ABSTRACT
STING (stimulator of interferon [IFN] genes) initiates type I IFN responses in mammalian cells through the detection of microbial nucleic acids. The membrane-bound obligate intracellular bacterium Chlamydia trachomatis induces a STING-dependent type I IFN response in infected cells, yet the IFN-inducing ligand remains unknown. In this report, we provide evidence that Chlamydia synthesizes cyclic di-AMP (c-di-AMP), a nucleic acid metabolite not previously identified in Gram-negative bacteria, and that this metabolite is a prominent ligand for STING-mediated activation of IFN responses during infection. We used primary mouse lung fibroblasts and HEK293T cells to compare IFN-β responses to Chlamydia infection, c-di-AMP, and other type I IFN-inducing stimuli. Chlamydia infection and c-di-AMP treatment induced type I IFN responses in cells expressing STING but not in cells expressing STING variants that cannot sense cyclic dinucleotides but still respond to cytoplasmic DNA. The failure to induce a type I IFN response to Chlamydia and c-di-AMP correlated with the inability of STING to relocalize from the endoplasmic reticulum to cytoplasmic punctate signaling complexes required for IFN activation. We conclude that Chlamydia induces STING-mediated IFN responses through the detection of c-di-AMP in the host cell cytosol and propose that c-di-AMP is the ligand predominantly responsible for inducing such a response in Chlamydia-infected cells.
IMPORTANCE
This study shows that the Gram-negative obligate pathogen Chlamydia trachomatis, a major cause of pelvic inflammatory disease and infertility, synthesizes cyclic di-AMP (c-di-AMP), a nucleic acid metabolite that thus far has been described only in Gram-positive bacteria. We further provide evidence that the host cell employs an endoplasmic reticulum (ER)-localized cytoplasmic sensor, STING (stimulator of interferon [IFN] genes), to detect c-di-AMP synthesized by Chlamydia and induce a protective IFN response. This detection occurs even though Chlamydia is confined to a membrane-bound vacuole. This raises the possibility that the ER, an organelle that innervates the entire cytoplasm, is equipped with pattern recognition receptors that can directly survey membrane-bound pathogen-containing vacuoles for leaking microbe-specific metabolites to mount type I IFN responses required to control microbial infections.
Introduction
The obligate intracellular pathogen Chlamydia trachomatis is the most common sexually transmitted bacterial infection and the leading cause of preventable blindness worldwide. As a consequence of inflammatory damage from repeated and chronic infections, severe sequelae can occur such as blinding trachoma, pelvic inflammatory disease, and infertility (1). Elementary bodies (EB), the infective form of C. trachomatis, attach to and invade epithelial cells of the conjunctiva and urogenital tract. After entry, the EB form transitions to the metabolically active reticulate body (RB), which replicates within a membrane-bound compartment known as the inclusion. Midway through the Chlamydia infectious cycle, RB replication becomes asynchronous, generating both RBs and EBs (2). EBs exit the cell through either lysis of the host cell or extrusion of the inclusion to infect neighboring cells (3).
Microbial compounds are recognized by surface-exposed and intracellular immune receptors, leading to the transcriptional activation of cytokines and other host defense genes (4). For instance, type I interferons (IFNs) are secreted in response to diverse viral and bacterial infections (5, 6). Although the antiviral properties of IFNs are fairly well characterized, the role that these cytokines play in controlling bacterial infections is less well understood, as they can both block and enhance pathogen replication in vivo (7–11). All cell types can initiate type I IFN responses through Toll-like receptor (TLR)-dependent and TLR-independent pathways. TLR-independent pathways include cytosolic sensing of microbial ligands such as cell wall fragments through NF-κB-activating NOD1/NOD2 receptors (12) and double-stranded DNA (dsDNA) or dsRNA-like molecules through the action of RNA polymerase III (13), RIG-I (14), MAVS (15), cGAS (16), and STING (stimulator of IFN genes; also known as Eris, MITA, and MPYS) (17–20).
C. trachomatis induces type I IFN responses in a variety of cell types. Previous work has provided evidence for (21) and against (22) a role for TLRs in the regulation of type I IFN responses to Chlamydia infection. Prantner and colleagues identified STING, a key mediator of responses to cytosolic DNA, RNA, cyclic dinucleotides, and membrane fusion (19, 23–25), as important for the induction of type I IFNs in Chlamydia-infected cells (22). STING is an endoplasmic reticulum (ER) integral membrane protein (26) that, upon exposure to cytoplasmic dsDNA, is redistributed to p62-positive punctate structures in the cytoplasm (“sequestosomes”) that colocalize with autophagy-related proteins (26, 27). Unlike sensing of dsDNA in the cytoplasm, which requires additional cofactors, STING can directly bind to bacterium-specific cyclic dinucleotides (24) and trigger potent type I IFN responses to intracellular pathogens such as Listeria monocytogenes and Legionella pneumophila (28–30).
Diadenylate cyclases (DACs) are found in a wide variety of Gram-positive bacteria and are required for the synthesis of cyclic di-AMP (c-di-AMP) (31). A related cyclic dinucleotide, c-di-GMP, is a well-established regulator of bacterial virulence, biofilm formation, motility, and gene expression (32). In contrast, the function of c-di-AMP is less well characterized. The DNA integrity-scanning protein DisA, found in Bacillus subtilis, synthesizes c-di-AMP in the presence of intact DNA to activate sporulation genes (33, 34). In addition, cell wall homeostasis is regulated by c-di-AMP, as elevated levels of this cyclic dinucleotide compensate for a lack of teichoic acids in Staphylococcus aureus (35), and enhances resistance to β-lactam antibiotics in B. subtilis by increasing peptidoglycan cross-linking (36). Overall, c-di-AMP appears to be important in Gram-positive bacteria for adaptations to membrane and cell wall stress.
In this study, we provide the first experimental evidence that C. trachomatis, a Gram-negative bacterium, synthesizes c-di-AMP during the late stages of infection and that the CT012 gene functions as a DAC. Furthermore, we show that c-di-AMP is largely responsible for driving type I IFN responses in Chlamydia-infected cells and that the sensing of this metabolite is mediated at the ER by the immune sensor STING.
RESULTS
The Chlamydia gene dacA (CT012) encodes a DAC.
The host cytosolic sensor STING could directly and indirectly sense RNA, DNA, and cyclic dinucleotides (17, 19, 24, 26, 37–41) to mediate IFN responses during Chlamydia infection (22, 42–47). Given recent evidence that STING senses cyclic dinucleotides directly (24), we were intrigued by the presence in C. trachomatis of an open reading frame (CT012) with sequence homology to the B. subtilis DAC domain-containing protein YbbP and the L. monocytogenes DAC DacA (Fig. 1A and B). B. subtilis contains three DAC domain-containing proteins (Fig. 1A), including the DNA integrity-scanning protein DisA (34). C. trachomatis CT012 has an N-terminal transmembrane (TM) domain and a putative catalytic domain with the conserved amino acids Asp-Gly-Ala (DGA) and Arg-His-Arg (RHR) found in bacterial DACs (Fig. 1B). On the basis of these similarities, we hypothesized that Chlamydia synthesizes c-di-AMP and that IFN responses may be mediated through the sensing of this metabolite in the host cytosol.
FIG 1 .
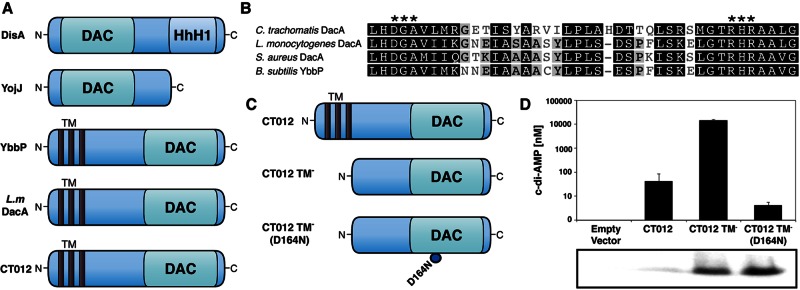
The C. trachomatis protein DacA (CT102) displays DAC activity. (A) Domain architecture of B. subtilis DAC domain-containing proteins DisA, YojJ, and Ybbp with L. monocytogenes DacA and the C. trachomatis putative DAC CT012. HhH1, helix-hairpin-helix motif; TM, three TM domains). (B) Amino acid sequence alignment of conserved domain regions of C. trachomatis DacA and DACs from c-di-AMP-producing bacteria with conserved residues critical for c-di-AMP synthesis indicated (asterisks). One hundred percent similarity, black; 80 to 100% similarity, dark gray; 60 to 80% similarity, light gray; <60% similarity, white. (C) Schematics of three C. trachomatis DacA recombinant variants used in this work to monitor c-di-AMP synthesis in E. coli. (D) UPLC-based quantification of c-di-AMP levels from nucleotide extracts of E. coli expressing full-length Chlamydia DacA, the DAC domain of DacA, or DacA with a D164N point mutation in the putative active site with corresponding immunoblot assays showing levels of DacA in soluble protein extracts. The data shown are means ± standard deviations of two independent experiments. Commercial c-di-AMP (Biolog) was used for calibration and quantification of nucleotide extracts.
We first determined if the putative Chlamydia DAC (CT012) is capable of synthesizing c-di-AMP. We expressed various hexahistidine-tagged forms of CT012 in Escherichia coli, which lacks any known DACs, including full-length CT012, a version lacking the TM domain (TM−) and a TM− CT012 with the aspartate in the DGA motif mutated to asparagine (Fig. 1C). Using ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS), we detected high levels of c-di-AMP in E. coli strains expressing either full-length or TM− CT012 but only minor amounts in strains expressing a D164N mutant form of TM− CT012 (Fig. 1D). Expression levels of these TM− variants of CT012 proteins were found to be similar by Western blotting (Fig. 1D). It is possible that, on a per-protein basis, full-length DacA is more active than the truncated forms. However, without a means to assess properly folded protein in this E. coli overexpression system, we cannot make any firm statement to that effect. Nonetheless, these findings indicate that CT012 has DAC activity and thus we have renamed the gene encoding this protein dacA.
C. trachomatis synthesizes c-di-AMP during infection.
To determine if Chlamydia synthesizes c-di-AMP during infection, nucleic acids were isolated from HeLa cells infected with C. trachomatis at various time points postinfection and analyzed by UPLC-MS/MS under conditions optimized for the separation and detection of cyclic dinucleotides. Synthetic c-di-AMP was used as the standard for the identification and quantification of c-di-AMP within extracts. c-di-AMP was detected in Chlamydia-infected cells as early as 12 h postinfection (hpi) and continued to increase throughout the infectious cycle (Fig. 2A). Peak c-di-AMP levels were observed at 36 hpi. It is unlikely that Chlamydia synthesizes c-di-GMP, a metabolite known to induce IFN responses, as c-di-GMP was undetectable in infected samples (data not shown) and Chlamydia lacks any proteins with GGDEF or EAL domains. We also generated DacA-specific antiserum that detected two immunoreactive bands consistent with the predicted DacA monomer and dimer molecular weights (Fig. 2A), consistent with recent observations with B. subtilis YbbP/CdaA (48), the closest homologue of Chlamydia DacA. The levels of DacA protein expression correlated with the abundance of c-di-AMP during infection and type I IFN responses in Chlamydia-infected murine embryonic fibroblasts (MEFs) (Fig. 2B).
FIG 2 .
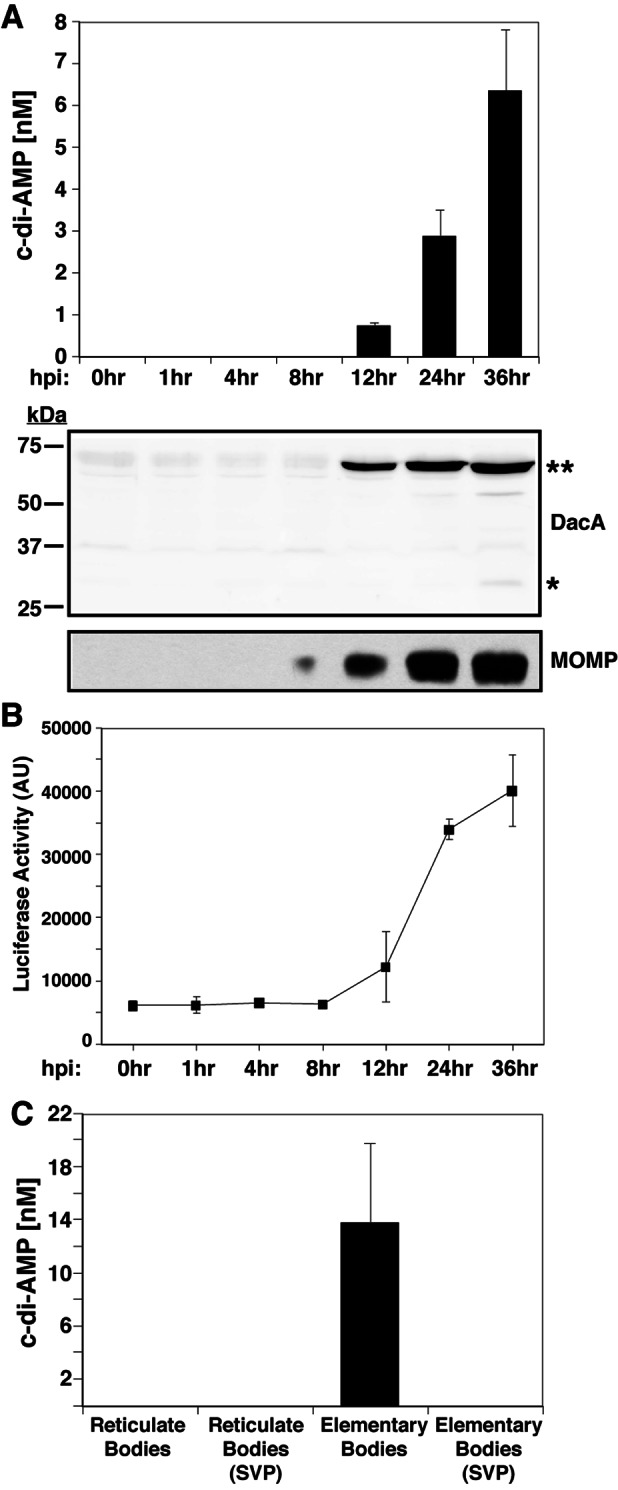
c-di-AMP is present in nucleic acid extracts from cells infected with C. trachomatis. (A) c-di-AMP levels in extracts of HeLa cells infected with C. trachomatis were quantified by UPLC at the indicated time points. The corresponding immunoblot shows Chlamydia DacA and MOMP expression levels. Asterisks indicate monomeric (*) and dimeric (**) forms of DacA. (B) MEFs induce type I IFN in response to Chlamydia infection. MEFs stably expressing an ISRE fused to a luciferase reporter gene were infected with C. trachomatis for the indicated times and lysed, and luciferase levels were assessed by an enzymatic assay. AU, arbitrary units. (C) c-di-AMP levels in lysates of density gradient-purified RBs and EBs. The signal for c-di-AMP was sensitive to SVP. The data shown are means ± standard deviations of two technical replicates.
We next compared the levels of c-di-AMP in infectious EBs with those in replicative RBs. Nucleic acids were extracted from density gradient-purified EBs and RBs and analyzed by UPLC-MS/MS. c-di-AMP was detected in EBs but not in RBs (Fig. 2C). The purity of gradient-purified EBs and RBs was assessed by their morphology (data not shown). These results are consistent with DNA microarray results (49) and proteomic data sets showing that DacA is expressed in EBs but not in RBs (50). EB lysates treated with snake venom phosphodiesterase (SVP) lost the peak corresponding to c-di-AMP, indicating that the peaks detected contain 3′-5′ phosphate linkages. Overall, these results indicate that Chlamydia synthesizes c-di-AMP during infection and that this metabolite accumulates primarily within EBs.
STING is required for type I IFN responses to C. trachomatis and cell autonomous control of bacterial replication.
Chlamydia induces type I IFN responses in a variety of cell types, and evidence from RNA interference-mediated gene silencing suggests that this response requires STING (22). We extended these findings to primary cells derived from STING-deficient animals. First, we determined that primary mouse lung fibroblasts (MLFs) induced significant levels of type I IFN as detected by the induction of luciferase expression in a reporter cell line containing an ISRE-luc transgene and by measuring secreted IFN-β by enzyme-linked immunosorbent assay (ELISA) (Fig. 2B and 3A and B). These MLFs also induced type I IFN in response to the delivery of the dsRNA analogue poly(I-C), dsDNA, and c-di-AMP to the cytosol (Fig. 3A), indicating that the sensing machinery for nucleic acids in the cytoplasm is intact in these cells.
FIG 3 .
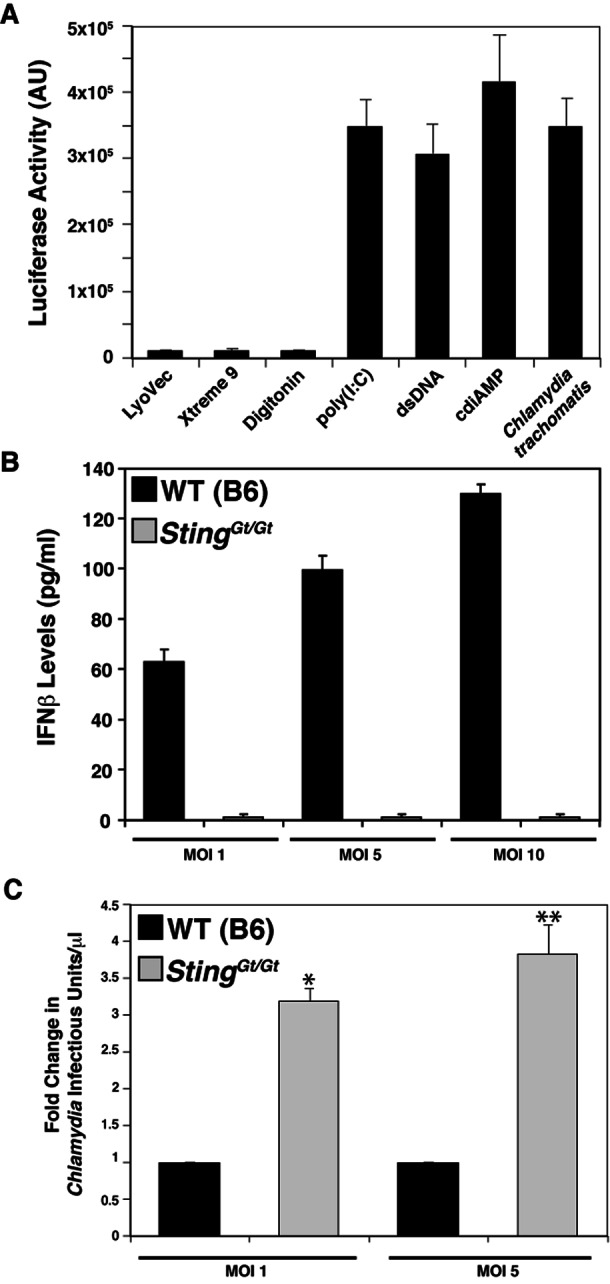
STING is required for the expression of type I IFN in mouse MLFs during C. trachomatis infection. (A) MLFs induce type I IFNs in response to various microbial-signal-like signals. MLFs were transfected with dsDNA or poly(I-C), loaded with c-di-AMP by digitonin permeabilization, or infected with C. trachomatis for 30 h. Cells treated with transfection (LuoVec and Xtreme9) or cell permeabilization reagents alone were included as controls. Levels of secreted type I IFNs in cell supernatants were assessed with a reporter cell line stably expressing an ISRE fused to a luciferase reporter gene and monitoring of luciferase enzymatic activity. AU, arbitrary units. (B) IFN-β responses to C. trachomatis infection requires STING. MLFs from wild-type (WT) and STINGGt/Gt (Goldenticket) mice were infected with C. trachomatis at the indicated MOIs for 30 h, and the levels of IFN-β in cell supernatants was determined by ELISA. (C) STING-deficient (STINGGt/Gt) MLFs are permissive for C. trachomatis replication. Wild-type and STINGGt/Gt MLFs were infected with C. trachomatis at the indicated MOIs, and the yield of infectious units was determined at 36 hpi. Fold change represents the ratio of the number of infectious units/μl observed in Goldenticket fibroblasts to that observed in wild-type fibroblasts. All data are means ± standard deviations of three independent samples ×, P < 0.001; ∗∗, P <0.001 (one-way analysis of variance and Newman-Keuls multiple-comparison test).
By forward genetic screening of N-ethyl-N-nitrosourea-mutagenized C57BL/6 mice, Sauer and colleagues identified a mouse strain with impaired type I IFN responses to L. monocytogenes (51). The loss of responsiveness in these mice (Goldenticket) was due to a destabilizing point mutation in STING. We isolated fibroblasts from wild-type and Goldenticket mouse lungs and tested their abilities to respond to Chlamydia infection. Goldenticket MLFs were unable to mount a type I IFN response to C. trachomatis at increasing multiplicities of infection (MOIs) (Fig. 3B), confirming STING’s essential role in mediating type I IFN responses to Chlamydia. Furthermore, we observed that wild-type MLFs produced fewer Chlamydia infectious units than did Goldenticket MLFs (Fig. 3C), indicating a protective role for type I IFNs in cell autonomous control of Chlamydia infection, as has been previously reported (46).
Chlamydia-induced type I IFN responses are mediated predominantly by c-di-AMP.
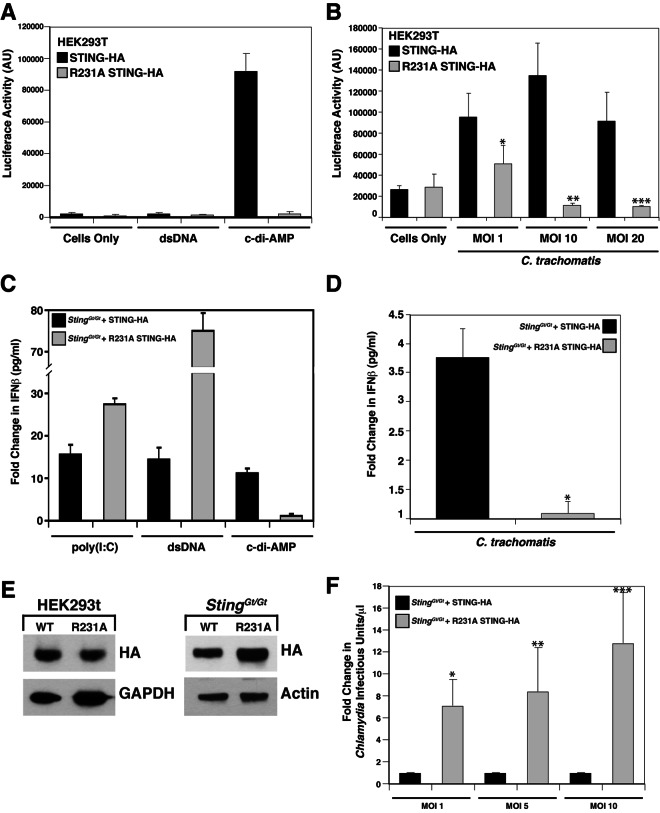
Because type I IFN responses to Chlamydia require STING (22), we postulated that sensing of Chlamydia is likely due to the detection of bacterial dsDNA, RNA, or c-di-AMP in the host cytoplasm. Burdette and colleagues recently identified a mutation (R231A) that renders STING incapable of responding to cyclic dinucleotides while retaining the ability to respond to dsDNA (24). To test the contribution of c-di-AMP to the triggering of type I IFN responses during Chlamydia infection, we stably expressed wild-type STING-hemagglutinin (HA) or STINGR231A-HA by retrovirus-mediated transduction of expression constructs into HEK293T cells. HEK293T cells do not express detectable levels of STING (24) or TLRs (52), and exogenously expressed STING is sufficient to recapitulate IFN responses to cyclic dinucleotides but not dsDNA (24) (Fig. 4A). Chlamydia infection triggered the expression of type I IFNs in HEK293T cells expressing wild-type STING-HA but not c-di-AMP-blind STINGR231A-HA (Fig. 4B). At high MOIs, we observed increased cytopathic effects (data not shown), which likely explains the apparent decrease in type I IFN responses. Overall, these findings indicate that c-di-AMP sensing during Chlamydia infection is a relevant inducer of IFN responses.
FIG 4 .
c-di-AMP is the main ligand responsible for Chlamydia-mediated type I IFN responses. (A and B) STING expression is sufficient for the induction of IFN responses during C. trachomatis infection. HEK293T cells stably expressing an ISRE-luciferase reporter were transduced with a wild-type STING-HA (black) or a STINGR231A-HA (gray) retroviral vector and treated with dsDNA or c-di-AMP (A) or infected with C. trachomatis for 30 h at the MOIs indicated (B). ×, P < 0.05; **, P < 0.001; ***, P < 0.001 (one-way analysis of variance and Newman-Keuls multiple-comparison test). Cells expressing mutant STINGR231A cannot induce type I IFNs in response to c-di-AMP. AU, arbitrary units. (C to F) MLFs expressing STING variants that cannot signal in response to cyclic dinucleotides fail to induce type I IFNs during C. trachomatis infection. Goldenticket MLFs transduced with a wild-type STING-HA (black) or a STINGR231A-HA (gray) construct were treated with poly(I-C), dsDNA, or c-di-AMP (C) or infected with C. trachomatis for 30 h (D). *, P < 0.05 (one-way analysis of variance and Newman-Keuls multiple-comparison test). Supernatants were tested for IFN-β levels by ELISA. Fold changes represent the ratios of total IFN-β detected by ELISA (in pg/ml) in cells treated with stimulants to that in untreated cells (C) and that in infected cells to that in uninfected cells (D). (E) Immunoblot analysis with anti-HA (STING) antibody of protein lysates from HEK293T cells and Goldenticket MLFs expressing various STING constructs indicates that comparable levels of STING were expressed in these cell lines. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin served as loading controls. WT, wild type. (F) Goldenticket MLFs expressing wild-type STING-HA (black) or STINGR231A-HA were infected with C. trachomatis, and infectious units were assessed as described in the legend to Fig. 3C. Note that Goldenticket MLF expressing the STING variant that cannot recognize ci-di-AMP did not induce IFN-β and control C. trachomatis replication. *, P < 0.05; **, P < 0.05; ***, P < 0.001 (one-way analysis of variance and Newman-Keuls multiple-comparison test). Fold changes represent the ratios of infectious units/μl observed in Goldenticket MLF complemented with STING-HAR231A-HA to those in wild-type STING-HA. The data shown are means ± standard deviations of three independent samples.
We could not directly address the role of Chlamydia DNA sensing in the induction of IFN responses in these cells because HEK293T cells do not express cytoplasmic dsDNA sensors that signal through STING (16, 24, 53). To address this problem, we complemented T-antigen/telomerase-immortalized (54) Goldenticket MLFs with a wild-type STING-HA or STINGR231A-HA expression construct and assessed the expression of type I IFNs during infection. We observed significant variability in IFN responses among batches of primary cells (data not shown) and thus chose to complement a clonal, immortalized Goldenticket MLF line with STING-HA constructs to minimize cell-to-cell variation. STINGR231A-HA MLFs secreted IFN in response to transfected cytosolic poly(I-C) and dsDNA (Fig. 4C) but did not respond to c-di-AMP (Fig. 4C) or C. trachomatis infection (Fig. 4D; see Fig. S2 in the supplemental material), indicating that c-di-AMP is the predominant Chlamydia-derived molecule recognized by the host cytoplasmic sensors in fibroblasts. While the expression of wild-type STING-HA and STINGR231A-HA, as assessed by immunoblotting, was comparable among HEK293T cells, STINGR231A-HA expression was slightly elevated compared to that of wild-type STING-HA in Goldenticket MLFs (Fig. 4E), which likely accounts for the observed elevated type I IFN responses to poly(I-C) and dsDNA (Fig. 4C). Finally, consistent with the role of type I IFNs in cell autonomous control of intracellular pathogens (46), C. trachomatis replication was significantly enhanced in Goldenticket MLFs expressing STINGR231A-HA compared to that in MLFs expressing wild-type STING-HA (Fig. 4F).
STING translocation from the ER to cytoplasmic signaling complexes upon c-di-AMP treatment correlates with the onset of type I IFN responses.
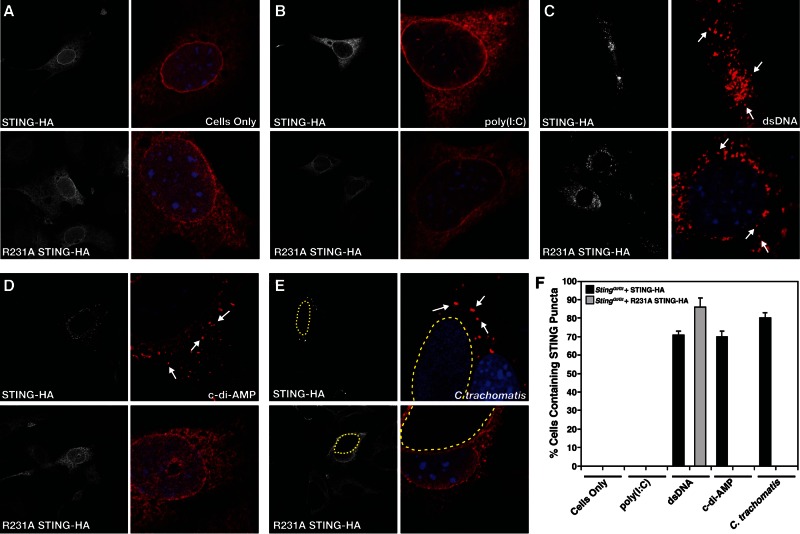
Upon the recognition of dsDNA in the cytoplasm, STING is redistributed from its steady-state localization at the ER to the Golgi apparatus and p62/SQSTSM-containing cytoplasmic structures (26). Exit from the ER is required for type I IFN activation in response to dsDNA (17), leading to the recruitment of serine/threonine-protein Tank-binding kinase 1 (TBK1), which phosphorylates the transcriptional factor IRF-3 (55). We tested if c-di-AMP is sufficient to mediate the relocalization of STING. At steady state, HA-tagged wild-type STING and STINGR231A localized to the ER-like structures in MLFs (see Fig. 6A). Goldenticket MLFs expressing HA-tagged wild-type STING, but not MLFs expressing STINGR231A, relocalized from the ER to punctate structures as early as 30 min posttreatment with c-di-AMP (Fig. 5). Although STINGR231A variants bind cyclic dinucleotides (24), we failed to see any STING translocation at the time points examined. In contrast, wild-type STING and STINGR231A were readily translocated to p62/SQSTSM-positive puncta upon transfection with dsDNA but not poly(I-C) (Fig. 6B and C; see Fig. S3 in the supplemental material). Neither c-di-AMP treatment (Fig. 6D) nor C. trachomatis infection (Fig. 6E) induced the translocation of STINGR231A. The percentages of cells containing STING-positive cytoplasmic punctate structures under all of the conditions tested are shown in Fig. 6F.
FIG 5 .
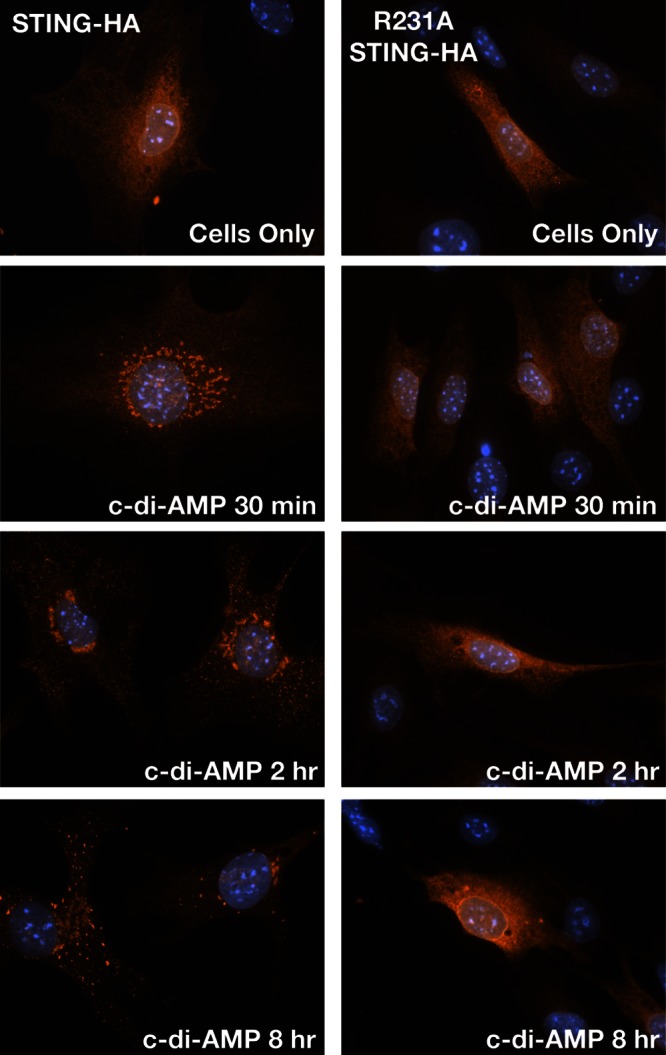
STING translocates to cytoplasmic structures in response to c-di-AMP. Goldenticket MLFs transduced with wild-type STING-HA (top) or STINGR231A-HA (bottom) were treated with c-di-AMP for the indicated times. Cells were immunostained with anti-HA (STING) antibody and imaged by epifluorescence microscopy.
FIG 6 .
(A) A STING variant (STINGR231A) that does not signal in response to c-di-AMP fails to translocate to cytoplasmic complexes upon Chlamydia infection. Immortalized Goldenticket MLFs expressing a wild-type STING-HA or a cyclic dinucleotide-blind STINGR231A-HA construct were treated with digitonin alone (A), transfected with the dsRNA analogue poly(I-C) (B) or dsDNA (C) for 24 h, loaded with c-di-AMP after digitonin permeabilization for 8 h (D), or infected with C. trachomatis for 30 h (E). Cells were fixed and stained with anti-HA (STING) antibody and imaged by confocal laser scanning microscopy. Note that STINGR231A cannot translocate to cytoplasmic punctate structures (arrows) in response to C. trachomatis infection or c-di-AMP stimulation but is responsive to dsDNA treatment. (F) Quantification of the percentage of cells displaying STING in cytoplasmic puncta after treatment with various inducers of type I IFNs. Cells displaying puncta (arrows) were scored as positive for STING translocation (n = 150).
Overall, these findings strongly suggest that c-di-AMP is the most prominent microbial ligand driving type I IFN responses during Chlamydia infection.
DISCUSSION
The Chlamydia-derived ligand(s) driving type I IFN responses and the corresponding sensing mechanism(s) are unclear. In this study, we provide evidence that Chlamydia synthesizes c-di-AMP and that this metabolite is a prominent driver of Chlamydia-induced IFN responses. To our knowledge, C. trachomatis is the first Gram-negative pathogen shown to synthesize c-di-AMP. Vibrio cholerae synthesizes a hybrid cyclic di-AMP-GMP molecule (56) via a protein lacking a canonical DAC domain, but whether this nucleic acid can induce IFN responses during infection is not known.
The role type I IFNs play in the control of Chlamydia infections in vivo is unclear. Mice lacking the type I IFN receptor gene (IFNAR−/−) cleared Chlamydia muridarum infections earlier and displayed reduced pathology in the oviducts (8), suggesting a detrimental role for type I IFNs in the outcome of genital tract infections with C. muridarum. However, type I IFNs can also induce cell autonomous defense mechanism with antichlamydial activities (46).
Both murine and human cell lines sense Chlamydia infections and mount type I IFN responses (45, 58–61). Some studies have implicated TLR3 in Chlamydia-mediated IFN responses in murine oviduct epithelial cells (21, 62), while others point to a role for STING in human cells (22). Here we provide genetic evidence supporting a prominent role for STING in the sensing of Chlamydia ligands by murine cells. MLFs derived from STING-deficient (Goldenticket) mice did not induce type I IFNs in response to C. trachomatis infection (Fig. 3B). Importantly, this defect in IFN expression by Goldenticket MLFs can be complemented with a wild-type copy of STING but not with a mutant form of STING (STINGR231A) that binds cyclic dinucleotides but cannot signal in response to them (24) (Fig. 4D). Because STINGR231A can still initiate IFN expression in response to cytoplasmic DNA, we conclude that chlamydial dsDNA in the cytoplasm is unlikely to be a major contributor to type I IFN responses during Chlamydia infection. A recent study suggested that the dsDNA sensor DDX41 (40) can also directly sense cyclic dinucleotides and requires STING as an adaptor protein (63). Whether sensing of c-di-AMP occurs directly through DDX41 or STING, the lack of IFN responses in cells expressing a STING variant blind to cyclic dinucleotide-dependent signaling argues that c-di-AMP, and not dsDNA, is the major ligand driving Chlamydia IFN responses. Similarly, Chlamydia-derived RNAs are unlikely to be a major inducer of IFN, as the expression of IFN-β in Chlamydia-infected cells is RIG-I and MAVS independent (22).
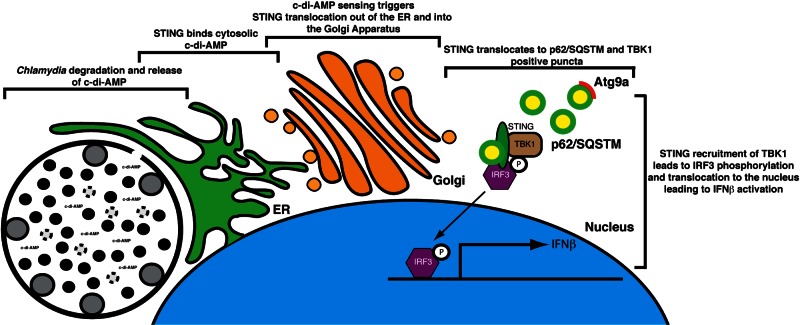
Unlike the cytosolic pathogen L. monocytogenes (64), where secreted metabolites are directly accessible to immune sensors in the cytoplasm, Chlamydia is surrounded by a membrane vacuole that is impermeable to small charged molecules >500 Da in size (65). This permeability barrier should limit the detection of chlamydial metabolites to microbial pattern recognition receptors. However, the inclusion is intimately associated with ER membranes, which has been interpreted as a pathogen-mediated process evolved to acquire lipids from the host (66–70). An alternative interpretation is that cells envelope large, abnormal vacuolar structures with ER membranes to increase the chance that sensors like STING can encounter microbe-derived compounds. In such a scenario, STING in the ER, an organelle that innervates the entire cytoplasm, has a greater likelihood of sensing chlamydial c-di-AMP and translocating to signaling platforms together with TBK1 and IRF3, as previously shown (29, 41, 71) (Fig. 7).
FIG 7 .
Model of activation of STING-dependent IFN-β responses in Chlamydia-infected cells. The bacterial metabolite c-di-AMP is synthesized by Chlamydia and sensed by STING at ER membranes enveloping the inclusion. STING is transported out of the ER (26) to the Golgi apparatus and eventually to p62/SQSTSM cytoplasmic complexes (see Fig. S3 in the supplemental material). As previously shown, the recruitment of TBK1 and IRF3 to these signaling platforms leads to IRF3 phosphorylation and translocation to the nucleus and expression of type I IFN genes (29, 71).
Upon sensing cytoplasmic dsDNA, STING relocalizes from the ER to the Golgi apparatus and cytoplasmic p62/SQSTSM-positive puncta (26). STING transport out of the ER upon the sensing of dsDNA is critical to the initiation of type I IFN responses (17). This intracellular transport event also occurs in response to cyclic dinucleotides, even in HEK293T cells, where cytoplasmic dsDNA sensors are not expressed (Fig. 4A) (24), and is important for immune signaling, as type I IFN induction in response to c-di-AMP is blocked by the fungal metabolite brefeldin A (unpublished data), an inhibitor of ER-to-Golgi apparatus transport (17). The molecular mechanisms regulating STING relocalization to cytoplasmic signaling complexes are unknown. Autophagy-related proteins have been implicated in the turnover of STING-positive signaling complexes, as Atg9a knockout cells show enhanced accumulation of STING and TBK1 in punctate structures and increased IFN-β production (26). Autophagy markers such as LC3 and Atg9a also colocalize with STING, yet STING-positive structures lack double-membrane-bound morphological characteristics of autophagosomes (26).
Unlike c-di-GMP, which has well-defined roles in bacterial virulence, including motility, biofilm formation, and virulence gene expression (72), the function of c-di-AMP in bacteria is less well understood. c-di-AMP was first identified in B. subtilis as a second messenger involved in signaling DNA integrity to regulate sporulation (33, 34). In S. aureus, c-di-AMP regulates peptidoglycan cross-linking and bacterial survival in the absence of lipoteichoic acids (35). Chlamydia contains a single gene for the synthesis of c-di-AMP, dacA, and a single putative phosphodiesterase gene with homology to proteins that cleave or inactivate c-di-AMP (73). While the role of c-di-AMP in C. trachomatis is unknown, the timing of c-di-AMP detection during infection mimics that of conversion of RB to EB forms (~15 hpi) and a quantitative proteomic study identified DacA only in purified EBs and not RBs (50). Given the prominent role of cyclic dinucleotides like c-di-GMP in the regulation of protein function, gene expression, and the posttranscriptional regulation of mRNAs, we speculate that c-di-AMP may have similar regulatory functions, especially during the transition from metabolically active RBs to infectious EBs.
As in other bacterial systems (29, 48), c-di-AMP may turn out to be an essential molecule in Chlamydia. Indeed, in a recent screen for essential Chlamydia genes, we have been unable to identify loss-of-function mutations in dacA (unpublished data). It is apt that an immune sensing mechanism would have evolved to detect conserved, essential bacterial metabolites.
MATERIALS AND METHODS
Organisms and cell culture.
C. trachomatis serovar LGV-L2 (strain 434/Bu) was grown in HeLa cells, MLFs, or HEK293T cells maintained in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum (FBS).
Primary LF isolation.
Primary lung fibroblasts (LFs) were isolated from Sting-deficient Goldenticket (51) and parental wild-type C57BL/6 mouse lungs. Briefly, lung tissues were minced and transferred into a sterile beaker containing a stir bar and 10 ml of Dulbecco’s modified Eagle’s medium (DMEM) with 0.14 Wunsch unit/ml Liberase Blendzyme 3 (Roche) and 1× antibiotic-antimycotic (penicillin-streptomycin and amphotericin B; Gibco). Samples were incubated at 37°C with slow stirring for 60 min, washed three times with 10 ml DMEM with 15% FBS and 1× antibiotic-antimycotic. Tubes were spun at 524 × g in a swinging-bucket centrifuge for 5 min, and the cell pellet was resuspended twice in 10 ml DMEM–15% FBS–1× antibiotic-antimycotic and transferred to a 10-cm dish. Cells were incubated at 37°C in a humidified 5% CO2 atmosphere. Primary LFs were stored in frozen aliquots or immortalized by retroviral delivery of simian virus 40 T antigen and human telomerase reverse transcriptase as previously described (54).
Plasmid constructs and virus production.
STING expression constructs (24) were delivered into HEK293T cells and immortalized LFs, and positive transductants were sorted on a FACSVantage sorter (BD) on the basis of the levels of enhanced green fluorescent protein (EGFP) expression. For stable expression of STING-HA, viruses were generated in 293T cells seeded into 10-cm dishes at 1 × 106 cells/dish and transfected with STING-HA retroviral constructs and packaging plasmid pCL10A1 at a 1:1 ratio with FuGENE6 (Promega). At 48 h posttransfection, the cell supernatant was collected, filtered over 0.44-µm filters (Pall), and added to immortalized Goldenticket LFs. Stable transductants were sorted on the basis of EGFP expression levels.
CT012 (dacA) and CT102 lacking the TM domain (minus amino acids 1 to 86) was inserted into pET28a for overexpression in BL21-CodonPlus-RIL cells (Stratagene). Mutations in the putative active site of CT012 were generated with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s guidelines.
Generation of anti-DacA antibodies.
Hexahistidine-tagged DacA was generated by cloning a fragment of CT012 lacking the TM domain into pET28a. Recombinant protein was produced in BL21-CodonPlus-RIL cells (Stratagene) and purified by affinity chromatography on nickel-charged resin (Qiagen). Purified, recombinant DAC was used to immunize a rabbit. The antiserum was precleared of cross-reactive antibodies by incubation with crude HeLa cell lysate at 4°C for 2 h prior to use in immunoblotting.
Analysis of DacA expression.
HeLa cell monolayers were infected with serovar LGV-L2 at an MOI of ~1. Infections were synchronized at 4°C, and total protein was collected at 0, 1, 4, 8, 12, 24, and 36 hpi by lysis in hot sample buffer (50 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 0.02% bromophenol blue), followed by sonication. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). Blots were blocked in 5% nonfat milk diluted in Tris-buffered saline (0.5 M Tris, 1.5 M NaCl, pH 7.4) with 0.1% Tween 20 (TBST) and incubated overnight with primary anti-Dac antibody diluted in 5% milk–TBST. The blots were washed in TBST and incubated with a horseradish peroxidase-linked anti-rabbit secondary antibody (GE Healthcare). Bound antibodies were detected with the SuperSignal West Dura detection kit (Thermo).
Cell stimulation.
Cells were infected with serovar LGV-L2 for 30 h or transfected with 250 ng/µl poly(I-C)/LyoVec (Invivogene) or 5 µg plasmid dsDNA (Xtreme 9; Roche) for 24 h. For c-di-AMP delivery, cells were incubated with 10 µg/ml digitonin (Calbiochem) and 10 µM c-di-AMP (Biolog) in permeabilization buffer (50 mM HEPES [pH 7], 100 mM KCl, 3 mM MgCl2, 85 mM sucrose, 0.2% bovine serum albumin, 0.1 mM dithiothreitol, 1 mM ATP, 0.1 mM GTP) for 30 min at 37°C in 5% CO2. The permeabilization buffer was removed and replaced with DMEM–10% FBS, and cells were incubated at 37°C in 5% CO2 for 8 h. Controls included digitonin permeabilization without c-di-AMP.
Type I IFN expression.
Expression of type IFNs was assessed either with reporter cell lines or by ELISA. For reporter cell line construction, lentiviruses containing an IFN-stimulated response element (ISRE)-luciferase reporter (SABiosciences) was used to stably transduce MEFs. These cells were seeded into 96-well plates in triplicate, and supernatants from cells treated with various IFN-inducing stimuli were added. Poststimulation, the medium was removed and cells were washed once with phosphate-buffered saline (PBS). Cells were lysed in 50 µl of britelite plus reagent per well (PerkinElmer), and the samples were transferred to a white OptiPlate (PerkinElmer) for measurement of luminescence on an EnSpire 2300 Multilabel Plate Reader (PerkinElmer).
For IFN-β ELISAs, cells were seeded into 96-well plates in triplicate at 1 × 104/well. Poststimulation, medium was collected and analyzed for IFN-β levels according to the manufacturer’s protocol (VeriKine Mouse IFN-β ELISA kit; Pestka Biomedical Laboratories). For MLFs expressing STING-HA constructs, results were normalized to the GFP expression of adherent cells as measured by an EnSpire 2300 Multilabel Reader (PerkinElmer).
c-di-AMP extraction.
E. coli BL21-CodonPlus-RIL (Stratagene) cells were used to overexpress Chlamydia DacA constructs. Overnight cultures were diluted in 100 ml minimal medium and incubated at 37°C for 3 h. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and cells were incubated at 37°C for 4 h (OD600 of 0.4). Cultures were spun at 5,100 rpm for 30 min at 4°C, and the bacterial pellets were resuspended in 250 µl extraction buffer (40:40:20 methanol-acetonitrile-H2O plus 0.1 N formic acid). Samples were incubated at −20°C for 30 min, and then insoluble material was pelleted by centrifugation at 13,000 rpm for 5 min at 4°C. A 200-µl volume of the supernatant was collected and neutralized with NH4HCO3 (0.6% final concentration). Samples were stored at −80°C until UPLC analysis. For c-di-AMP extraction from infected cells, HeLa cells were seeded into six-well plates and infected with C. trachomatis at an MOI of 1. Infections were synchronized by centrifugation at 3,000 rpm for 30 min at 4°C and then transferred to a 37°C 5% CO2 humidified incubator. At the indicated time points, infected cells were harvested and nucleotides were extracted from the pellet as described above.
UPLC-MS/MS analysis.
c-di-AMP was quantified by UPLC-MS/MS as previously described (74). Immediately before analysis, 100 µl of each sample was centrifuged under a vacuum for 2 h to remove the extraction buffer and the remaining pellet was then resuspended in 100 µl water. A 10-µl volume was analyzed on a Quattro Premier XE mass spectrometer (Waters) coupled with an Acquity Ultra Performance LC system (Waters) alongside an eight-point standard curve of purified c-di-AMP (Biolog) ranging from 250 to 1.95 nM. The limit of detection of cyclic dinucleotides was >1 nM. c-di-AMP was detected in all infected cell lines.
Isolation of EB and RB forms of Chlamydia.
Purification of C. trachomatis EBs and RBs was performed by density gradient centrifugation as previously described (50). To ensure the purity of serovar LGV-L2, stocks were plaque purified twice before generating EB and RB forms for c-di-AMP analysis. For c-di-AMP extraction, final pellets were resuspended in 250 µl extraction buffer and processed as described for E. coli.
Inclusion-forming unit assay.
Wild-type and Goldenticket MLFs were seeded at 1 × 104/well into a 96-well plate. Infections were conducted in triplicate at various MOIs. At 40 hpi, cells were lysed in H2O and SPG (3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM l-glutamic acid, 218 mM sucrose, pH 7.4) was added to a final concentration of 1×. Serial dilutions were used to infect HeLa cell monolayers for 24 h, at which point, cells were fixed with methanol and processed for immunofluorescence microscopy with anti-major outer membrane protein (anti-MOMP) antibodies. The number of inclusions per well was determined with a Cellomics High Content imaging system (Thermo Scientific).
Immunofluorescence assays for STING localization.
For routine immunofluorescence microscopy, cells were fixed with 3% formaldehyde–0.025% glutaraldehyde for 20 min, permeabilized with 0.1% Triton X-100–PBS for 20 min, and then blocked with 5% bovine serum albumin in PBS. The primary antibodies used were rat anti-HA (1:500; Roche) and rabbit anti-p62/SQSTSM1 (1:1,000; Sigma) antibodies. Primary antibodies diluted in PBS were incubated with permeabilized cells at 4°C for 1 h; this was followed by staining with fluorophore-conjugated secondary antibodies (Molecular Probes) for an additional 1 h. DNA was stained with Hoechst (Molecular Probes) at a concentration of 1:10,000 in PBS. Cells were mounted on slides with Mowiol (Sigma) and stored at 4°C. Cells were imaged by confocal laser scanning microscopy on a Zeiss 510 inverted confocal microscope.
SUPPLEMENTAL MATERIAL
c-di-AMP is present in nucleic acid extracts from HEK293T cells and MLFs infected with C. trachomatis. c-di-AMP levels in nucleic acid extracts of HEK293T cells (A) or MLFs (B) infected with C. trachomatis for 36 h were quantified by UPLC-MS. The data shown are means ± standard deviations of two independent experiments. (C) Representative chromatograph from UPLC-MS experiments highlighting detectable c-di-AMP peaks during the infection of HEK293T cells. Download
Expression of type I IFN during Chlamydia infection is dependent on the ability of STING to sense cyclic dinucleotides. MLFs were infected with C. trachomatis for 30 h. Levels of secreted type I IFNs in cell supernatants were assessed with a reporter cell line stably expressing an ISRE fused to a luciferase reporter gene and monitoring of luciferase enzymatic activity. Download
c-di-AMP and C. trachomatis induce STING translocation from the ER to p62/SQSTM vesicles. MLFs stably expressing STING-HA were treated with digitonin only (A) or c-di-AMP (B) for 8 h or infected with C. trachomatis for 30 h (C) and immunostained with anti-HA and anti-p62/SQSTM antibodies with 4ʹ,6-diamidino-2-phenylindole (DAPI) to detect STING-containing vesicles. Cells were imaged by confocal laser scanning microscopy. Download
ACKNOWLEDGMENTS
This work was supported by NIH award 1RO1-AI081694 to R.H.V. and NIH NRSA fellowships AI096866-02 to J.R.B. and AI091100 to D.L.B. Additional funding includes NIH grants AI063302 and AI080749 to R.E.V. and 2-U54-AI-057153 to C.M.W. R.H.V. and R.E.V. are Burroughs Welcome Investigators in the pathogenesis of Infectious Diseases.
We acknowledge the Michigan State University mass spectrometry facility for assistance in quantifying c-di-AMP.
Footnotes
Citation Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. 2013. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4(3):00018-13. doi:10.1128/mBio.00018-13.
REFERENCES
- 1. Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. 2010. Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 201:S134–S155 [DOI] [PubMed] [Google Scholar]
- 2. Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 [DOI] [PubMed] [Google Scholar]
- 3. Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820 [DOI] [PubMed] [Google Scholar]
- 5. Monroe KM, McWhirter SM, Vance RE. 2010. Induction of type I interferons by bacteria. Cell. Microbiol. 12:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kawai T, Akira S. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141:137–145 [DOI] [PubMed] [Google Scholar]
- 7. Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. 2012. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13:954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagarajan UM, Prantner D, Sikes JD, Andrews CW, Jr, Goodwin AM, Nagarajan S, Darville T. 2008. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect. Immun. 76:4642–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200:527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li XD, Knapp S, Kovarik P. 2011. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathog. 7:e1001345 http://dx.doi.10.1371/journal.ppat.1001345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 200:437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Núñez G. 2007. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 178:2380–2386 [DOI] [PubMed] [Google Scholar]
- 13. Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54–65 [DOI] [PubMed] [Google Scholar]
- 15. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 16. Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550 [DOI] [PubMed] [Google Scholar]
- 19. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 106:8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. 2008. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28:5014–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derbigny WA, Johnson RM, Toomey KS, Ofner S, Jayarapu K. 2010. The Chlamydia muridarum-induced IFN-beta response is TLR3-dependent in murine oviduct epithelial cells. J. Immunol. 185:6689–6697 [DOI] [PubMed] [Google Scholar]
- 22. Prantner D, Darville T, Nagarajan UM. 2010. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J. Immunol. 184:2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishikawa H, Barber GN. 2011. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell. Mol. Life Sci. 68:1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, Rixon FJ, Herold BC, Fitzgerald KA, Paludan SR. 2012. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 13:737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 106:20842–20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. 2012. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect. Immun. 80:1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abdul-Sater AA, Grajkowski A, Erdjument-Bromage H, Plumlee C, Levi A, Schreiber MT, Lee C, Shuman H, Beaucage SL, Schindler C. 2012. The overlapping host responses to bacterial cyclic dinucleotides. Microbes Infect. 14:188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Römling U. 2008. Great times for small molecules: c-di-AMP, a second messenger candidate in bacteria and Archaea. Sci. Signal. 1:e39 http://dx.doi.10.1126/scisignal.133pe39 [DOI] [PubMed] [Google Scholar]
- 32. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Witte G, Hartung S, Büttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30:167–178 [DOI] [PubMed] [Google Scholar]
- 35. Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7:e1002217 http://dx.doi.10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 83:623–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. 2012. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46:735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, Tasaka-Fujita M, Asahina Y, Yoneyama M, Fujita T, Watanabe M. 2013. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type-I interferon-dependent innate immunity. Hepatology 57:46–58 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. 2011. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 187:2595–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Devitt A, Lund PA, Morris AG, Pearce JH. 1996. Induction of alpha/beta interferon and dependent nitric oxide synthesis during Chlamydia trachomatis infection of McCoy cells in the absence of exogenous cytokine. Infect. Immun. 64:3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rödel J, Groh A, Vogelsang H, Lehmann M, Hartmann M, Straube E. 1998. Beta interferon is produced by Chlamydia trachomatis-infected fibroblast-like synoviocytes and inhibits gamma interferon-induced HLA-DR expression. Infect. Immun. 66:4491–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rödel J, Vogelsang H, Prager K, Hartmann M, Schmidt KH, Straube E. 2002. Role of interferon-stimulated gene factor 3gamma and beta interferon in HLA class I enhancement in synovial fibroblasts upon infection with Chlamydia trachomatis. Infect. Immun. 70:6140–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. 2005. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J. Immunol. 174:7186–7193 [DOI] [PubMed] [Google Scholar]
- 46. Vignola MJ, Kashatus DF, Taylor GA, Counter CM, Valdivia RH. 2010. cPLA2 regulates the expression of type I interferons and intracellular immunity to Chlamydia trachomatis. J. Biol. Chem. 285:21625–21635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Derbigny WA, Hong SC, Kerr MS, Temkit M, Johnson RM. 2007. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect. Immun. 75:1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. 2012. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high-level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 288:2004–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol. Microbiol. 82:1185–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, Endres S, Hartmann G. 2002. Quantitative expression of Toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168:4531–4537 [DOI] [PubMed] [Google Scholar]
- 53. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O’Hayer KM, Counter CM. 2006. A genetically defined normal human somatic cell system to study ras oncogenesis in vivo and in vitro. Methods Enzymol. 407:637–647 [DOI] [PubMed] [Google Scholar]
- 55. Burdette DL, Vance RE. 2013. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 14:19–26 [DOI] [PubMed] [Google Scholar]
- 56. Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard JC, Taylor GA, Dietrich WF, Starnbach MN. 2008. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J. Immunol. 180:6237–6245 [DOI] [PubMed] [Google Scholar]
- 58. Rödel J, Assefa S, Prochnau D, Woytas M, Hartmann M, Groh A, Straube E. 2001. Interferon-beta induction by Chlamydia pneumoniae in human smooth muscle cells. FEMS Immunol. Med. Microbiol. 32:9–15 [DOI] [PubMed] [Google Scholar]
- 59. Nagarajan UM, Ojcius DM, Stahl L, Rank RG, Darville T. 2005. Chlamydia trachomatis induces expression of IFN-gamma-inducible protein 10 and IFN-beta independent of TLR2 and TLR4, but largely dependent on MyD88. J. Immunol. 175:450–460 [DOI] [PubMed] [Google Scholar]
- 60. Ishihara T, Aga M, Hino K, Ushio C, Taniguchi M, Iwaki K, Ikeda M, Kurimoto M. 2005. Inhibition of Chlamydia trachomatis growth by human interferon-alpha: mechanisms and synergistic effect with interferon-gamma and tumor necrosis factor-alpha. Biomed. Res. 26:179–185 [DOI] [PubMed] [Google Scholar]
- 61. Derbigny WA, Kerr MS, Johnson RM. 2005. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J. Immunol. 175:6065–6075 [DOI] [PubMed] [Google Scholar]
- 62. Derbigny WA, Shobe LR, Kamran JC, Toomey KS, Ofner S. 2012. Identifying a role for Toll-like receptor 3 in the innate immune response to Chlamydia muridarum infection in murine oviduct epithelial cells. Infect. Immun. 80:254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. 2012. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13:1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Böttcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. 2012. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31:4153–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heinzen RA, Hackstadt T. 1997. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect. Immun. 65:1088–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651 [DOI] [PubMed] [Google Scholar]
- 68. Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. 2011. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 7:e1002198 http://dx.doi.10.1371/journal.ppat.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Derré I, Swiss R, Agaisse H. 2011. The lipid transfer protein CERT interacts with the chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 7:e1002092 http://dx.doi.10.1371/journal.ppat.1002092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dumoux M, Clare DK, Saibil HR, Hayward RD. 2012. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic 13:1612–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20 http://dx.doi.10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U. S. A. 109:12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
c-di-AMP is present in nucleic acid extracts from HEK293T cells and MLFs infected with C. trachomatis. c-di-AMP levels in nucleic acid extracts of HEK293T cells (A) or MLFs (B) infected with C. trachomatis for 36 h were quantified by UPLC-MS. The data shown are means ± standard deviations of two independent experiments. (C) Representative chromatograph from UPLC-MS experiments highlighting detectable c-di-AMP peaks during the infection of HEK293T cells. Download
Expression of type I IFN during Chlamydia infection is dependent on the ability of STING to sense cyclic dinucleotides. MLFs were infected with C. trachomatis for 30 h. Levels of secreted type I IFNs in cell supernatants were assessed with a reporter cell line stably expressing an ISRE fused to a luciferase reporter gene and monitoring of luciferase enzymatic activity. Download
c-di-AMP and C. trachomatis induce STING translocation from the ER to p62/SQSTM vesicles. MLFs stably expressing STING-HA were treated with digitonin only (A) or c-di-AMP (B) for 8 h or infected with C. trachomatis for 30 h (C) and immunostained with anti-HA and anti-p62/SQSTM antibodies with 4ʹ,6-diamidino-2-phenylindole (DAPI) to detect STING-containing vesicles. Cells were imaged by confocal laser scanning microscopy. Download