Abstract
Hydroxyproline plays a major role in stabilizing collagenous domains in eukaryotic organisms. Lack of this modification is associated with significant lowering in thermal stability of the collagen triple helix and may also affect fibrillogenesis and folding of the peptide chains. In contrast, even though bacterial collagens lack hydroxyproline, their thermal stability is comparable to fibrillar collagen. This has been attributed to the high frequency of charged amino acids found in bacterial collagen. Here we report a thermally stable hydroxyproline-free ABC heterotrimeric collagen mimetic system composed of decapositive and decanegative peptides and a zwitterionic peptide. None of the peptides contain hydroxyproline and furthermore the zwitterionic peptide does not even contain proline. The heterotrimer is electrostatically stabilized via multiple interpeptide lysine-aspartate and lysine-glutamate salt-bridges and maintains good thermal stability with a melting temperature of 37 °C. The ternary peptide mixture also populates a single composition ABC heterotrimer as confirmed by circular dichroism (CD) and Nuclear Magnetic Resonance (NMR) spectroscopy. This system illustrates the power of axial salt-bridges to direct and stabilize the self-assembly of a triple helix and may be useful in analogous designs in expression systems where the incorporation of hydroxyproline is challenging.
Stability of the collagen triple helix in phylogenetically distant organisms such as mammals and bacteria is determined by a combination of enthalpic and entropic factors that depend on the amino acids occupying the X and Y positions of the canonical Xaa-Yaa-Gly triplet repeat.1 For example, the cyclic amino acids proline (Pro, P) and hydroxyproline (Hyp, O) found predominantly in the collagenous domains of eukaryotic organisms,2 restrict the backbone dihedral angles close to the native folded state3,4 providing entropic stabilization. The stereoelectronic and inductive effect of the electronegative hydroxyl substituent provides additional stabilization by pre-organizing the Hyp ring pucker5,6 and lowering the barrier to cis-trans isomerization of hydroxyprolyl amide bond.7,8 Therefore, lowering the Hyp content is often associated with significant loss in thermal stability of the triple helix9,10 that can also prevent fibrillogenesis in some native collagen types.11
Bacteria lack a crucial enzyme, prolyl hydroxylase, required for post-translational hydroxylation of proline, however, the consequent lack of hydroxyproline does not significantly affect the thermal stability of their collagenous domains.12,13 For example, the melting temperature of the collagenous domains of Scl1 and Scl2 bacterial proteins in Streptococcus Pyogenes have been shown to be comparable to that of vertebrate fibrillar collagen.14 These organisms have evolved alternate mechanisms to compensate for the lack of hydroxyproline.15 In silico genomic analysis of a variety of bacteria and bacteriophages has indicated a high occurrence of threonine in the Y position where its hydroxyl group can form hydrogen bonds to the backbone carbonyl.16 However, organisms that lack a predominance of threonine as well as proline residues contain an unusually high frequency of the charged amino acids lysine, arginine, glutamate and aspartate. Although, these residues generally destabilize the triple helix,17 it has been demonstrated that the loss in stability is significantly compensated if the acidic and basic residues form specific charge pairs. 18-20
The stability of previously reported collagen mimetic systems containing salt-bridge interactions had sizable contribution from proline and hydroxyproline.21,22 In contrast, collagen constructs that contained no hydroxyproline were stabilized by terminally fused trimerization domains such as T4 fibritin foldon23 or cysteine knots.24
Here we report a hydroxyproline free ABC heterotrimeric collagen triple helix composed of decacharged positive and negative peptides and a zwitterionic peptide that contains neither proline nor hydroxyproline. Multiple lysine-glutamate and lysine-aspartate axial charge pairs electrostatically stabilize the heterotrimer. To our knowledge, this is the first report of a collagen mimetic system that self-assembles into a stable triple helix despite the complete lack of cyclic amino acids in one of the peptide chains.
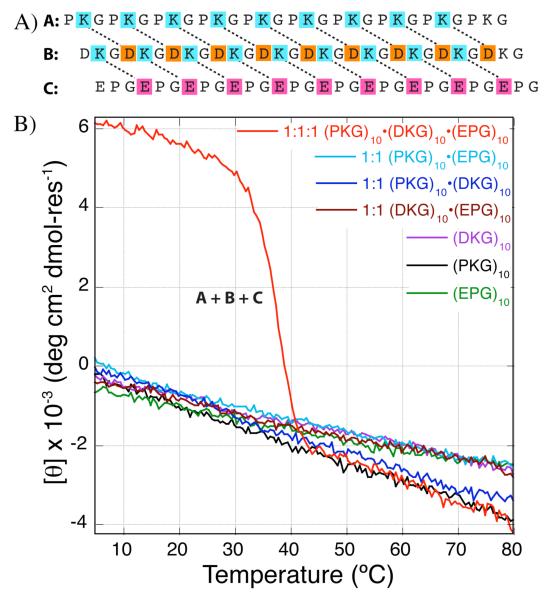
Using salt-bridges rationally engineered into a 3•(POG)10 template, we have previously reported AAB and ABC heterotrimeric collagen mimetic systems that show controlled composition and/or register.25-27 Of particular interest is the system composed of (PKG)10, (EOG)10 and (POG)10 peptides that self-assemble into an ABC heterotrimer with a melting temperature of 54 °C.21 The primary sources of stabilization in this heterotrimer are the multiple lysine-glutamate charge pairs and the high content of cyclic amino acids. In order to design a hydroxyproline-free ABC heterotrimer, we made two modifications to this system: first the 10 hydroxyproline residues in (EOG)10 were substituted with prolines and second, the neutral (POG)10 chain was substituted with a zwitterionic peptide (DKG)10. This eliminates twenty hydroxyproline residues and ten proline residues replacing them with ten lysine, ten glutamate and ten aspartate residues, all of which are known to destabilize a triple helix unless they form axial charge pairs (Figure 1A).1,21
Figure 1.
(A) Schematic illustration of hydroxyproline-free ABC heterotrimer composed of (PKG)10, (DKG)10 and (EPG)10 peptides electrostatically stabilized through multiple lysine-aspartate and lysine-glutamate salt-bridges. All three peptides contain an N-terminal YG label to assist in determination of concentration. (B) Thermal melting profile of the individual, binary and ternary mixtures of (PKG)10, (DKG)10 and (EPG)10 peptide. The monomers as well as binary mixture were monitored at 225.0 nm and the ternary mixture was monitored at 222.4 nm.
All peptides synthesized in this study include an N-terminal tyrosine for spectrophotometric determination of concentration. The Gly17 residue is 15N-isotopically enriched for NMR spectroscopic study. YG labels are omitted in further discussion of the peptides for brevity. Amino acid residues are abbreviated by their single letter code followed by an XYG subscript indicating their peptide chains.
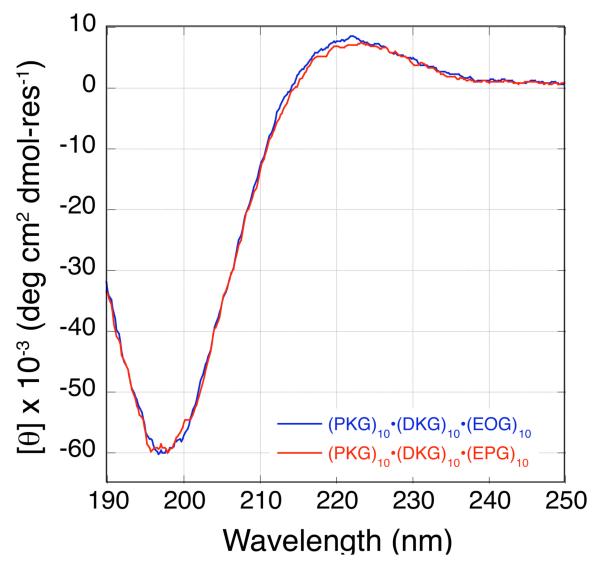
The CD spectrograph of a mixture of (PKG)10, (EPG)10 and (DKG)10 peptides in Figure 1B shows a single cooperative unfolding transition at 37 °C. All monomers and possible binary mixture of the three peptides show a linear decrease in molar residue ellipticity (MRE) indicating the absence of well folded states. The energetic compensation from fewer charge pairs in the competing AAB heterotrimeric and homotrimeric states is not sufficient to offset the loss in stability due to lower cyclic amino acid content. The homologous ABC heterotrimer, containing (EOG)10 instead of (EPG)10, unfolds at a higher melting temperature of 43 °C (Figure S2.C) while the two ABC heterotrimers have nearly identical CD spectra at 5 °C (Figure 2). Interestingly, the mixture of (PKG)10 and (EOG)10 unfolds at 39 °C (Figure S2.B) but a mixture of (PKG)10 and (EPG)10 does not form a triple helix. The lack of triple helices formed by binary mixtures of peptides in the (EPG)10 containing system means that its ABC system has no compositional competitors, unlike the (EOG)10 containing system, and therefore the (PKG)10•(DKG)10•(EPG)10 triple helix self-assembles more cleanly.
Figure 2.
CD wavelength scan of (PKG)10•(DKG)10•(EOG)10 in blue and (PKG)10•(DKG)10•(EPG)10 in red recorded at 5 °C.
Although the 225 nm maximum observed in the wavelength scan of the ternary peptide mixture (Figure 2) is indicative of a polyproline type II (PPII) helix,28,29 it provides little information on the association state of the heterotrimer. The highly cooperative nature of the thermal unfolding data (Figure 1b) is strongly suggestive of the formation of a triple helical structure. In order to further confirm the formation of a triple helix, we performed analytical ultracentrifugation (AUC) sedimentation velocity experiments over a large range of sample concentrations. These experiments showed that both (PKG)10•(DKG)10• (EPG)10 and (PKG)10•(DKG)10•(EOG)10 heterotrimers assembled into a major species with a molecular weight consistent only with the trimer. Furthermore the observed frictional ratios were fully consistent with an extended helical structure. Detailed description of the AUC data is presented in the supporting information.
After conclusively determining the oligomerization state to be three, as is expected for a collagen triple helix, establishing the triple helical topology of the assembly is particularly important in this case because of the lack of hydroxyproline and low percentage of proline residues. Further, one chain of this helix, (DKG)10, lacks cyclic amino acids all together. These characteristics might be expected to significantly perturb the backbone dihedral angles and therefore the fold of the triple helical assembly. To assess this we use NMR.
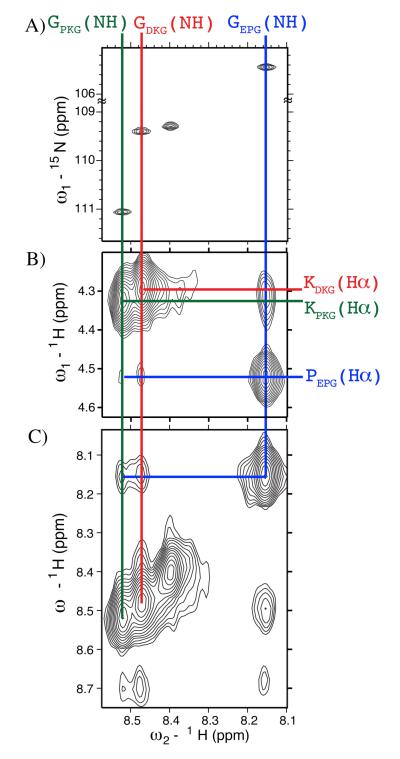
In theory, a mixture of three peptides can populate ten unique triple helices of different compositions corresponding to the A3, B3 and C3 homotrimers and A2B, AB2, A2C, AC2, B2C, BC2 and ABC heterotrimers. Detection of these competing states is difficult in CD, especially if they represent minor components. 1H,15N-Heteronuclear Single Quantum Coherence (HSQC) provides higher sensitivity and greater resolution of competing states. The HSQC spectra of a mixture of (PKG)10, (EPG)10 and (DKG)10 (Figure 3A) obtained at 20 °C shows a monomer cross peak and three additional cross peaks of equal intensity that correspond to the 15N-isotopically enriched Gly present in each chain of the triple helix. The presence of only three triple helical cross peaks in the HSQC spectra and a single thermal transition observed in CD suggest that, within the limit of detection, the mixture of (PKG)10, (EPG)10 and (DKG)10 populates a single composition ABC heterotrimer.
Figure 3.
1H,15N-HSQC spectrum (A) and two-dimensional plane of a three-dimensional 1H,15N-NOESY-HSQC (B and C) spectrum of a mixture of (PKG)10, (DKG)10 and (EPG)10 peptides.
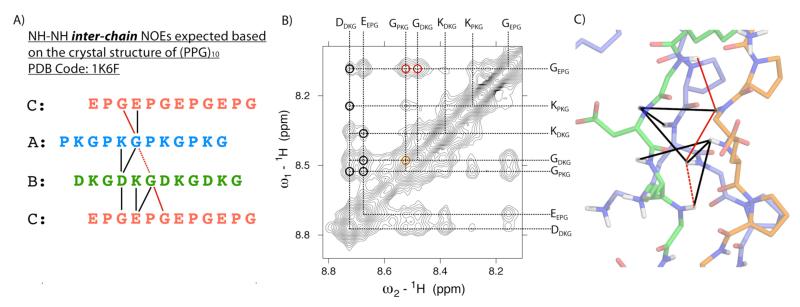
The supramolecular topology of the triple helix is characterized by the PPII conformation of the peptide chains and their single residue stagger with respect to each other. The definitive proof for this structure comes from our NMR data which shows that the three chains, (PKG)10, (DKG)10 and (EPG)10, form contacts that are within 5 Å of one another (Figure 3b,c and 4). Further, the NOEs observed are those expected for a canonical triple helix. The crystal structure of (PPG)10 30 shows that nine backbone NH-NH NOE correlations should be observed, of these we can definitively assign eight while the ninth may exist but is obscured by spectral overlap (Figure 4). See supporting information for a detailed discussion of the NMR assignment.
Figure 4.
(A) NH-NH inter-chain NOE expected based on the crystal structure of (PPG)10 collagen triple helix30 (PDB code: 1K6F). (B) 1H,1H-NOESY spectrum of a mixture of (PKG)10, (EPG)10 and (DKG)10 showing the NH-NH NOE cross peaks. G(NH)-G(NH) correlations are indicated in black and red. The GDKG(NH)-GPKG(NH) correlation shown in orange/dashed could not be definitively assigned due to spectral overlap. (C) Model of the ABC heterotrimer composed of (PKG)10, (EPG)10 and (DKG)10 chains indicating observed NH-NH inter-chain correlations.
The single residue stagger of peptide chains in the ABC heterotrimer leads to a unique situation where six different triple helices, called registers, are possible for the permutation of A, B and C chains in leading, middle and lagging positions. The number of potential axial charge pairs varies for each register and populating some registers may be more energetically favorable compared to others (Scheme S1). In our case, the six registers can be divided into two subsets of three registers each. Registers within a subset are symmetry related and will generated nearly identical NOE correlation map. Also, as shown in Figure S4, registers from one subset form nearly twice as many charge pairs as those from the other. 1H,1H-NOESY data shows that registers that contain fewer charge pairs and are therefore energetically unfavorable, are not populated while one or more of the more favorable register are formed (see Supporting Information for the discussion on determination of registers).
Chemically synthesized short peptides are most commonly used to study the structure-function relationship of native collagen. While these systems have been very useful, as we move from refining our knowledge of this relationship to obtaining insights into fibrillogenesis, disease modeling, protein binding and the design of a robust and functional extracellular biomimetic scaffold, the need for longer polypeptides with multiple role-specific sub-domains becomes indispensable. Being effectively inaccessible via solid phase synthesis due to their size, these will need to be obtained by recombinant expression in various hosts. Although eukaryotic expression systems capable of various post-translational modifications have been developed,31,32 due to the difficulty of isolation and purification, protein translation in bacterial expression systems remains lucrative. In this article, we have successfully demonstrated that hydroxyproline can be substituted with charge pairs with moderate loss in thermal stability of the resulting collagen triple helix. This strategy could be used to design hydroxyproline-free collagen constructs that can be expressed in bacterial hosts such as E.Coli. Furthermore, the specificity of salt-bridge interaction can provide a level of control in designing composition and register specific collagen heterotrimers that is inaccessible with hydroxyproline. This is reflected in the single composition ABC heterotrimer populated by the ternary peptide mixture. The designed hydroxyproline free heterotrimer might also be used as an autonomous trimerization domain for designing composition specific heterotrimeric collagenous or non-collagenous fusion proteins such as the coiled-coil collagenous motif found in type IX collagen.
Supplementary Material
ACKNOWLEDGMENT
This work was funded in part by National Science Foundation (DMR-0645474 and DMR-1206899) and the Robert A. Welch Foundation (Grant No. C1557). The development of the UltraScan software is supported by the National Institutes of Health through grant RR022200 (to BD). Supercomputer time allocations were provided through National Science Foundation grant TG-MCB070038 (to BD). BD acknowledges the support of the San Antonio Cancer Institute grant P30 CA054174 for the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio.
Footnotes
ASSOCIATED CONTENT
Synthesis and characterization of peptides, preparation of CD and NMR samples, experimental parameters and additional NMR and analytical ultracentrifugation experiments and analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- (1).Persikov AV, Ramshaw JA, Kirkpatrick A, Brodsky B. Biochemistry. 2000;39:14960. doi: 10.1021/bi001560d. [DOI] [PubMed] [Google Scholar]
- (2).Ramshaw JA, Shah NK, Brodsky B. J. Struct. Biol. 1998;122:86. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- (3).Okuyama K. Conn. Tiss. Res. 2008;49:299. doi: 10.1080/03008200802325110. [DOI] [PubMed] [Google Scholar]
- (4).Bella J. J. Struct. Biol. 2010;170:377. doi: 10.1016/j.jsb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- (5).Shoulders MD, Satyshur KA, Forest KT, Raines RT. Proc. Nat. Acad. Sci. USA. 2010;107:559. doi: 10.1073/pnas.0909592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- (7).Eberhardt ES, Panisik N, Raines PR. J. Am. Chem. Soc. 1996;118:12261. doi: 10.1021/ja9623119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Reimer U, Fischer G. Biophys. Chem. 2002;96:203. doi: 10.1016/s0301-4622(02)00013-3. [DOI] [PubMed] [Google Scholar]
- (9).Berg RA, Prockop DJ. Biochem. Biophys. Res. Comm. 1973;52:115. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- (10).Jimenez S, Harsch M, Rosenblo J. Biochem. Biophys. Res. Comm. 1973;52:106. doi: 10.1016/0006-291x(73)90960-1. [DOI] [PubMed] [Google Scholar]
- (11).Perret S, Merle C, Bernocco S, Berland P, Garrone R, Hulmes DJS, Theisen M, Ruggiero F. J. Biol. Chem. 2001;276:43693. doi: 10.1074/jbc.M105507200. [DOI] [PubMed] [Google Scholar]
- (12).Boydston JA, Chen P, Steichen CT, Turnbough CL. J. Bacter. 2005;187:5310. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Han RL, Zwiefka A, Caswell CC, Xu Y, Keene DR, Lukomska E, Zhao ZH, Hook M, Lukomski S. Appl. Microbiol. Biotech. 2006;72:109. doi: 10.1007/s00253-006-0387-5. [DOI] [PubMed] [Google Scholar]
- (14).Xu Y, Keene DR, Bujnicki JM, Hook M, Lukomski S. J. Biol. Chem. 2002;277:27312. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- (15).Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. J. Biol. Chem. 2007;282:29757. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- (16).Rasmussen M, Jacobsson M, Bjorck L. J. Biol. Chem. 2003;278:32313. doi: 10.1074/jbc.M304709200. [DOI] [PubMed] [Google Scholar]
- (17).Chan VC, Ramshaw JA, Kirkpatrick A, Beck K, Brodsky B. J. Biol. Chem. 1997;272:31441. doi: 10.1074/jbc.272.50.31441. [DOI] [PubMed] [Google Scholar]
- (18).Gurry T, Nerenberg PS, Stultz CM. Biophys. J. 2010;98:2634. doi: 10.1016/j.bpj.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. J. Mol. Biol. 2002;316:385. doi: 10.1006/jmbi.2001.5342. [DOI] [PubMed] [Google Scholar]
- (20).Gauba V, Hartgerink JD. J. Am. Chem. Soc. 2007;129:2683. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- (21).Gauba V, Hartgerink JD. J. Am. Chem. Soc. 2007;129:15034. doi: 10.1021/ja075854z. [DOI] [PubMed] [Google Scholar]
- (22).Xu F, Zahid S, Silva T, Nanda V. J. Am. Chem. Soc. 2011;133:15260. doi: 10.1021/ja205597g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bachinger HP, Engel J. J. Mol. Biol. 2001;308:1081. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- (24).Krishna OD, Kiick KL. Biomacromolecules. 2009;10:2626. doi: 10.1021/bm900551c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).O’Leary LE, Fallas JA, Hartgerink JD. J. Am. Chem. Soc. 2011;133:5432. doi: 10.1021/ja111239r. [DOI] [PubMed] [Google Scholar]
- (26).Fallas JA, Lee MA, Jalan AA, Hartgerink JD. J. Am. Chem. Soc. 2012;134:1430. doi: 10.1021/ja209669u. [DOI] [PubMed] [Google Scholar]
- (27).Jalan AA, Hartgerink JD. Biomacromolecules. 2012;14:179. doi: 10.1021/bm3015818. [DOI] [PubMed] [Google Scholar]
- (28).Brown AM, Zondlo NJ. Biochemistry. 2012;51:5041. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- (29).Ramani R, Hanski S, Laiho A, Tuma R, Kilpelainen S, Tuomisto F, Ruokolainen J, Ikkala O. Biomacromolecules. 2008;9:1390. doi: 10.1021/bm7012845. [DOI] [PubMed] [Google Scholar]
- (30).Berisio R, Vitagliano L, Mazzarella L, Zagari A. Protein Sci. 2002;11:262. doi: 10.1110/ps.32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bulleid NJ, John DCA, Kadler KE. Biochem. Soc. Trans. 2000;28:350. [PubMed] [Google Scholar]
- (32).Pinkas DM, Ding S, Raines RT, Barron AE. ACS Chem. Biol. 2011;6:320. doi: 10.1021/cb100298r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.