ABSTRACT
Pseudomonas aeruginosa is a ubiquitous microorganism and the most common Gram-negative bacterium associated with nosocomial pneumonia, which is a leading cause of mortality among critically ill patients. Although many virulence factors have been identified in this pathogen, little is known about the bacterial components required to initiate infection in the host. Here, we identified a unique trimethyl lysine posttranslational modification of elongation factor Tu as a previously unrecognized bacterial ligand involved in early host colonization by P. aeruginosa. This modification is carried out by a novel methyltransferase, here named elongation factor Tu-modifying enzyme, resulting in a motif that is a structural mimic of the phosphorylcholine present in platelet-activating factor. This novel motif mediates bacterial attachment to airway respiratory cells through platelet-activating factor receptor and is a major virulence factor, expression of which is a prerequisite to pneumonia in a murine model of respiratory infection.
IMPORTANCE
Phosphorylcholine is an interesting molecule from the microbiological and immunological point of view. It is a crucial epitope for the virulence of many important human pathogens, modulates the host immune response, and is involved in a wide number of processes ranging from allergy to inflammation. Our current work identifies a novel bacterial surface epitope structurally and functionally similar to phosphorylcholine. This novel epitope is crucial for initial colonization of the respiratory tract by Pseudomonas aeruginosa and for development of pneumonia. This opens up new targets for the development of novel drugs to prevent P. aeruginosa pneumonia, which is particularly important given the frequent emergence of multidrug-resistant strains.
Introduction
Pseudomonas aeruginosa is the most common Gram-negative organism associated with nosocomial pneumonia and is the leading cause of mortality among critically ill patients with airways damaged from mechanical ventilation, trauma, or antecedent viral infection (1). The majority of these infections result from aspiration of the microorganism colonizing the mucosal surfaces of the oropharyngeal airways (1). Understanding the early molecular events that facilitate infection is key for the identification of effective prevention and treatment strategies for P. aeruginosa pneumonia.
Phosphorylcholine (ChoP) is a common epitope present on the surface of major pathogenic bacterial species, including two major human pathogens of the respiratory tract, Streptococcus pneumoniae and Haemophilus influenzae (2, 3). In these pathogens, ChoP acts as a mimic of the eukaryotic platelet-activating factor (PAF). The interaction of ChoP with PAF receptor (PAFR) on the host cells is a crucial step for the virulence of these pathogens (4–6). In previous work, utilizing monoclonal antibodies (MAbs) specific for ChoP, we detected this posttranslational modification on the elongation factor Tu (EF-Tu) on the surface of P. aeruginosa (7). Although generally considered to be a cytoplasmic protein, EF-Tu can also be found surface exposed in many microorganisms, including P. aeruginosa, where it mediates the binding of factor H, conferring resistance to human complement (8).
In all 92 P. aeruginosa clinical isolates studied, the modification of EF-Tu was detected at higher levels at environmental temperatures than at 37°C (7), suggesting a role at early stages of P. aeruginosa infection.
In the present study, through a combination of approaches, including mass spectrometry of purified modified or unmodified EF-Tu, site-directed mutagenesis of key residues, and genetic loss-of-function/gain-of-function studies, we demonstrate that P. aeruginosa mimics ChoP by the transfer of three methyl groups to a lysine on EF-Tu, resulting in a chemical structure similar to that of ChoP. Furthermore, binding and invasion experiments with cultured human airway cells treated with PAFR antagonist or with specific PAFR small interfering RNA (siRNA) demonstrate that this modification of EF-Tu mediates the attachment of P. aeruginosa to host cells. Thus, this motif represents a novel bacterial strategy to mimic the structure of the natural ligand of PAFR that P. aeruginosa exploits to initiate colonization of the respiratory tract and cause pneumonia.
RESULTS
Identification of a unique gene involved in EF-Tu modification.
In order to identify the gene(s) responsible for the posttranslational modification of EF-Tu, we screened by Western blot analysis the PA14 mutants from the transposon mutant library of genes potentially involved in the synthesis of ChoP that had been identified in a number of different bacteria (see Table S1 in the supplemental material). None of the P. aeruginosa PA14 transposon mutants in the homologs of these genes showed altered ChoP expression, indicating that the mechanism responsible for EF-Tu modification differs from previously characterized pathways.
A systematic analysis of the ordered transposon mutant library in P. aeruginosa PA14 was undertaken; whole-cell lysates of 5,514 PA14 mutants were individually screened for the lack of modified EF-Tu by Western blotting. Only one mutant, PA14_09870::MAR2xT7, with the transposon located in a gene encoding a hypothetical methyltransferase, showed no reactivity with the ChoP-specific MAb TEPC-15 at either 25°C or 37°C (Fig. 1A). We refer to this gene as eftM (EF-Tu-modifying enzyme). To confirm these results, we used allelic exchange to delete a 100-bp fragment within the locus of the homologous PAO1 gene, PA4178, to derive the mutant PAO1ΔeftM from PAO1. In contrast to the wild-type (WT) strain, PAO1ΔeftM did not modify EF-Tu (Fig. 1A), while the level of EF-Tu was the same (Fig. 1B). Wild-type strain PAO1 and the mutant showed similar growth rates with nonsignificant differences at 25°C or 37°C; doubling time in the mid-log phase ranged from 33 min to 36 min at 37°C and from 92 to 96 min at 25°C. Furthermore, the total amounts of protein per bacterium of the two strains were similar (PAO1, 1,100 ± 80 fg/CFU; PAO1ΔeftM, 1,000 ± 10 fg/CFU).
FIG 1 .
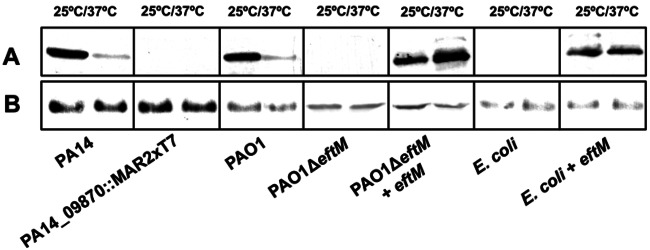
Western blot analysis of EF-Tu modification in different strains. Proteins of whole-cell extracts from different strains grown at 25°C or 37°C were subjected to electrophoresis and immunoblot analysis using antibodies specific for ChoP (MAb TEPC-15) (A) or specific for EF-Tu as a loading control (B).
The EftM protein is conserved in all completed genomes of P. aeruginosa strains present at http://www.pseudomonas.com (9), with one exception, P. aeruginosa DK2 (a longitudinal isolate from a cystic fibrosis [CF] patient that is known to have genomic deletions [10, 11]). Of these P. aeruginosa strains, PA7 has the lowest sequence identity (87%), while all other strains have >99% sequence identity, to PAO1 EftM. Homologs of EftM are also present in many other Pseudomonas species, including P. mendocina, P. stutzeri, P. fulva, and P. syringae. BLAST analysis using the EftM protein sequence indicates the broad distribution of homologs within other gammaproteobacteria (including Shewanella sp. and Vibrio sp.), as well as within the Firmicutes.
EftM trimethylates EF-Tu lysine residue 5.
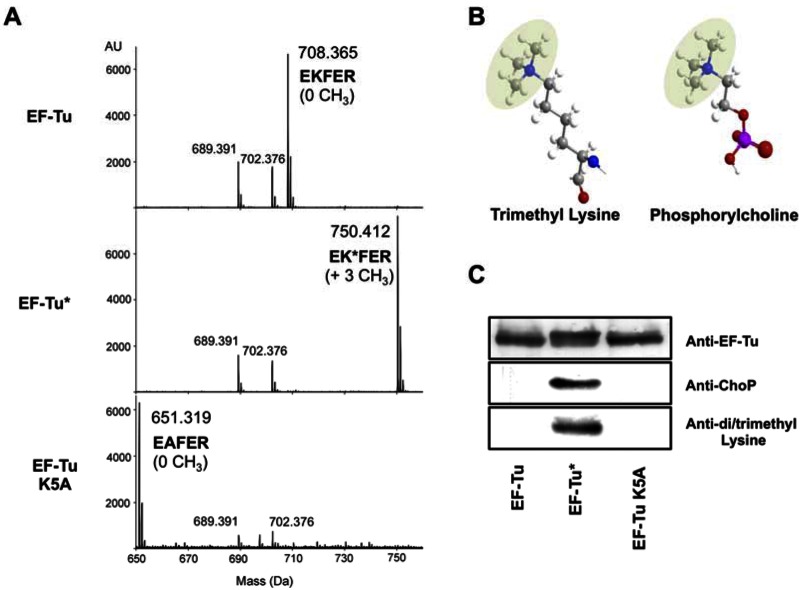
A tandem mass spectrometry approach was chosen to characterize the modification of EF-Tu. Analysis of purified EF-Tu from PAO1ΔeftM digested with trypsin identified a peak with a mass of 708.365 Da corresponding to a peptide with the residues 4 to 8 (EKFER) of P. aeruginosa EF-Tu (Fig. 2A, top). Similar analysis of EF-Tu purified from PAO1 grown at 25°C identified a peak with a mass of 750.412 Da (Fig. 2A, middle). Surprisingly, this increase corresponded to the addition of three methyl groups (42.047 Da) to lysine residue 5, resulting in a chemical structure similar to the ChoP epitope present in PAF (Fig. 2B). Analysis of purified EF-Tu carrying a point mutation replacing the conserved lysine residue 5 of the protein with alanine (EF-Tu K5A) showed the absence of the methyl groups (Fig. 2A, bottom). Purified EF-Tu from PAO1 grown at 25°C (referred to as EF-Tu*) reacted with the MAb TEPC-15 and with an anti-di/trimethyl lysine-specific antibody, unlike purified EF-Tu from PAO1ΔeftM or EF-Tu K5A (Fig. 2C). These data indicate that the posttranslational modification of EF-Tu detected by the anti-ChoP antibody is in fact a trimethylation of EF-Tu lysine residue 5.
FIG 2 .
Trimethylation of EF-Tu by the elongation factor Tu-modifying enzyme (EftM). (A) Tandem mass spectrometry analysis of purified EF-Tu from the EftM-deficient mutant PAO1ΔeftM (EF-Tu, top), from the WT strain PAO1 (EF-Tu*, middle), and from PAO1 harboring the plasmid containing a K5A mutation in the tufB gene (EF-Tu K5A, bottom) digested with trypsin. (B) Molecular structure of trimethyl lysine and phosphorylcholine with the methyl groups shadowed on both molecules. Atoms of hydrogen, carbon, nitrogen, oxygen, and phosphorus are colored in white, gray, blue, red, and pink, respectively. (C) A Western blot of recombinant wild-type EF-Tu isolated from the EftM-deficient mutant (EF-Tu) and from the WT strain PAO1 (EF-Tu*) and recombinant EF-Tu harboring a K5A mutation in the tufB gene isolated from PAO1 (EF-Tu K5A) was probed with anti-EF-Tu, anti-ChoP (MAb TEPC-15), and anti-di/trimethyl lysine antibodies.
To further demonstrate the activity of EftM, we transferred a plasmid expressing the eftM gene to PAO1ΔeftM as well as to Escherichia coli DH5α, which lacks this gene in its genome. Western blot analysis of EF-Tu from all strains harboring this plasmid demonstrated that eftM is sufficient to modify EF-Tu even in a heterologous system such as E. coli (Fig. 1A). Moreover, mass spectrometry analysis of P. aeruginosa EF-Tu purified from E. coli expressing eftM identified the same modification as that observed in PAO1 grown at 25°C (see Fig. S1 in the supplemental material). Altogether, these results indicate that eftM encodes a novel methyltransferase that modifies lysine 5 of EF-Tu, resulting in a chemical structure similar to that of ChoP present in PAF.
Trimethylation of EF-Tu mimics PAF to interact with PAFR.
The effect of EF-Tu modification with three methyl groups as a structural mimic of PAF with respect to its interaction with epithelial cells was evaluated. Both oropharyngeal epithelial cells and bronchoepithelial cells bound purified methylated EF-Tu (EF-Tu*) almost 2-fold more efficiently than nonmodified EF-Tu (Fig. 3A). Accordingly, the ability of PAO1ΔeftM to enter both types of airway epithelial cells was impaired compared with that of PAO1 or the mutant complemented with eftM (Fig. 3B). To investigate whether trimethylated EF-Tu could interact with PAFR on the airway epithelial cells and thereby affect adherence and uptake of P. aeruginosa, we performed bacterial invasion assays with cells treated with a PAFR antagonist or with PAFR-specific small interfering RNA (siRNA). PAFR antagonist decreased the internalization of PAO1 and PAO1ΔeftM complemented with eftM in a dose-dependent manner but had no effect on the internalization rate of PAO1ΔeftM (see Fig. S2 in the supplemental material). To confirm this result, we reduced the PAFR cellular expression by more than 90% using PAFR siRNA (Fig. 3C). Control cells were transfected with a scrambled siRNA sequence. PAFR knockdown reduced the internalization of PAO1 to ~60% of the levels seen with the control cells, whereas the internalization of the mutant PAO1ΔeftM was not affected by the absence of PAFR (Fig. 3D).
FIG 3 .
Effect of trimethylation of EF-Tu on the interaction with the host cells. (A) Purified nonmethylated EF-Tu (EF-Tu) and methylated EF-Tu (EF-Tu*) were labeled with IRDye 800CW and incubated either with oropharyngeal cells or with bronchoepithelial cells. Binding of each fluorescent protein to each cell line was measured after exhaustive washing of the cells. (B) Internalization of P. aeruginosa by oropharyngeal cells or bronchoepithelial cells. Monolayers of cells were incubated with the indicated P. aeruginosa strains grown at 25°C, and the amount of bacterial internalization was determined. (C) Whole-cell lysates from oropharyngeal and bronchoepithelial cells that had been treated with either a scrambled siRNA sequence or PAFR-specific siRNA were subjected to electrophoresis and immunoblot analysis with PAFR-specific antibodies or β-actin-specific antibodies as loading control. Specific siRNA treatment reduced the level of PAFR by >90%. (D) Scrambled or PAFR-specific siRNA-treated cells were incubated with the indicated P. aeruginosa strains, and the amount of bacterial internalization was determined. The internalization of P. aeruginosa by cells treated with PAFR-specific siRNA is plotted as a percentage of that obtained for scrambled siRNA-treated cells. Three independent experiments, each done in triplicate, were performed. Error bars represent standard errors of the means. Statistical analyses were done with Student’s unpaired two-tailed t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
EF-Tu trimethylation is a prerequisite to cause pneumonia.
To gain insight into the potential impact of the EF-Tu modification in the development of P. aeruginosa respiratory infection, we tested the ability of the EftM-deficient mutant to cause pneumonia and fatal infection in a murine model. In order to simulate the initial phase of the infection from an exogenous source and given that EF-Tu modification is maximal at 25°C (Fig. 1) (12), animals were infected intranasally with 2 × 107 CFU of PAO1 or PAO1ΔeftM grown at 25°C. Analysis of survival indicated that mutation of eftM impaired the virulence of P. aeruginosa (Fig. 4A). The 50% lethal dose (LD50) of the mutant was higher (1.99 × 107 CFU) than that of the wild-type strain (2.25 × 106 CFU), and the mean time to death for an infection dose of 2 × 107 bacteria was shorter for PAO1 (26.75 h) than for PAO1ΔeftM (80 h). After 24 h, mice infected with PAO1 had significantly higher levels of bacteria in their nasal cavities and in their lungs compared to levels in mice infected with PAO1ΔeftM (Fig. 4B), although those differences were detectable 12 h postinfection (see Fig. S3 in the supplemental material), which indicated a role for trimethylation of EF-Tu in the early stages of the infection.
FIG 4 .

Effect of trimethylation of EF-Tu on P. aeruginosa respiratory infection. (A) Survival curves over 96 h of mice infected intranasally with 2 × 107 CFU of P. aeruginosa strain PAO1 (n = 6) or its derived isogenic mutant PAO1ΔeftM (n = 6). Hazard ratio = 23.52; 95% confidence interval (CI), 4 to 138; P = 0.005. (B) Bacterial load in the nasal wash and in the lungs of mice (n = 19 for each strain) 24 h after infection with the indicated P. aeruginosa strains. Error bars represent standard errors of the means. *, P < 0.05, and ***, P < 0.001, by Student’s unpaired two-tailed t test.
To establish the in vivo role of the methylated EF-Tu–PAFR interaction in P. aeruginosa virulence, we tested a PAFR antagonist for its ability to reduce, transiently, the bacterial load recovered from the nasal cavities and lungs of mice infected with PAO1 or PAO1ΔeftM grown at room temperature. Administration of a single dose of the antagonist to the nostrils of mice just prior to infection resulted in a 20-fold decrease in bacterial burden in the nasal wash and lungs of mice infected with PAO1 compared with those of mice treated with saline. In contrast, the treatment with the antagonist had no effect on the bacterial load of the mutant from the airway tract (Fig. 5A), suggesting that the interaction of modified EF-Tu with PAFR is specific and critical in the early stages of the infection.
FIG 5 .

In vivo role of methylated EF-Tu–PAFR interaction in a murine P. aeruginosa respiratory infection. (A) Mice (n = 8 per group) were treated with saline or PAFR antagonist (50 nM) and, after 15 min, infected intranasally with 2 × 107 CFU of the indicated P. aeruginosa strains. Bacterial loads in the nasal wash and in the lung were determined 12 h after the infection. (B) Twenty-four hours after intranasal inoculation with 2 × 107 CFU of the indicated P. aeruginosa strains, the bacterial loads in the nasal wash and in the lung were determined in WT BALB/c and PAFR−/− mice (n = 19 WT, 8 PAFR−/−). Error bars represent standard errors of the means. *, P < 0.05, and ***, P < 0.001, by Student’s unpaired two-tailed t test.
Consistently, PAFR−/− mice were as resistant as wild-type mice to the infection by the mutant PAO1ΔeftM (Fig. 5B). However, as previously described (13), 24 h postinfection, PAFR−/− mice were more susceptible to the infection by PAO1 than were the wild-type mice, likely due to defects in phagocytosis. Thus, PAFR contributes to resistance to P. aeruginosa respiratory infection by recognition of trimethylated EF-Tu, a posttranslational modification which is drastically reduced at 37°C.
DISCUSSION
The results described in this report show that trimethylation represents a novel strategy to structurally mimic PAF that P. aeruginosa exploits to decorate EF-Tu and interact with PAFR to promote respiratory infection.
EF-Tu is a highly conserved protein that plays a major role in mRNA translation by positioning the incoming aminoacyl-tRNAs (aa-tRNAs) in the ribosomal A site. EF-Tu accounts for 5 to 9% of total bacterial cell protein and thus is one of the most abundant proteins in bacteria. EF-Tu is generally considered to be a cytoplasmic protein. However, EF-Tu has also been described to be surface exposed in many microorganisms such as Acinetobacter baumannii (14), Mycoplasma pneumoniae (15), or Mycobacterium tuberculosis (16). In these pathogens, EF-Tu mediates the interaction between the microorganism and specific cellular and humoral host components. P. aeruginosa also exposes EF-Tu on the cell surface (7, 8) and mediates the binding of factor H conferring resistance to human complement attack (8). Furthermore, using specific antibodies against ChoP, we also detected the presence of this motif on the P. aeruginosa surface (7). In this work, we demonstrate that the posttranslational modification of EF-Tu detected by the anti-ChoP antibody is in fact a trimethylation of lysine residue 5 present in the N terminus of EF-Tu, which, in combination with our previous results, indicates that this region of the protein is surface exposed and may be involved in the binding of the complement regulator component.
To date, ChoP is the most common bacterial epitope that mimics PAF (2, 3, 6, 17). Thus, trimethylation of EF-Tu lysine 5 represents a novel bacterial strategy to mimic the structure of the natural ligand of the PAFR. Our findings indicate that trimethylation of EF-Tu mediates the interaction of P. aeruginosa with human epithelial cells via PAFR but does not confer increased susceptibility to the bactericidal effect of the complement (see Fig. S4 in the supplemental material), as occurs with ChoP in other microorganisms (6, 18), suggesting that trimethylation differs from ChoP in some critical aspects of bacterium-host interactions. It would be interesting to know whether other pathogens in which the presence of ChoP has been detected linked to surface proteins only by using specific antibodies (18) contain the same motif to mimic this epitope. On the other hand, it is likely that other pathogens which present trimethylated surface proteins, such as Rickettsia prowazekii (19), use a strategy similar to that described for P. aeruginosa in this study to impact their virulence.
Previous studies indicate that EF-Tu methylation regulates translation in other microorganisms (20). Comparisons of P. aeruginosa EF-Tu structure to EF-Tu structures from other microorganisms and three-dimensional (3D) modeling indicate that lysine residue 5 is located on the surface-exposed portion of the protein distant from the GTP/GDP or tRNA binding domains of the protein that are crucial for its activity (21). However, methylation of EF-Tu could potentially alter the accommodation of the amino acyl-tRNA in the ribosomal A site. For this reason, although growth curves and the total amounts of protein per bacterium of the WT strain PAO1 and the EftM-deficient mutant were similar, we cannot exclude the possibility that the results obtained in our mouse model of respiratory infection could be partially due to specific protein expression differences between strains expressing or not expressing modified EF-Tu. Nevertheless, with our experiments using a PAFR antagonist, specific PAFR siRNA treatment of the epithelial cells, or PAFR−/− mice, we demonstrate that one of the major effects of EF-Tu trimethylation on P. aeruginosa virulence resides in its ability to interact with PAFR.
Our findings indicate that the EF-Tu modification is maximal at room temperature and that it is quickly reduced at body temperature, suggesting that trimethylation of EF-Tu acts as a ligand of P. aeruginosa for PAFR on the airway epithelium at the initial stages of lung infection. It will be of interest to determine whether this EF-Tu–PAFR interaction is also crucial to initiate P. aeruginosa infections following burns or eye injury. Further, it remains to be determined how these initial interactions alter epithelial cell responses and the overall host response to infection. It is likely that modified EF-Tu stimulates downstream signaling events originating at the PAFR promoting P. aeruginosa infection. A similar process occurs with S. pneumoniae, where the interaction of ChoP with PAFR mediates the invasion of the endothelium and allows the bacteria to reach the subarachnoid space to cause meningitis (22).
A thorough understanding of these steps will be critical in order to develop drugs to block this bacterial ligand-host cell interaction or inhibit the activity of this new methyltransferase, EftM.
MATERIALS AND METHODS
Bacteria and cell lines.
P. aeruginosa reference strains PAO1 and PA14 and the nonredundant transposon insertion mutant library of PA14 (23) were used in this study. E. coli strains XL1-Blue, DH5α, and S17-1 were used in the cloning experiments. Bacteria were grown at 37°C or 25°C in Luria-Bertani (LB) broth or on LB solidified with 1.5% agar.
Human bronchoepithelial immortalized cells (16HBE14o−) and human dysplastic oral keratinocytes (DOK) (European Collection of Cell Cultures 94122104; Salisbury, Wiltshire, United Kingdom) were used in this study. The cells were propagated in Earle’s minimal essential medium plus 1% l-glutamine culture medium or in Dulbecco’s modified Eagle’s medium, respectively. Both media were supplemented with 10% fetal calf serum and 1% penicillin and streptomycin. Before use in experiments, cells were incubated in tissue culture plates at 37°C and 5% CO2, until confluence was reached.
Western blot analysis.
To study the expression of EF-Tu and its posttranslational modification, equivalent numbers of bacterial cells were resuspended in Laemmli buffer, boiled for 5 min, and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Separated proteins were transferred onto Immobilon-P membranes which were blocked for 2 h at room temperature with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) and incubated overnight at 4°C with the following primary antibodies: mouse MAb TEPC-15 (Sigma-Aldrich, St. Louis, MO) specific for the ChoP epitope; a rabbit polyclonal antibody, anti-EF-Tu (7); or a rabbit polyclonal pan anti-di/trimethyl lysine antibody (Upstate, Temecula, CA). The membranes were subsequently washed three times with PBS and incubated for 1 h at room temperature with alkaline phosphatase- or peroxidase-conjugated (Sigma-Aldrich, St. Louis, MO) secondary antibody. Finally, membranes were treated with the Fast 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium kit (Sigma-Aldrich, St. Louis, MO) or enhanced chemiluminescence (ECL) Western blotting substrate (Thermo Scientific, Rockford, IL).
To evaluate the expression of PAFR, epithelial cell proteins were extracted using RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA) and the concentration of solubilized protein samples was measured using a Coomassie blue staining protein assay kit according to the manufacturer’s instructions (Bio-Rad, Hercules, CA). The solubilized proteins were then boiled for 5 min in Laemmli buffer; each lane was loaded with 20 µg of protein before separation, transfer, and detection as described above. PAFR and β-actin (used as a loading control) were detected using a rabbit polyclonal anti-PAFR antibody (Cayman, Ann Arbor, MI) and with mouse monoclonal anti-β-actin (Sigma-Aldrich, St. Louis, MO), respectively. Alkaline phosphatase-conjugated secondary antibodies were used for detection and developed as described above.
Construction of mutants and plasmids.
We partially deleted eftM in the strain PAO1 using the procedure described by Quénée et al. (24) for gene deletion and antibiotic resistance marker recycling in P. aeruginosa using the cre-lox system. Briefly, upstream and downstream PCR products (see Table S2 in the supplemental material) of eftM were digested with either BamHI or EcoRI and HindIII and then cloned by three-way ligation into pEX100Tlink deleted for the HindIII site and digested with EcoRI and BamHI to produce plasmid pEXeftM, which was transformed into E. coli strain XL1-Blue. Transformants were selected on LB agar plates with 30 µg/ml ampicillin. The lox-flanked gentamicin resistance cassette (aac1), obtained from the HindIII-digested plasmid pUCGmlox, was cloned into the plasmid pEXeftM digested with the same enzyme, producing plasmid pEXeftMGm, which was transformed into E. coli XL1-Blue. Transformants were selected on LB agar plates with 30 µg/ml ampicillin and 5 µg/ml gentamicin. These plasmids were then transformed into the E. coli helper strain S17-1. Plasmid pEXeftMGm was transferred from E. coli S17-1 to PAO1, and selection for double recombinants using LB agar plates with 5% sucrose and 30 µg/ml gentamicin was performed to produce the PAO1∆eftMGm strain. Double crossovers were first screened for susceptibility to carbenicillin (200 µg/ml) and by PCR amplification using primers eftM-F-ErI and eftM-R-BhI (see Table S2). For the removal of the gentamicin resistance cassette (to yield the PAO1ΔeftM mutant), plasmid pCM157 was electroporated into the mutant. Transformants were selected on LB agar plates with 250 µg/ml tetracycline. One transformant was grown overnight in LB broth with 250 µg/ml tetracycline in order to allow the expression of the cre recombinase. Plasmid pCM157 was then cured from the strains by three successive passages in LB broth. Selected colonies were then screened for susceptibility to tetracycline (250 µg/ml) and gentamicin (30 µg/ml) and were checked by PCR amplification and DNA sequencing.
To purify P. aeruginosa EF-Tu, we constructed the shuttle expression vector pUCP18ApGw(tufB), which encodes His-tagged EF-Tu, using the directions provided in the Invitrogen Gateway system (Life Technologies, Carlsbad, CA) and primers described in Table S2 in the supplemental material. Essentially, tufB was amplified by PCR from the genome of PAO1 using primers tufBF and tufBR and cloned into the Entry vector, pENTR/D-TOPO. His-tagged EF-Tu was transferred to the Gateway compatible expression vector pUCP18ApGw, which was derived from pUCP18 (25) by cloning reading frame A from the Gateway Vector Conversion System (Life Technologies, Carlsbad, CA) into the SmaI site, using LR clonase. The resulting plasmid was named pUCP18ApGw(tufB).
To generate the mutant protein EF-Tu K5A, we used the protocol described above, but the primer tufBF was replaced with tufBFK5A, where the lysine 5-encoding codon of tufB is replaced by an alanine-encoding codon. The resulting plasmid was named pUCP18ApGw(tufBK5A).
eftM was also cloned into the shuttle expression vector pUCP18ApGw using the protocol described above and primers PA4178F and PA4178R (see Table S2 in the supplemental material) with PAO1 genomic DNA as a template. The resulting plasmid was named pUCP18ApGw(eftM).
All molecular biology techniques were performed according to standard protocols as described previously (26).
Purification and labeling of EF-Tu.
P. aeruginosa EF-Tu was purified from strain PAO1, PAO1ΔeftM, or E. coli DH5α harboring plasmid pUCP18ApGw(tufB). For the purification of the mutated EF-Tu K5A, PAO1 harboring the plasmid pUCP18ApGw(tufBK5A) was used. All purifications were performed after growing the strains at 25°C and using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Briefly, bacterial cells were suspended in lysis buffer consisting of 50 mM sodium phosphate (pH 8), 300 mM NaCl, 10 mM imidazole, and lysozyme (1 mg/ml) (Sigma-Aldrich, St. Louis, MO). The cells were disrupted by sonication (10 sets of 30-s pulses), and the resulting homogenate was centrifuged at 12,000 × g to remove cellular debris. Ni-NTA (5 ml in a 50% slurry) was added to the resulting supernatant, and the suspension was swirled at 4°C at 100 rpm for 1 h on a rotary shaker. The Ni-NTA resin was washed three times with 20 ml of wash buffer consisting of 50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, and 20 mM imidazole. The His-tagged proteins were eluted with 1.5 ml of 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl and 250 mM imidazole. Purity (>97%) was confirmed by SDS-PAGE analysis.
Proteins were labeled with the infrared dye 800CW using the IRDye 800CW protein labeling kit (Li-Cor, Lincoln, NE) according to the manufacturer’s instructions.
Tandem mass spectrometry analysis.
Proteins (~30 µg) were digested with trypsin (0.2 mg) (Sigma-Aldrich, St. Louis, MO) at 37°C overnight. Digested proteins were placed on a polished steel target (Bruker Daltonics, Bremen, Germany), mixed with 1 µl of matrix (2,5-dihydroxybenzoic acid in 70/30 acetonitrile/water with 0.1% trifluoroacetic acid), allowed to air dry, and analyzed with an Autoflex III matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with a 200-Hz Smartbeam laser. Spectra were recorded in the reflector, positive mode, at a laser frequency of 200 Hz within a mass range from 500 to 4,300 Da. The IS1 voltage was 19 kV, the IS2 voltage was maintained at 16.65 kV, the lens voltage was 8.30 kV, the reflector voltage was 21 kV, and the reflector 2 voltage was 9.7 kV. The spectra were calibrated using a peptide calibration standard (Bruker Daltonics, Bremen, Germany) or autolysis trypsin peaks.
Cell culture assays.
Monolayers of 16HBE14o− or DOK epithelial cells were grown to confluence in 24-well tissue culture plates (~5 × 105 cells per well) as mentioned above and used for standard invasion assays as described previously (27). The cells were infected with bacteria grown at 25°C at a multiplicity of infection (MOI) of 50:1 for 60 min.
In some experiments, cells were treated with different concentrations of the PAFR antagonist 1-O-hexadecyl-2-acetyl-sn-glycerol-3-phospho-(N,N,N-trimethyl)-hexanolamine (Calbiochem, Darmstadt, Germany) for 30 min prior to infection. This antagonist is a structural analog of PAF that interacts with the PAFR and potentially blocks the interaction of other PAFR binding molecules. This antagonist shows inhibitory activity on platelet aggregation and phospholipid turnover (28).
siRNA treatment.
Sixteen HBE14o− and DOK epithelial cells were plated at 2.5 × 105 cells/well in 6-well tissue culture plates. After 24 h, cells were transfected with 20 nM specific anti-PAFR siRNAs or scrambled nonspecific siRNAs as a control. siRNAs were diluted in 1 ml Opti-MEM medium containing a 1:500 dilution of Oligofectamine according to the instructions of the manufacturer. After 24 h, cells were used in standard internalization assays as described above. No changes in the cell proliferation were observed due to the treatment of the cells with anti-PAFR siRNAs. All reagents were obtained from Life Technologies, Carlsbad, CA.
Murine model of acute respiratory infection.
pafr-deficient mice of BALB/c background were kindly provided by Elaine Tuomanen (St. Jude Children’s Hospital, Memphis, TN) (29). WT BALB/c (Harlan, Chicago, IL) and pafr-deficient mice of matching age and sex were used in this study. Mice were anesthetized with a ketamine (65 mg/kg of body weight) and xylazine (13 mg/kg) mixture and infected by intranasal administration with 2 × 107 CFU. In some experiments, 10 µl of saline or saline containing PAFR antagonist (50 nM) was administered in each nostril 15 min before the infection. At the time points indicated, animals were sacrificed and nasal washes and lung homogenates were aseptically obtained and plated for quantitative bacterial cultures. In the survival studies, mice were infected with doses ranging from 5 × 106 to 5 × 107 CFU and monitored daily during a period of 7 days. LD50 was determined using the Reed-Muench method (30). All animal experiments were performed according to institutional and national guidelines and were approved by the Animal Care and Use Committees of the institutions.
Statistical analysis.
Two-tailed unpaired Student t tests were used to compare the different conditions assayed during in vitro models of infection and to compare bacterial loads in vivo. Survival curves were compared using a log rank test. Analysis was conducted using GraphPad Prism 5.01 software.
Serum resistance assays.
Complement-mediated serum bactericidal activity was determined as previously described (31).
SUPPLEMENTAL MATERIAL
Tandem mass spectrometry analysis of P. aeruginosa EF-Tu purified from E. coli harboring a plasmid expressing eftM and digested with trypsin. The figure shows the EF-Tu-derived peptide that is modified with three methyl groups due to P. aeruginosa EftM. Download
Effect of a PAFR antagonist on the interaction of P. aeruginosa with bronchoepithelial cells. Monolayers of cells treated with different concentrations of the antagonist [1-O-hexadecyl-2-acetyl-sn-glycerol-3-phospho-(N,N,N-trimethyl)-hexanolamine] or with saline (control) were incubated with the indicated P. aeruginosa strains grown at 25°C, and the amount of bacterial internalization was determined. The internalization of P. aeruginosa by cells treated with the antagonist is plotted as a percentage of that obtained for untreated cells. Three independent experiments, each done in triplicate, were performed. Error bars represent standard errors of the means (SEMs). Statistical analyses were done with Student’s unpaired two-tailed t test: **, P < 0.01; ***, P < 0.001. Download
Time course analysis of P. aeruginosa respiratory tract infection. Mice (n > 4 per group) were infected intranasally with 2 × 107 CFU of PAO1 or PAO1ΔeftM and euthanized at different times to determine the bacterial load in the nasal wash and in the lung. Error bars represent standard errors of the means (SEMs). Statistical analyses were done with Student’s unpaired two-tailed t test: *, P < 0.05; ***, P < 0.001. Download
Resistance of PAO1 and PAO1ΔeftM to complement-mediated killing. PAO1 and PAO1ΔeftM were grown to mid-log phase at 25°C or 37°C and treated for 60 min in 20% normal human serum. The percentage of survival is the number of CFU remaining compared with controls in which complement activity was heat inactivated. Three independent experiments each done in triplicate were performed. Error bars represent standard errors of the means (SEMs). Download
List of P. aeruginosa genes homologous to genes involved in ChoP biosynthesis, expression, or metabolism in other microorganisms.
Primers used in this study.
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Economia y Competitividad and Instituto de Salud Carlos III, Spanish Network for the Research in Infectious Diseases grants SAF2012-38426 and REIPI C03/14 and RD06/0008 to S.A., and a Research Grant from the Cystic Fibrosis Foundation (GOLDBE10G0) to J.B.G.
Footnotes
Citation Barbier M, Owings JP, Martínez-Ramos I, Damron FH, Gomila R, Blázquez J, Goldberg JB, Albertí S. 2013. Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. mBio 4(3):e00207-13. doi:10.1128/mBio.00207-13.
REFERENCES
- 1. Sadikot RT, Blackwell TS, Christman JW, Prince AS. 2005. Pathogen host-interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mosser JL, Tomasz A. 1970. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem. 245:287–298 [PubMed] [Google Scholar]
- 3. Weiser JN, Shchepetov M, Chong ST. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435–438 [DOI] [PubMed] [Google Scholar]
- 5. Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13–27 [DOI] [PubMed] [Google Scholar]
- 6. Clark SE, Weiser JN. 2013. Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect. Immun. 81:392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barbier M, Oliver A, Rao J, Hanna SL, Goldberg JB, Albertí S. 2008. Novel phosphorylcholine-containing protein of Pseudomonas aeruginosa chronic infection isolates interacts with airway epithelial cells. J. Infect. Dis. 197:465–473 [DOI] [PubMed] [Google Scholar]
- 8. Kunert A, Losse J, Gruszin C, Hühn M, Kaendler K, Mikkat S, Volke D, Hoffmann R, Jokiranta TS, Seeberger H, Moellmann U, Hellwage J, Zipfel PF. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 179:2979–2988 [DOI] [PubMed] [Google Scholar]
- 9. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas genome database: improved comparative analysis and population genomic capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 http://dx.doi.org/10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. 108:7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rau MH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L. 2012. Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ. Microbiol. 14:2200–2211 [DOI] [PubMed] [Google Scholar]
- 12. Weiser JN, Goldberg JB, Pan N, Wilson L, Virji M. 1998. The phosphorylcholine epitope undergoes phase variation on a 43-kilodalton protein in Pseudomonas aeruginosa and on pili of Neisseria meningitidis and Neisseria gonorrhoeae. Infect. Immun. 66:4263–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Zoelen MA, Florquin S, Meijers JC, de Beer R, de Vos AF, de Boer OJ, van der Poll T. 2008. Platelet-activating factor receptor contributes to host defense against Pseudomonas aeruginosa pneumonia but is not essential for the accompanying inflammatory and procoagulant response. J. Immunol. 180:3357–3365 [DOI] [PubMed] [Google Scholar]
- 14. Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, Ye JY, Weitao T. 2012. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal 2012:128705 http://dx.doi.org/10.1100/2012/128705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041–1051 [DOI] [PubMed] [Google Scholar]
- 16. Xolalpa W, Vallecillo AJ, Lara M, Mendoza-Hernandez G, Comini M, Spallek R, Singh M, Espitia C. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332–3341 [DOI] [PubMed] [Google Scholar]
- 17. Serino L, Virji M. 2000. Phosphorylcholine decoration of lipopolysaccharide differentiates commensal Neisseriae from pathogenic strains: identification of licA-type genes in commensal Neisseriae. Mol. Microbiol. 35:1550–1559 [DOI] [PubMed] [Google Scholar]
- 18. Smani Y, Docobo-Pérez F, López-Rojas R, Domínguez-Herrera J, Ibáñez-Martínez J, Pachón J. 2012. Platelet-activating factor receptor initiates contact of Acinetobacter baumannii expressing phosphorylcholine with host cells. J. Biol. Chem. 287:26901–26910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bechah Y, El Karkouri K, Mediannikov O, Leroy Q, Pelletier N, Robert C, Médigue C, Mege JL, Raoult D. 2010. Genomic, proteomic, and transcriptomic analysis of virulent and avirulent Rickettsia prowazekii reveals its adaptive mutation capabilities. Genome Res. 20:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kraal B, Lippmann C, Kleanthous C. 1999. Translational regulation by modifications of the elongation factor Tu. Folia Microbiol. (Praha) 44:131–141 [DOI] [PubMed] [Google Scholar]
- 21. Roy H, Becker HD, Mazauric MH, Kern D. 2007. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 35:3420–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Radin JN, Orihuela CJ, Murti G, Guglielmo C, Murray PJ, Tuomanen EI. 2005. Beta-arrestin 1 participates in platelet-activating factor receptor-mediated endocytosis of Streptococcus pneumoniae. Infect. Immun. 73:7827–7835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quénée L, Lamotte D, Polack B. 2005. Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. Biotechniques 38:63–67 [DOI] [PubMed] [Google Scholar]
- 25. Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Kowalski MP, Pier GB. 2004. Localization of cystic fibrosis transmembrane conductance regulator to lipid rafts of epithelial cells is required for Pseudomonas aeruginosa-induced cellular activation. J. Immunol. 172:418–425 [DOI] [PubMed] [Google Scholar]
- 28. Grigoriadis G, Stewart AG. 1991. 1-O-hexadecyl-2-acetyl-sn-glycero-3-phospho (N,N,N trimethyl) hexanolamine: an analogue of platelet-activating factor with partial agonist activity. Br. J. Pharmacol. 104:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishii S, Shimizu T. 2000. Platelet-activating factor (PAF) receptor and genetically engineered PAF receptor mutant mice. Prog. Lipid Res. 39:41–82 [DOI] [PubMed] [Google Scholar]
- 30. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 31. Weiser JN, Pan N, McGowan KL, Musher D, Martin A, Richards J. 1997. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tandem mass spectrometry analysis of P. aeruginosa EF-Tu purified from E. coli harboring a plasmid expressing eftM and digested with trypsin. The figure shows the EF-Tu-derived peptide that is modified with three methyl groups due to P. aeruginosa EftM. Download
Effect of a PAFR antagonist on the interaction of P. aeruginosa with bronchoepithelial cells. Monolayers of cells treated with different concentrations of the antagonist [1-O-hexadecyl-2-acetyl-sn-glycerol-3-phospho-(N,N,N-trimethyl)-hexanolamine] or with saline (control) were incubated with the indicated P. aeruginosa strains grown at 25°C, and the amount of bacterial internalization was determined. The internalization of P. aeruginosa by cells treated with the antagonist is plotted as a percentage of that obtained for untreated cells. Three independent experiments, each done in triplicate, were performed. Error bars represent standard errors of the means (SEMs). Statistical analyses were done with Student’s unpaired two-tailed t test: **, P < 0.01; ***, P < 0.001. Download
Time course analysis of P. aeruginosa respiratory tract infection. Mice (n > 4 per group) were infected intranasally with 2 × 107 CFU of PAO1 or PAO1ΔeftM and euthanized at different times to determine the bacterial load in the nasal wash and in the lung. Error bars represent standard errors of the means (SEMs). Statistical analyses were done with Student’s unpaired two-tailed t test: *, P < 0.05; ***, P < 0.001. Download
Resistance of PAO1 and PAO1ΔeftM to complement-mediated killing. PAO1 and PAO1ΔeftM were grown to mid-log phase at 25°C or 37°C and treated for 60 min in 20% normal human serum. The percentage of survival is the number of CFU remaining compared with controls in which complement activity was heat inactivated. Three independent experiments each done in triplicate were performed. Error bars represent standard errors of the means (SEMs). Download
List of P. aeruginosa genes homologous to genes involved in ChoP biosynthesis, expression, or metabolism in other microorganisms.
Primers used in this study.