Abstract
Background and Objective
The rapid development of molecular biology techniques allows for the introduction of real-time polymerase chain reaction (PCR) methods with a limit of mutation detection at 1 % in a background of wild-type DNA. Analysis of KRAS mutations in codons 12, 13, and 61, together with analysis of BRAF mutations in codon 600, are predictive biomarkers for anti-epidermal growth factor receptor (EGFR) treatment in colorectal cancer. Our aim was to compare PCR methods for KRAS mutations and BRAF mutation analysis using DNA isolated from tissue samples previously evaluated for presence of tumor cells using a quantitative scale and the percentage of tumor cells (PTC) scale. We addressed the question of whether a low number of tumor cells can be qualified for somatic mutation testing.
Results
Our study showed that PTC as low as 10 % was good enough to detect KRAS G12D, G13D, and Q61L mutations in formalin-fixed paraffin-embedded (FFPE) material. Furthermore, our results indicate that up to 20 % of colorectal cancer may carry mutations in the KRAS codon 61 and BRAF codon 600, which suggests the value of these mutation analyses because patients carrying them are unlikely to respond to cetuximab or panitumumab. A low level of KRAS somatic mutation detection has not been studied in depth in the context of clinical outcomes in patients; therefore, we compared new PCR methods, (KRBR-RT 50 Entrogen; ViennaLab StripAssay) and re-evaluated KRAS and BRAF status in patients with relapse after targeted therapy.
Conclusions
The importance of molecular results was confirmed by clinical observation of a patient with relapse who had qualified for targeted therapy with KRAS WT status (but was diagnosed by less sensitive single-stranded conformation polymorphisms method). Interestingly, during anti-EGFR treatment, it came to the selection of cells with KRAS G12C mutation which were present from the beginning in the tumor but at a low level (detected by PCR methods) only and led consequently to the metastasis. Taking into consideration the limit of detection, labor time, and assay cost, the real-time PCR method seems to be very promising especially for FFPE material with the PTC below 15 %.
Introduction
Thirty years ago the KRAS gene was proposed as an oncogene in human cancer [1], and today we already know that it is one of the most frequently activated oncogenes with 17–25 % of all human tumors [2]. Since that time we have learned that mutations in KRAS arise after the loss of APC (the tumor suppressor gene) during colon cancer progression in humans [3]. Moreover, the expression of KRAS with the most common G12D mutation in the colon epithelium stimulates hyperproliferation in an MEK-dependent manner [4]. Experiments in mice carrying a mutation in the APC revealed that mutant KRAS promotes tumor progression [4], while human colon cancer cells (DLD-1), expressing KRAS mutated in codon 13 (G13D), are not sensitized to the inhibition of MEK but when exposed to small molecule inhibitors of RAF (AZ628 and sorafenib) the cells were hypersensitive to RAF inhibition [4]. Interestingly, transcriptomes data from colorectal cell lines with mutations in KRAS or BRAF demonstrated that glucose deprivation may drive the acquisition of KRAS pathway mutation in tumors, and that glycolysis inhibitors preferentially suppress the growth of those tumor cell lines [5].
Since the late 1980s, KRAS mutations (mostly in codon 12) have been observed in a broad range of solid tumors: lung adenocarcinoma [6], pancreatic adenocarcinoma [7], stomach cancer [8], and colorectal cancer [9]. Moreover, evaluation of KRAS status in hyperplastic colorectal polyps showed the presence of a mutation in codons 12 and 13 in 47 % of samples, indicating that hyperplastic colorectal polyps may actually be true premalignant lesions [10].
The next step to understanding tumorigenesis was BRAF mutation analysis in colorectal tumors. Rajagopalan et al. [11], in the systematic evaluation of KRAS and BRAF mutations, demonstrated that both genes are mutated at a similar phase of tumorigenesis: after initiation but before malignant conversion. Additionally, BRAF tumorigenesis action occurs in tumors that do not carry mutations in a KRAS gene [11]. The study emphasized the role of repair processes in tumor formation. Further assessment of BRAF c. 1799T>A transversion in exon 15 (V600E) in papillary thyroid cancer showed significant correlation with distant metastases suggesting that the mutation status of BRAF could be used as a potential marker of patients with advanced thyroid cancer [12]. Moreover, a study with MAPK/ERK kinase inhibitors revealed that the BRAF mutation is associated with the enhanced and selective sensitivity to MEK inhibition when compared to cell lines with the KRAS mutation [13]. Also, in tumors with the V600E mutation, ERK signaling is inhibited in cells exposed to RAF inhibitors; therefore, RAF inhibitors can be effective in those patients suffering from a tumor with the BRAF mutation [14].
In the current clinical settings, broad knowledge regarding the KRAS and BRAF genes has been used to personalize treatment. Because mutations in the KRAS gene result in activation of the epidermal growth factor receptor (EGFR) signaling pathway, only metastatic colorectal cancer patients with wild-type KRAS tumors qualify for anti-EGFR targeted therapies with cetuximab or panitumumab [15]. Also, a clinical trial with the BRAF inhibitor (PLX4032) demonstrated antitumor activity in tumors with the BRAF V600E mutation and confirmed that V600E BRAF is a valid therapeutic target in human cancer [16].
Therefore, recent guidelines for anti-EGFR therapies has forced assessment for KRAS mutation in codons 12 and 13 in colorectal cancer. Likewise, codon 61 has been included in many diagnostic tests because the KRAS mutation in codon 61 predicts resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer [17]. Moreover, the presence of the V600E mutation in the BRAF gene is more and more frequently taken into consideration before patient-targeted therapy qualification, because wild-type BRAF is required for a response to panitumumab or cetuximab [18, 19]. The right strategy to assess the KRAS and BRAF mutation status is crucial for the pathologist and diagnostician, especially due to the fact that not only formalin-fixed paraffin-embedded (FFPE) samples are taken for analysis but also fresh biopsy and cytology samples [20]. Old strategies such as Sanger sequencing, the gold standard in mutation testing, or single-stranded conformation polymorphisms (SSCPs) have been used in diagnostics in the past 20 years. Both methods are simple tools to detect germline mutation; however, their detection of somatic mutations depends on the percentage of mutated cells in the studied material with a limit of detection ranging from 10 to 30 %. Therefore, we have compared two polymerase chain reaction (PCR)-based KRAS and V600E BRAF detection kits, including novel colorectal adenocarcinoma samples and old samples previously diagnosed as KRAS-wild type by SSCP, where patients did not respond to anti-EGFR therapy.
Methods
Study Samples and Patient Characteristics
We reviewed a cohort of 40 consecutive specimens of colorectal carcinoma obtained from the Department of Tumor Pathology and Pathomorphology, The Franciszek Lukaszczyk Oncology Center in Bydgoszcz, Poland. Informed consent for mutation testing was obtained from all patients. In each case, a representative of the carcinoma tissue area, an FFPE block, was identified by the pathomorphologist and scraped for DNA isolation. Each individual case was evaluated for V600E BRAF and KRAS mutations using two different methods. The first was a real-time PCR mutation assay based on mutation-specific PCR, which detects the 11 most common KRAS mutations in codons 12, 13, and 61 and the single V600E BRAF mutation (EntroGen). The second method tested was the KRAS-BRAF StripAssay (ViennaLab), which detects the 10 most common KRAS mutations in codons 12 and 13 and the V600E BRAF mutation. Two additional interesting retrospective cases were added to our study to repeat the mutation status analysis using more sensitive PCR methods. These samples were found not to carry mutations in KRAS codons 12 and 13; however, the patients, after cetuximab treatment and a short remission (2 months), had new metastasis.
Finally, three additional controls were added to our study: the HT29 cell line was included as an internal control for the V600E BRAF mutation and KRAS WT. Then two KRAS controls (Ctrl 8 with the G12D mutation and Ctrl 9 with the G13D mutation found previously by the TheraScreen®: K-RAS Mutation Kit for the detection of seven mutations (by DxS Diagnostic Innovations) [21]. All of the colorectal carcinoma patients were examined for age, sex, number of metastatic sites, and chemotherapy (Table 1).
Table 1.
Patient characteristics
| Characteristics | Study population (n = 40) | KRAS and BRAF | |
|---|---|---|---|
| WT | Mutated | ||
| Mean age and range (years) | 56 (23–76) | 55 (29–72) | 57 (23–76) |
| Sex (male/female) | 25/15 | 10/6 | 15/9 |
| Tumor site | |||
| Colon | 30 | 12 | 18 |
| Rectum | 10 | 4 | 6 |
|
ECOG PS | |||
| 1 | 38 | 15 | 23 |
| 2 | 1 | 0 | 1 |
| 3 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 |
| Number of metastatic sites | |||
| 0 | 3 | 2 | 1 |
| 1 | 7 | 1 | 6 |
| 2 | 5 | 3 | 2 |
| >2 | 25 | 10 | 15 |
| Sites of metastasis | |||
| Liver | 20 | 6 | 14 |
| Lung | 8 | 4 | 4 |
| Lymph nodes, peritoneum, local infiltration | 15 | 8 | 7 |
| Other | 5 | 2 | 3 |
| Treatment | |||
| Adjuvant chemotherapy or chemoradiotherapy | 3 | 2 | 1 |
| Metastatic chemotherapy after adjuvant chemotherapy | 19 | 9 | 10 |
| Primary metastatic chemotherapy | 18 | 5 | 13 |
| Treatment anti-EGFR | |||
| CHT for primary metastatic disease, CHT due to relapse after adjuvant | 4 | 3 | 1* |
KRAS status was analyzed using real-time PCR methods. ECOG PS analysis was performed for 39 patients, clinical data for 1 patient was not available
* Patients first diagnosed as KRAS-WT using the SSCP method and then, after cetuximab treatment and disease progression—KRAS status was analyzed using real-time PCR method
Histological Evaluation and DNA Isolation
One slide with one section per tissue sample, stained with hematoxylin and eosin (H&E), was evaluated by the pathomorphologist for the quantitative presence of a tumor. In each tissue section, the extent of the tumor pattern (quantitative scale, QS) and percentage of tumor cells (PTC) were estimated. The quantitative scale of histological grading (H) shows the presence of a tumor within a tissue sample, based on the percentage of the neoplastic area of the section assessed microscopically and is as follows: QS H1+, over 1 % up to 20 % tumor presence; H2+, over 20 % up to 40 %; H3+, over 40 % up to 60 %; H4+, over 60 % up to 80 %; H5+, over 80 % up to 100 %.
The QS characterized the tissue section as being representative for a tumor while the PTC allowed one to assess a share of neoplastic cell nuclei. The latter parameter was analyzed based on the number of tumor cells compared to all nucleated cells, using a subjective method of counting microscopically ten neighboring cells as a smallest virtual “decimal cell group,” dispersed uniquely within each tissue section. Next, it was tenfolded to the “hundredth” and “thousandth” cell groups in the next two steps, obtaining “tenth,” “hundredth,” or “thousandth” cell measures, respectively, to an approximate estimation of the cell number, if needed, with a method accuracy of approximately 10 %. To increase the extent of neoplastic patterns, a tissue section with a low representation of cancer was subjected to a macrodissection procedure during which non-neoplastic tissue, marked by the pathomorphologist, was cut out from paraffin blocks. Subsequently, slides with the new section stained with H&E were evaluated again.
DNA isolation was also performed after the macrodissection of a region indicated by the pathomorphologist, from FFPE. Genomic DNA was derived from FFPE colon cancer tissue using Qiagen’s QIAamp FFPE Mini Kit, according to the manufacturer’s instructions with the following modification: in order to lyse all materials, a resuspended pellet in 180 μl of Tissue Lysis Buffer with 20 μl of proteinase K was vortexed and continuously shaken at short intervals during overnight incubation at 56 °C. The DNA quality and quantity was measured by Nanodrop and a total of 650 ng was used for real-time PCR.
Evaluation of 11 KRAS and V600E BRAF Mutations Using Real-Time Assay
Evaluation of KRAS and V600E BRAF status using real-time PCR assay was performed using the K-Ras/B-Raf Mutation Analysis Panel Kit for real-time PCR (EntroGen, Tarzana, CA, USA) (Figs. 1, 2), which employs allele-specific primers that are 100 % complementary to mutant variants of the KRAS and BRAF genes. Detection of the amplification products was performed with the use of fluorescent hydrolysis probes. Probes tagged with the FAM fluorophore were complementary to the KRAS and BRAF genes and the following mutations localized in codon 12: G12S, G12D, G12V, G12C, G12A, G12R; in codon 13: G13D; and in codon 61: Q61H (61CAA>CAT), Q61L, Q61R, Q61H (61CAA>CAC) of the KRAS gene and the V600E mutation in the BRAF gene. The internal control gene’s probe tagged with a VIC fluorophore allowed for controlled analysis of the DNA template in the reaction (Fig. 1b). Reagent preparation, 50 ng of the tested DNA per reaction, LC480 set up (Roche), and data analysis using absolute quantification/second derivative maximum method were completed according to manufacturer’s instructions. The assay can detect a 1 % mutation in a background of wild-type DNA.
Fig. 1.
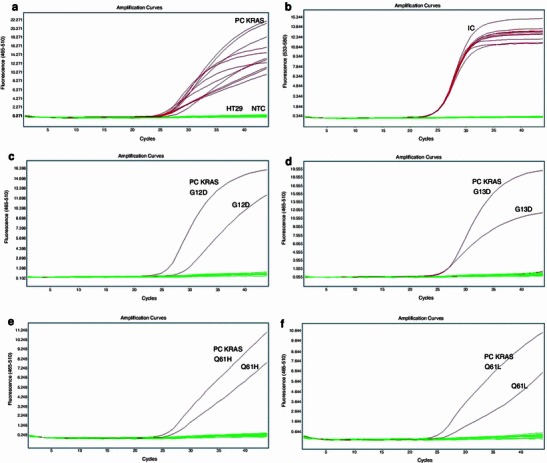
KRAS mutation analysis in codons 12, 13, and 61. Each curve represents the time course of the real-time PCR assay. a Amplification plots for positive controls (PC KRAS) and KRAS wild-type HT29 cell line (flat lines). Amplification curves represent following positive controls in order: G12S, G12D, G12V, G12C, G12A, G12R, G13D, Q61H (61CAA>CAT), Q61L, Q61R, and Q61H (61CAA>CAC) (amplification curves for PC in range: C p 24.02–27.34). b Amplification plots for VIC/yellow reporter for HT29 cell line to ensure that each reaction was properly loaded. Amplification curves represent internal controls for each of 11 reactions (amplification curves in range: C p 25.07–25.50), flat lines represent NTC. c Representative sample with KRAS G12D mutation’s detection (amplification curves of PC with C p 27 and evaluated sample with C p 28.85). d Representative sample with G13D mutation’s detection (amplification curves for PC with C p 26.64 and evaluated sample with C p 25.51). e Representative sample with Q61H mutation’s detection (amplification curves for PC with C p 25.91 and evaluated sample with C p 26.69). f Representative sample with Q61L mutation’s detection (amplification curves for PC with C p 25.17 and evaluated sample with C p 26.04)
Fig. 2.
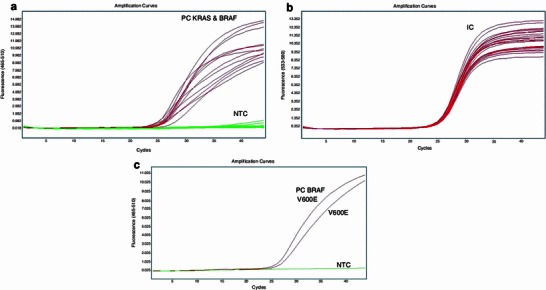
KRAS and BRAF V600E mutation analysis. One-step analysis with 11 probes for KRAS and 1 probe for V600E analysis a filter: FAM 465-510; amplification curves are for PC only, flat baseline represents no amplification for the KRAS WT, BRAF WT sample. b Internal controls for PC and analyzed KRAS wild-type and BRAF V600 wild-type sample; lines represent internal controls: all have C p in range: 25.17–25.66; filter 533–680. c Two-step analysis: first, WT KRAS samples were determined; and next those tested for V600 BRAF status: representative sample with V600E BRAF mutation’s detection (amplification curves for PC with C p 26.15 and evaluated sample with C p 26.79)
Evaluation of 10 KRAS and V600E BRAF Mutations Using PCR with Reverse Hybridization
Ten mutations in the KRAS gene (codons 12 and 13) and one mutation in BRAF (V600E) were evaluated using the KRAS-BRAF StripAssay (ViennaLab Diagnostics, Austria) via PCR amplification with biotinylated primers and hybridization of the amplification products to test strips containing allele-specific oligonucleotide probes, according to the manufacturer's instructions. The presence or absence of a mutation was detected thanks to the binding of biotinylated fragments to streptavidin-alkaline phosphatase complexes and the use of color substrates. Interpretation of the results was based on a naked-eye analysis of colorized test strips and always started from the interpretation of the control line, which indicates the correct function of the conjugate solution and color developer and was followed by the positive/negative reactions of the PCR positive and negative controls.
Results
A total of 40 FFPE samples and one HT29 cell line were screened for 11 KRAS and single V600E BRAF mutations using two different methods: (1) real-time PCR, which detects 11 KRAS mutations in codons 12, 13, and 61 and an additional V600E in BRAF; and (2) a PCR-based method with reverse hybridization, which detects 10 KRAS mutations in codons 12 and 13 and an additional V600E as well. In this studied cohort, two colon carcinoma samples (numbers 37 and 38) were previously tested using TheraScreen (DxS, Manchester, UK) and were selected as additional positive controls (for mutated KRAS codons 12 and 13) [21]. A further two samples (numbers 39 and 40) were previously determined using SSCP as wild-type KRAS and were included in the study.
Patient and Specimen Characteristics
Clinical and pathological characteristics of this cohort are summarized in Table 1. We qualified FFPE colorectal cancer material for molecular analysis using a quantitative scale (QS) and PTC. Our study shows that QS = H2+ with a PTC as low as 10 % is good enough to detect a mutation in codon 61 (Q61L; Table 2). The more common mutation, G13D, was also detected in FFPE material qualified as QS = H2+ and with PTC 15 %.
Table 2.
Clinical and pathological characteristics of 41 samples and comparison of KRAS (codons 12, 13, 61) and BRAF mutation analysis of PCR-based methods
| Sample | Real-time PCR EntroGen (KRAS 11 targets) | PCR + StripAssay ViennaLab (KRAS 10 targets) | EntroGen + ViennaLab BRAF V600E | Tumor volume | Tumor (%) |
|---|---|---|---|---|---|
| 1 | WT | WT | WT | H4+ | 50 |
| 2 | WT | WT | WT | H3+ | 20 |
| 3 | G12C | G12C | WT | H5+ | 30 |
| 4 | G12V | G12V | WT | H4+ | 20 |
| 5 | WT | WT | WT | H4.5+ | 50 |
| 6 | Q61L | WT | WT | H2+ | 10 |
| 7 | WT | WT | WT | H4+ | 50 |
| 8 | G12D | G12D | WT | H4+ | 60 |
| 9 | G12D | G12D | WT | H4+ | 20 |
| 10 | G13D | G13D | WT | H3+ | 10 |
| 11 | WT | WT | WT | H3.5+ | 20 |
| 12 | WT | WT | WT | H3.5+ | 15 |
| 13 | WT | WT | WT | H3.5+ | 20 |
| 14 | WT | WT | WT | H4+ | 60 |
| 15 | WT | WT | WT | H5+ | 60 |
| 16 | WT | WT | WT | H4+ | 25 |
| 17 | WT | WT | WT | H2.5+ | 15 |
| 18 | G12D | G12D | WT | H3+ | 10 |
| 19 | G12D | G12D | WT | H3+ | 20 |
| 20 | G13D | G13D | WT | H2+ | 15 |
| 21 | G12V | G12V | WT | H3+ | 20 |
| 22 | Q61H | WT | WT | H4+ | 25 |
| 23 | WT | WT | WT | H3+ | 10 |
| 24 | WT | WT | V600E | H3+ | 35 |
| 25 | G12V | G12V | WT | H4+ | 40 |
| 26 | G12D | G12D | WT | H2+ | 15 |
| 27 | G12C | G12C | WT | H4+ | 20 |
| 28 | WT | WT | WT | H3.5+ | 20 |
| 29 | G12V | G12V | WT | H4+ | 30 |
| 30 | WT | WT | WT | H3+ | 20 |
| 31 | G13D | G13D | WT | H4+ | 25 |
| 32 | WT | WT | WT | H3+ | 25 |
| 33 | WT | WT | WT | H3.5+ | 15 |
| 34 | G12V | G12V | WT | H4+ | 20 |
| 35 | G13D | G13D | WT | H4+ | 30 |
| 36 | WT | WT | V600E | H3+ | 15 |
| 37 Ctrl (DxS: G12D) | G12D | G12D | WT | H3+ | 30 |
| 38 Ctrl (DxS: G13D) | G13D | G13D | WT | U | U |
| 39 SSCP: KRAS WT | G12C | G12C | WT | H3+ | 15 |
| 40 SSCP:KRAS WT | WT | WT | WT | H2+ | 5 |
| Ht29 cell line | WT | WT | V600E | N/A | N/A |
N/A nonapplicable, U unknown
To illustrate the practical application of real-time PCR methods and the clinical benefit from KRAS mutation detection at low PTC, we carefully evaluated the clinical and molecular data of four patients who received targeted therapy. Two of them had short-term improvement and then progression as detailed below. The first patient (no. 39) was a 55-year-old woman who was diagnosed in 1997 with colon adenocarcinoma (G2pT3N2) and had radical surgery followed by 1 year of adjuvant chemotherapy. Two years later, abdominal recurrence (tumor and retroperitoneal lymph node metastases) was diagnosed. This prompted a 4-month course of chemotherapy, followed by radical surgery. Nine years later the patient experienced an inoperable recurrence of cancer in the abdomen and pelvic region. After third-line palliative chemotherapy, the patient was referred for genetic evaluation. SSCP analysis indicated KRAS WT status, which qualified her (the patient) for concomitant targeted therapy (cetuximab: first dose, 400 mg/m2; subsequent doses, 250 mg/m2) and chemotherapy (5-fluorouracil 400 mg/m2 bolus and then 600 mg/m2 over 22 h for day 1; leucovorin 200 mg/m2 before 5-fluorouracil). After 4 months of treatment, stabilization of the metastatic lesions within the pelvis was observed, but two new metastatic lesions within the skin layers had occurred. Fine needle aspiration of the new metastatic lesions indicated adenocarcinomas, which were tested for KRAS and V600E BRAF mutation using the real-time PCR method (KRBR-RT50, Entrogen). Interestingly, KRAS G12C mutation was diagnosed in these new lesions. This encouraged us to retest FFPE from recurrence for KRAS status and to check the first diagnosis using the real-time PCR method, which has a limit of detection of 1 %. We confirmed the presence of KRAS G12C mutation in this specimen. After the progression described above, the patient started the next line of chemotherapy and is still alive at the time of writing.
The second patient (no. 40) was a 64-year-old man who was diagnosed in 2011 with local adenocarcinoma of the rectum (G2 pT3N2). The patient received short radiation therapy followed by radical surgery and then adjuvant chemotherapy. During this chemotherapy, a CT scan of the abdomen was performed and metastatic disease was diagnosed. The patient was referred for genetic KRAS mutation evaluation. SSCP analysis indicated KRAS WT status, which qualified him for concomitant targeted therapy (cetuximab: first dose, 400 mg/m2 day 1; subsequent doses: 250 mg/m2) and chemotherapy (irinotecan 180 mg/m2 day 1, 5-fluorouracil 400 mg/m2 bolus and then 2,400 mg/m2 over 48 h; leucovorin 200 mg/m2 before 5-fluorouracil). At first, partial remission was observed, but therapy was stopped after seven cycles due to clinical and radiological progression of the disease. As further genetic analysis showed, the patient did not have mutations in KRAS (codons 12, 13, and 61) or V600E BRAF.
Comparison of the Mutational Status of KRAS Codons 12 and 13 and BRAF Codon 600 Determined by Real-Time PCR Methods and PCR with Reverse Hybridization
Before the novel real-time PCR and strip analysis was evaluated for the detection of somatic mutations in colorectal carcinoma with unknown KRAS and BRAF status, the methods were tested using tumor samples known to harbor KRAS mutations, detected previously using the TheraScreen K-Ras Mutation Kit (DxS). KRAS control 8 carries the most common mutation in primary and metastatic colorectal cancer, G12D [22], whereas KRAS control 9 carries a mutation in codon 13 (G13D) [21]. The novel methods for the evaluation of BRAF mutation and wild-type KRAS status involved the use of human colon adenocarcinoma cell line (HT29), which is known to carry wild-type KRAS and the V600E BRAF mutation (Fig. 1a, b).
Among the 40 patients of the study cohort, KRAS codon 12 was mutated in 14 cases, codon 13 in 5 cases, and codon 61 in 2 cases. We used the same isolated DNA for real-time PCR with allele-specific oligonucleotides (EntroGen) and PCR with a hybridization step to a strip test containing allele-specific probes. The same final results for codons 12 and 13 were evaluated using both kits for all tested samples: mutations G12D (Fig. 1c) and G12V were detected in 6 and 5 separate cases for each mutation, respectively, G12C was detected in 3 samples (Fig. 3), and G13D was detected in 5 samples (Fig. 1d), including positive controls (numbers 37, 38). The BRAF mutation in codon 600 was detected in 2 cases by both methods (Fig. 2). No mismatch between real-time PCR (EntroGen) and StripAssay was observed in the detection of mutations in KRAS codons 12 and 13, and BRAF codon 600.
Fig. 3.
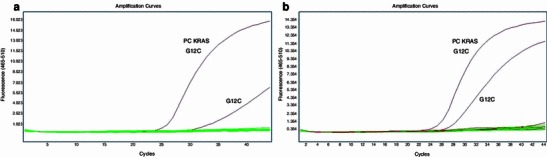
Retrospective KRAS and V600 BRAF mutation status analysis in sample no. 39 (previously determined as wild-type KRAS using the SSCP method). a Detection of KRAS G12C mutation in sample no. 39 (amplification plots for G12C positive control with C p 25.71 and sample no. 39 with C p 30.95, in baselines determining BRAF V600 wild type and no other mutations in KRAS codons 12, 13, and 61). b Detection of the same KRAS G12C mutation in the metastatic tumor cells (amplification plots for G12C positive control with C p 25.71 and sample no. 39 with C p 27.34)
Detection of Mutation in KRAS Codon 61 by Real-Time PCR Method
The mutation analysis of KRAS in codon 61 of exon 3 was performed using the real-time PCR method only (EntroGen). Positive FAM signals were detected in two cases, in which probes complementary to Q61H (Fig. 1e) and Q61L (Fig. 1f) hybridized to the sequence of interest and yielded amplicons with C p = 26.
Low Level of KRAS Mutation Detection in Codon 12 in the Context of Clinical Outcomes in Patients Previously Evaluated with KRAS WT Status by the Less Sensitive SSCP Method
To determine whether patients—those who did not respond well to targeted therapy—have the mutation in KRAS codon 61 or BRAF V600E, we re-evaluated two samples (numbers 39, 40) previously tested by the SSCP method and evaluated as KRAS wild-type in codons 12 and 13. Sample no. 40 did not carry any KRAS mutation in codons 12, 13, or 61, and no V600E BRAF mutation was detected (Table 2). Sample no. 39 was retested and the KRAS G12C mutation was detected in codon 12 by both methods. Each analysis was repeated at least twice by the real-time PCR method and by the KRAS StripAssay, producing the same results (Fig. 3). Interestingly, this sample (no. 39) was derived from a patient who had taken cetuximab in combination with LF4 (5-fluorouracil + leucovorin) for 4 months as fourth-line chemotherapy.
Discussion
Real-time PCR methods are becoming increasingly popular in molecular diagnostics and may replace the gold standard of Sanger sequencing or other methods with an insufficient limit of somatic mutation detection in the present quantitative PCR era. Allele-specific quantitative PCR with hydrolysis probes has been reported to surpass the 1 % sensitivity level [15]. In addition, a recent study involving serial dilution of DNA carrying a KRAS c.34G>T or a c.35G>A mutation demonstrated that minimum DNA input is 32 pg, the equivalent of 4–6 cells of high molecular DNA, to give a C p value <35 [23]. The K-Ras/B-Raf Mutation Analysis Panel Kit for real-time PCR (EntroGen) uses allele-specific quantitative PCR with hydrolysis probes to detect common mutations not only in V600E BRAF and KRAS codons 12 and 13, but, as opposed to the KRAS-BRAF StripAssay, also in codon 61. In cohort two of this study, the samples carried the following mutations in codon 61: Q61H and Q61L. Those mutations were not detected by the StripAssay due to the lack of allele-specific probes for mutations in codon 61 of the KRAS gene in the tested assay. The KRAS codon 61 mutation has been observed in 10.5 % of patients with KRAS codons 12 and 13 wild-type and was associated with a lower response rate and a worse progression-free survival [17]. Our results are in accordance with those data. We observed 10.5 % of samples with a mutation in KRAS codon 61 and another 10.5 % with V600E BRAF mutation in 19 samples with KRAS wild-type status in codons 12 and 13 (Figs. 1e, f, 2). Therefore, this mutation analysis should be recommended in routine diagnostics because the presence of KRAS mutation in codon 61 or BRAF in codon 600 can help the oncologist optimize the treatment, especially when patients with those mutations probably will not respond to anti-EGFR monoclonal antibodies [17].
Previous mutation analysis revealed that the V600E mutation in BRAF and KRAS mutations are mutually exclusive in colorectal [11] and gastric cancer [24]. Our data are consistent with those observations: both V600E BRAF mutations were observed in the sample that had KRAS wild-type status in codons 12, 13, and 61.
In SSCP analysis, the limit of mutation detection is as low as 10 %—and is lower than the gold standard (direct sequencing) where the limit of detection is in the range of 10–30 %, depending on the system and the quality of DNA [25]. Another evaluation of molecular methods for somatic mutation analysis performed at Johns Hopkins Medical Institution showed that the limit of detection for Sanger sequencing, pyrosequencing, and melting curve analysis of KRAS mutation was approximately 15–20 %, 5 %, and 10 % mutant alleles, respectively [26]. A different comparison of peptide nucleic acid PCR clamping and direct sequencing also showed the inability of the gold standard method to identify low levels of mutation-bearing colorectal cancer cells [27]. Macrodissection or laser microdissection could help increase the percentage of tumor in the specimen for DNA isolation, but also adds labor time to the pathomorphologist’s schedule. In the FFPE samples containing 10 % tumor cells, 1 % of mutant cell detection corresponds to genetic heterogeneity within the same tumor, as previously reported for KRAS in CRCs [7, 28]. Therefore, quantitative PCR methods will increase mutation detection as was demonstrated in a diagnostic case with 1 % of neoplastic cells in the entire section and around 10 % in the macrodissected area [28]. Mutation was not detected in DNA isolated from the whole section while G12D mutation was identified in the same macrodissected sample, taking into consideration that in both cases DNA quality was unfavorable [28]. On the other hand, the results obtained by Tsiatis et al. demonstrated that the percentage of the KRAS mutant cells was not universally lower, but sometimes greater than that predicted based on tumor evaluation by the pathologists or individual tumor cells harboring extra copies of mutant KRAS alleles when two samples in the series had tumor percentages estimated to be <20 % (<10 % predicted mutant alleles), but with >45 % KRAS mutant alleles identified using pyrosequencing [26].
Although those molecular diagnostics results increased the importance of molecular methods with best limit of detection and minimum false negative and false positive results, the clinical significance of low-level mutants in relation to their prognostic and therapeutic benefit has not been fully understood [26]. Our results demonstrate for the first time that patients with a very low content of tumor cells harboring mutations in KRAS codon 12 were previously diagnosed as carrying wild-type KRAS due to the limit of detection of approximately 10 % in the SSCP method.
Patients with the KRAS G12C mutation, treated with cetuximab (Erbitux) had stabilization for a short period, and then novel metastatic foci occurred with KRAS mutated in the same codon 12. Therefore, we might conclude that outcome of personalized treatment is highly associated with tumor heterogeneity observed in patients because the detected mutation was probably present in only 5 % of tumor cells. In the case of patient no. 39, during the targeted anti-EGFR treatment, it came to the selection of KRAS G12C cells (which were present from the beginning in the tumor but at a low level of around 5 %, Fig. 3a) and in consequence to the metastasis (Fig. 3b).
On the other hand, patient no. 40 did not respond to cetuximab—we even confirmed KRAS WT and BRAF WT status by real-time PCR methods. This reminds us that the patient could have a KRAS mutation in codon 146 [17] or another molecular mechanism is involved in his lack of response to cetuximab.
Therefore, when the molecular method is chosen for somatic mutation screening, not only costs but also several other factors should be taken into consideration such as the best limit of detection, sensitivity (false negatives) and specificity (false positives), reproducibility, and reasonable turnaround time (Table 3) [26]. SSCP and KRAS-BRAF StripAssay differ in cost of reagents and the limit of detection but both have the longest turnaround time, while the real-time PCR method (K-Ras/B-Raf Mutation Analysis Panel Kit for real-time PCR, EntroGen) has prices comparable with those of StripAssay, but its turnaround time is minimal with the same limit of detection (Table 3).
Table 3.
Comparison of estimated assay cost, turnover time and limit of detection without DNA isolation
| Method | SSCP | EntroGen (KRAS-BRAF real-time) | ViennaLab (KRAS-BRAF StripAssay) |
|---|---|---|---|
| Reagent costs | Low | High | High |
| Labor time | High | Low | High |
| Turnover time (h) | 5 | 2.5 | 5–6 |
| Limit of detection | 10 % [25] | 1 % (instructions for use v.2.6) | 1 % (instructions for use 03/2010) |
Conclusions
Our comparison of two sensitive methods with a limit of detection of 1 % mutant DNA indicated the same level of mutation detection reliability. However, labor time indicates the real-time PCR method as preferred. Our results confirm that somatic hotspot mutations can be reliably detected using allele-specific quantitative PCR with hydrolysis probes [23, 29]. We identified in DNA isolated from colorectal cancer 10 % mutation outside of KRAS codons 12/13 and another 10 % in BRAF V600 codon. All mutations were detected in material with a minimum 10 % of tumor cells (PTC). Our clinical sample (patient no. 39) with a low content of tumor cells may suggest preponderance of PCR methods in detecting KRAS mutation over the SSCP, as well as the importance of this in clinical practice.
Acknowledgments
We thank Dr. Z. Pawlowicz for generating the conditions advantageous for our research. This work is supported by a Foundation for Polish Science, cofinanced from European Union, Regional Development Fund (HOMING PLUS/2010-2/7).
Conflict of interest
The authors have no conflicts of interest that are directly relevant to the content of this article.
References
- 1.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA. 1982;79(11):3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kranenburg O. The KRAS oncogene: past, present, and future. Biochimica et biophysica acta. 2005;1756(2):81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 4.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA, Jr, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325(5947):1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317(15):929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 7.Andreyev HJ, Tilsed JV, Cunningham D, Sampson SA, Norman AR, Schneider HJ, Clarke PA. K-ras mutations in patients with early colorectal cancers. Gut. 1997;41(3):323–329. doi: 10.1136/gut.41.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KH, Lee JS, Suh C, Kim SW, Kim SB, Lee JH, Lee MS, Park MY, Sun HS, Kim SH. Clinicopathologic significance of the K-ras gene codon 12 point mutation in stomach cancer. An analysis of 140 cases. Cancer. 1995;75(12):2794–2801. doi: 10.1002/1097-0142(19950615)75:12<2794::AID-CNCR2820751203>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256(5053):102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 10.Otori K, Oda Y, Sugiyama K, Hasebe T, Mukai K, Fujii T, Tajiri H, Yoshida S, Fukushima S, Esumi H. High frequency of K-ras mutations in human colorectal hyperplastic polyps. Gut. 1997;40(5):660–663. doi: 10.1136/gut.40.5.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418(6901):934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 12.Namba H, Nakashima M, Hayashi T, Hayashida N, Maeda S, Rogounovitch TI, Ohtsuru A, Saenko VA, Kanematsu T, Yamashita S. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab. 2003;88(9):4393–4397. doi: 10.1210/jc.2003-030305. [DOI] [PubMed] [Google Scholar]
- 13.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, Solit DB. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Krieken JH, Jung A, Kirchner T, Carneiro F, Seruca R, Bosman FT, Quirke P, Flejou JF, Plato Hansen T, de Hertogh G, Jares P, Langner C, Hoefler G, Ligtenberg M, Tiniakos D, Tejpar S, Bevilacqua G, Ensari A. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453(5):417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 19.Santini D, Spoto C, Loupakis F, Vincenzi B, Silvestris N, Cremolini C, Canestrari E, Graziano F, Galluccio N, Salvatore L, Caraglia M, Zito FA, Colucci G, Falcone A, Tonini G, Ruzzo A. High concordance of BRAF status between primary colorectal tumours and related metastatic sites: implications for clinical practice. Ann Oncol Off J Eur Soc Med Oncol. 2010;21(7):1565. doi: 10.1093/annonc/mdq318. [DOI] [PubMed] [Google Scholar]
- 20.Krol LC, Hart NA, Methorst N, Knol AJ, Prinsen C, Boers JE. Concordance in KRAS and BRAF mutations in endoscopic biopsy samples and resection specimens of colorectal adenocarcinoma. Eur J Cancer. 2012;48(7):1108–1115. doi: 10.1016/j.ejca.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Lewandowska MA, Jóźwicki W, Żurawski B, Starzyński J, Rhone P. Can we believe in CE-IVD tests for KRAS mutation analysis? Oncology as a challenge for modern medicine. Biała Podlaska: PSW; 2012. [Google Scholar]
- 22.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O’Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Eijk R, Licht J, Schrumpf M, Talebian Yazdi M, Ruano D, Forte GI, Nederlof PM, Veselic M, Rabe KF, Annema JT, Smit V, Morreau H, van Wezel T. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 2011;6(3):e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim IJ, Park JH, Kang HC, Shin Y, Park HW, Park HR, Ku JL, Lim SB, Park JG. Mutational analysis of BRAF and K-ras in gastric cancers: absence of BRAF mutations in gastric cancers. Hum Genet. 2003;114(1):118–120. doi: 10.1007/s00439-003-1027-0. [DOI] [PubMed] [Google Scholar]
- 25.Sivertsson A, Platz A, Hansson J, Lundeberg J. Pyrosequencing as an alternative to single-strand conformation polymorphism analysis for detection of N-ras mutations in human melanoma metastases. Clin Chem. 2002;48(12):2164–2170. [PubMed] [Google Scholar]
- 26.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12(4):425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207(12):762–768. doi: 10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Kotoula V, Charalambous E, Biesmans B, Malousi A, Vrettou E, Fountzilas G, Karkavelas G. Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics. PLoS One. 2009;4(11):e7746. doi: 10.1371/journal.pone.0007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewandowska MA, Jóźwicki W, Starzynski J, Kowalewski J. Analysis of EGFR mutation frequency and coexistence of KRAS and EGFR mutations using RT-PCR in lung adenocarcinoma: May a clinical and pathological model of a patient’s qualification for targeted therapy have an impact on time to obtain genetic results? Pol J Cardio-Thorac Surg. 2012;9(4):443–451. doi: 10.5114/kitp.2012.32682. [DOI] [Google Scholar]


