Abstract
Optogenetics with microbial opsin genes, and pharmacogenetics with designer receptors, represent potent and versatile experimental modalities that can be integrated with each other as well as with a rich diversity of synergistic methods to provide fundamental opportunities in neuroscience research. Since initial steps were taken with these approaches less than 10 years ago, we are witnessing a rapid rise in publications (Fig. 1). The 7th Annual Brain Research Meeting in New Orleans in October 2012, Optogenetics and Pharmacogenetics in Neuronal Function and Dysfunction, brought together leading researchers that have developed and used these tools to explore a wide range of questions in nervous system function and dysfunction. This special issue of Brain Research includes articles by speakers in this meeting and others, which together synthesize and summarize the state of the art for optogenetics and designer receptors.
Keywords: Optogenetics, Pharmacogenetics, Channelrhodopsin, DREADD, Designer receptor, Viral transduction, Neurotransmitter signalling, Motivation/cognition, Autonomic function, Sensory Function, Translational neuroscience
Introduction
Optogenetics involves the introduction of genes encoding light-sensitive transmembrane ion conductance regulators (most commonly microbial opsins) to enable excitation or inhibition of targeted cells, with the biological effect depending upon the specific opsin employed (Fig. 2). The broad field of microbial opsin biophysics (Stoeckenius and Oesterhelt, 1971; Matsuno-yagi and Mukohata, 1977; Nagel et al., 2002) helped set the stage for the first expression of microbial opsins in neurons by Deisseroth and colleagues at Stanford (Boyden et al., 2005; Zhang et al., 2011). Now many opsins with distinct properties are employed for optogenetics, with the most widely applied being a mammalian codon-optimized channelrhodopsin (ChR2) and a third-generation halorhodopsin optimized for membrane trafficking in mammalian cells (eNpHR3.0), though there are many other examples and more being developed or modified regularly (Mattis et al., 2011). As described in other papers in this issue and elsewhere (Fenno et al., 2011; Tye and Deisseroth, 2012), opsins can be introduced into neurons by using transgenic animals that express these proteins, or by using viral vectors to introduce opsin genes into neurons through microinjection into the parenchyma.
Figure 2.
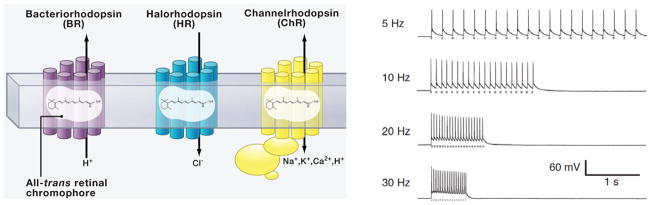
Left, BRs and related proteins pump protons from the cytoplasm to the extracellular medium, and HRs pump chloride into the cytoplasm; both hyperpolarize the cell. ChRs conduct cations across the membrane in both directions but always along the electrochemical gradient of the transported ions, and typically depolarize cells. Adapted from Zhang et al, 2011. Right, brief photostimuli at different frequencies (gray dashes below each spike trace) reliably elicit action potential in hippocampal neurons. Adapted from Boyden et al., 2005.
Pharmacogenetics in this context refers to the use of designer receptors to provide a lock-and-key approach to selectively modulate neuronal function by pharmacological means. The DREADDs (Designer Receptors Exclusively Activated by Designer Drugs, developed by Bryan Roth and colleagues at the University of North Carolina; Armbruster et al., 2007), are G-protein coupled receptors (GPCRs) engineered from muscarinic receptors, and include Gs, Gq and Gi varieties. When expressed in neurons, DREADDs are not sensitive to endogenous ligands and show little or no constitutive activity, but are very sensitive to the orally available ligand clozapine N-oxide (CNO) which is otherwise pharmacologically inert (Fig. 3). As with opsins discussed above, DREADDs and other designer receptors (e.g., RASSLs; see Gaven et al. this issue) are typically introduced into neurons by viral vectors. By including promoters for specific neuronal phenotypes, these receptors can be expressed in a cell-type specific manner in vivo, allowing behavioral and physiological studies of specific brain circuits with simple systemic administration of CNO. A recent review of DREADD technology is found in (Rogan and Roth, 2011), and contributions by Farrell and Roth, Gaven et al., Nair et al., and Vazey and Aston-Jones (summarized below) provide additional information on designer receptor technology.
Figure 3.
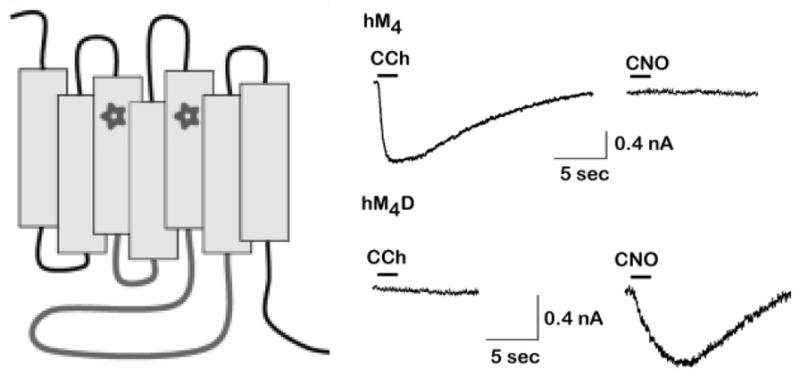
Left, DREADDs are mutant muscarinic receptors formed by mutations in two transmembrane regions (stars correspond to Y149C and A239G in hM3).The Gs-coupled DREADD also has the second and third intracellular loops (gray) of the β1-AR replacing those of the original M3 muscarinic receptor. Adapted form Rogan and Roth (2011). Right, Current induced in HEK293 cells that express native muscarinic receptors (hM4) or the hM4Di DREADD (hM4D), in response to the muscarinic agonist carbachol (CCh) or the DREADD agonist CNO. Adapted from (Armbruster et al., 2007).
Here, we organize and summarize the papers in this issue according to five focus areas: techniques, motivation and cognition, autonomic function, sensory analysis, and clinical application.
Techniques
Techniques for both optogenetics and designer receptors have developed rapidly over the last several years, and continue to provide exciting new tools and applications that take advantage of these approaches. In this issue, Farrell and Roth (Roth and colleagues originated the DREADD method) describe the properties of DREADDs and their significance for pharmacological research with translational implications. They describe several strengths and applications of this technology, and propose a new term, “pharmacosynthetics”, to describe the science of synthetic ligand-GPCR pairs (e.g., DREADDs) for selective pharmacological manipulation of neurons and intracellular signaling pathways.
Next, recent developments in optogenetics methods and applications are described in papers by Kravitz et al. and Kahn et al. Kravitz et al. take on the challenge of using ChR2-mediated spiking to identify distinct cell types in electrophysiological recordings from awake behaving animals. They expressed ChR2 in the neurons of the direct or indirect pathways of striatum and performed electrical recordings together with optical stimulation using an implanted microwire array with integrated optical fiber. They describe and address multiple challenges and confounds that can arise with this potentially extremely useful class of experiment and provide a Matlab analysis tool for general application.
Beginning in 2010, optogenetics has been integrated with fMRI (ofMRI; Lee et al., 2010) in an approach that promises both to provide a means to globally map changes in brain activity elicited by defined neural populations under different conditions, and to help determine the contributions of different classes of circuit element activity to the blood oxygenation level-dependent (BOLD) signal. Here Kahn et al., address this second question, combining optogenetic drive of cortical neurons with rodent fMRI. Across stimulus patterns and analyses, the strongest correlation was observed between spiking activity and the BOLD response, with transfer functions generated based on local spiking (but not those generated by local field potential or LFP) providing accurate estimation of the measured BOLD response.
Neurotransmitter-specific signaling and function
Neurotransmitter functions have been studied conventionally using pharmacological approaches, taking advantage of receptors associated with specific transmitter molecules. Although these methods have produced a wealth of knowledge about these systems, applications have suffered from either or both of limitations in the specificity of drugs for specific receptors, or in access to the brain specific pharmacological agents. Optogenetics and pharmacogenetics circumvent many of these limitations, and allow much more selective manipulation of specific circuit elements. Janak and Steinberg review the use of optogenetics to manipulate and study brain dopamine neurons in a specific manner, including the use of transgenic tyrosine hydroxylasecre rats combined with floxed ChR2 vectors. They discuss both advances and opportunities afforded by optogenetics and caveats that should be kept in mind when using these methods to study DA or other brain neurons. Gaven et al. focus on another class of synthetic receptors, termed RASSLs (Receptors Activated Solely by Synthetic Ligands). In this paper, they specifically describe the development and properties of 5HT4 RASSLs, and identify the first biased agonists for wildtype 5HT4 receptors that allow analysis of Gs vs Gq signaling by this receptor. These are just two examples of the powerful ways in which optogenetics and designer receptors can be used to understand signaling, physiology, and behavioral functions relevant to identified neurotransmitter systems in the brain.
Motivation/Cognition
Some of the most powerful applications of optogenetics and pharmacogenetics are in behavioral studies, affording selective manipulation of systems and circuits associated with a variety of complex behavioral functions. Although many papers in this special issue describe use of these methods to study behavioral function, three in particular focus on studies of motivation and cognitive processes. First, Nieh et al provide a comprehensive review of insights that optogenetics has provided into brain circuits that mediate emotional and motivational processes. They review studies from their own lab and from others that use opsins in several different brain levels (mesolimbic dopamine system, striatum, hypothalamus or amygdala) to better define and distinguish functional subcircuits within each of these areas, and how these different neural circuits are involved in defining emotional valence and mediating motivational responses. Second, Nair et al. review results from studies using classical methods, as well as their own recent studies using designer receptors, into functions of the lateral habenula, a brain region that has received considerable interest recently (in part for its projections that regulate midbrain dopamine neurons). They describe how the designer receptors provide significant advantages over classical methods and are helping to define the roles of the habenula in a network of subcortical nuclei that regulate aversive learning, motivation and stress responses. Third, Smith and Graybiel extend these types of behavioral studies to the neural substrates of habit. They first review classical studies that laid the groundwork for current understanding of habit circuitry, and then describe their own recent study using optogenetics to extend that understanding. They find that optogenetic inhibition of infralimbic cortex in rats reveals a surprisingly complex role of this area in regulating habitual behavior, perhaps involving rapid neuroplasticity during or after optogenetic inhibition of IL neurons.
Autonomic
Optogenetics and pharmacogenetics are well suited for a wide variety of behavioral studies, including autonomic function. Papers by Ray et al., and Guyenet et al., both focus on advances in understanding the neural substrates of respiratory function through designer receptor- or opsin-regulation of activities in specific neuronal populations. Ray et al. used combinatorial genetics to express the hM4Di DREADD in neurons that express the early growth response 2 transcription factor (Egr2), rendering these cells specifically sensitive to inhibition by the DREADD agonist CNO. This approach allowed the investigators to circumvent the lethality of prenatal lesions of these neurons, so that they could be silenced in adults in which they had developed normally. These studies showed that Egr2 neurons play an important role in adult respiratory function. Guyenet et al. review a series of studies using classical, optogenetic and pharmacogenetic methods, that examine neural substrates of respiratory control. Results with classical methods (lesions, electrophysiology) are extended with optogenetics by this group and others showing that the retrotrapezoid nucleus, raphe serotonergic neurons, and nearby glial cells are critically involved in the control of respiration. In particular, these cells appear to be important for respiratory response to elevated CO2.
Sensory
Sensory functions are also amenable to study with optogenetics or pharmacogenetics. One paper by Shimano et al in this issue uses optogenetic manipulations of dorsal cochlear neurons to establish some basic labeling and physiological responses to photo-stimulation or –inhibtion in these auditory neurons, establishing baseline data that will be useful for future studies of anatomical or physiological properties of circuits involved in auditory processing.
Translational
Optogenetics and pharmacogenetics are powerful techniques for investigating the neural substrates of clinical disorders and helping develop new therapeutics. Vazey and Aston-Jones provide an overview of both methods, and specifically describe exciting recent studies with these methods that have led to a better understanding the etiology of motor dysfunctions in Parkinson’s disease. They also discuss future potential applications of these methods for non-dopamine systems (e.g., locus coeruleus) to investigate and perhaps some day even treat motor or non-motor parkinsonian dysfunctions.
Conclusions
As revealed by the presentations at the 7th Annual Brain Research meeting, and in this special issue of Brain Research, optogenetics and pharmacogenetics are powerful methodologies that are finding application in a wide range of neuroscience studies, and that share important features. A major strength of both approaches is the use of modern genetics and viral vector techniques to introduce non-native proteins into neurons, that function as channels, pumps or receptors. In both cases, these novel proteins are not sensitive to endogenous compounds, and are sensitive only to exogenous non-native stimuli (light or CNO) that otherwise have no effect on neural tissue. Therefore, both approaches provide a striking increase in selectivity for neuronal manipulations, allowing causal analyses of the roles of neural circuits in defined functions.
Although these methods share important features, they also have different but complementary strengths and limitations, so that together they provide an especially wide range of applications. Optogenetics allows millisecond-scale temporal accuracy in manipulating neuronal activity, important for circuit or behavioral functions wherein the rate or timing of neural activity may be important. However, this method requires implantation of an optical fiber for most behaving-animal work, and due to light scattering the amount of tissue that can be photostimulated in vivo by a single fiber is limited (although the most light-sensitizing of the recently-engineered opsins can recruit even deep brain structures in mice without implanted hardware; Yizhar et al., 2011). Designer receptors, on the other hand, have the advantage that they are sensitive to an orally available agonist (CNO) so that no optical devices are needed and DREADDs that are widely distributed in brain can be stimulated by a simple systemic CNO administration. However, their action is slow, on the order of minutes, so that their utility in analyzing neural processes that rely on rate or timing of neural activity is quite limited (although this property may be ideal for certain studies such as tonic inactivation of motivational circuits, or stimulation of remaining neurons to compensate for loss in neurodegenerative disorders).
Figure 4 lists some of the technical advances with optogenetics and pharmacogenetics that were highlighted at the 7th Annual Brain Research meeting, as well as many of the applications that were presented. As shown, optogenetics and pharmacogenetics have spurred a great deal of technical development, and have now entered into use in many areas of neuroscience research. Notably, this is only the beginning of what is sure to be an expanding list of applications, as the power of these exciting technologies continues to develop.
Figure 4.
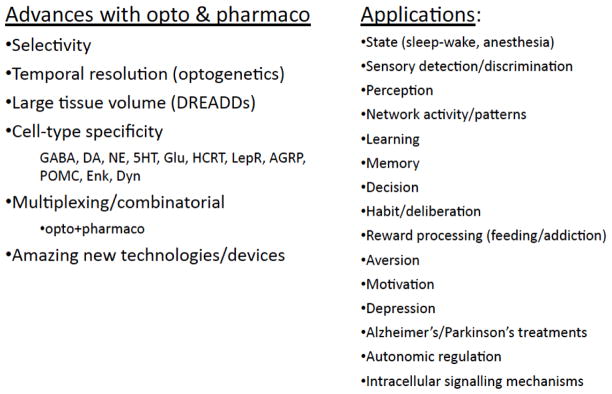
Advances reported with optogenetics or pharmacogenetics, and applications described for these methods, at the recent 7th Annual Brain Research meeting in New Orleans.
Figure 1.

Numbers of articles published over the past 10 years that include optogenetics or designer receptors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–8. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2010;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–92. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno-Yagi A, Mukohata Y. Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun. 1977;78:237–43. doi: 10.1016/0006-291x(77)91245-1. [DOI] [PubMed] [Google Scholar]
- Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature Methods. 2011;9:159–72. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–8. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–52. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacological Reviews. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Deisseroth K. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nature Reviews Neuroscience. 2012;13:251–66. doi: 10.1038/nrn3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, et al. Neocortical excitation-inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–8. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–57. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


