Abstract
The majority of people in the developed world spend more than 90% of their lives indoors. Here, we examine our understanding of the bacteria that co-inhabit our artificial world and how they might influence human health.
The need for culture-independent analysis of microbial diversity
The global trend towards industrialization and urbanization has led to ever more people living and working indoors. Some studies estimate that humans in industrialized countries spend as much as 90% of their lives inside [1,2]. They are born in hospitals, raised in homes or apartments, work in office buildings or factories and move to nursing homes in their old age. This trend towards indoor living is also occurring in developing nations. Thirty years ago, the Chinese city of Shenzhen was a small city of 300,000 inhabitants. Since then, it has grown into a massive industrial megalopolis housing over 10 million people, many of whom have moved there from rural areas [3]. Thus, for billions of humans, the built environment (BE) now represents the modern ecological habitat of Homo sapiens sapiens.
In the process of creating BEs, we have also inadvertently created an incredible diversity of microhabitats that have been colonized by thousands of different microorganisms. Indeed, BEs contain many potential novel habitats for microbial life that have chemical and physical properties not found in the natural world. Humans convey a large fraction of the colonizing microbes into these habitats by shedding them from their bodies or transporting them by means of their clothes and shoes, whereas the rest originate from water or other environmental sources (for example, soil) [4-7]. Depending on the environment, microbes can also be transported indoors by pets, insects and other animals [8].
While culture-based studies have found microbes to be ubiquitous in the BE, recent culture-independent molecular studies have shown that our understanding of indoor microbial diversity remains extremely sparse [9-13]. These molecular methods are also an integral part of the tool kit for expanding our understanding of what species of microorganisms live in these ecosystems, how they function and how they migrate among humans, physical surfaces and other environments [6,14,15].
The most commonly used molecular approach for exploring microbial diversity is based on the PCR amplification and sequencing of genes encoding small subunit ribosomal RNA (16S rRNA) directly from environmental sources [16]. In this approach, DNA is directly extracted from all the microbial cells present in a sample and 'universal' PCR primers designed from conserved regions of the 16S rRNA gene are used to amplify this gene from all the microbial genomes in the sample [16,17]. This approach readily determines several orders of magnitude more microbial diversity than culture-based methods, often discovering novel organisms with no known growth conditions, and has dramatically altered our understanding of global microbial diversity [14]. Although powerful, there are two obvious sources of bias in this method. First, different DNA extraction techniques allow access to different parts of the microbial diversity to varying extents, through uneven cell lysis [18,19]. Second, there are no truly universal PCR primers, with even the most comprehensive pair covering approximately 85% of the known taxa [20]. Despite these limitations, this technology, when combined with a rigorous experimental design, is extremely useful for characterizing the dynamics and ecological patterns of microbial ecosystems [21,22]. The initial 16S rRNA diversity studies performed in the BE cloned PCR amplicons into plasmid vectors before transformation, colony isolation and sequencing [9,10,12]. However, the application of direct emulsion-PCR-based sequencing in 2006 [23] dramatically increased the depth and breadth of analysis; similar technologies now enable the simultaneous processing of thousands of samples at a depth of hundreds of thousands of sequence reads each [24,25].
Here, we review the literature on culture-independent analysis of microbial diversity in the BE and explore what it tells us about the dynamics and distribution of bacteria and fungi in these environments. So, why is the BE even worth exploring? First, as already stated, the BE is the primary habitat for a large and growing percentage of humanity, whose health might depend upon our firm comprehension of the interaction between humans and the microbiology of buildings. Second, the climate, materials and design of artificial environments could have unexpected and interesting consequences for the selection and growth of microbes that might help us to design healthier buildings. Finally, a deeper understanding of indoor microbial diversity should help inform public health policy, particularly in settings with many immune-compromised individuals (for example, hospitals, intensive care units and nursing homes). The renewed interest in microbial ecology, and the application of next-generation sequencing technology, presents exciting opportunities to uncover hidden diversity and interactions in these environments. As we will show, these methods have already revealed the BE to be a surprisingly complex and highly dynamic microbial ecosystem that collects and selects for a remarkable array of novel diversity.
Unnatural selection
It goes without saying that the BE contains chemical compounds and physical conditions entirely unlike the natural world where microbes evolved. Drywall, floor wax, polyester, carpeting, constant temperature and humidity, novel hydrocarbons and polymers - these are just some of the many features found in BEs that potentially provide unusual growth conditions and niches for bacteria and fungi. While natural microbial evolution might not have originally selected for phenotypes able to exploit these conditions or materials, the incredible diversity and adaptability of the microbial world means that there probably exists a microbe that will survive on almost any BE surface or condition. Moreover, the short generation times and rapid evolutionary mechanisms (for example, horizontal gene transfer) found in the microbial world mean that many microbes might have already adapted to the BE over the past few hundred years.
Culture-based studies have repeatedly discovered conditions in the BE that enrich for particular bacteria and fungi [26-28]. However, while culture isolation focuses on the presence of particular bacteria or fungi, molecular studies of BEs typically uncover polymicrobial communities comprising hundreds or thousands of uncultured microbes. When conditions in the BE enrich strongly for particular traits, these polymicrobial communities tend to be dominated by a few microbial groups (Table 1). For instance, an analysis of bacterial 16S rRNA sequence diversity in hospital therapy pools and showerheads found they enriched for diverse, primarily uncultured, soil-dwelling Mycobacteria, as well as many Proteobacteria and other, rarer groups [9,29]. Shower curtains tend to be dominated by dozens of different Sphingomonas and Methylobacterium species, many of them uncultured [10]. The latter are particularly interesting as some Methylobacterium species have been shown to grow on an enormous diversity of different carbon compounds (that is, C2, C3 and C4 compounds) [30], and some species thrive on the decay products of human skin cells [31]. Studies of showerheads and hospital shower stalls find them, like therapy pools, to be hospitable environments for Actinobacteria and Proteobacteria [29,32]. Other highly selective environments include the tubing of catheters [33,34], which maintain a significantly different microbial community internally compared with externally, and NASA clean-rooms used for spacecraft assembly [35,36]. In the latter case, the extensive sealing and sterilization protocols appeared to select for only the hardiest of microbes that also survive space journeys [35].
Table 1.
Signature microbes of various built environments
| Site | Bacterial species |
|---|---|
| Office | Streptococcus spp. |
| Corynebacterium spp. | |
| Flavimonas spp. | |
| Lactobacillus spp. | |
| Burkholderia spp. | |
| Bacillus spp. | |
| Bradyrhizobium spp. | |
| Neonatal intensive care unit | Propionibacterium spp. |
| Enterobacter spp. | |
| Neisseria spp. | |
| Pseudomonas spp. | |
| Streptococcus spp. | |
| Staphylococcus spp. | |
| Aquarium filter | Nictrospumilus spp. (archaea) |
| Nitrosospira spp. | |
| Nitrosomonas spp. | |
| Hospital air | Kytococcus sedentarius |
| Staphylococcus epidermidis | |
| S. haemolyticus | |
| Ralstonia pickettii | |
| Enterobacter spp. | |
| Kocuria rhizophila | |
| Micrococcus luteus | |
| Microcystis aeruginosa | |
| Prochlorococcus marinus | |
| Methylocella silvestris | |
| Methylobacterium extorquens | |
| Catheter | Pseudomonas spp. |
| Staphylococcus spidermidis | |
| Enterococcus faecalis | |
| Klebsiella spp. | |
| Restroom | Propionibacterium spp. |
| Corynebacterium spp. | |
| Micrococcaceae | |
| Streptococcus spp. | |
| Staphylococcus spp. | |
| Bacteriodaceae | |
| Shower curtain | Methylobacterium spp. |
| Sphingomonas spp. | |
| Showerhead | Mycobacterium spp. |
| Pseudomonas aeruginosa | |
| Legionella spp. | |
| Therapy pool | Mycobacterium spp. |
| Sphingomonas spp. | |
| Classroom | Propionibacterineae |
| Xanthomonadaceae | |
| Micrococcineae | |
| Sphingomonas spp. | |
| Caenibacterium | |
| Staphylococcus spp. | |
| Enterobacteriaceae | |
| Corynebacterineae | |
Colonization and dispersal within the built environment
Molecular studies have also enhanced our understanding of how indoor microbes colonize surfaces and are transmitted between microenvironments. Studies of keyboards [37], restrooms [4] and offices [5] show that humans are the main source of the microbial diversity in many indoor environments and are also its principle vector. Perhaps unsurprisingly, a great number of surface microbes can be traced back to human skin, although gut and nasal/oral environments also contribute [5,6]. Molecular studies of bacterial diversity in offices [5,12], hospitals [11], airplanes [7,38], kitchens [39] and restrooms [4] have consistently found a predominance of human skin-associated, and gastrointestinal or urogenital-associated, bacteria on most physical surfaces. In fact, Tringe and colleagues showed that the bacteria living on the skin of individuals inhabiting a particular space define the microbial community observed in air samples taken from that same space [13]. Given that the average human sheds approximately 1.5 million skin cells an hour, carrying with them approximately 15 million bacterial cells [40], humans clearly contribute substantially, and perhaps predominantly, to indoor microbial diversity.
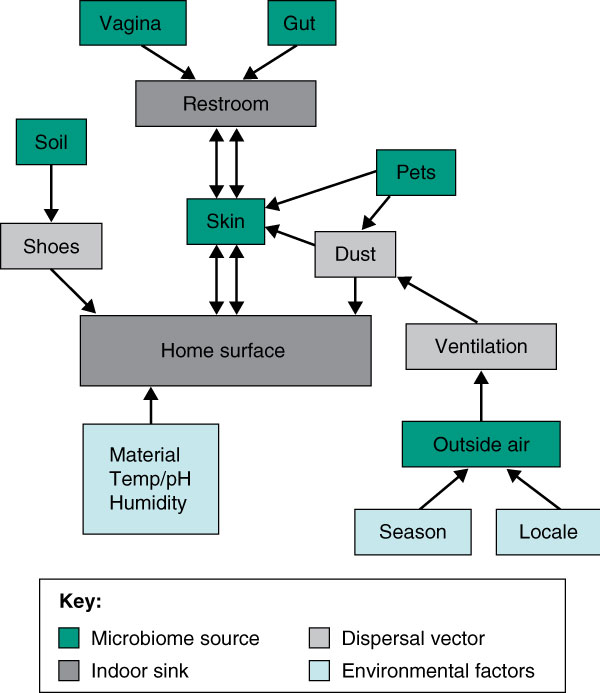
Direct transfer of skin microbiota through surface contact has also been shown to have a considerable and area-specific impact. Depending on the surface, skin contact can transfer millions of microbial cells in each event [41], leading to the rapid and continual inoculation and re-inoculation of surfaces touched repeatedly, such as light switches, door handles, keyboards, soap dispensers and toilet seats [4,5,37]. Furthermore, a number of studies have demonstrated that the dominant microbiota associated with human skin differs dramatically among individuals [42-46]. Indeed, it might be possible to identify people based on the unique microbial signature they leave behind in a room or on a surface, which has significant potential applications in forensics. Figure 1 diagrams various sources and transmission routes of hypothetical BE microbial communities and indicates how they might contribute to the microbial ecology of indoor surfaces.
Figure 1.
Illustration showing the various sources and transmission routes of hypothetical built environment microbial communities. Also indicated is how these communities might contribute to the microbial ecology of indoor surfaces, as well as various abiotic factors (for example, temperature) that may influence microbial diversity.
Despite the evidence suggesting that humans interact microbially with their indoor spaces, the mechanistic relationship enabling this interaction remains poorly characterized. As the BE can also be a source of microbes that colonize people, it is essential that we investigate 'who lives where' in the BE. Thus far, the literature has primarily explored the phenomenon of fungal contamination on damp surfaces [40,47-49]; the role of hygiene in removing microbial communities [46,50]; and the length of time microbes can survive on surfaces [51]. The microbial diversity of dust has also been explored [12,52,53], as has the diversity of indoor air [11,13,54]. One study investigated the temporal succession of microbial communities performed on indoor dust and found building-specific seasonal patterns that were a combination of skin cells shed from inhabitants within the buildings and seasonally defined microbiota from the external environment [12].
In addition to human skin, other aspects of the human microbiome can also contribute significantly to the BE microbiome, although the contribution tends to be more situation specific. As might be expected, human gut and vaginal flora play a major role in shaping the microbial communities associated with surfaces in restrooms [4], daycare centers [55] and neonatal intensive care units [56]. Although few molecular studies have directly investigated the impact of pets, it is expected that animal skin, hair, fecal material (for example, cat litter), saliva and possibly fleas all contribute significantly to microbial communities associated with indoor surfaces and air [1,8]. The impact of fish tanks on the BE microbiome might also be worth considering as these environments have a unique microbial consortia [57,58]. Understanding the contribution of animal inhabitants might be particularly helpful in understanding the causes of childhood asthma [59].
Most indoor settings also obtain a considerable fraction of their microbiota from environmental sources. Dust samples in offices consistently uncovered bacteria associated with soil and the plant rhizosphere [5]. The amount and type appears to depend on the degree of outdoor ventilation and the local climate. Kembel and colleagues found significant differences in the contribution of natural environmental bacteria in buildings that were well ventilated versus those with sealed windows [11]. Rintala and co-workers showed dramatic changes in dust microflora according to different seasons [12], while Hewitt and colleagues found a strong climatic signal in the office dust of buildings in Tucson, Arizona (desert soil bacteria), compared with dust in New York and San Francisco [5]. The restroom study of Flores et al. also found soil bacteria over the restroom floor that was probably transferred from shoes [4].
Clearly, BE microbial communities comprise microbes from a diverse array of human and environmental habitats. The ability to compare vast metagenomic datasets of either ribosomal sequences or whole microbial genome fragments from BE sinks with environmental, animal and human source communities, and the application of bioinformatic tools that estimate the relative contribution of various sources to communities (for example, SourceTracker [6]), will be instrumental in understanding microbial colonization and transmission in the BE.
Open questions and future directions
The molecular studies cited in this review have just begun to inform and advance the understanding of microbial diversity in the BE. As the methods improve in terms of cost, speed and depth, the experimental design possibilities grow proportionally. Analyzing thousands of samples simultaneously provides a much broader assessment of the role humans and environmental sources play in the development and persistence of the indoor microbiome. These 'faster, better, cheaper' methods will also allow more sophisticated statistical design and lengthy time-series analyses.
Proper study designs and increased sequencing power will make it possible to understand how BEs are colonized, how microbes spread through these environments, and the rate at which microbial communities change in the BE. For example, a recent study funded by the Alfred P Sloan Foundation plans to investigate all these factors in a newly built hospital as it becomes occupied with staff and patients [60]. The relatively low cost of sample processing and sequencing means that this study is examining >15,000 surfaces (both human and built), with daily (35 samples a day) collection for an entire year, providing unparalleled resolution for investigating colonization and transmission of bacteria, fungi and viruses in the hospital infrastructure. This study aims to identify the microbial streets and highways that enable rapid transmission of bacteria through this ecosystem, and it is expected that pathogens will also use these routes. Because the human microbiome contributes the majority of the BE microbes, and humans also probably vector a significant proportion of soil and other environmental microbes through shoes, hands and clothes (for example, see [60]), future studies need to follow and sample the human inhabitants as they enter and move through the BE over time, but to do this it will be necessary to define the rate of change in the microbiome to sample at the appropriate temporal and spatial resolution.
A number of other key questions concerning the BE will need to be addressed, such as the role of surface materials, ventilation, local climatic conditions and the physical conditions of the building in shaping the microbial diversity. Answers to these questions will require thorough and systematic sampling, rigorous statistical design and collaboration with experts in building science and engineering. The issue of ventilation and airflow appears to deserve special attention as these affect the rate and direction of bioaerosol transmission to sensitive respiratory mucosal surfaces. Studies by Kembel et al. [11] and Poza et al. [61] provide tantalizing evidence that the amount of external air makes a significant difference in the microbial diversity in a hospital setting, perhaps even affecting overall pathogen loads, and the usage of different environments (for example, operating room versus waiting room) also impacts the microbial profiles of these spaces. Collaborations between microbiologists and building scientists will be especially important for understanding the impact of materials and machine-aided ventilation, especially given their variability and complexity between different BEs.
Thus far, the molecular studies of the BE have primarily investigated bacterial diversity through the sequencing of community 16S rRNA gene amplicons. The advantages of using this gene for surveying microbial diversity are well known [62]. However, the limits of amplicon sequencing for both diversity and functional studies are also familiar, including PCR bias [63], variable copy numbers per genome [64], limited taxonomic resolution [65] and indirect functional knowledge (that is, inference of community function through phylogenetic relationships). Higher throughput and sensitivity (for example, lower DNA concentrations required to generate sequence data), lower sequencing costs and advances in computation should enable the future application of shotgun metagenomics (that is, sequencing of all genomic DNA in a given sample) and whole-genome sequencing in the indoor environment [66,67]. This will allow the simultaneous analysis of microbial diversity, including deeper taxonomic resolution (strains and subspecies) and more direct functional analysis at the genomic level. For example, the Hospital Microbiome Project is implementing Illumina Nextera library generation to sequencing thousands of microbial and viral metagenomes. Deeper taxonomic resolution and determination of gene function will be important for verifying pathogens in indoor settings by allowing the differentiation between commensals and pathogens (for example, Escherichia coli K12 versus E. coli OH157:H7) and for detecting pathogenic islands or antibiotic resistance markers on chromosomal and extra-chromosomal elements. Table 2 compares and contrasts shotgun metagenomics and other methods (amplicon sequencing, metatranscriptomics, metaproteomics and metabolomics) used in microbial community research that could potentially be applied to the BE.
Table 2.
Comparison of different techniques, including pros and cons, for analysis of microbial communities
| Technique | Description | Advantages | Disadvantages |
|---|---|---|---|
| Amplicon sequencing | The amplification and sequencing of a single gene from a broad selection of the microbiome. Traditionally applied to 16S rRNA for bacteria, but now being applied to a wide range of targets | Primarily cost and depth of analysis. Amplicon sequencing is very cheap and enables a rapid and deep characterization of the varying structure of microbial life under changing environmental gradients | This approach provides a narrow field of view, targeting a single gene, be it taxonomic or functionally informative, and only gives information about that one gene. It also can be affected by primer and amplification biases |
| Genomic sequencing | Sequencing of the genome of representatives of a community, ideally resulting in a single sequence, but more often resulting in 100s of genomic fragments | The genome enables a defined link between potential function and phylogeny, so that one can deduce that species x performs process y. When linked to a cultured cell, it can also be used to define gene function through targeted biochemical tests | Throughput is a problem. Sequencing the genome of isolated organisms has become rudimentary, but few can be isolated. Screening sorted cells from a community, followed by sequencing, is becoming viable, but often results in limited coverage of the genome owing to amplification bias |
| Metagenomic sequencing | Sequencing of a random sample of the genomic DNA from the cells of a microbial consortium | This technique enables broad observation of the taxonomic and functional genes from an entire community, without the bias associated with amplicon sequencing. With sufficiently deep sequencing, it is also possible to reassemble microbial genomes and other genetic elements | Current sequencing platforms require extensive starting material, although this is changing. Cost can be prohibitive, leading to only shallow characterization of the most dominant microbial taxa. The output only describes potential function, and it is often difficult to link function and phylogeny definitively. Also has the potential to sequence DNA from dead cells |
| Metatranscriptomic sequencing | Random sequencing of the messenger, small and other RNAs from a microbial community that define the mechanism and response of microbial gene expression | As with metagenomics, this technique enables broad taxonomic and functional characterization, but of expressed genes, which enables deeper analysis of the community and targets the active members of the community. Sequence data can be mapped to known genomes to help identify phylogenetic-specific functional responses | Cost is prohibitive as the steps required to remove the 90-95% of ribosomal RNA to enable deeper characterization of the mRNA are expensive and time consuming, which also limits throughput. RNA is sensitive to degradation, and the half-life of mRNA is very short, which creates biases from sampling the community. |
| Metaproteomic sequencing | Random sequencing of the amino acid sequences that represent the protein material in a microbial community | The primary advantage is the ability to identify proteins that have not only been expressed as mRNA but have also been folded and have potentially formed active proteins - for example, enzymes - in a cell. When combined with genomics and metatranscriptomics, it is possible to define protein isoforms and map protein function to phylogeny | The primary disadvantage of cost and throughput, which is still higher than metagenomics or metatranscriptomics. By itself, it is complicated to assign taxonomy |
| Metabolomic sequencing | Random characterization of the metabolic products present in a sample that might have been generated by a microbial community | This is the zenith of microbial activity, and when compared with that of other samples, the relative change in metabolite concentration can explain a lot about the functional consequence of genomic potential, or transcript and protein abundance | As with proteomics, metabolomics is limited by cost and throughput but also by identification of products. It also currently has a limit of detection, with very rare metabolites being hard to detect |
Methods such as shotgun metagenomics will also be necessary to analyze the diversity of viruses (bacteriophage and eukaryotic viral diversity, also known as the 'virome' [68]) in the BE, which is very poorly understood. Bacteriophage, in particular, play a major role in the dynamics of natural microbial populations (predator-prey relationships) [69] and also facilitate the horizontal transfer of crucial genes (for example, toxin and metabolic genes) among bacteria [70]. Models of BE microbial ecology will certainly be incomplete without a parallel understanding of viral community diversity, and understanding the contribution of bacteriophage to bacterial pathogenicity in places such as hospitals could be an extremely important contribution to public health. Recent studies have used deep-sequencing approaches to characterize viral diversity and gene content for bacteriophage [71,72] and eukaryotic viruses [73] in the human microbiome, and similar approaches could be applied in the BE. In fact, we ourselves have been funded by the Sloan Foundation to study viral metagenomics in the BE. Although we have found evidence of viral-like particles (VLPs) in all of our samples, the low biomass on indoor surface biofilms presents a significant challenge to this work, particularly with short-lived and low-abundance eukaryotic viruses.
Molecular studies of fungal community diversity, clearly of great interest in the BE, have also been limited by choice of marker gene and taxonomic breadth of primer amplification [53,74,75]. However, the recent development of novel primers targeting the intergenic spacer region between small- and large-subunit ribosomal RNA genes is being applied to amplify a broader diversity of fungal sequences that could help expand our understanding of BE fungal diversity [75]. Dollive and colleagues have also developed a sorely needed software pipeline for the analysis of eukaryotic 18S rRNA gene sequences [76]. The combination of primers that capture a greater taxonomic diversity and better software tools for analysis should allow greater insight not only into BE fungal diversity but also for a deeper understanding of the interactions between BE fungal and bacterial communities.
One criticism of environmental molecular studies, which is of particular relevance for the BE, is the issue of whether a microbial DNA signature comes from a viable organism. Identification of a microbe through DNA sequencing only verifies that the particular organism was present at some time, but it might no longer be viable, especially considering the extreme conditions present on most surfaces (for example, extremely low humidity, limited nutrients, UV exposure). Understanding which of the microbes in a community persist in a viable state will be especially important for verifying their pathogenic potential. While culturing remains the standard for establishing viability, it is limited in that most organisms are recalcitrant to culture. Therefore, it is essential that we develop methods for both the in situ radiolabeling of active organisms and the direct sequencing of rRNA and mRNA from microbial communities. One of the common problems in bacteria transcriptomics is the removal of ribosomal RNA, which comprises 90 to 95% of the total RNA. In eukaryotic cells, the mRNA has a poly-A tail, which can be used to selectively isolate these sequences. Therefore, since 2008, many methods have been developed to remove rRNA and improve the enrichment of bacterial mRNA in community samples (for example, [77,78]). Indeed, the development of rapid rRNA removal and mRNA enrichment tools has led to these methods being applied even in low-biomass environments, such as subsections of human intestinal epithelium tissue, which now put these techniques in reach of researchers working with BE samples [79]. RNA is rapidly degraded in the environment, and its presence suggests the existence of active cells [77]. However, the low biomass on most surfaces will require innovative strategies to reach the nucleic acid quantities required for most sequencing platforms (that is, 0.5 to 1 µg). The development of novel extraction techniques (for example, MoBio power kits) and sequencing library preparations (for example, Nextera-Illumina, which requires 1 to 50 ng of starting material) means that a solution is potentially imminent.
Concluding remarks
Studies of the unseen majority inside the BE were initiated to determine how bacteria and fungi might affect human health long after it was realized that spending time indoors, away from fresh air, might increase rates of acute and chronic illness. In recent years, it has become apparent that studying the BE might also allow us to better interrogate the ecological interactions between microorganisms. This means that our artificial environments, in and of themselves, have become a new ecosystem worthy of significant investigation. Research focused on the BE has also allowed us to elucidate how microbes interact in this space, and it is, in turn, enabling us with the help of new technologies to understand better how humans interact with the microbiology of indoor spaces. Recent discoveries in the microbiome of the BE, facilitated by enlightened investment, show us that we share our living spaces with an immense diversity of microbial life, some potentially bad, others good, and most just along for the ride. As the resource and personnel costs of performing high-throughput microbiological studies decrease, it will be possible to perform ever-more-detailed characterization of the rate of transmission of microbes between the BE and ourselves and of the metabolic consequences of sharing our living and working spaces with the invisible biosphere. Without a doubt, this new field of research will lead to discoveries that will change the way that we build, clean and live in our new ecosystem.
Competing interests
The authors declare that they have no competing interests
Contributor Information
Scott T Kelley, Email: skelley@mail.sdsu.edu.
Jack A Gilbert, Email: gilbertjack@anl.gov.
Acknowledgements
This work was supported in part by a grant from the Alfred P Sloan Foundation and the US Department of Energy under Contract DE-AC02-06CH11357.
References
- Custovic A, Taggart SC, Woodcock A. House dust mite and cat allergen in different indoor environments. Clin Exp Allergy. 1994;24:1164–1168. doi: 10.1111/j.1365-2222.1994.tb03323.x. [DOI] [PubMed] [Google Scholar]
- Hoppe P, Martinac I. Indoor climate and air quality. Review of current and future topics in the field of ISB study group 10. Int J Biometeorol. 1998;42:1–7. doi: 10.1007/s004840050075. [DOI] [PubMed] [Google Scholar]
- Shen J. Urban Growth and Sustainable Development in Shenzhen City. Open Environ J. 2008;2:71–79. [Google Scholar]
- Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. Microbial biogeography of public restroom surfaces. PLoS ONE. 2011;6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. Office space bacterial abundance and diversity in three metropolitan areas. PLoS ONE. 2012;7:e37849. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, Piceno YM, Tom LM, Desantis TZ, Jones BW, Andersen GL, Hwang GM. Bacterial communities in commercial aircraft high-efficiency particulate air (HEPA) filters assessed by PhyloChip analysis. Indoor Air. 2013;23:50–61. doi: 10.1111/j.1600-0668.2012.00787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, Zoratti EM, Woodcroft KJ, Bobbitt KR, Wegienka G, Boushey HA, Lynch SV. Man's best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126:410–412. doi: 10.1016/j.jaci.2010.05.042. 412 e411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent LT, Kelley ST, St Amand A, Pace NR, Hernandez MT. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc Natl Acad Sci USA. 2005;102:4860–4865. doi: 10.1073/pnas.0501235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley ST, Theisen U, Angenent LT, St Amand A, Pace NR. Molecular analysis of shower curtain biofilm microbes. Appl Environ Microbiol. 2004;70:4187–4192. doi: 10.1128/AEM.70.7.4187-4192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Jones E, Kilne J, Northcutt D, Stenson J, Womack AM, Bohannan BJM, Brown GZ, Green JL. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala H, Pitkaeranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Zhang T, Liu X, Yu Y, Lee WH, Yap J, Yao F, Suan ST, Ing SK, Haynes M, Rohwer F, Wei CL, Tan P, Bristow J, Rubin EM, Ruan Y. The airborne metagenome in an indoor urban environment. PLoS ONE. 2008;3:e1862. doi: 10.1371/journal.pone.0001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA, Le Paslier D, Simone P, Vogel TM. Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J. 2012;6:1677–1687. doi: 10.1038/ismej.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, Hirsch PR, Vogel TM. Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol. 2011;77:1315–1324. doi: 10.1128/AEM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Bizzoco RW, Ellis DG, Lipson DA, Poole AW, Levine R, Kelley ST. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl Environ Microbiol. 2007;73:2612–2623. doi: 10.1128/AEM.02567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, Bailey MJ, Gordon JI, Kowalchuk GA, Gilbert JA. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaissie EJ, Penzak SR, Dignani MC. The hospital water supply as a source of nosocomial infections: a plea for action. Arch Intern Med. 2002;162:1483–1492. doi: 10.1001/archinte.162.13.1483. [DOI] [PubMed] [Google Scholar]
- Casini B, Valentini P, Baggiani A, Torracca F, Frateschi S, Nelli LC, Privitera G. Molecular epidemiology of Legionella pneumophila serogroup 1 isolates following long-term chlorine dioxide treatment in a university hospital water system. J Hosp Infect. 2008;69:141–147. doi: 10.1016/j.jhin.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Szewzyk U, Szewzyk R, Manz W, Schleifer KH. Microbiological safety of drinking water. Annu Rev Microbiol. 2000;54:81–127. doi: 10.1146/annurev.micro.54.1.81. [DOI] [PubMed] [Google Scholar]
- Perkins SD, Mayfield J, Fraser V, Angenent LT. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl Environ Microbiol. 2009;75:5363–5372. doi: 10.1128/AEM.00658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom ME, Chistoserdova L. Plants in the pink: cytokinin production by methylobacterium. J Bacteriol. 2002;184:1818. doi: 10.1128/JB.184.7.1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti V, Vohra J, Goonetilleka S, McDonald IR, Straubler B, Stackebrandt E, Kelly DP, Wood AP. Molecular detection and isolation of facultatively methylotrophic bacteria, including Methylobacterium podarium sp. nov., from the human foot microflora. Environ Microbiol. 2004;6:820–830. doi: 10.1111/j.1462-2920.2004.00623.x. [DOI] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci USA. 2009;106:16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Wilson SS, St Amand AL, Pace NR. Culture-independent microbiological analysis of foley urinary catheter biofilms. PLoS ONE. 2009;4:e7811. doi: 10.1371/journal.pone.0007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sriprakash KS, McMillan D, Gowardman JR, Patel B, Rickard CM. Microbiological pattern of arterial catheters in the intensive care unit. BMC Microbiol. 2010;10:266. doi: 10.1186/1471-2180-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Duc MT, Vaishampayan P, Nilsson HR, Torok T, Venkateswaran K. Pyrosequencing-derived bacterial, archaeal, and fungal diversity of spacecraft hardware destined for Mars. Appl Environ Microbiol. 2012;78:5912–5922. doi: 10.1128/AEM.01435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73:2600–2611. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Kelley ST. Molecular survey of aeroplane bacterial contamination. J Appl Microbiol. 2005;99:502–508. doi: 10.1111/j.1365-2672.2005.02651.x. [DOI] [PubMed] [Google Scholar]
- Flores GE, Bates ST, Caporaso JG, Lauber CL, Leff JW, Knight R, Fierer N. Diversity, distribution and sources of bacteria in residential kitchens. Environ Microbiol. 2012;15:588–596. doi: 10.1111/1462-2920.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffal AA, Banat IM, ElMogheth AA, Nsanze H, Bener A, Ameen AS. Residential indoor airborne microbial populations in the United Arab Emirates. Environment International. 1997;23:529–533. doi: 10.1016/S0160-4120(97)00055-X. [DOI] [Google Scholar]
- Dawson P, Han I, Cox M, Black C, Simmons L. Residence time and food contact time effects on transfer of Salmonella Typhimurium from tile, wood and carpet: testing the five-second rule. J Appl Microbiol. 2007;102:945–953. doi: 10.1111/j.1365-2672.2006.03171.x. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, Suematsu M, Benno Y. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–1238. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC. NISC Comparative Sequencing Program. Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvarinen A, Meklin T, Vepsalainen A, Nevalainen A. Fungi and actinobacteria in moisture-damaged building materials-concentrations and diversity. Internal Biodeter Biodegrad. 2002;49:27–37. doi: 10.1016/S0964-8305(01)00103-2. [DOI] [Google Scholar]
- Nevalainen A, Seuri M. Of microbes and men. Indoor Air. 2005;15:58–64. doi: 10.1111/j.1600-0668.2005.00344.x. [DOI] [PubMed] [Google Scholar]
- Lignell U, Meklin T, Rintala H, Hyvarinen A, Vepsalainen A, Pekkanen J, Nevalainen A. Evaluation of quantitative PCR and culture methods for detection of house dust fungi and streptomycetes in relation to moisture damage of the house. Lett Appl Microbiol. 2008;47:303–308. doi: 10.1111/j.1472-765X.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- Bright KR, Boone SA, Gerba CP. Occurrence of bacteria and viruses on elementary classroom surfaces and the potential role of classroom hygiene in the spread of infectious diseases. J Sch Nurs. 2010;26:33–41. doi: 10.1177/1059840509354383. [DOI] [PubMed] [Google Scholar]
- Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian A, Larsson L. Characterization of the microbial community in indoor environments: a chemical-analytical approach. Appl Environ Microbiol. 2003;69:3103–3109. doi: 10.1128/AEM.69.6.3103-3109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkaranta M, Meklin T, Hyvarinen A, Paulin L, Auvinen P, Nevalainen A, Rintala H. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl Environ Microbiol. 2008;74:233–244. doi: 10.1128/AEM.00692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen K, Rintala H, Hirvonen MR, Vepsalainen A, Hyvarinen A, Meklin T, Toivola M, Nevalainen A. Indoor air particles and bioaerosols before and after renovation of moisture-damaged buildings: The effect on biological activity and microbial flora. Environ Res. 2008;107:291–298. doi: 10.1016/j.envres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Lee L, Tin S, Kelley ST. Culture-independent analysis of bacterial diversity in a child-care facility. BMC Microbiol. 2007;7:27. doi: 10.1186/1471-2180-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt KM, Mannino FL, Gonzalez A, Chase JH, Caporaso JG, Knight R, Kelley ST. Bacterial diversity in two neonatal intensive care units (NICUs). PLoS ONE. 2013;8:e54703. doi: 10.1371/journal.pone.0054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakawa H, Tajima Y, Numata Y, Tsuneda S. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol. 2008;74:894–900. doi: 10.1128/AEM.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS ONE. 2011;6:e23281. doi: 10.1371/journal.pone.0023281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Palmer MW, Andersen GL, Halonen MJ, Josephson KC, Maier RS, Martinez FD, Neilson JW, Stern DA, Vercelli D, Wright AL. Environmental determinants of and impact on childhood asthma by the bacterial community in household dust. Appl Environ Microbiol. 2010;76:2663–2667. doi: 10.1128/AEM.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital Microbiome. http://www.hospitalmicrobiome.com
- Poza M, Gayoso C, Gomez MJ, Rumbo-Feal S, Tomas M, Aranda J, Fernandez A, Bou G. Exploring bacterial diversity in hospital environments by GS-FLX Titanium pyrosequencing. PLoS ONE. 2012;7:e44105. doi: 10.1371/journal.pone.0044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, Bailey MJ, Gordon JI, Kowalchuk GA, Gilbert JA. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahllof I, Baillie H, Kjelleberg S. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl Environ Microbiol. pp. 3376–3380. [DOI] [PMC free article] [PubMed]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP Jr, Rohwer F, Haynes M. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrmark K, Bodeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandstrom-Durling M, Clemmensen KE, Lindahl BD. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–677. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, Nabel CS, Hill DA, Artis D, Bachman MA, Custers-Allen R, Grunberg S, Wu GD, Lewis JD, Bushman FD. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, Joint I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS ONE. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13:R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]