An analytical approach is presented to identify and resolve several distinct structural species dynamically mixed in multiple crystallographic data sets.
Keywords: dynamic crystallography, structural heterogeneity
Abstract
Dynamic behavior of proteins is critical to their function. X-ray crystallography, a powerful yet mostly static technique, faces inherent challenges in acquiring dynamic information despite decades of effort. Dynamic ‘structural changes’ are often indirectly inferred from ‘structural differences’ by comparing related static structures. In contrast, the direct observation of dynamic structural changes requires the initiation of a biochemical reaction or process in a crystal. Both the direct and the indirect approaches share a common challenge in analysis: how to interpret the structural heterogeneity intrinsic to all dynamic processes. This paper presents a real-space approach to this challenge, in which a suite of analytical methods and tools to identify and refine the mixed structural species present in multiple crystallographic data sets have been developed. These methods have been applied to representative scenarios in dynamic crystallography, and reveal structural information that is otherwise difficult to interpret or inaccessible using conventional methods.
1. Introduction
Structural differences introduced into a protein structure resting in crystal lattices can be observed with the aim of testing hypothesized mechanisms of its function. Comparison of the captured structures before and after a controlled perturbation at atomic resolution provides the major source of dynamic structural information. However, structural heterogeneity is intrinsic to any dynamic process. The presence of mixed structural species, such as those associated with substrate, intermediate and product complexes, often complicates structural interpretation and thus casts doubt on the mechanisms.
This common practice in protein crystallography involves multiple data sets acquired from crystals of chemically modified protein. For example, site-directed mutagenesis, cocrystallization and soaking of native crystals with various reagents (cofactor, substrate, inhibitor or product) are commonly used techniques. In a temperature-scanning cryocrystallographic experiment, many difference electron-density maps as a function of temperature are required (Yang et al., 2011 ▶). Despite different experimental approaches, a common theme emerges: the same data-collection protocol is repeated while a single parameter is varied to perturb the molecular structure in the crystals from its resting state. Applying the term ‘dynamic’ here refers in a broader sense to structural changes that occur in response to a single controlled external stimulus. Many other forms of stimuli may be applied to induce structural changes in crystals, e.g. gases (O2, CO or NO), pH, redox potential, pressure, chemical modifications including mutations or electromagnetic fields, X-rays or other radiation, most notably light. Even an uncontrolled accidental variable may yield informative structural differences. The different molecular environments arising from crystal packing, alternative crystal lattices and noncrystallographic symmetry (NCS) also provide structural variants that may reveal important structural dynamics (Ren et al., 2012 ▶), although these differences are often dismissed as uninteresting artifacts of crystallization. Despite the differing natures of the external stimuli, structural differences or changes take two distinct forms (which are most often observed in combination): changes in structural conformation arising from atomic motion and changes in the relative proportions of several distinct static conformations. In this post-structural genomics era, the introduction of an extra dimension in addition to the three spatial dimensions expands this traditionally static technique into dynamic crystallography. In this technique, a desired control parameter is varied to initiate a reaction, to stimulate a response or to perturb a structure from its resting state. Meanwhile, a sequence of ever-evolving structures is monitored in order to obtain dynamic insights into the working machinery of a protein. In this sense, dynamic crystallography aims at feasible experimental approaches that include three integral parts: (i) the introduction of a controlled stimulus to a responsive macromolecular system, (ii) the repeated collection of crystalloraphic data sets as the condition varies and (iii) the extraction of structural and population changes as function of the control parameter. The majority of data-collection techniques (ii) will remain identical to everyday static cryocrystallography, while experimental control (i) and joint data analysis (iii) require innovative approaches. A recent study of DNA polymerase η serves as a good example of our definition of dynamic crystallography (Nakamura et al., 2012 ▶).
The key to a successful experiment in dynamic crystallography is to generate and accumulate desired structural species to a detectable concentration. A common difficulty is structural heterogeneity (Fig. 1 ▶ and Supplementary Movie 11). It is noteworthy that experimentally achieving an effectively homogeneous structural species other than the resting state in a dynamic experiment is seldom practical or feasible. Nor is it necessary if the following technique becomes available: the ability to isolate data pertaining to a single structural species from a mixture, a process referred to as analytical trapping (Rajagopal, Kostov et al., 2004 ▶). The limited ability to isolate a single structural species from a mixture has hindered structural dynamics studies of many biologically important systems by crystallography. The root cause of the current limitation is the tradition of one structure or one ensemble per data set in crystallography. Structural refinement has been strictly on a per-data-set basis. We describe a new methodology, along with its underlying theoretical background and computational algorithms, that identifies and refines several structures simultaneously against a much larger number of data sets. The new capability to characterize common structural species from multiple data sets allows us to numerically deconvolute structural constituents in an evolving mixture, none of which ever attains a population large enough to be observed by direct measurement of a nearly homogeneous state. We apply this methodology to several experimental scenarios and quantitatively analyze the structural heterogeneity of electron density using a suitable strategy for each unique case.
Figure 1.
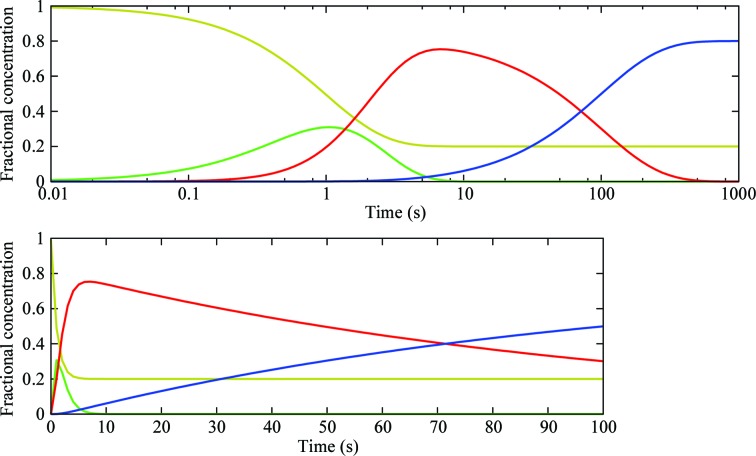
Fractional concentrations of various states in a hypothetical reaction. A hypothetical reaction starts from a ground state (gold), 80% of which enters the reaction at time 0 and first converts to an intermediate state (green) at a unit rate. This intermediate then converts to a second intermediate state (red) at a slower rate of 0.9 s−1. Finally, the product (blue) forms at a much slower rate of 0.01 s−1, which allows the intermediate (red) to accumulate to a high concentration. At the end of the reaction, there is still 20% of ground state that never enters the reaction. In reality, this percentage could be very large despite best efforts owing to practical reasons in reaction initiation in crystals (Fig. 3 ▶ b). Fractional concentrations as function of time are plotted on logarithmic and linear time scales.
2. Methods
This central section of this article describes a computational procedure that aims to extract a small set of distinct structures to simultaneously satisfy a large set of experimental electron-density maps. We recognize that this computational problem posed in dynamic crystallography, as formulated in §2.1, does not have a unique mathematical solution. We apply singular value decomposition (SVD) to jointly analyze the large and diverse data sets observed in a dynamic crystallographic experiment. §2.2 provides a brief summary of SVD and presents several goals that can be achieved by application of SVD to our dynamic data sets. However, the lack of a unique algebraic solution first requires the incorporation of structural knowledge into the procedure, which is facilitated by our choice of the real-space refinement approach presented in §2.3. Secondly, the subspace with much reduced dimensionality obtained by SVD provides a good guideline for the search in real-space refinement (§2.4). These combined efforts usually lead to either a unique set of structural solutions that fits the data sets or a small number of equally good fits from which one must choose. A closely related topic of minimal parameterization of protein structures is briefly mentioned in §2.5, which greatly eases the actual implementation of real-space refinement. Finally, several possible evaluations of real-space fitting are discussed in §2.6. An overview of the entire procedure is summarized in the scheme in Fig. 2 ▶.
Figure 2.
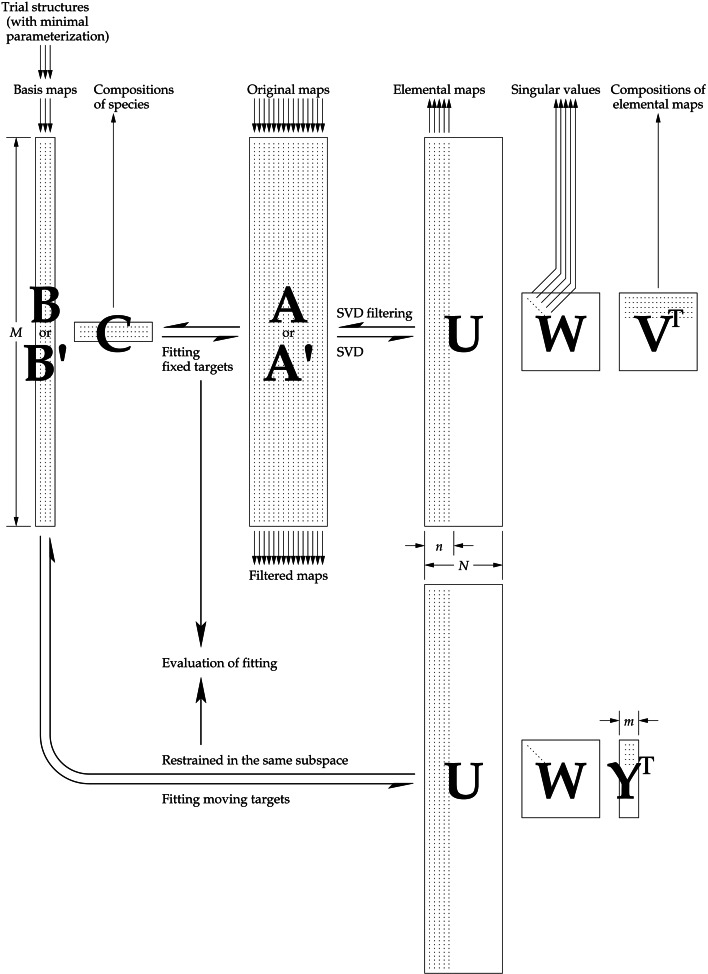
Scheme of the computational procedure. The observed original maps are first linearized and arranged in a large rectangular matrix A. SVD is performed on the matrix to produce factorized matrices U, W and V T. Judging by the ranking of singular values in W and the map features, several outstanding elemental maps and their corresponding compositions are selected from U and V T. Multiplication of the selected singular triplets results in noise-filtered maps in matrix A′. On the other hand, initial basis maps are calculated from manually built structures that are heterogeneously mixed in the observations. Each trial structure is minimally parameterized to lessen the requirement for stereochemical restraints. Least-squares fitting of linear combinations of the basis maps to all maps in A′ results in the coefficient matrix C. Meanwhile, each basis map should also be a linear combination of the accepted elemental maps. The residual of fitting is minimized while the trial structures are altered according to the next simplex downhill iteration. At the convergence of the fitting, map correlation coefficients are evaluated. The final trial structures are considered to be the refined heterogeneous structures. The final coefficient matrix C contains the compositions of the mixtures in matrix A.
2.1. Data matrix in real space
We adopt a real-space approach to take advantage of its unique feature of locality. Firstly, structural signals are often localized in real space, e.g. near the active site of an enzyme or the chromophore in a photoreceptor, but they are distributed across the entire reciprocal (diffraction) space. However, experimental noise is evenly distributed in both real space and reciprocal space. Secondly, local structural adjustment affects only a small subset of nearby grid points in an electron-density map in real space, but affects the entire data set in reciprocal space. If a known reference structure such as a native state is available in the isomorphous form of the perturbed structures, difference density maps are usually preferred because of their sensitivity to small structural changes (Fig. 3 ▶ b). However, see example 3 for a non-isomorphous case. A set of (difference) electron-density maps are arranged to form a large rectangular data matrix A (Fig. 2 ▶), in which each column represents a map at all grid points and each row represents a density value at a specific grid point as a function of the controlled variable. Matrix A may contain between hundreds and millions of grid points or rows, denoted M, and between a few and hundreds of conditions or columns, denoted N. A typical data matrix A is therefore very elongated, with M >> N.
Figure 3.
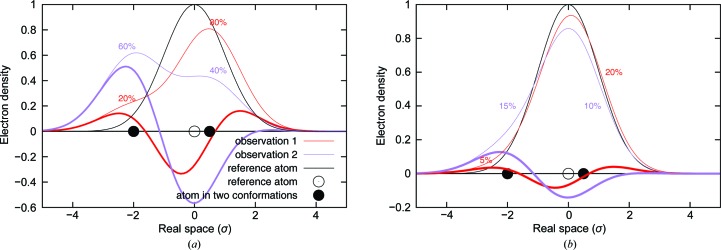
One-dimensional simulation of difference electron density in two observations. One atom moves from its starting position (open circle) to two distinct new conformations (black dots) by different distances. The electron density of the atom at the reference position is represented by a Gaussian curve with a peak amplitude of 1 and a deviation of σ, shown as a thin line in black. The electron densities resulting from the two displaced conformations present in different proportions are distinctly different, as shown by thin lines in red and purple. The thick lines are the difference electron density obtained by subtracting the initial reference electron density. Observation 1 in red shows two equally strong positive peaks separated by a negative peak, despite the fact that one conformation is four times more populated than the other. Observation 2 in purple shows only one pair of large positive–negative peaks. The minor conformation on the right, although at 40%, barely generates any positive density owing to its much smaller displacement. (a) represents an ideal scenario in which all molecules in the initial state have entered a reaction pathway and thus the reference structure is not present in the mixture. Although easier to understand, this case is rarely found in practice. The red observation 1 results from a mixture at a 20:80% = 1:4 ratio of the two displaced conformations and the purple one is a 60:40% = 3:2 mixture. (b) illustrates a more realistic scenario in which the reference structure contributes 75% to both observations. That is to say, only the remaining 25% of molecules have entered the reaction. The red observation contains 5:20% = 1:4 of the displaced conformations, while the purple observation contains 15:10% = 3:2. In this case, the electron-density maps (thin lines) are all very similar, except for differences in the tails of some peaks. In contrast, the difference maps (thick lines) are more revealing in representing structural changes. In addition, the two thin red lines in (a) and (b) differ in shape, as do the two thin purple lines. This illustrates the fact that the electron-density maps are also affected by the occupancy of the reference structure. However, the difference maps (thick lines) are only sensitive to the composition of the displaced conformations. The shapes of the difference maps, including the points of zero crossing, are not affected by the total extent of reaction initiation. This demonstrates another advantage of the difference map in describing structural changes in addition to its sensitivity.
Assume that each of the N observed maps in matrix A is a linear combination of a small number of m maps, in which each of the m maps results from a homogeneous chemically meaningful structural species. The jth observation (where j = 1, …, N) can therefore be written as
B 1–B m denote m basis maps, each of which represents a distinct structural species. Together, they account for all of the maps in matrix A. cij is the ith coefficient of the linear combination for the jth observation.
Our goal is to obtain this small set of m distinct structures that would produce the corresponding basis maps B 1–B m and to concomitantly solve the coefficient sets C j = (c 1j, …, cmj) for all N observations. Since both B i and C j are to be determined, this presents a large nonlinear fitting problem with MN data points and m(M + N) unknowns. Since M >> N, the data-to-parameter ratio is approximately N/m, a practical limit of which is presented below. This purely algebraic problem posed in dynamic crystallography has no unique solution if no structural knowledge enforces B 1–B m being images of chemically sensible structures. A complete and direct solution may be difficult. Here, we propose and implement a strategy aimed at a satisfactory solution to this problem through iterative refinement, under the assumption that m is small. If this assumption does not hold, that is too many structural conformations coexist to allow clean resolution, a different strategy is required (example 2).
2.2. Singular value decomposition
The first questions are as follows. Do the observed maps in A represent structural change? If so, do their differences depend on the controlled variable or on some other factors? Does the variation arise from a simple linear transition from a beginning to an end state? If not, how many distinct structures coexist in the observation matrix A? Obtaining a correct estimate of m alone is important in many projects. We apply SVD to the data matrix A.
The SVD approach in linear algebra is a robust process to factorize a rectangular matrix. It has been widely used for signal processing (Henry & Hofrichter, 1992 ▶) in many disciplines of science and has been successfully adapted to analyze crystallographic data (Schmidt et al., 2003 ▶; Rajagopal, Schmidt et al., 2004 ▶). If an M × N data matrix A, where M >> N, is factorized by SVD (Fig. 2 ▶), then
The matrix U has the same M × N shape as A. Each column of U retains the format of a (difference) electron-density map, but has been decomposed such that it cannot be represented by any linear combination of maps in the other columns. The N maps in U are denoted ‘elemental maps’, which are independent of one another or, in algebraic terms, orthonormal. Notice that these elemental maps differ from the basis maps B 1–B m in (1). Basis maps are images of distinct chemical structures that are related by a reaction scheme, while an elemental map usually does not reflect a single chemically meaningful structure. Unlike elemental maps, basis maps are not necessarily orthonormal to one another. See example 3 for examples of elemental maps, which may feature differences in spatial resolution, mutations or space group or differences between subunits related by NCS. The square N × N matrix W contains all zeros except for N positive values on its major diagonal, known as the singular values. The magnitude of a singular value can be thought of as a weight or significance of its corresponding elemental map. Matrix V is also N × N in shape, but its transpose V T must be used in multiplication with the other matrices. Each column of V, that is each row of V T, contains the relative composition of an elemental map as a function of the controlled variable, e.g. a time course or a temperature variation. This function also reveals whether the presence of an elemental map is correlated with factors other than the controlled variable, which is used as a powerful means of identifying systematic variations or errors such as radiation damage (example 3). A singular triplet denotes (i) an elemental map U k, (ii) its singular value wk and (iii) the composition function V k corresponding to the elemental map, where k = 1, …, N. U k and V k are also referred to as the left and right singular vectors, respectively.
Singular triplets are often sorted in descending order of their singular values wk. The trailing insignificant singular triplets can be eliminated with little loss of signal but with very effective noise reduction. The number n of the remaining significant singular triplets is the effective rank of the data matrix A, where n < N. All possible linear combinations of these n significant elemental maps define the range of matrix A. It can be shown that in a least-squares sense a linear combination of the n elemental maps in matrix U would satisfactorily reproduce the major features in each and every observed map in matrix A and yield a noise-filtered matrix A′. The jth column of matrix A′, that is the jth noise-filtered map, is
where vjk is an item of V k, the relative composition of the kth elemental map in the jth observation. The singular value wk behaves as a weight in forming the coefficient wkvjk for the linear combination.
It is also guaranteed that any possible map of a homogeneous structural solution cannot exceed the range of matrix A and must also be a linear combination of the n significant elemental maps. Thus, a close representation of a basis map B i can be written as
where yik is the relative composition of the kth elemental map in the ith basis map. These quantities are not derived from SVD analysis: rather, they describe the desired solutions. Again, the singular value wk functions as a weight in forming the coefficient wkyik for the linear combination. These coefficients of the elemental maps determine the qualitative features of a basis map. Combining equations (1–4) above, we obtain an important relationship,
where Y k = (y 1k, …, ymk). The dot product of two solution vectors C j and Y k is constrained by the result of SVD analysis, vjk. In other words, the qualitative features of the observed maps characterized by V k and the basis-map features determined by Y k are related by the concise relationship (5) for j = 1, …, N and k = 1, …, n. Each Y k can be easily obtained given trial vectors C 1–C N during refinement. Since the data-to-parameter ratio of the solution is given by N/m, where N > m, solving m independent structures from N observed maps is a substantially overdetermined linear problem.
In summary, SVD analysis firstly acts as an effective noise filter, secondly determines n ≥ m, the upper bound of coexisting structural species in the observation matrix A, thirdly identifies and removes systematic errors (example 3) and fourthly provides guidance for the search range of subsequent analyses including structural refinement, which takes the form of a concise restraining relationship between two solution vectors (5).
2.3. Real-space refinement against multiple difference maps
Since a direct solution of B i and C j in (1) is not straightforward, it would require initial trial structures for all heterogeneously mixed species in the observed maps in matrix A. Trial basis maps B i are then calculated from the trial structures (Fig. 2 ▶). The subsequent fitting of linear combinations of basis maps with observed maps is essentially a real-space refinement. The difficulty in this real-space refinement of multiple conformations, each with an unknown occupancy in C j, against multiple difference maps in matrix A is illustrated by two simulations containing only one atom in each of two distinct conformations (Fig. 3 ▶). The simulated one-dimensional difference ‘maps’ with different occupancies for each conformation demonstrate the combined effect of occupancy and atomic displacement. This simulation clearly suggests firstly that refinement of displacements and their occupancies against multiple observations must be carried out simultaneously in order to explain all experimental difference maps, secondly that difference maps are far more sensitive to small structural changes than electron-density maps (Henderson & Moffat, 1971 ▶) and thirdly that the features in a difference map are independent of the total occupancy of all conformations displaced from the initial reference state. Here, the term ‘small structural change’ refers to small atomic displacements occurring in only a small portion of a larger structure and to only a small fraction of all molecules. When one or more of these small quantities approaches zero, ‘small structural change’ becomes undetectable. The threshold of detectability is dictated by the magnitude of noise in matrix A.
Computationally, this real-space refinement protocol is a multi-parameter minimization between the observed difference maps in matrix A or the SVD-filtered maps in A′ (3) and linear combinations of the calculated difference maps of the distinct structures. This least-squares procedure is identical to conventional curve fitting except that it takes place in multiple three-dimensional real spaces simultaneously. This minimization process adjusts both trial conformations and their coefficients in linear combinations in order to synthesize difference maps that are as close to the observed maps as possible. When all observations are accounted for at the convergence of the minimization, refined structures are achieved in a least-squares sense and their compositions are determined. Many options are available for effective implementation of this multi-parameter minimization. However, this work has not yet included such efforts. For a proof-of-concept implementation, we take advantage of the simplicity of the downhill simplex method (Nelder & Mead, 1965 ▶).
Our approach fundamentally differs from the commonly used alternative conformations and other models of conformational polymorphism (Gros et al., 1990 ▶; Levin et al., 2007 ▶; Lang et al., 2010 ▶) by increasing the number of observed data points instead of the number of parameters to fit in the multi-parameter minimization. Previous approaches have attempted to extract conformational polymorphism or to account for thermal motions from a single data set obtained under a fixed condition. Thus, an isolated static ensemble of structures is no more than a time-averaged snapshot of a dynamic system and does not display a trend of structural change, nor does it suggest the cause of the change. To address the challenge of structure refinement in dynamic crystallography, here we replace the one structure or one ensemble to one data set tradition in structural refinement by our implementation of m structures to N data sets, where N > m (example 1).
2.4. Moving-target refinement
The real-space refinement that we propose replaces the need for direct solution of B i and C j from the large nonlinear system presented in (1). However, in practice, the multi-parameter minimization is also difficult to perform. The central problem offered by the large nonlinear system in (1) is how to simultaneously solve for both a small set of m distinct structural species that correspond to basis maps B 1–B m and coefficient vectors C 1–C N for all observed difference maps A 1–A N. A possible mathematical (but not chemically plausible, and thus false) solution to this system is to allow the ith structural species to become arbitrary by adjusting all of the ith coefficients in C 1–C N to near zero. That is to say, if a structural species makes a negligible contribution to all observed maps in matrix A, this species can adopt an arbitrary structure. In practice such arbitrary solutions occur frequently unless we follow the guidance given by the SVD result (5).
One workable technique to avoid arbitrary solutions is to synthesize trial basis maps from n elemental maps according to the current chemically plausible trial structures and to minimize differences between the trial basis maps and the trial structures in the upcoming refinement cycle by improving the trial structures. That is to say, the refinement target includes not only the experimental observations but also some trial basis maps synthesized on-the-fly (Fig. 2 ▶). Unlike the fixed experimental maps, a synthesized map is a moving target: it evolves with the trial structures as refinement proceeds. However, this moving target must be a linear combination of the elemental maps and therefore must remain within the range of matrix A. This moving-target strategy incorporates the SVD result into the minimization procedure and serves as a powerful constraint to prevent arbitrary solutions.
2.5. Minimal parameterization of macromolecular structures
This topic lies largely outside the scope of this article. However, it is a necessity to render the multi-structural refinements presented here computationally more feasible. We briefly describe the approach of minimal parameterization and will present it elsewhere in greater detail.
All structure-refinement programs require stereochemical restraints to maintain a nearly ideal geometry for macromolecular structures during refinement (Adams et al., 2010 ▶; Afonine et al., 2012 ▶). The stringency of these restraints (Grosse-Kunstleve et al., 2004 ▶) suggests that three Cartesian coordinates per atom overparameterize a macromolecular structure. Overparameterization and stereochemical restraints are two conflicting forces in complex structural refinement: reducing one would lessen the requirement for the other. We adopt the commonly used concept of minimal, or near-minimal, parameterization of macromolecular structure (Rice & Brünger, 1994 ▶), in which we seek to maintain the ideal geometry of a structure and to simplify or even eliminate stereochemical restraints. ‘Minimal parameterization’ has to be interpreted in relation to the spatial resolution of the crystal structure. Because crystal structures in a typical resolution range, say 1.5–3 Å, exhibit very narrow distributions of covalent bond lengths and angles, many degrees of freedom can be eliminated (at least in the early stages of refinement).
In the scheme of minimal parameterization, the most fundamental parameter is the rotation of a chemical group around a defined axis. At the termini of a main chain or the end of a side chain this angular parameter is simply the torsion angle about a single chemical bond. In the middle of a chain, a limited rotation is defined about an axis connecting two atoms separated by one or more other atoms in the chain, not about a chemical bond. For example, a peptide plane is rotated slightly about an axis connecting two consecutive Cα atoms. To move a Cα atom, two peptides are rotated slightly about the axis connecting the first and the third Cα atoms. Minimal parameterization maintains the planarity of aromatic rings, e.g. phenyl, imidazole, indole and pyrrole rings, as well as guanidinium and peptide groups, thus requiring no separate planarity restraints on these groups. On average, each protein side chain requires only 1.75 torsion angles, in contrast to 13 Cartesian coordinates, to describe all possible conformations of non-H atoms. Each residue needs 25 Cartesian coordinates, but less than four angular parameters. The minimal parameterization of a protein structure in combination with the locality of real-space refinement (§2.1) potentially confines the numerical process of minimization to a small scale.
2.6. Goodness of fit in real-space refinement
Several statistical measures are employed to evaluate the goodness of fit in real space. The commonly used root-mean-squared deviation (r.m.s.d.) between the observed and modeled values is normalized in R nrmsd. The normalized mean modulus deviation R nmmd is sometimes favored and is well known as the R factor. These are defined in real-space refinement as
and
where the summations are over all grid points of all maps involved in the refinement for an overall measure or over each map for individual evaluation. Δρo and Δρc are observed and calculated difference electron densities, respectively. Specifically, Δρo comes from either the entire matrix A or individual columns of matrix A. More often, the SVD-filtered matrix A′ is used (3). Δρc is synthesized from m calculated maps from the refined structures using the refined coefficient sets C j (1). kj is a scale factor between observed and calculated maps, which is usually specific to each jth observed map in matrix A.
It is obvious that fitting a difference electron-density map results in a far greater normalized residual than fitting an electron-density map. Because difference electron densities are in the summation in the denominators of (6) and (7), these R factors typically have much greater numerical values than the equivalent R factors with electron densities in the denominators. In addition, the scale factors kj certainly affect these R factors. Alternatively, the real-space correlation coefficient (CC) between Δρo and Δρc is often a better indicator of the quality of a fit given the noisy nature of difference signals. The acceptable CC critically depends on the mask of electron-density maps. In our experience, a CC in the range 0.5–0.7 indicates a good fit for a difference map within 10 Å around the active site (examples 1 and 2), while a CC of >0.9 is expected for reasonable fitting of an individual side chain against a 2F o − F c map (example 3).
In summary, we integrate four key technical components in our implementation of the joint real-space analysis of a large number of heterogeneous maps: (i) SVD identifies principal components and reduces experimental noises in a large unified data matrix A, (ii) multiple structures are simultaneously refined in real space against all observed maps and (iii) the incorporation of synthesized moving-target trial maps guides the refinement within a valid search range. Finally, (iv) minimal parameterization of the real-space structures (Rice & Brünger, 1994 ▶) serves to significantly ease the requirement of stereochemical restraints during this large-scale structural refinement. We now apply this approach to examples of dynamic crystallographic data acquired by different experimental techniques.
3. Results
3.1. Example 1: temperature-scanning cryocrystallography
At a sufficiently low temperature a reaction may effectively stall at a certain intermediate step, which allows certain structural intermediates to accumulate at observable concentrations. Since the rates of individual reaction steps are affected differently by temperature, different mixtures of intermediates may be trapped at different temperatures. To capture short-lived intermediates during the photoconversion reaction in Pseudomonas aeruginosa bacteriophytochrome (PaBphP), a ‘trap–pump–trap–probe’ strategy was employed to collect light and dark data sets from photoactive crystals of the PAS-GAF-PHY photosensory core at cryogenic temperatures ranging from 100 to 180 K (Yang et al., 2011 ▶). This temperature scan generates a total of 112 independent F light − F dark difference maps derived from 14 light and six corresponding dark data sets and from eight monomers in the asymmetric unit related by NCS. In all maps difference densities are tightly concentrated near the bilin chromophore. It is immediately obvious that the features in the observed difference maps vary continuously with pump temperature and that photoreaction proceeds from the far-red-absorbing Pfr ground state towards the red-absorbing Pr product state (Yang et al., 2011 ▶) as the pump temperature rises (Fig. 4 ▶). We may also conclude from the smoothly varying maps that the number of distinct structural species is far less than the total number of maps, that each difference map contains a mixture of multiple structural species and that the relative compositions of structural species vary with the pump temperature.
Figure 4.
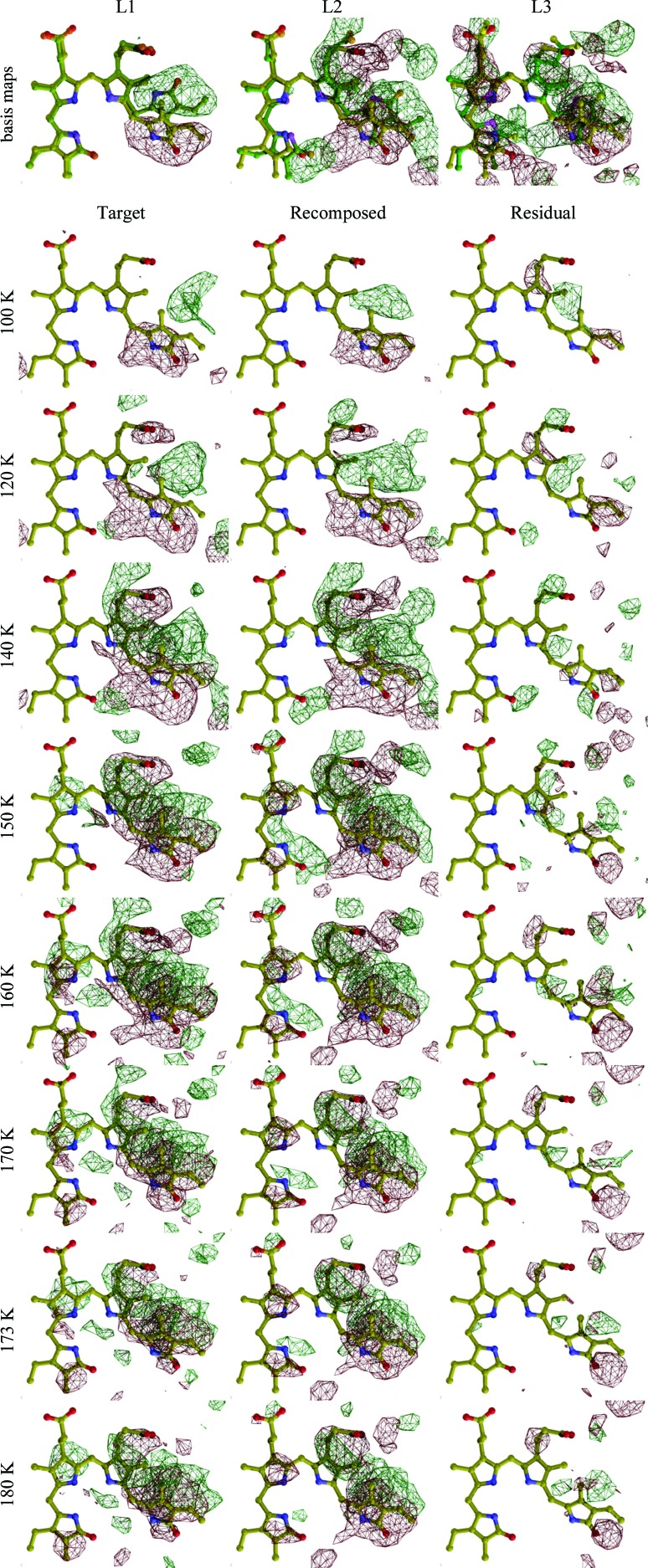
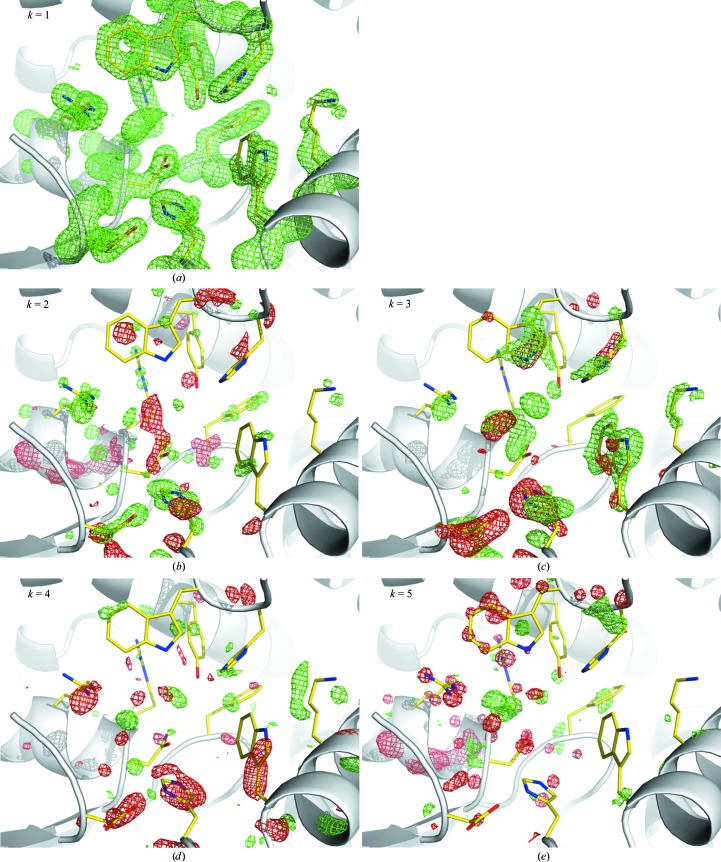
Difference electron-density maps near the PaBphP chromophore. Positive and negative electron densities relative to the dark structure are shown in green and red, respectively. The three maps in the top row are calculated from three refined structural species: L1, L2 and L3, respectively. Each other row denotes a different temperature. The ball-and-stick model in yellow shows the chromophore in its dark Pfr state. The models in green show the refined chromophore structures. Surrounding protein side chains also undergo conformation changes in concert with the chromophore, but are omitted here for clarity. The array of maps contains eight representative temperature settings out of the 14 in total. Only the dark-state chromophore is shown. The first column of maps show the NCS-averaged maps, which are the target maps in real-space refinement. The second column of maps are linear combinations of the three basis maps shown in the top row. The coefficients used in the composition are presented in Fig. 5 ▶. The third column shows the residual of fitting.
To demonstrate the effectiveness of our joint analysis of the heterogeneous mixture in real space, we mask the densities to consider only the volume within a 3 Å radius of a region that contains the chromophore and some adjacent key protein residues and subject the masked difference maps to SVD analysis (§2.2). The data matrix A consists of 53 923 grid points in each masked difference map and 14 NCS-averaged maps (Fig. 4 ▶) at various pump temperatures. Applying SVD to matrix A reveals several significant singular values, of which the top three stand out, indicating three major structures (Fig. 5 ▶). We manually built three trial structures denoted L1, L2 and L3 into the maps in matrix A (Yang et al., 2011 ▶), followed by real-space refinement of a partial structure that includes the biliverdin chromophore and its surrounding protein residues. Additional constraints are applied during refinement according to plausible interactions between the protein and chromophore in each structure. At the convergence of least-squares fitting, both atomic positions and the fractional concentration of each structure are determined. Structural heterogeneity is thereby resolved (Fig. 5 ▶).
Figure 5.
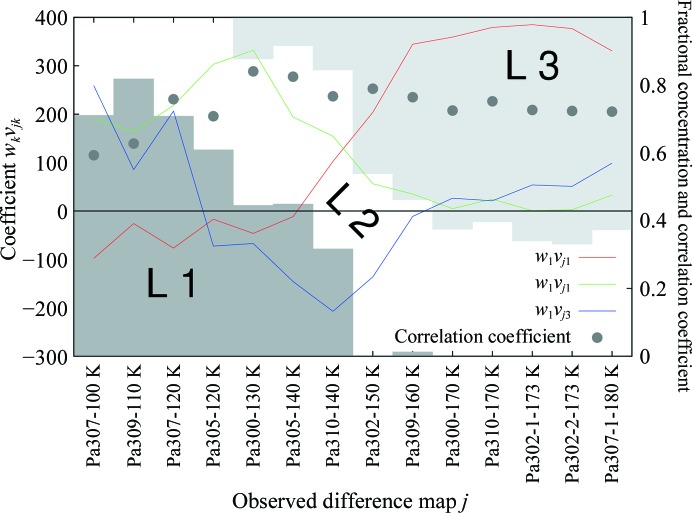
SVD and real-space refinement of PaBphP. The linear combination coefficients wkvjk for the three most significant elemental maps k = 1, 2 and 3 are shown as red, green and blue curves, respectively, as a function of the observed difference maps j. The labels on the horizontal axis are in the format crystal ID-temperature. The red curve indicates that the top elemental map k = 1 mostly exhibits features at temperatures of 160 K and higher. During the transition period from 140 to 160 K, low-temperature features represented by the elemental map k = 2 are gradually replaced by high-temperature features in the elemental map k = 1. At temperature settings lower than 140 K, structural changes are mostly described by elemental maps k = 2 and 3. The white and two gray areas show fractional concentrations of three refined structural species in all difference maps. The darker gray represents L1. Its population falls to nearly 0 at temperatures above 140 K. The lighter gray area is L3, which starts to appear at 130 K and reaches its maximum of more than 60% at high temperatures. The white area in between shows the population of L2, which is always present and is mixed with other species at all temperatures. Its maximum concentration exceeds 50% at 140–150 K. The fractional concentration expressed here is with respect to the total molecules that ever underwent photoconversion; thus, the sum over the three fractions is always 1. In this sense, fractional concentration differs from the occupancy as commonly used in the PDB. The black dots show real-space correlation coefficients between the target and the recomposed maps (Fig. 4 ▶) as an indicator of the goodness of fit.
Since no single data set arises from a single homogeneous structure in the temperature-scanning experiments, existing refinement-software tools are not capable of refining several structures common to all data sets simultaneously. Conventional refinement in reciprocal space would seek to refine four alternative conformations (the ground state and three intermediate structures L1, L2 and L3) against each of the 14 data sets, with a total of 56 structures, which is clearly a case of overfitting. There is no effective way to constrain the 14 ground-state structures to remain identical and refine a common structure; nor can this be achieved for three intermediates. As an example, using the conventional refinement technique, each soaking time and pH setting resulted in an independently refined structure in the study of the DNA polymerase (Nakamura et al., 2012 ▶). More seriously, owing to the limited extent of photolysis and reaction initiation in large crystals, the occupancy of the dark state remains high in the illuminated crystals. The total occupancy of these three intermediates is estimated to be 15–30% in the various temperature-dependent data sets. Conventional refinement approaches are very insensitive to occupancy and it is even harder to determine the conformations of species with low occupancies. Low occupancy is also a common difficulty in diffusion-based experiments under reasonable soaking times and concentrations. Early stages of drug-screening experiments often suffer from low-occupancy binding caused by the poor affinity of drug leads yet to be optimized.
A practical limit on m is related to how structurally distinct the m species are. An intermediate structure often differs only slightly from its predecessor and successor. In order to isolate these closely similar structural species, it is important to ensure that the number of observations N > m. In this example, N/m is nearly 5, which thus far sets a practical lower limit. However, when the number of distinct species m becomes large, increasing the number of observation data sets N even more may result in a large unmanageable numerical problem and thus is not always practical. In the next example, we refine an entire molecule but analyze each data set with a unique single structure.
3.2. Example 2: a transient asymmetric state of a dimeric hemoglobin
In a time-resolved experiment, diffraction data are collected while a reaction progresses at near-physiological temperature following rapid reaction initiation in the crystal. Nevertheless, as in any kinetic experiment, crystallographic data at a given time point capture a mixture of structural intermediates. The composition of the mixture evolves with time in an ever-changing process (Fig. 1 ▶ and Supplementary Movie 1).
To study cooperative changes in ligand-binding affinity upon photodissociation of CO in the homodimeric hemoglobin HbI from Scapharca inaequivalvis, a sequence of Laue diffraction data sets were collected at 1.6 Å resolution at various time delays ranging from 100 ps to 50 µs after a short 35 ps laser pulse had photodissociated the CO ligand (see Knapp et al., 2006 ▶, for the experimental protocol). Although the largest difference signals resulted from a rapid response to CO dissociation and cluster near the heme region of both subunits, additional difference densities are distributed extensively throughout several more distant helices in the globin (see Knapp et al., 2006 ▶, and Ren et al., 2012 ▶, for an overview of the difference maps and SVD analysis).
When applying our real-space analysis protocol, we find that the assumption in (1) does not hold for the entire dimeric molecule. That is, the number m of distinct dimer structures is too large and exceeds the capability of the algorithm. However, if we restrict our real-space analysis to a local region such as the heme-binding pocket of one subunit, we observe that the conformational changes occur in several discrete steps throughout the time course. The number of these distinct local structures is small enough to be refined simultaneously by our protocol. The same is true for the heme-binding pocket of the other subunit and for other local regions. Real-space refinement of several structures against a set of many difference maps, each confined to a local region, has been demonstrated in example 1 for a bacteriophytochrome. Therefore, we do not repeat this example. The results for this hemoglobin example show that the structural transitions in different local regions are asynchronous. This temporal mismatch between structural events in different local regions generates a large number of intermediate dimer structures throughout the probed time course. An alternative strategy considers an entire dimer as a region but analyzes each time point as a unique single structure. Both strategies have been tested. The results of the two strategies must be merged to form a more complete picture describing the local and global structural changes throughout the entire time course, which will be reported elsewhere.
This alternative strategy is particularly applicable to the earliest time point of 100 ps since it appears that at this time the photoproduct is still relatively homogeneous in structure, in contrast to later time points in the sequence. At 100 ps it is clear that CO has been dissociated and is positioned in the distal-pocket docking site. The difference map (Fig. 6 ▶ b) calculated from a photoproduct model that reflects such a change reproduces the strong signal associated with CO in the observed difference map (Fig. 6 ▶ a). However, owing to the achievable experimental signal-to-noise ratio, the observed difference signal associated with the heme motion is weaker and disconnected, although the distribution of positive and negative densities flanks the heme group as expected. To execute real-space refinement, the heme group is parameterized under the minimal principle, in which the individual pyrrole rings remain planar while the angles between their normals are allowed to vary. After convergence via least-squares fitting, the heme group is shifted towards the distal site. The difference map calculated from the refined structure (Fig. 6 ▶ b) shows continuous positive and negative densities; continuity is by-and-large absent in the observed difference map (Fig. 6 ▶ a). The correlation coefficient between the observed and calculated maps is 0.53 around the heme group. To achieve a higher correlation coefficient, the connectivity of the calculated map would have to be broken and this in turn would require sacrificing the planarity of the pyrrole rings. Our minimal parameterization protocol does not allow experimental errors to artificially distort the pyrrole rings even in the absence of explicit stereochemical restraints (Grosse-Kunstleve et al., 2004 ▶).
Figure 6.
Light–dark difference electron-density maps of dimeric S. inaequivalvis hemoglobin upon photolysis of the CO form. Positive and negative electron densities relative to the reference dark structure are shown in green and red, respectively. The heme region in subunit B is shown. The ball-and-stick model in yellow is the dark state and the green model is the refined photoproduct at 100 ps. (a) Observed difference map at 1.6 Å resolution contoured at ±4σ. (b) Difference map calculated from the refined structure.
This real-space protocol enables us to confirm light-induced CO dissociation and the general features of the structural response at the heme group. It also reveals global structural features in a transient asymmetric state that are directly relevant to cooperative ligand binding in this homodimeric hemoglobin (Ren et al., 2012 ▶). Although this refinement task falls into the one structure to one data set tradition, these scattered signals are usually too subtle to be confidently modeled and refined using existing tools.
3.3. Example 3: mixed conformations in fluoroacetate dehalogenases
A third example concerns a representative case in structure–function studies that often involve a large number of mutant and complex structures, where isomorphism is not always available. The numerical resolution of heterogeneous structural species unevenly distributed in multiple data sets provides a means to jointly analyze many static data sets from commonly practiced mutation studies and inhibitor screening in drug discovery. The techniques of cocrystallization and post-crystallization soaking in static crystallography generate drug-induced structural changes and collectively provide dynamic insights into protein function if analyzed from the perspective of dynamic crystallography.
In the previous two examples, we analyzed difference electron-density maps in which density values are distributed symmetrically around zero. However, such an isomorphous difference Fourier technique, which is very sensitive to weak signal, is not always possible. In this example, we show that our algorithms are equally applicable to electron-density maps including 2F o − F c maps or F o − F c omit maps, in which density values are distributed asymmetrically around zero and structural signals are only associated with positive density values. This approach could be applied to the DNA polymerase case (Nakamura et al., 2012 ▶).
Fluoroacetate dehalogenases catalyze the hydrolysis of fluoroacetate into glycolate. To establish the structural and dynamic basis for cleaving the carbon–fluorine bond, which is the strongest covalent bond in organic chemistry, many high-resolution static structures of the fluoroacetate dehalogenase enzyme RPA1163 from Rhodopseudomonas palustris CGA009 have been determined (Chan et al., 2011 ▶). Direct inspection of the electron-density maps in the active site of the enzyme reveals multiple conformations for several key residues and the substrate or product present in the active site. However, the detail seems complex and it is difficult to refine the occupancy of each conformation. Although the real-space strategy presented in this paper is motivated by challenges in analyzing time-resolved and temperature-scan data sets, we here show that it can also apply to deconvolute the structural heterogeneity present in an array of static structures.
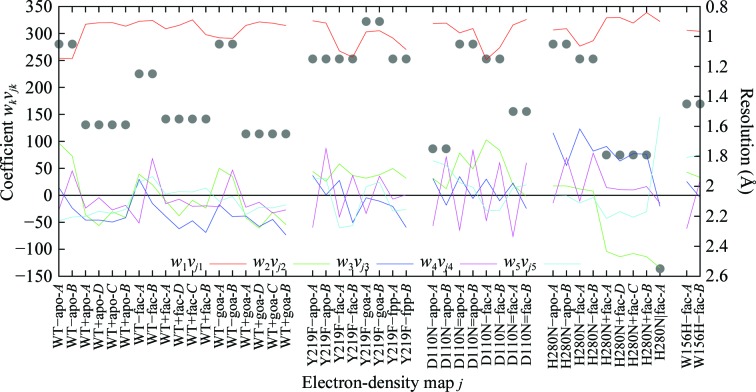
From 19 structures of the fluoroacetate dehalogenase enzyme determined at resolutions between 2.5 and 0.9 Å, we assembled a total of 45 independently measured monomeric structures (Chan et al., 2011 ▶). These structures include the wild type, mutants and various complexes with substrate or products in monoclinic, orthorhombic and tetragonal space groups. To remove prior model bias, we calculated simulated-annealing F o − F c omit maps (SAOMs) that omit several key residues around the active site (Adams et al., 2010 ▶). The SAOMs from all 45 monomers are aligned according to their active-site structures. These yield a data matrix A of 136 199 grid points (M) from 45 maps (N) around the active site to be subjected to SVD analysis (§2.2). SVD yields seven significant singular triplets. Interpretation of these elemental maps is usually difficult, since they are not images of structurally homogeneous species. The top elemental map (k = 1) can be considered as an average map (Fig. 7 ▶ a). Its composition (red curve in Fig. 8 ▶) stands out well above all other components, suggesting that overall the active-site structures agree well despite differences in resolution, space group, mutations and ligand-binding status. However, unlike the analysis of data from the examples described above, several singular triplets in this example are associated with systematic differences that originate in the different sources of data. The elemental map k = 2 (Fig. 7 ▶ b) presents the distinction between high- and low-resolution maps, as shown by the fact that the green composition curve w 2 vj 2 (Fig. 8 ▶) largely correlates with the resolution of each map (black dots). The elemental maps k = 3 and k = 5 (Figs. 7 ▶ c and 7 ▶ e) single out the unique features of the mutant structure H280N and data obtained in the tetragonal space group, respectively. It is most interesting that the elemental map k = 4 (Fig. 7 ▶ d) identifies a systematic difference between chain A and chain B in the monoclinic space group and to a lesser extent between chains A/C and chains D/B in the orthorhombic space group. This difference is apparently derived from NCS, but may have functional implications (Ren et al., 2012 ▶).
Figure 7.
Elemental maps of fluoroacetate dehalogenase structures. Green and red meshes are contoured at ±3σ, respectively, except in (a) k = 1, where the only green contour is at 1.5σ. The top elemental map k = 1 is an average of all 45 simulated-annealing omit maps and contains few areas of negative density.
Figure 8.
SVD analysis of multiple crystal structures of the fluoroacetate dehalogenase enzyme from R. palustris. The linear combination coefficients wkvjk for the five most significant elemental maps k = 1, 2, 3, 4 and 5 are shown in red, green, blue, magenta and cyan, respectively, as a function of the jth simulated-annealing omit map. The other 40 less significant components are omitted for clarity. The labels on the horizontal axis indicate wild type (WT) or mutant (Y219F, D110N, H280N and W156H), apoenzyme (apo) or ligand complex (fac, fluoroacetate; goa, glycolate; fpp, 2-fluoropropanoate) and chain ID (A, B, C and D). The symbols – and = denote monoclinic space groups under different soaking conditions; + and | denote orthorhombic and tetragonal space groups, respectively. The structures in the monoclinic space group contain A and B chains in the asymmetric unit, those in the orthorhombic space group contain A, D and C, B chains (which correspond to two copies of the monoclinic A and B chains) and those in the tetragonal space group contain only the A symmetry-related chains (one of its symmetry-related chains corresponds to the monoclinic B chain). The black dots show the resolution limits of each structure.
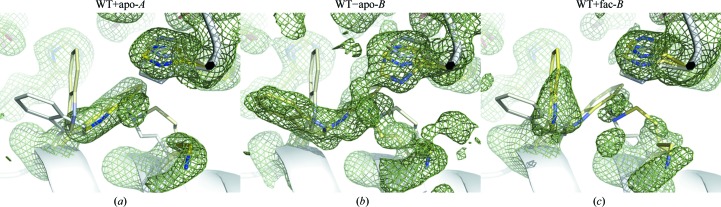
Three distinct conformations in the active site are jointly refined during the simultaneous fitting of 18 SAOMs of wild-type monomers (Fig. 9 ▶). Their occupancies in each of the 18 monomeric structures are also refined. As shown in Table 1 ▶, the three active-site conformations are populated very differently in the 18 maps depending on the space group and NCS. For example, conformation 2 is more populated in the orthorhombic chain B and conformation 3 is only populated in the monoclinic chain B. This analysis helps to unify aspects of a seemingly complex structural interpretation and identifies a consistent structural distinction between chains A and B.
Figure 9.
Representative simulated-annealing omit maps of fluoroacetate dehalogenase. Three jointly refined conformations are shown as stick models in yellow. The color saturation level is determined by the refined fractional concentration (Table 1 ▶). Fainter colors denote lower concentrations. Only His155, Lys181 and Trp185 are shown in the foreground. The green mesh is contoured at 1.5σ. See the legend to Fig. 8 ▶ for the format of the map labels.
Table 1. Refined fractional concentrations of three coexisting conformations.
See the legend to Fig. 8 ▶ for the format of the map labels.
| Conformation | ||||
|---|---|---|---|---|
| SAOM | 1 | 2 | 3 | Correlation coefficient |
| WT−apo-A | 0.712 | 0.096 | 0.192 | 0.863 |
| WT−apo-B (Fig. 9 ▶ b) | 0.455 | 0.141 | 0.404 | 0.855 |
| WT+apo-A (Fig. 9 ▶ a) | 0.752 | 0.216 | 0.033 | 0.906 |
| WT+apo-D | 0.684 | 0.261 | 0.054 | 0.903 |
| WT+apo-C | 0.707 | 0.230 | 0.063 | 0.914 |
| WT+apo-B | 0.646 | 0.321 | 0.034 | 0.919 |
| WT−fac-A | 0.925 | 0.075 | 0.000 | 0.907 |
| WT−fac-B | 0.822 | 0.179 | 0.000 | 0.890 |
| WT+fac-A | 0.707 | 0.213 | 0.080 | 0.927 |
| WT+fac-D | 0.520 | 0.352 | 0.128 | 0.918 |
| WT+fac-C | 0.650 | 0.260 | 0.090 | 0.930 |
| WT+fac-B (Fig. 9 ▶ c) | 0.408 | 0.509 | 0.083 | 0.916 |
| WT−goa-A | 0.576 | 0.126 | 0.298 | 0.898 |
| WT−goa-B | 0.452 | 0.144 | 0.404 | 0.863 |
| WT+goa-A | 0.787 | 0.191 | 0.022 | 0.913 |
| WT+goa-D | 0.646 | 0.306 | 0.048 | 0.901 |
| WT+goa-C | 0.765 | 0.198 | 0.037 | 0.909 |
| WT+goa-B | 0.467 | 0.469 | 0.064 | 0.907 |
4. Discussion
Protein conformation evolves in discrete steps along a reaction pathway. However, the population of each distinct conformation varies continuously as the reaction proceeds (Fig. 1 ▶ and Supplementary Movie 1). Observations from such dynamic processes inevitably involve heterogeneous structural species. Our basic strategy is to analyze multiple observations jointly in order to identify, extract and refine several constituents that are constant in conformation but vary in population. Both the constants and the variations are required to warrant a unique solution to the problem (1). If the intermediate constituent structures are largely disordered or adopt a wide distribution in the conformational space, this strategy will be less effective. At the opposite extreme, where little variation is observed in map composition, redundant and correlated measurements, although heterogeneous, do not contribute to the numerical resolution of a mixture, which can be understood as no unique solution to a singular linear system. A cryo-trapping experiment at a single temperature may be difficult to interpret in detail if heterogeneous structures are present, because such an isolated observation lacks the ability to reveal a trend. Thus, coexisting features cannot be assigned with high confidence to different structural species. Resolution of this difficulty provides the fundamental rationale behind temperature-scanning cryocrystallography (Yang et al., 2011 ▶).
Conventional structural refinement in macromolecular crystallography is now largely carried out in reciprocal space (Adams et al., 2010 ▶; Afonine et al., 2012 ▶), although real-space refinement was more common in the early years (Diamond, 1971 ▶). Small-scale real-space refinement (Chapman, 1995 ▶) is often performed in conjunction with model fitting. It is a rather straightforward technique in which atoms need to be moved into areas of higher electron density in target maps such as 2F o − F c or F o − F c omit maps while maintaining stereochemical restraints. Our approach of real-space refinement in dynamic crystallography extends this significantly: multiple structures and their compositions are refined concomitantly against many (difference) electron-density maps simultaneously. Although a difference refinement protocol in reciprocal space can refine structures such as mutants and ligand-bound forms (Terwilliger & Berendzen, 1995 ▶, 1996 ▶), it is not effective for simultaneous refinement of multiple conformations, especially when the occupancy of each conformation is much lower than that of the coexisting parent conformation. Furthermore, the difference refinement protocol is not capable of refining one or more common conformations against multiple data sets (example 1). Whether our goal of refinement in dynamic crystallography can be achieved in reciprocal space and whether it carries greater advantage remain to be explored.
The difference refinement protocol (Terwilliger & Berendzen, 1995 ▶, 1996 ▶) and many other treatments of structural polymorphism strictly hold the one structure or one ensemble to one data set tradition in static crystallography. For example, discrete alternative conformations are commonly used to model a small number of coexisting conformations with high occupancy for certain residues or structural segments. The atomic displacement parameters are also used to model atomic disorder and could suggest functionally important motions (Lu et al., 2005 ▶). Possible thermal motions could be included in structural refinement of an ensemble to improve agreement with measured diffraction data (Gros et al., 1990 ▶). Recent advances, such as ensemble refinement (Levin et al., 2007 ▶), may explain disordered electron densities even better and help to visualize conformational polymorphism (Lang et al., 2010 ▶). However, it is questionable whether this additional structural information depicts functional dynamics of proteins or merely reflects the uncertainty in the data (Terwilliger et al., 2007 ▶). Table 2 ▶ summarizes crystallographic techniques that acquire or model dynamic structural information.
Table 2. Crystallographic techniques that acquire or model dynamic structural information.
| Technique | Information | Drawbacks |
|---|---|---|
| Atomic displacement parameters | Thermal vibration | Little relevance to function (Lu et al., 2005 ▶) |
| Alternative conformations | Coexisting conformations | Static |
| Ensemble refinement (Gros et al., 1990 ▶; Levin et al., 2007 ▶) | Conformational polymorphism (Lang et al., 2010 ▶) | More parameters and data limitation (Terwilliger et al., 2007 ▶) |
| Structural comparison | Structural differences | Stable structures only |
| Chemical trapping | Intermediate, especially after rate-limiting step† | Difficulty in design |
| Cryotrapping | Intermediate structure | May alter reaction pathway |
| Time-resolved crystallography | Intermediate and reaction mechanism | Difficulty in rapid reaction initiation |
To capture an intermediate after a natural rate-limiting step that cannot accumulate to a sufficient concentration, chemical trapping is the only approach possible to slow down or to stall the next step so that the original step is no longer rate-limiting.
Supplementary Material
Simulation of structural heterogeneity in a crystal: simulation of the hypothetical reaction described in Fig. 1. The array of dots represent protein molecules in a crystal lattice. Their colors indicate the distinct structural states. Fractional concentrations of various states are displayed in the color bar. This simulation shows that molecules convert from one state to the next in a random thermodynamic fashion independently of one another, even when the reaction of each molecule is synchronously initiated (Moffat, 2001) at the same time point 0. The rates of the reaction steps govern when the concentration of each state will rise, peak and fall, and what concentration each state can reach. The reaction in one molecule is not tied to those in any other molecules by any force. Heterogeneity is intrinsic during any dynamic processes. This is true in both crystalline and solution states. A non-ideal experimental initiation of the reaction can only contribute further to heterogeneity.. DOI: 10.1107/S0907444913003454/lv5027sup1.gif
Acknowledgments
We thank S. L. Perry for critical discussions and reading of the manuscript. BioCARS is supported by the National Institutes of Health, National Center for Research Resources under grant No. RR007707 and by the National Institute of General Medical Sciences under grant No. 8P41GM103543. Part of this research was supported by the Natural Sciences and Engineering Research Council of Canada through a graduate scholarship (PWYC) and an operating grant (EFP) as well as by the Canada Research Chairs Program (EFP). We acknowledge the following software components used in this work: CCP4 (http://ccp4.ac.uk), dynamiX (ZR), gnuplot (http://gnuplot.info), PHENIX (http://phenix-online.org), Precognition (Renz Research Inc., Westmont, Illinois, USA), PyMOL (http://pymol.org), Python (http://python.org), RAVE (http://xray.bmc.uu.se/usf) and SciPy (http://scipy.org). ZR designed the research, developed the analytical methods and algorithms, implemented the dynamiX computer software, analyzed the data and wrote the paper. PWYC, KM, EFP, WER, VŠ and XY provided the study cases, interpreted the results and commented on the paper. The authors declare no competing financial interests.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: MV5027).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Chan, P. W. Y., Yakunin, A. F., Edwards, E. A. & Pai, E. F. (2011). J. Am. Chem. Soc. 133, 7461–7468. [DOI] [PMC free article] [PubMed]
- Chapman, M. S. (1995). Acta Cryst. A51, 69–80.
- Diamond, R. (1971). Acta Cryst. A27, 436–452.
- Gros, P., van Gunsteren, W. F. & Hol, W. G. J. (1990). Science, 249, 1149–1152. [DOI] [PubMed]
- Grosse-Kunstleve, R. W., Afonine, P. V. & Adams, L. W. (2004). IUCr Comm. Crystallogr. Comput. Newsl. 4, 19–36.
- Henderson, R. & Moffat, J. K. (1971). Acta Cryst. B27, 1414–1420.
- Henry, E. R. & Hofrichter, J. (1992). Methods Enzymol. 210, 129–192.
- Knapp, J. E., Pahl, R., Šrajer, V. & Royer, W. E. (2006). Proc. Natl Acad. Sci. USA, 103, 7649–7654. [DOI] [PMC free article] [PubMed]
- Lang, P. T., Ng, H.-L., Fraser, J. S., Corn, J. E., Echols, N., Sales, M., Holton, J. M. & Alber, T. (2010). Protein Sci. 19, 1420–1431. [DOI] [PMC free article] [PubMed]
- Levin, E. J., Kondrashov, D. A., Wesenberg, G. E. & Phillips, G. N. Jr (2007). Structure, 15, 1040–1052. [DOI] [PMC free article] [PubMed]
- Lu, W., Wang, C., Yu, E. & Ho, K. (2005). Proteins, 62, 152–158. [DOI] [PMC free article] [PubMed]
- Moffat, K. (2001). Chem. Rev. 101, 1569–1581. [DOI] [PubMed]
- Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y. & Yang, W. (2012). Nature (London), 487, 196–201. [DOI] [PMC free article] [PubMed]
- Nelder, J. A. & Mead, R. (1965). Comput. J. 7, 308–313.
- Rajagopal, S., Kostov, K. S. & Moffat, K. (2004). J. Struct. Biol. 147, 211–222. [DOI] [PubMed]
- Rajagopal, S., Schmidt, M., Anderson, S., Ihee, H. & Moffat, K. (2004). Acta Cryst. D60, 860–871. [DOI] [PubMed]
- Ren, Z., Šrajer, V., Knapp, J. E. & Royer, W. E. (2012). Proc. Natl Acad. Sci. USA, 109, 107–112. [DOI] [PMC free article] [PubMed]
- Rice, L. M. & Brünger, A. T. (1994). Proteins, 19, 277–290. [DOI] [PubMed]
- Schmidt, M., Rajagopal, S., Ren, Z. & Moffat, K. (2003). Biophys. J. 84, 2112–2129. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1995). Acta Cryst. D51, 609–618. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1996). Acta Cryst. D52, 1004–1011. [DOI] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Adams, P. D., Moriarty, N. W., Zwart, P., Read, R. J., Turk, D. & Hung, L.-W. (2007). Acta Cryst. D63, 597–610. [DOI] [PMC free article] [PubMed]
- Yang, X., Ren, Z., Kuk, J. & Moffat, K. (2011). Nature (London), 479, 428–432. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Simulation of structural heterogeneity in a crystal: simulation of the hypothetical reaction described in Fig. 1. The array of dots represent protein molecules in a crystal lattice. Their colors indicate the distinct structural states. Fractional concentrations of various states are displayed in the color bar. This simulation shows that molecules convert from one state to the next in a random thermodynamic fashion independently of one another, even when the reaction of each molecule is synchronously initiated (Moffat, 2001) at the same time point 0. The rates of the reaction steps govern when the concentration of each state will rise, peak and fall, and what concentration each state can reach. The reaction in one molecule is not tied to those in any other molecules by any force. Heterogeneity is intrinsic during any dynamic processes. This is true in both crystalline and solution states. A non-ideal experimental initiation of the reaction can only contribute further to heterogeneity.. DOI: 10.1107/S0907444913003454/lv5027sup1.gif