Abstract
A recent study investigates the in vitro DNA binding behavior of PRDM9, a zinc finger protein involved in the localization of recombination hotspots in mammals.
Please see related research article: http://genomebiology.com/2013/14/4/R35
Keywords: DNA binding, hotspots, meiotic recombination, PRDM9, zinc finger
During meiosis, homologous chromosomes need to be paired and to segregate properly in order to ensure the transmission of a correct number of chromosomes per gamete. The search for homology is initiated by the deliberate placement of DNA double-strand breaks along the chromosomes, followed by the rejoining of DNA segments. This process, referred to as meiotic recombination, is both a key source of genetic variation, creating new combination of alleles in the population, as well as an important source of genomic instability, responsible for numerous human diseases. Understanding how the locations of double-strand breaks are determined and how they lead to the shuffling of DNA therefore carries important implications for molecular and evolutionary biology, as well as for medical genetics.
In humans, as in many mammals, recombination events tend to be clustered in small regions of the genome called hotspots. Unexpectedly, given the importance of this process, the location and intensity of hotspots has been shown to vary among humans and between mice strains. Furthermore, hotspots are not shared between closely related species such as humans and chimpanzees, indicating a rapid turnover in their locations.
In the last few years, exciting work from a variety of disciplines has uncovered the central role of the PR domain zinc finger protein 9 (PRDM9) in specifying the location of most hotspots genome-wide. This protein binds specific DNA sequences through its zinc finger domain and tri-methylates histone H3 on lysine 4 (H3K4me3) through its SET domain, which somehow results in the recruitment of the recombination machinery. The PRDM9 zinc finger array is highly variable within and between species, both in terms of the number and identity of its zinc fingers; in humans and mice, this variability leads to differences in the locations and intensity of hotspots. How specific DNA sequences are recognized by the zinc finger domain of PRDM9 is therefore a central question for our understanding of how recombination events are specified. The relationship between PRDM9 variants and the bound motifs appears to be rather complex, however, and much remains to be understood about exactly how DNA recognition is achieved. A recent study by Billings et al. [1] addressed these questions experimentally by expressing PRDM9 in Escherichia coli. The protein retains its trimethylating activity in vitro, and its binding behavior seems to recapitulate in vivo activity in mice (that is, the protein variants bind in vitro only to the hotspots they are known to activate in vivo), therefore enabling a detailed dissection of the interaction between the mouse protein variants and the DNA sequence.
How many zinc fingers are required to bind DNA?
Billings et al. examined the minimum length of segments bound by PRDM9 at previously known hotspots using gel-shift assays, focusing on three hotspots bound by the PRDM9Cst variant (present in the CAST/Eij mouse strain) and one bound by the PRDM9Dom2 variant (present in the C57BL/6J mouse strain) [1]. PRDM9Cst has 11 fingers and therefore can bind up to 33 base pairs (bp), whereas PRDM9Dom2 has 12 fingers and hence can bind up to 36 bp. The authors found that the four hotspots (pre B cell leukemia homeobox 1 (Pbx1), H2.0-like homeobox (Hlx1), estrogen-related receptor gamma (Esrrg-1) and proteasome (prosome, macropain) subunit, beta type, 9 (Psmb9)) present a minimum-binding site between 30 and 34 bp, suggesting that PRDM9 uses all its zinc fingers to bind DNA. Importantly, as the authors note, the use of the full complement of PRDM9's zinc fingers for binding suggests that PRDM9 binds continuously to DNA for more than one helical turn, a conclusion further supported by the finding that binding is inhibited by the addition of Mg2+.
At first glance, this result seems to contrast with previous computational analyses that found a hotspot consensus motif matching only to the second half of PRDM9's zinc finger domain (Figure 1) [2-4]. However, in humans, it was reported that bases flanking the central motif and extending up to 50 bp are significantly over-represented in hotspots, relative to matched coldspots, and show a threefold periodicity, indicative of zinc finger binding [3]. This is consistent with the conclusion that all fingers of PRDM9 are used for binding, therefore recognizing a longer motif than the consensus one.
Figure 1.
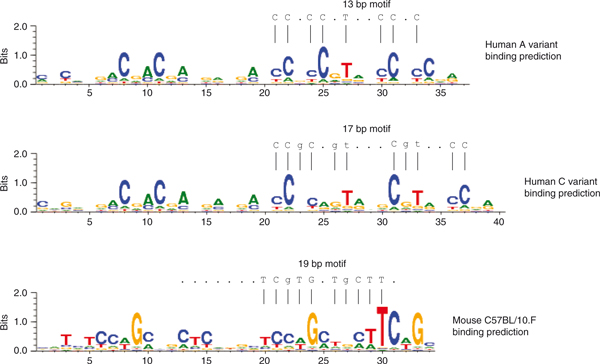
PRDM9 binding predictions and hotspot consensus motifs in humans (A and C variant [3,4]) and mice (C57BL/10.F strain [2]). Binding predictions were obtained from the polynomial model described by Persikov et al. [9].
For one hotspot (Hlx1), Billings et al. further mutated each of the 31 positions of the binding site. This revealed great variability in specificity among bases, with high specificity for those matching the first half of the zinc finger domain (especially the fingers 4 to 6). On that basis, they propose that the other, less specific fingers are used to stabilize the protein-DNA complex.
A mixture of different motifs?
Billings et al. then compare the sequences bound by PRDM9Cst at the three hotspots analyzed, finding that the three sequences have little in common, even though they are able to compete with each other for binding to PRDM9 in gel-shift assays. Furthermore, the few matches seem equally distributed among different fingers. One interpretation is that PRDM9 is able to bind to a mixture of different motifs, thus resolving the apparent paradox that PRDM9 is at the same time very permissive (in that it can bind to degenerate versions of the consensus motif) and highly sensitive (in that specific mutations can completely knock down hotspot activity [5]). This hypothesis would also explain why, in humans, PRDM9 was shown to influence both hotspots containing and lacking an exact match to the consensus motif [6,7], and would further suggest that there is a greater variety of target sequences in Western chimpanzees, where no consensus motif could be found [8], than in humans. More generally, these results raise the question of how many distinct motifs coexist for PRDM9 in humans and other species.
As the authors state, the intriguing results of this study raise as many questions as they answer. Notably, the study reports that instances of the same zinc fingers repeated along the protein do not share the same DNA specificity, and so the protein-DNA interaction seems to be highly context-dependent. The source of this dependence, however, remains unclear, highlighting that we still have a limited understanding of the behavior of long zinc fingers, or perhaps that PRDM9 presents unusual features.
To deepen our understanding of this enigmatic protein, we need to know where it binds in vivo genome-wide (for example, through PRDM9 ChIP-seq data), and to compare these locations with those of double-strand breaks (for example, as identified in mice through ChIP-seq of disrupted meiotic cDNA 1 homolog (DMC1) by Smagulova et al. [2]). It would also be helpful to characterize how PRDM9 binding is affected by chromatin organization or by the presence of other co-factors. Such analyses would help to understand why, even though there are hundreds of thousands of motifs for any version of PRDM9, only a small subset yields double-strand breaks. In turn, an answer to this question may help to understand what constraints restrict the state space of possible motifs to which PRDM9 could bind. In the longer term, the goal is to be able to predict, for a given variant, where double-strand breaks will occur. Such a complete understanding may be of practical use in engineering specific breaks in the genome and, regardless, will yield important insights into evolutionary biology and human genetics.
Abbreviations
bp: base pair; ChIP-seq: chromatin immunoprecipitation followed by high throughput sequencing; PRDM9: PR domain zinc finger protein 9.
Competing interests
The author declares that they have no competing interests.
Acknowledgements
I would like to thank Molly Przeworski for helpful discussions and comments on this manuscript.
References
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, Petkov PM. DNA binding specificities of the long zinc finger recombination protein PRDM9. Genome Biol. 2013;4:R35. doi: 10.1186/gb-2013-14-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, Aldrich MC, Ambrosone CB, Amos C, Bandera EV, Berndt SI, Bernstein L, Blot WJ, Bock CH, Boerwinkle E, Cai Q, Caporaso N, Casey G, Cupples LA, Deming SL, Diver WR, Divers J, Fornage M, Gillanders EM, Glessner J, Harris CC. et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci USA. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, Humburg P, Iqbal Z, Lunter G, Maller J, Hernandez RD, Melton C, Venkat A, Nobrega MA, Bontrop R, Myers S, Donnelly P, Przeworski M, McVean G. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Osada R, Singh M. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics. 2009;25:22–29. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]


