ABSTRACT
WspR is a hybrid response regulator-diguanylate cyclase that is phosphorylated by the Wsp signal transduction complex in response to growth of Pseudomonas aeruginosa on surfaces. Active WspR produces cyclic di-GMP (c-di-GMP), which in turn stimulates biofilm formation. In previous work, we found that when activated by phosphorylation, yellow fluorescent protein (YFP)-tagged WspR forms clusters that are visible in individual cells by fluorescence microscopy. Unphosphorylated WspR is diffuse in cells and not visible. Thus, cluster formation is an assay for WspR signal transduction. To understand how and why WspR forms subcellular clusters, we analyzed cluster formation and the enzymatic activities of six single amino acid variants of WspR. In general, increased cluster formation correlated with increased in vivo and in vitro diguanylate cyclase activities of the variants. In addition, WspR specific activity was strongly concentration dependent in vitro, and the effect of the protein concentration on diguanylate cyclase activity was magnified when WspR was treated with the phosphor analog beryllium fluoride. Cluster formation appears to be an intrinsic property of phosphorylated WspR (WspR-P). These results support a model in which the formation of WspR-P subcellular clusters in vivo in response to a surface stimulus is important for potentiating the diguanylate cyclase activity of WspR. Subcellular cluster formation appears to be an additional means by which the activity of a response regulator protein can be regulated.
IMPORTANCE
Bacterial sensor proteins often phosphorylate cognate response regulator proteins when stimulated by an environmental signal. Phosphorylated response regulators then mediate an appropriate adaptive cellular response. About 6% of response regulator proteins have an enzymatic domain that is involved in producing or degrading cyclic di-GMP (c-di-GMP), a molecule that stimulates bacterial biofilm formation. In this work, we examined the in vivo and in vitro behavior of the response regulator-diguanylate cyclase WspR. When phosphorylated in response to a signal associated with surface growth, WspR has a tendency to form oligomers that are visible in cells as subcellular clusters. Our results show that the formation of phosphorylated WspR (WspR-P) subcellular clusters is important for potentiating the diguanylate cyclase activity of WspR-P, making it more active in c-di-GMP production. We conclude that oligomer formation visualized as subcellular clusters is an additional mechanism by which the activities of response regulator-diguanylate cyclases can be regulated.
Introduction
The intracellular secondary messenger cyclic di-GMP (c-di-GMP) promotes biofilm growth in many gram-negative bacteria by stimulating exopolysaccharide (EPS) and adhesin production. c-di-GMP also inhibits flagellar motility. Multiple diguanylate cyclases with a characteristic GG(D/E)EF domain produce c-di-GMP in response to diverse environmental stimuli (1–4). One of the best-studied diguanylate cyclases is WspR from Pseudomonas species (5–8). In the opportunistic pathogen Pseudomonas aeruginosa, c-di-GMP production catalyzed by WspR stimulates synthesis of the EPS Pel, which is important for biofilm formation (9, 10).
WspR is the output response regulator of the Wsp signal transduction complex (Fig. 1A). The crystal structure of WspR revealed that it has a CheY-like receiver (response regulator) domain connected to a GGEEF diguanylate cyclase domain via a coiled-coil linker (Fig. 1B) (6). The in vitro diguanylate cyclase activity of WspR is stimulated by phosphorylation (7) and inhibited by c-di-GMP binding at a site of inhibition (I site) on its GGEEF domain (6, 11). In vivo, deletion of the wspF gene in the Wsp operon locks the Wsp signal transduction system into an “on” state so that phosphorylation of WspR is greatly increased and levels of intracellular c-di-GMP and biofilm formation increase dramatically (7). In a ΔwspF strain, yellow fluorescent protein (YFP)-tagged WspR (WspR-YFP) forms dynamic subcellular cytoplasmic clusters that are visible by fluorescence microscopy (12). In wild-type P. aeruginosa, YFP-tagged WspR does not form visible subcellular clusters in cells grown in broth but does form clusters in cells grown on an agar surface. Thus, we use WspR cluster formation as an assay for surface-stimulated Wsp signal transduction.
FIG 1 .
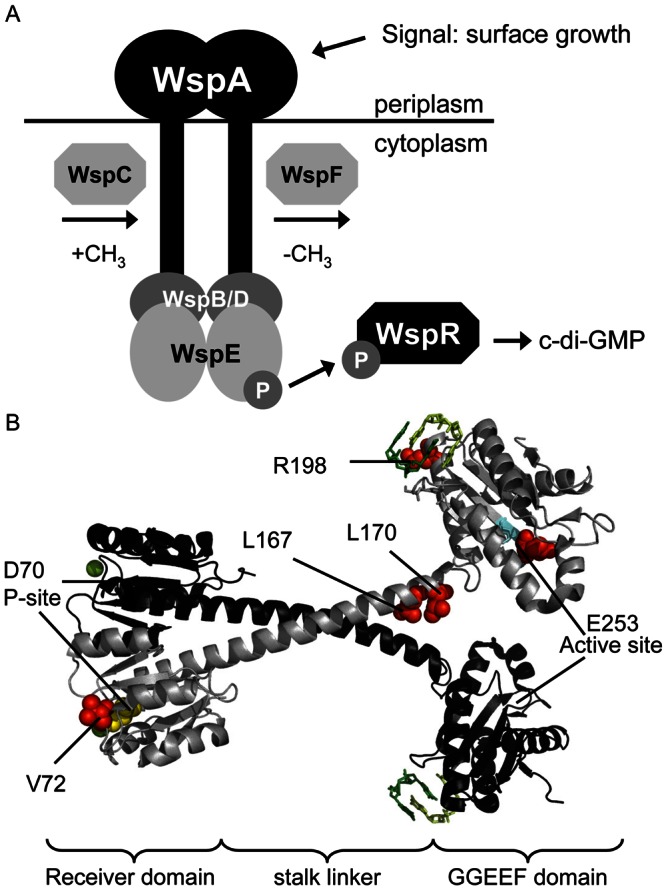
(A) A model for the Wsp signal transduction complex. WspA is a membrane-bound receptor protein which detects a signal associated with growth on a surface. The signal is communicated to the histidine kinase WspE, which catalyzes phosphotransfer to the response regulator-diguanylate cyclase WspR. Phosphorylated WspR produces the secondary messenger c-di-GMP. The methyltransferase WspC and the methylesterase WspF likely play a role in adaptation to the surface signal. WspB and WspD are scaffolding proteins important for function and proper localization of the Wsp complex (18). (B) Locations of WspR mutations examined in this study with respect to the published crystal structure of WspR (PDB 3BRE) rendered by the PyMOL program (v0.99rc6) (http://www.pymol.org). WspR is depicted as a dimer, one molecule in grey and the other in black. Red spheres show locations of the mutated residues; cyan indicates the conserved GGEEF motif of the cyclase active site; yellow indicates the conserved aspartate of the phosphorylation site; green balls are Mg2+ ions needed for phosphorylation. C-di-GMP dimers bound at the I sites are shown.
To understand the physiological significance of WspR subcellular cluster formation, we characterized six single-amino-acid variants of WspR. The data suggest a model in which the diguanylate cyclase activity of phosphorylated WspR (WspR-P) is potentiated by oligomerization, with oligomers becoming visible as subcellular clusters. Subcellular clustering appears to reflect an additional layer of regulation of a response regulator protein. Our data further suggest that cluster formation is an intrinsic property of WspR-P.
RESULTS
Description of WspR variants.
Two of the WspR variants that we analyzed, the D70A and V72D variants (WspRD70A and WspRV72D), are mutated in the response regulator domain, which we refer to here as the CheY-like receiver domain (Fig. 1B). The D70A substitution replaces the phosphorylatable aspartate of WspR and is similar to the D70N mutation, which renders WspR inactive and insensitive to phosphorylation (5, 12). The V72D variant originated with a random mutagenesis of wspR with screening for mutants that stimulated increased EPS production by P. aeruginosa (J. W. Hickman and C. S. Harwood, unpublished). Another two of the variants have substitutions in the GGEEF domain. WspR activity is inhibited by the binding of c-di-GMP to the I site in its GGEEF domain. The R198A change renders the I site unable to bind c-di-GMP, so that WspRR198A activity is not inhibited by c-di-GMP (6). E253A changes the active-site region from GGEEF to GGAEF and abolishes cyclase activity (8). In vitro work from the Sondermann laboratory (5) showed that unphosphorylated WspR is in equilibrium between dimeric and tetrameric species. The dimeric form of WspR has low basal levels of activity, while the tetrameric form of WspR has high levels of activity and is considered the active conformation (5). The mutations L167D and L170D in the linker stalk region affect WspR tetramerization. WspRL167D tends toward the monomeric species and is inactive, while WspRL170D stabilizes the tetrameric species of the protein and is very active (6). The activity of WspRL170D is also resistant to inhibition by c-di-GMP (6).
Effects of wspR mutations on subcellular clustering.
We fused the yfp gene to the various wspR mutant genes and to wild-type wspR, integrated the constructs into the attB site in the P. aeruginosa chromosome, and expressed them under the control of the arabinose promoter in a wspR deletion mutant. We have previously shown that WspR-YFP is active in cells (12). We estimate from immunoblotting experiments that the average P. aeruginosa PAO1 cell contains about 300 WspR molecules, equivalent to about a 1-μmol concentration (see Fig. S1 in the supplemental material). When the Wsp signal transduction system is active and WspR is phosphorylated, individual cells have between one and four visible clusters of WspR-YFP, suggesting that one cluster may comprise 75 to 300 molecules of WspR. Immunoblot analysis revealed that each of the variants was expressed at approximately the same level as wild-type wspR from its native promoter (data not shown). Immunoblot analysis also confirmed that the YFP tag remained fused to WspR and the WspR variants and was not cleaved off. We observed the clustering behavior of the YFP-tagged proteins using epifluorescence microscopy (Fig. 2) and quantified cluster formation as the percentage of cells in a population with at least one WspR-YFP cluster (Table 1). The variants WspRL170D, WspRR198A, and WspRE253A formed subcellular clusters in broth-grown cells (Table 1; Fig. 2A), a condition in which wild-type WspR (WspRwt) expressed in a wild-type background does not form clusters. However, surface growth further stimulated cluster formation (Table 1). Cells expressing WspRD70A and WspRV72D did not have significant levels of clusters when grown in liquid or on an agar surface (Table 1; Fig. 2B). Cells with WspRL167D expressed in a wild-type background had very low levels of clusters (4%) when grown on agar (Table 1; Fig. 2B)
FIG 2 .
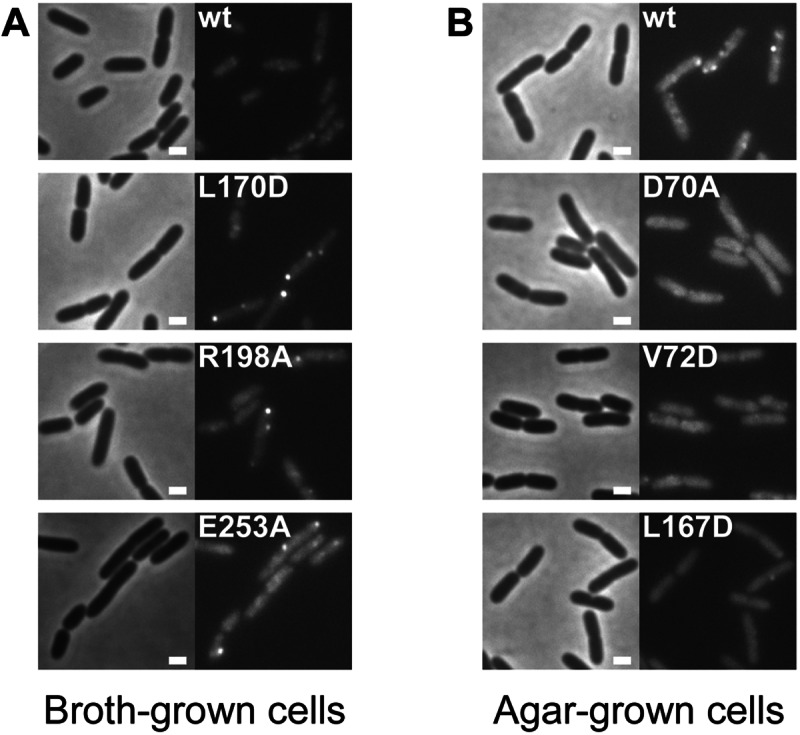
Fluorescence micrographs of strain PAO1 derivatives expressing the denoted wspR alleles fused with YFP. Left, phase-contrast images; right, fluorescent images. The scale bar represents 1 µm. (A) Broth-grown cells. (B) Agar-grown cells.
TABLE 1 .
Quantitative analysis of subcellular cluster formation in strains expressing different YFP-tagged WspR variants
| Mutation | Location | % cells with clusters (n) for genotypea |
|||||
|---|---|---|---|---|---|---|---|
| Broth grown |
Agar grown |
||||||
| ΔwspA | WT | ΔwspF | ΔwspA | WT | ΔwspF | ||
| None | 1 (715) | 1 (295) | 34 (493) | 0 (339) | 43 (411) | 77 (494) | |
| D70A | P site | 0 (250) | 0 (178) | 0 (138) | 0 (256) | 0 (190) | 0 (139) |
| V72D | D + 2b | 2 (228) | 2 (572) | 0 (256) | 0 (99) | 0 (599) | 1 (310) |
| L167D | Linker stalk | 0 (158) | 0 (506) | 23 (381) | 0 (288) | 4 (684) | 72 (632) |
| L170D | Linker stalk | 0 (604) | 29 (632) | 82 (222) | 1 (266) | 75 (468) | 72 (186) |
| R198A | I site | 0 (173) | 9 (265) | 36 (140) | 0 (128) | 38 (366) | 48 (261) |
| E253A | Cyclase active site | 0 (266) | 12 (145) | 75 (232) | 0 (194) | 68 (276) | 75 (130) |
Percentages represent the numbers of cells with at least one well-defined fluorescent spot divided by the total number of visualized cells (n), shown in parentheses. A well-defined fluorescent spot is defined by dividing the maximum pixel intensity by the average pixel intensity in the cell. All cells with a resultant number above an empirically determined threshold are considered cells with at least one cluster. WT, wild type.
Two residues downstream of conserved aspartate at the P-site.
As noted above, phosphorylation of WspR is greatly increased in a ΔwspF strain (7), and WspR forms subcellular clusters not only in cells grown on an agar surface but also in cells grown in shaken liquid culture (12). Consistent with this, WspR variants expressed in a ΔwspF background had increased cluster formation as long as the aspartate at position 70 was intact (Table 1). The one exception was WspRV72D, which did not form subcellular clusters in a ΔwspF background. As we show below, WspRV72D is nevertheless very active in vivo and in vitro. An examination of the in vitro activity of purified WspRV72D, described in the section on WspR concentration-dependent activity below, provides a possible explanation for its anomalous behavior.
None of the WspR variants formed clusters in more than 1 to 2% of cells of a strain deleted for the wspA receptor gene, a background in which phosphorylation is predicted not to occur because the Wsp system cannot form an active signal transduction complex (Table 1). Thus, phosphorylation appears to be essential for WspR subcellular cluster formation.
Comparison of in vivo clustering frequencies with in vivo diguanylate cyclase activities of WspR variants.
Because of limitations in the sensitivity of assays to quantitate c-di-GMP extracted from cells grown on complex media, we decided to use P. aeruginosa colony morphology as a way to roughly estimate relative levels of c-di-GMP in cells. High intracellular levels of c-di-GMP stimulate the production of EPS, resulting in “wrinkly” colony morphologies and the uptake of the Congo red dye (7, 13). As shown in Fig. 3, there is generally a correlation between the numbers of subcellular clusters formed by cells harboring various WspR variants and the degree of wrinkliness of their colonies. One exception is the WspRE253A variant, which has a smooth colony morphology because the E253A mutation completely abolishes diguanylate cyclase activity. WspRE253A is shown here as a negative control, but for these and subsequent studies we do not consider it when drawing a connection between cluster formation and activity. The other exception is the WspRV72D variant, which did not form subcellular clusters but did form very wrinkly colonies, indicating that it is highly active in vivo. A WspRD70A/V72D double mutant lacked diguanylate cyclase activity and formed smooth colonies (not shown), indicating that an intact aspartate at position 70 is required for the WspRV72D variant to be active.
FIG 3 .
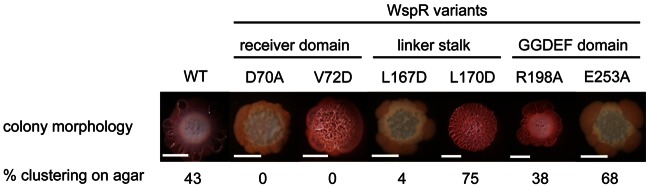
P. aeruginosa strain PAO1 expressing WspR variants spotted onto tryptone-Congo red agar plates with 1% arabinose. Bar = 5 mm. The native wspR gene is deleted, and the selected wspR allele is inserted into the neutral attB site under an arabinose-inducible PBAD promoter.
WspR diguanylate cyclase specific activity is concentration dependent.
The data presented so far show that with the exception of the WspRV72D and WspRE253A variants, the more active WspR variants have a greater propensity to form clusters. But does cluster formation stimulate the activity of a given variant? To test the hypothesis that WspR is most active when it is in a more concentrated form within a subcellular cluster, we looked into how the WspR concentration affected its in vitro activity.
When activated with the phosphor analog beryllium fluoride, WspRwt increased in specific activity as the concentration of the WspR protein was increased (Fig. 4). WspRwt that was not treated with BeF3− did not demonstrate concentration-dependent activity in the range of protein concentrations tested. We were unable to test higher concentrations of WspRwt because it started to precipitate from the assay buffer. This was true of several of the variants that we tested. WspRL170D was active enough to show a concentration-dependent increase in activity without BeF3− treatment, demonstrating that concentration-dependent activation does occur in the absence of phosphorylation. WspRL170D-BeF3− was about as active as WspRwt-BeF3−, suggesting that both have reached the maximum amount of activation by BeF3−. WspRD70A had little catalytic activity with or without BeF3− addition. The lack of activation of WspRD70A by BeF3− indicates that BeF3− activates WspRwt through mimicking phosphorylation at D70 and not through another mechanism.
FIG 4 .
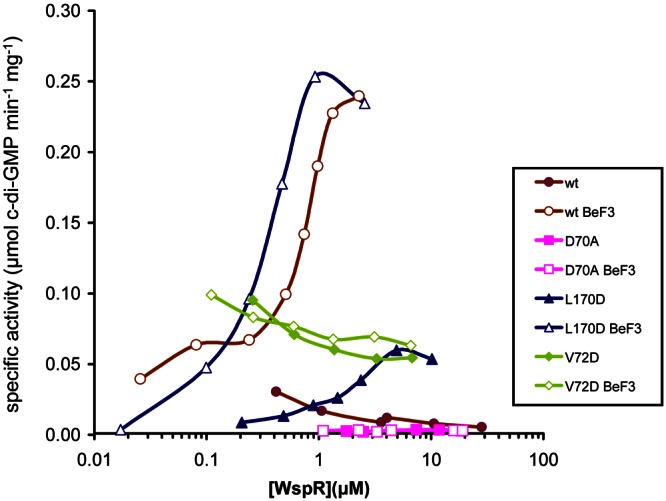
Specific activity of WspR as a function of the WspR concentration. The diguanylate cyclase activities of WspR proteins were assayed at concentrations ranging from 17 nM to 28 µM following their equilibration in assay buffer at 22°C for 16 to 24 h.
When we examined the concentration-dependent activity of WspRV72D, we found that it had high activity relative to those of untreated WspRwt and WspRL170D at low protein concentrations (Fig. 4). In addition WspRV72D displayed no concentration-dependent activity and was not activated by BeF3−. These observations raised the question of what the configuration of WspRV72D is. To look at this, we subjected WspRwt and WspRV72D to size exclusion chromatography. The elution profile of both proteins showed peak maxima at elution volumes of 12.3 and 13.8 ml, which roughly correspond to the tetramer and dimer peak maxima at 11.7 and 13.3 ml previously reported in the literature (6). We analyzed the diguanylate cyclase activities of the eluted fractions corresponding to each peak. Surprisingly, both dimer and tetramer fractions of WspRV72D showed an intermediate amount of activity (see Fig. S2 in the supplemental material). As shown previously (6), the tetramer fraction of WspRwt had high activity while the dimer fraction had very little activity.
Subcellular cluster formation appears to be an intrinsic property of WspR-P.
One hypothesis is that WspR associates with a c-di-GMP receptor protein and this stimulates WspR cluster formation. Although we cannot exclude this possibility, we have been unable to obtain evidence in its favor. We know that WspR affects Pel EPS biosynthesis. So the Pel EPS machinery, which is known to require c-di-GMP for activity, and the FleQ transcriptional regulator, which controls Pel gene expression in response to c-di-GMP, are two effectors with which WspR might interact. We have previously reported that WspR forms clusters in a Δpel Δpsl double mutant (12), suggesting that it does not nucleate around the Pel EPS biosynthetic machinery. Here we determined that WspR forms clusters in a ΔfleQ mutant, and we found no evidence for an interaction between FleQ and WspR in bacterial two-hybrid assays (not shown).
In considering other proteins around which WspR might nucleate, we know that WspE plays a role in initiating WspR-P clustering behavior because WspE is the protein that transphosphorylates WspR (Fig. 1A). However, WspR-P moves away from WspE, and it stays clustered when it does so. We have observed that WspR-YFP and cyan fluorescent protein (CFP)-tagged WspA (WspA-CFP) (which is in a complex with WspE) are sometimes colocalized in cells, but we do not usually see them in the same place (12). Another obvious protein that might play a role in WspR cluster formation is the cytoskeletal protein MreB, which has been shown to influence the localization of a number of bacterial proteins (14–16). When we treated cells with the drug A22 (17), MreB localization was disrupted but subcellular cluster formation by WspR-YFP in a ΔwspF strain background was not affected (data not shown). In short, while it is not possible to draw firm conclusions from negative data such as these, we were unable to obtain evidence that subcellular cluster formation is anything other than an intrinsic property of WspR-P.
DISCUSSION
Our current model is that a surface-associated signal sensed by the WspA receptor protein stimulates the autophosphorylation of WspE, which then transphosphorylates the WspR protein (Fig. 1A). Although WspR-P is active, our results suggest that the formation of WspR-P subcellular clusters potentiates the diguanylate cyclase activity of this protein (Fig. 5). This would have the effect of amplifying the surface signal that is detected by the Wsp system. We do not yet know the identity of the surface stimulus for Wsp. We have speculated that WspA senses changes in the physical environment of its transmembrane region or its periplasmic domain (18). We have tested strains carrying mutations in surface features, including flagella and pili, that have been implicated in surface sensing in other bacteria and did not see a major effect on subcellular clustering of WspR (12), although we have not tested the effects of different surface appendages in combination. Increasing the viscosity of liquid cultures also did not stimulate WspR clustering (12).
FIG 5 .
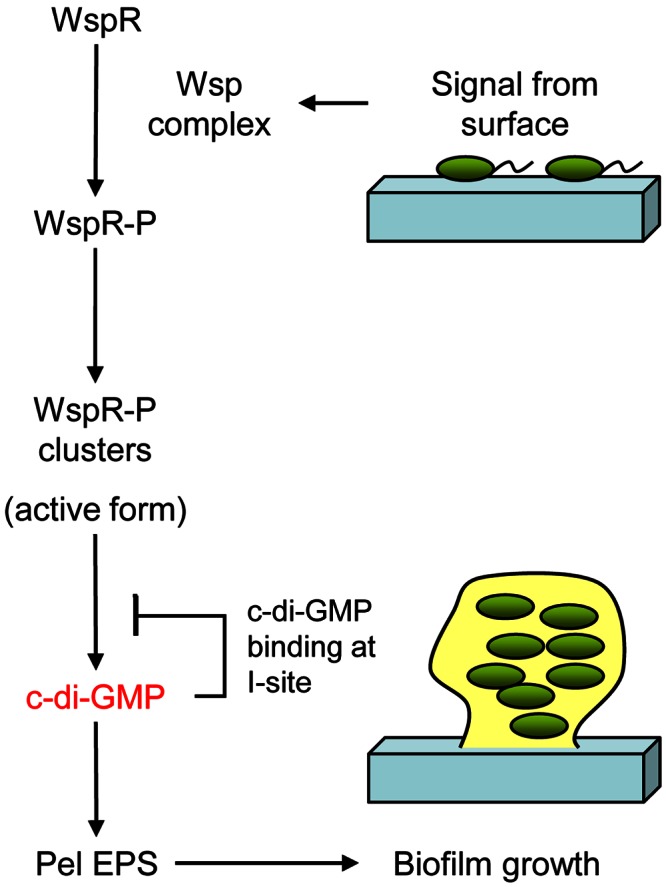
Model for WspR-P activation in the context of Wsp signal transduction and P. aeruginosa biofilm growth. A surface-associated signal activates the Wsp complex to phosphorylate WspR. WspR-P assumes a conformation that allows subcellular cluster formation. WspR-P subcellular clustering further stimulates its diguanylate cyclase activity. C-di-GMP bound to c-di-GMP effectors in the EPS biosynthetic machinery increases production of Pel EPS, resulting in biofilm growth of the P. aeruginosa cells.
In our model, WspR in its active conformation, WspR-P tetramers, tends to oligomerize (cluster), and the oligomerized form of WspR-P is even more active. We found that a concentrated and phosphorylated form of the WspR protein is the form that has the highest diguanylate cyclase specific activity in vitro. Phosphorylation seems to stimulate oligomerization. When WspR variants were not phosphorylated by WspA due to being expressed in a ΔwspA mutant background, they did not form subcellular clusters. The same was true of the unphosphorylatable variant WspRD70A. De and coworkers found that the WspRL167D variant is compromised in its ability to tetramerize and tends to form monomers and be inactive (6). Consistent with this, this variant formed very few subcellular clusters when expressed in wild-type cells grown on agar (Table 1). However, overphosphorylation of WspRL167D in a ΔwspF background restored cluster formation and c-di-GMP production as assessed by wrinkly colony formation.
The WspRV72D variant is highly active in vivo and in vitro but does not form visible subcellular clusters in P. aeruginosa. We found that WspRV72D differed from WspRwt and WspRL170D in that its specific activity did not increase as its concentration was increased and its activity also was not potentiated by treatment with beryllium fluoride (Fig. 4). In addition, we determined that the dimer form of WspRV72D is active, in contrast to WspRwt, which is active only as tetramers. Thus, WspRV72D behaves in vitro as we would predict from our observation that it does not form clusters in vivo.
It seems likely that WspR-P clusters can dissociate as well as form, and one hypothesis is that c-di-GMP stimulates cluster dissolution. Indeed, the active tetramer form of WspR is also the species that is subject to product inhibition by c-di-GMP, and there is evidence that the c-di-GMP-inhibited tetrameric form of WspR dissociates further into inactive elongated dimers (5). From this, one would predict that WspR variants that are resistant to c-di-GMP inhibition would be better cluster formers. The data in Table 1 support this. WspR variants WspRE253A, WspRL170D, and WspRR198A are all not subject to c-di-GMP inhibition in vivo, and all form clusters in wild-type cells grown in liquid. In contrast WspRWT forms almost no clusters under these conditions. If binding of c-di-GMP to WspR stimulates cluster dissolution, then one would predict that cells that are overexpressing a phosphodiesterase in trans would have greater numbers of WspR clusters. We carried out this experiment with the phosphodiesterase PA2133 and found that its expression had no effect on WspR-YFP clustering in P. aeruginosa. Also, WspR produces high levels of c-di-GMP when expressed in a ΔwspF background, as evidenced by the formation of wrinkly colonies, yet a high percentage of such cells have WspR-YFP clusters. It is possible that highly phosphorylated WspR is more resistant to c-di-GMP inhibition, and we also cannot rule out that WspR has a specific interaction with a phosphodiesterase. Thus, although we are unable to conclude at this point that c-di-GMP binding to WspR-P alters its ability to stay oligomerized and maintain clusters in cells, this is an intriguing possibility that warrants follow-up.
The concept that increasing the local concentration of a protein is a mechanism to stimulate its activity is not new. There are many examples of oligomeric proteins that increase in specific activity as their concentration increases, including NtrC, FtsZ, PleD, MinD, and IRE1 (19–23). The functional consequences of oligomerization differ depending on the protein. For example, phosphorylation of the response regulator domain of the AAA+ ATPase protein NtrC promotes its assembly into a hexameric ring structure that has ATPase activity necessary for remodeling a σ54-RNA polymerase closed complex into an open complex able to transcribe genes (24). PleD is a diguanylate cyclase from Caulobacter crescentus with two response regulator domains and resembles WspR in that it also forms subcellular clusters. PleD-P forms clusters at the pole of cells, and c-di-GMP that it releases at this location activates other polarly localized proteins to initiate a program of stalk morphogenesis (25–27). PleD is part of a protein interaction network that exists at the C. crescentus cell pole, and its example invites speculation that subcellular clusters of WspR may also be delivering c-di-GMP to a specific site in cells. We cannot say with certainty that WspR is not functioning this way or that it is not part of a protein interaction network. However, we have failed to identify proteins that interact with WspR, and WspR does not appear to localize to any particular place in cells (Fig. 2) (12).
How then does c-di-GMP produced by WspR specifically affect Pel EPS synthesis but not Psl synthesis or swimming and swarming motility (9, 10; V. Huangyutitham and C. S. Harwood, unpublished)? There are two related possibilities. We know that the depletion in total intracellular c-di-GMP due to a wspR mutation is small. Thus, phenotypes that are very dominant in P. aeruginosa may not be much affected by this small change. Such could be the case for Psl-mediated phenotypes. This is the EPS that is mostly responsible for attachment and biofilm formation in P. aeruginosa strain PAO1 (28). A related possibility raised by recently reported work with Salmonella (29) is that the differential effects of WspR on c-di-GMP-sensitive processes is due to differences in the affinities of receptor proteins for c-di-GMP. PelD and FleQ, two proteins that regulate Pel synthesis in response to c-di-GMP, have relatively low affinities for c-di-GMP (15 µM and 2 µM, respectively) (30, 31), while other known c-di-GMP receptors have affinities in the nanomolar range (32). So a slight change in c-di-GMP caused by WspR DGC activity could affect FleQ and PelD activities but not the activities of high-affinity receptors that remain saturated with c-di-GMP.
The work presented here suggests that active WspR-P tetramers oligomerize to form a more active protein conformation in cells. This oligomerized conformation probably contains on the order of 20 tetramers, and a WspR-YFP tagged oligomer is visible in cells as a cluster. About 6% of all response regulators are involved in c-di-GMP metabolism (24), and it is possible that oligomerization potentiates the activities of many of these. In future work, it will be interesting to determine the exact configuration of the oligomeric form of WspR-P and the mechanistic basis for how this stimulates activity. It will also be important to examine the kinetics of WspR cluster formation and dissolution in cells. Cluster dissolution would be a logical way for cells to adapt to a surface stimulus.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains used in this work are listed in Table 2. Cells were routinely grown in LB medium (10 g liter−1 tryptone, 5 g liter−1 NaCl, and 5 g liter−1 yeast extract) at 37°C unless otherwise noted. Media were supplemented with the following antibiotics: for P. aeruginosa, tetracycline (100 µg ml−1) and gentamicin (50 µg ml−1); for E. coli, tetracycline (2 to 20 µg ml−1), gentamicin (10 µg ml−1), ampicillin (50 to 100 µg ml−1), chloramphenicol (34 µg ml−1), or kanamycin (30 to 50 µg ml−1).
TABLE 2 .
Strains used in this study
| Strain | Source or reference |
|---|---|
| Pseudomonas aeruginosa | |
| PAO1 | 35 |
| PAO1 wspR-yfp | 12 |
| PAO1 ΔwspR | 7 |
| PAO1 ΔwspFR | 7 |
| PAO1 ΔwspAR | 12 |
| PAO1 ΔwspR attB::miniCTX-wspR-yfpa | This study |
| PAO1 ΔwspR attB::miniCTX-wspRD70A-yfp | This study |
| PAO1 ΔwspR attB::miniCTX-wspRV72D-yfp | This study |
| PAO1 ΔwspR attB::miniCTX-wspRL167D-yfp | This study |
| PAO1 ΔwspR attB::miniCTX-wspRL170D-yfp | This study |
| PAO1 ΔwspR attB::miniCTX-wspRR198A-yfp | This study |
| PAO1 ΔwspR attB::miniCTX-wspRE253A-yfp | This study |
| PAO1 ΔwspF wspR-yfp | 12 |
| PAO1 ΔwspFR attB::miniCTX-wspR-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRD70A-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRV72D-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRL167D-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRL170D-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRR198A-yfp | This study |
| PAO1 ΔwspFR attB::miniCTX-wspRE253A-yfp | This study |
| PAO1 ΔwspA wspR-yfp | 12 |
| PAO1 ΔwspAR attB::miniCTX-wspR-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRD70A-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRV72D-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRL167D-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRL170D-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRR198A-yfp | This study |
| PAO1 ΔwspAR attB::miniCTX-wspRE253A-yfp | This study |
| Escherichia coli | |
| DH5α | Gibco-BRL |
| S17-1 | American Type Culture Collection |
| Rosetta 2 | Novagen |
The miniCTX backbone of pTG142 was integrated in the chromosome alongside wspR. See Materials and Methods and reference 34.
Construction of strains with expression of wspR under arabinose-inducible control.
wspR was translationally fused to yfp with a PVPVAT linker and a KpnI site between the two genes, as previously described (12). A blunt fragment containing a ribosome binding site (RBS) and wspR-yfp was created by cloning wspR-yfp into the EcoRI and XbaI sites of pGFP-C (12), excising with NheI and XbaI, and treating with T4 DNA polymerase. This fragment was ligated at the SmaI site of pSW196 (33), which was cured of its KpnI and EcoRI sites. The resultant plasmid, pTG142, contains an arabinose-controllable promoter upstream of its multiple cloning site and can integrate into the attB site on the P. aeruginosa chromosome. Variant alleles of wspR were amplified using PCR from sources noted in Table 2 and swapped with the wild-type wspR gene between the EcoRI and KpnI restriction sites of pTG142. The resulting plasmids were introduced into the appropriate P. aeruginosa strains via conjugation from E. coli S17-1 to integrate into the chromosome at the attB site (34). Transconjugants were selected on LB plates containing tetracycline (100 µg ml−1) and chloramphenicol (10 µg ml−1). Integration into the attB site on the chromosome was checked using PCR with the primer Pser-up (34) and a primer internal to wspR (wspR1; 5′ GACTACCTGGTCAAGCTGCCGGACG 3′).
Fluorescence microscopy.
Sample preparation and microscopy were performed as previously described (12). To analyze liquid-grown cells, cells were grown while shaking to an optical density at 600 nm (OD600) of 0.3 to 0.5 in LB broth at 37°C, back diluted to an OD600 of 0.1 in fresh LB with 1% l-arabinose, and incubated with shaking at 25°C for 3 h for induction of wspR. To remove interfering fluorescence from LB medium, cells were washed by centrifuging for 5 min at 10,000 × g and resuspended in phosphate-buffered saline (PBS) (pH 7.4). The resuspended cell preparation (3 µl) was spotted onto a 0.8% agarose PBS pad on a microscope slide and then covered with a coverslip. To analyze surface-grown cells, liquid-grown cells were diluted to an OD600 of 0.01, and 100 µl was spread on LB 2.5% agar plates with 1% l-arabinose and incubated at 22 to 25°C for 20 h. Cotton swabs were used to transfer surface-grown cells to PBS agarose pads for imaging.
Fluorescence intensity measurements and analysis.
Images were analyzed as previously described (12). Briefly, Metamorph software (6.3r2) was used to detect and create regions corresponding to isolated single cells. Average pixel intensity and maximum pixel intensity were acquired for each cell. The ratio of maximum pixel intensity to average pixel intensity was calculated and used to determine which cells had at least one subcellular cluster. The threshold ratio of 1.74 was determined by eye to lower the incidence of false positives in the wspR negative-control strain.
Congo red colony morphology assay.
Broth-grown cells were diluted in LB to an OD600 of 0.005, and 2 µl of the dilution was spotted on tryptone-Congo red plates (7). The cells were allowed to grow at room temperature for 6 days. Images of surface-illuminated colonies were captured on the 6th day using a digital camera mounted on a dissection microscope (SZX-ILLK100; Olympus).
Protein expression and purification.
Alleles of wspR were amplified by PCR from sources listed in Table 2 and cloned into the pET29 or pETDuet vector (Novagen), the latter with the phosphodiesterase PA2133 cloned into the secondary cloning site. The protein was overexpressed in the E. coli strain Rosetta 2 (Novagen). Cells were grown to mid-log phase in LB at 37°C, subcultured at an OD600 of 0.01 in terrific broth (12 g liter−1 tryptone, 24 g liter−1 yeast extract, 0.4% glycerol, 72 mM K2HPO4, and 17 mM KH2PO4), and grown at 30°C to an OD600 of 0.5 to 1.0. Media were supplemented with appropriate antibiotics. The cells were equilibrated to 16 to 18°C for 30 min and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h. Cells were pelleted at 11,000 × g at 4°C for 15 min and frozen at −20°C. Cells were resuspended in wash buffer (25 mM Tris, 0.3 M NaCl, 10 mM imidazole, pH 7.3) and lysed using a French press. Cell debris was removed by spinning at 11,000 × g and then at 12,000 × g for 15 min (each) and then filtered through 0.45-µm and 0.22-µm filters. His-tagged WspR proteins were isolated using affinity chromatography with HisPur cobalt columns (Pierce) and eluted with elution buffer (25 mM Tris, 0.3 M NaCl, 150 mM imidazole, pH 7.3). Arginine (0.1 M) was added to prevent precipitation. Protein was concentrated using Amicon Ultra filter units and stored at 4°C for no longer than 24 h before assaying. When WspR was diluted from a concentrated stock, its specific activity remained relatively high for a period of a few hours. Activity then decreased gradually to a constant level after about 20 h. We used these “equilibrated” preparations of WspR for the experiments shown in Fig. 4.
Calculation of WspR cellular concentration.
Rabbit antisera were raised against purified WspR (Covance Research Products, Denver, PA). Bands in immunoblots were quantified using ImageQuant 5.1 software. A standard curve of pixel intensity versus ng of WspR was generated using pure WspR. The pixel intensity of the WspR blot in cell lysate samples was then converted to ng of WspR. The amount of WspR was converted to molecules and divided by the amount (in µg) of cell lysate loaded. PAO1 cells were grown to an OD600 of 0.5, cells per ml were determined from these samples, cells were harvested, and cell lysates were prepared. Total protein in cell lysates was determined and calculated as mg of total protein per ml of original culture. From this, we determined a value of 3.52 × 106 cells per µg ⋅ cell lysate protein. Replicates (11) across 4 independent experiments give an average of 283 (standard deviation, 67) molecules of WspR per cell.
Diguanylate cyclase activity assay.
Assays were carried out using the Enzchek pyrophosphate assay kit (Invitrogen). The concentration of MgCl2 was increased to 2 mM, GTP (USB) was at 0.5 mM, and the pyrophosphatase from the kit was replaced with 0.4 U ml−1 pyrophosphatase (inorganic from E. coli; Sigma). To assay WspR activity, WspR was equilibrated in reaction buffer overnight, and then enzymes from the kit and pyrophosphatase were added to reaction mixtures before starting the reaction with GTP. The accumulation of phosphate was monitored over time at OD360.
Beryllium fluoride treatment.
Beryllium fluoride was synthesized in situ from beryllium chloride and sodium fluoride (Sigma). Various amounts of WspR (0.25 µM to 10 µM) were equilibrated in reaction buffer (50 mM Tris-Cl, 10 mM MgCl2, pH 7.5) overnight before activation with 0.1 mM BeCl2 and 10 mM NaF for 5 min at room temperature. Each reaction was run through a PD-10 desalting column to remove extra BeF3−, BeCl2, and NaF from the treated WspR protein in order to prevent inhibition of the pyrophosphatase in the Enzchek assay. Diguanylate cyclase activity of WspR-BeF3− was assayed immediately after BeF3− treatment and desalting.
Size exclusion chromatography.
Protein (60 µM) was injected into a Superdex 200 10/300 GL column (GE). The buffer used consisted of 25 mM Tris (pH 7.5), 100 mM NaCl, 1 mM dithiothreitol (DTT), and 2 mM MgCl2. Chosen fractions were directly assayed for their diguanylate cyclase activity immediately after elution from the column.
SUPPLEMENTAL MATERIAL
Immunoblots of WspR used to estimate the amount of WspR in a P. aeruginosa strain PAO1 cell. Cell lysates of PAO1 and PAO1 ΔwspR were run next to a standard curve of purified protein. This immunoblot is representative of four immunoblots used to estimate the cellular concentration of WspR. Download
Size exclusion chromatography of WspRwt and WspRV72D and the specific activities of fractions. The dashed lines denote the elution volumes corresponding to the fractions analyzed. Download
ACKNOWLEDGMENTS
Public Health Service grant GM56665 from the National Institute of General Medical Sciences supported this work.
We thank Jason Hickman, Holger Sondermann, Matthew Parsek, and Richard Siehnel for strains. We are grateful to Ruth Silversmith for helpful discussions and for the beryllium fluoride protocol. We thank Kimberly Cowles and Zemer Gitai for A22 and the protocol for treating MreB, and we thank Seemay Chou and Joseph Mougous for help with size exclusion chromatography.
Footnotes
Citation Huangyutitham V, Güvener ZT, Harwood CS. 2013. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4(3):e00242-13. doi:10.1128/mBio.00242-13.
REFERENCES
- 1. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 2. Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 21:929–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sondermann H, Shikuma NJ, Yildiz FH. 2012. You’ve come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 15:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 5. De N, Navarro MV, Raghavan RV, Sondermann H. 2009. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6:e67 http://dx.doi.org/10.1371/journal.pbio.0060067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malone JG, Williams R, Christen M, Jenal U, Spiers AJ, Rainey PB. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology (Reading, Engl.) 153:980–994 [DOI] [PubMed] [Google Scholar]
- 9. Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264 http://dx.doi.org/10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′′-5′′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015–32024 [DOI] [PubMed] [Google Scholar]
- 12. Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66:1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shaevitz JW, Gitai Z. 2010. The structure and function of bacterial actin homologs. Cold Spring Harb. Perspect. Biol. 2:a000364 http://dx.doi.org/10.1101/cshperspect.a000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White CL, Kitich A, Gober JW. 2010. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76:616–633 [DOI] [PubMed] [Google Scholar]
- 17. Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol. Microbiol. 86:720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Z, Lutkenhaus J. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337–1343 [DOI] [PubMed] [Google Scholar]
- 20. Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature 457:687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mettke I, Fiedler U, Weiss V. 1995. Mechanism of activation of a response regulator: interaction of NtrC-P dimers induces ATPase activity. J. Bacteriol. 177:5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 282:29170–29177 [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Lutkenhaus J. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9:435–442 [DOI] [PubMed] [Google Scholar]
- 24. Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lam H, Matroule JY, Jacobs-Wagner C. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell 5:149–159 [DOI] [PubMed] [Google Scholar]
- 27. Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14:1913–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol. Microbiol. 86:1424–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitney JC, Colvin KM, Marmont LS, Robinson H, Parsek MR, Howell PL. 2012. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287:23582–23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 35. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblots of WspR used to estimate the amount of WspR in a P. aeruginosa strain PAO1 cell. Cell lysates of PAO1 and PAO1 ΔwspR were run next to a standard curve of purified protein. This immunoblot is representative of four immunoblots used to estimate the cellular concentration of WspR. Download
Size exclusion chromatography of WspRwt and WspRV72D and the specific activities of fractions. The dashed lines denote the elution volumes corresponding to the fractions analyzed. Download


