Abstract
Examining NFAT transcription factor dynamics in single cells during signal propagation from cell membrane to the nucleus reveals unique modes of operation for protein isoforms.
Keywords: Transcription factors, nuclear dynamics, signaling, NFAT, p53, NF-κB
Extracellular signals can trigger intracellular signaling pathways that frequently involve the translocation of transcription factors from the cytoplasm, where they are sequestered, into the nucleus, where they elicit gene activation. The study of translocation dynamics and their effect on gene expression has many unanswered questions (Box 1) but has benefited in recent years from the development of live-cell techniques for the study of biological systems. Traditional transcriptional analyses often provide an averaged output measured from a cell population [1], which tends to mask the behavior of individual cells. Using novel approaches, however, it has become possible to focus on transcription factor dynamics in individual living cells. In a study recently published in Molecular Cell, Yissachar et al. [2] use the nuclear factor of activated T-cells (NFAT) signaling system pathway employed by the immune system to examine the dynamic properties of NFAT transcription factor translocation to the nucleus in response to extracellular cues in rat cells, and discover striking differences in comparison to the previously characterized p53 and nuclear factor-κB (NF-κB) signaling systems.
Transcription factors can exhibit nucleo-cytoplasmic oscillations during unperturbed and stress conditions
Temporal and spatial information extracted from time-lapse movies of cells expressing fluorescently tagged transcription factors shows that nucleo-cytoplasmic oscillations are common. For example, DNA damage leads to increased levels of the tumor suppressor p53, a transcription factor, and its negative regulator, transformed Mouse double minute 2 homolog (Mdm2), in the nucleus. p53 accumulation follows an oscillating mode of approximately 6 hours for each pulse, while Mdm2 makes a delayed nuclear appearance but follows a similar dynamic pattern [3]. As damage increases, more pulses are observed, yet their intensity remains unmodified (Figure 1a). The basal dynamics of p53 under non-stressed conditions show similar pulses; however, these pulses do not culminate in gene activation, due to an absence of the post-translational modifications required to activate p53 [4]. Similar pulsation phenomena are observed for the NF-κB transcription factor after tumor necrosis factor α (TNF-α) stimulation. Asynchronous nucleo-cytoplasmic 2-hour oscillations begin as a response to TNF-α and continue for more than 20 hours [5], with the frequency of NF-κB oscillations acting as a control switch for the expression of different gene sets. Similar experiments performed under physiological TNF-α levels show comparable 2-hour pulses, although the response is substantially delayed, less synchronized and occurs in a smaller fraction of the cell population (Figure 1b), implying a mechanism of threshold activation [6].
Figure 1.
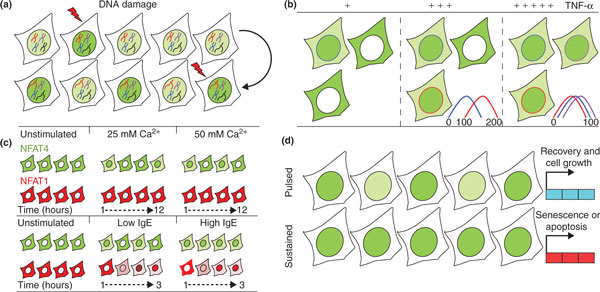
Transcription factor dynamics in single living cells. (a) DNA damage affects transcription factor dynamics. p53 (green) exhibits more frequent nuclear pulses as damage levels increase (disrupted black and red chromosomes). (b) Increasing doses of extracellular stimulation (plus signs) affects the synchronization of the initial cytoplasm-to-nucleus translocation time. High levels of TNF-α increase the overall population response, observed as elevated nuclear NF-κB (green) and increased synchronization. Colored plots represent the initial NF-κB nuclear translocation times (minutes) for each depicted cell. (c) NFAT isoforms respond differently to stimuli. Top: calcium levels modulate the frequency of NFAT4 nuclear localization. A rise in calcium levels increases the frequency of NFAT4 nuclear localization (green) bursts, which last for up to 12 hours, while NFAT1 remains cytoplasmic (red). Bottom: immunoglobulin E (IgE) mediates activation of NFAT1 and NFAT4. In response to low antigen levels, NFAT4 exhibits nuclear bursts that become sustained nuclear localization at high antigen levels. NFAT1 shows a gradual response to both low and high antigen levels, but with higher amplitude at high levels. (d) Unique transcription factor dynamics control gene expression patterns. Pulsed or sustained p53 nuclear accumulation (green) under DNA damage conditions determines cell fate by driving expression of distinct sets of genes. Pulsed p53 leads to cell recovery and proliferation, whereas sustained p53 ends in activation of senescence or apoptosis pathways.
Transcription factor isoforms: redundancy or unique functionality?
Yissachar et al. [2] have now used NFAT transcription factors as a basis for investigating how activation dynamics can vary between isoforms within a transcription factor family. The NFAT family consists of five protein members (NFAT1 to NFAT5), four of which are regulated by calcium signaling. All contain a DNA-binding domain, and have an NFAT homology region, which harbors a transactivation domain. NFAT1, 2 and 4 are transcription factors that function in T cells, and have been considered redundant owing to their high sequence and structural similarity. Indeed, knockout mice lacking a single NFAT gene exhibit only minor alterations in immune response, and it is only when additional NFAT genes are knocked out simultaneously that an acute immune response emerges. However, accumulating evidence now suggests that the transcriptional response can be differentially mediated by each isoform, either by way of varying expression levels in different tissues or by interactions with different co-activators. Since 'isoform redundancy' is nowadays a term less frequently used to excuse our limited understanding of protein diversity, we should assume that the distinctness of each isoform lies at intricate functional levels yet to be unraveled.
Consistent with this view, the study by Yissachar et al. [2] demonstrates that NFAT isoforms harbor functional uniqueness. Using an elegant set-up for monitoring transcription factors in living cells, the authors examined the dynamics of the transcriptional response produced by two NFAT isoforms, NFAT1 and NFAT4, in mast cells. The authors dissected the temporal and spatial elements of NFAT transcriptional dynamics in response to various cues, such as an increase in extracellular calcium or an immune response. Their results reveal the true colors of NFAT1 and NFAT4 signaling traits and, importantly, the subtle yet biologically significant differences in transcriptional dynamics.
Signaling in real-time: from extracellular calcium to nuclear transcription factor accumulation
Immunofluorescent staining of endogenous NFAT1 and NFAT4 reveals a cytoplasmic localization in mast cells under unperturbed conditions. Upon calcium stimulation, NFAT4 translocates to the nucleus in a subpopulation of cells, while NFAT1 remains uniformly cytoplasmatic across the cell population. Using two-color live-cell time-lapse imaging, performed on mast cells stably co-expressing GFP-NFAT1 or GFP-NFAT4, together with a marker of the cell nucleus, NFAT4 is observed shuttling in and out of the nucleus, while GFP-NFAT1 does not translocate (Figure 1c). The oscillating behavior of NFAT4 persists for more than 12 hours and consists of irregularly dispersed 10-minute periods of nuclear localization. Halving the calcium concentration did not reduce the duration of NFAT4 nuclear localization or its nuclear intensity levels. Nevertheless, low calcium levels did reduce the frequency of the bursts, as well as the percentage of cells that actually responded to the signal (Figure 1c). Altogether, these findings demonstrate that an intensification of the extracellular calcium signal strength increases both the responsiveness of the cell population and the number of translocation events within a time period. Interestingly, once the initial signal had been relayed, nuclear NFAT4 molecules shifted to a dynamic behavior free from calcium level influence. This dynamic behavior is probably governed by unidentified kinases or binding proteins.
Merging of different extracellular signals on the same signaling pathway results in diverse nuclear outputs
The calcium-stimulated nuclear translocation of NFAT transcription factors is mediated by their dephosphorylation at the hands of calcineurin phosphatase. Given that the same pathway is activated during an immune response, the authors tested the effect of immunoglobulin E (IgE) activation on NFAT translocation. Even though NFAT4 exhibited nuclear localization bursts in only a fraction of the cell population under these conditions, as observed with calcium treatment, its translocation behavior was notably different in that oscillations ceased after only 2 hours or so. Interestingly, an increase in antigen concentration brought about a long-lasting nuclear NFAT4 localization time (2 to 3 hours) in place of the bursting phenotype (10-minute bursts).
As with NFAT4, NFAT1 behavior also differed between calcium and IgE stimulations. Whereas calcium did not perturb NFAT1's uniform cytoplasmic distribution, a gradual nuclear accumulation occurred over the course of several hours. This redistribution was then reversed by a slow clearing out from the nucleus (Figure 1c).
Comparative analysis of NFAT isoform translocation dynamics
Exhaustive comparative image analysis of NFAT1 and NFAT4 behavior in hundreds of cells revealed inherent differences in the dynamic performance of transcription factor isoforms, even when acting in the same cells and responding to the same extracellular signals. First, the difference in response times showed a unique behavior for each isoform. Under high antigen levels, the NFAT4 response was highly synchronized in the responding cells and reached 20% of its maximum amplitude after 1 minute or so. Decreased antigen levels led to a loss of synchronization within the population, but notably, even though some cells had a delayed response, the whole population finally responded. In contrast, the response time for NFAT1 was somewhat slower, averaging over 3 to 4 minutes, and was unaffected by antigen levels. Second, although increased antigen levels led to the doubling of nuclear duration times for both NFAT1 and NFAT4, the magnitude of NFAT4 nuclear accumulation remained constant, whereas NFAT1 exhibited a dose-dependent increase in nuclear localization.
Autocorrelation analysis of the NFAT data attributed two kinetic populations to NFAT4 nuclear dynamics. A slow component (55 minutes) determines the overall response duration and is common to both NFAT4 and NFAT1, while a fast component unique to NFAT4 (1.5 minutes) is responsible for nuclear bursts. The rapidity of this response substantially differs from the prolonged p53 and NF-κB responses [3,5], even though NF-κB can respond to the same Fc receptor signaling pathway.
A pulsing signal received by the immune system
IgE-mediated activation triggers signaling through the constant presence of the signaling molecule in the extracellular medium. However, immune cells traveling in between cells and tissues are most likely to encounter brief and pulsatile signals, as observed during T-cell activation and calcium signaling [7]. To simulate such a situation in culture, Yissachar et al. [2] used antigen pulses of varying durations and varying signal strength. They measured a 60-second nuclear import rate for NFAT4, which was considerably faster than the 5 minutes observed for NFAT1. Correspondingly, nuclear export rates of NFAT4 were 3 minutes and 32 minutes for NFAT1. When the NFAT dephosphorylation signal was terminated with cyclosporine, a calcineurin phosphatase inhibitor, similar (fast) export times were observed for both isoforms. This observation is consistent with NFAT molecules constantly shuttling between the nucleus and cytoplasm during signal activation, with the net accumulation of NFAT molecules in the nucleus determining the amplitude of the response.
Finally, Yissachar et al. established that NFAT1 accumulation intensifies in response to an increase in either the activating signal or the pulse duration. By contrast, the same magnitude of response was observed for NFAT4 irrespective of treatment, although each cell responded in an all-or-none fashion. For low antigen levels and short pulses, only 10% of the cell population responded, whereas long pulses increased this proportion to 65%, owing to more cells having an opportunity to respond when the signal persisted. Another recent study similarly showed a differential outcome on the activation of gene transcription between pulsed and sustained signaling of p53 [8]. In that study, short p53 pulses delayed the activation of genes regulating cell senescence and proliferation, while prolonged pulses accelerated the expression of senescence- and apoptosis-related genes (Figure 1d).
Perspectives
Yissachar et al. demonstrate that the mode of transcriptional activation can vary according to a transcription factor's nucleo-cytoplasmic oscillations, and to its sensitivity to the intensity and duration of an activating signal. Having established that these variations occur even between highly similar transcription factor isoforms, leading them to respond quite differently to the same signal transduction pathway disseminating through the cell, it now remains to be seen how these diverse outputs translate to changes in the dynamic make-up of the transcriptome. For instance, do transcription factor bursts in the nucleus result in stochastic gene expression events? Is the number, frequency or length of transcription factor pulses exploited by the cell to increase mRNA expression levels, or does a gene perhaps become desensitized to the signal after its initial expression takes place? By combining live-cell systems for examining transcription factor dynamics with those that are capable of tracking transcriptional output in real time [9,10], it will be possible to comprehensively visualize and analyze the complete signaling transcription pathway.
Box 1. Transcription factor dynamics: unanswered questions
Crucial questions regarding transcription factor dynamics in the context of signaling pathways remain unanswered, even after decades of biochemical analysis. Questions to be explored include:
• Is a single stimulation pulse sufficient for the activation of a particular signaling pathway and for the realization of the ultimate transcriptional response, or are several repeated inputs required?
• Are cells sensitive to different signal intensities, or are they instead responsive to the frequencies at which signals arrive?
If we are to understand how cells integrate the information arriving at the cell surface into a physiologically meaningful transcriptional response, we will need to decipher the kinetics of the signaling response. A fruitful approach to addressing these questions is proving to be the visualization of transcription factors in action at the single-cell level.
Abbreviations
Fc: crystallisable fragment (of an antibody); GFP: green fluorescent protein; IgE: immunoglobulin E; Mdm2: Mouse double minute 2 homolog; NFAT: nuclear factor of activated T-cells; NF-κB: nuclear factor-κB; TNF: tumor necrosis factor.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Alon Kalo, Email: alonkalo@gmail.com.
Yaron Shav-Tal, Email: Yaron.Shav-Tal@biu.ac.il.
Acknowledgements
This work was supported by a European Research Council grant and a Union for International Cancer Control grant to YST.
References
- Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;13:4–6. doi: 10.1016/S0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- Yissachar N, Sharar Fischler T, Cohen AA, Reich-Zeliger S, Russ D, Shifrut E, Porat Z, Friedman N. Dynamic Response Diversity of NFAT Isoforms in Individual Living Cells. Mol Cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Oscillations in NF-κB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Turner DA, Paszek P, Woodcock DJ, Nelson DE, Horton CA, Wang Y, Spiller DG, Rand DA, White MR, Harper CV. Physiological levels of TNFα stimulation induce stochastic dynamics of NF-κB responses in single living cells. J Cell Sci. 2010;123:2834–2843. doi: 10.1242/jcs.069641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzer M, Schafer A, Borgmann S, Grabbe S, Zanker KS, Brocker EB, Kampgen E, Friedl P. Antigen presentation in extracellular matrix: interactions of T cells with dendritic cells are dynamic, short lived, and sequential. Immunity. 2000;13:323–332. doi: 10.1016/S1074-7613(00)00032-7. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7:631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]


