The B-box domains of BBX25 physically interact with the bZIP domain of HY5, and BBX24 and BBX25 inhibit the activation of BBX22 expression by HY5, probably by forming inactive heterodimers. In contrast with their role during deetiolation, BBX24 and BBX25 can switch roles and function independently of HY5 in the hypocotyl shade avoidance response.
Abstract
ELONGATED HYPOCOTYL5 (HY5) is a basic domain/leucine zipper (bZIP) transcription factor, central for the regulation of seedling photomorphogenesis. Here, we identified a B-BOX (BBX)–containing protein, BBX25/SALT TOLERANCE HOMOLOG, as an interacting partner of HY5, which has been previously found to physically interact with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1). BBX25 physically interacts with HY5 both in vitro and in vivo. By physiological and genetic approaches, we showed that BBX25 is a negative regulator of seedling photomorphogenesis. BBX25 and its homolog BBX24 regulate deetiolation processes and hypocotyl shade avoidance response in an additive manner. Moreover, genetic relationships of bbx25 and bbx24 with hy5 and cop1 revealed that BBX25 and BBX24 additively enhance COP1 and suppress HY5 functions. BBX25 accumulates in a light-dependent manner and undergoes COP1-mediated degradation in dark and light conditions. Furthermore, a protoplast cotransfection assay showed that BBX24 and BBX25 repress BBX22 expression by interfering with HY5 transcriptional activity. As HY5 binds to the BBX22 promoter and promotes its expression, our results identify a direct mechanism through which the expression of BBX22 is regulated. We suggest that BBX25 and BBX24 function as transcriptional corepressors, probably by forming inactive heterodimers with HY5, downregulating BBX22 expression for the fine-tuning of light-mediated seedling development.
INTRODUCTION
Plants have evolved a network of sophisticated and highly complex mechanisms to cope with fluctuating abiotic (e.g., light and temperature) and biotic (e.g., pathogens and insects) conditions. To withstand the constant diurnal and seasonal variations in quality, quantity, duration, and direction of light, plants have a battery of photoreceptors to perceive and integrate light signals to optimize growth and development. In Arabidopsis thaliana, red:far-red (R:FR) light is perceived by phytochromes (phyA to phyE), blue/UV-A light is perceived by cryptochromes (cry1 and cry2) and phototropins (phot1 and phot2), and UV-B is perceived by UVR8 photoreceptor (Casal, 2013).
Genetic, molecular, and biochemical studies have led to the identification of several light signaling intermediates, which act downstream of photoreceptors (Jiao et al., 2007; Chen and Chory, 2011). CONSTITUTIVE PHOTOMORPHOGENIC (COP)/DE-ETIOLATED/FUSCA function downstream of multiple photoreceptors and act as suppressors of photomorphogenesis. COP1 is an E3 ubiquitin ligase, which suppresses photomorphogenesis in dark conditions by targeting many photomorphogenesis-promoting factors, such as ELONGATED HYPOCOTYL5 (HY5), HY5 HOMOLOG, LONG AFTER FAR-RED LIGHT1, LONG HYPOCOTYL IN FAR- RED1 (HFR1) and B-Box22 (BBX22) for degradation (Lau and Deng, 2012). In light-grown seedlings, multiple photoreceptors repress COP1 activity by inactivating and relocating COP1 to the cytosol (Osterlund and Deng, 1998), thus stabilizing photomorphogenesis-promoting factors in the nucleus. SUPPRESSOR OF PHYTOCHROME A (SPA1 to SPA4) proteins act redundantly and can physically interact with COP1 to suppress photomorphogenic growth in the dark (Seo et al., 2003; Laubinger et al., 2004; Zhu et al., 2008). Furthermore, COP1 promotes the hypocotyl shade avoidance syndrome (SAS) response by regulating early shade transcription factors (Crocco et al., 2010). The COP1/SPA ubiquitylation complex targets HFR1 but not HY5 to promote hypocotyl and leaf petiole elongation in response to low R:FR light, suggesting that different COP1 signaling pathways mediate deetiolation and SAS (Rolauffs et al., 2012). Phytochrome interacting factors (PIFs) collaborate redundantly with COP1 to suppress photomorphogenesis in the dark and promote SAS (Lorrain et al., 2008; Leivar et al., 2009). In light conditions, PIFs physically interact with Pfr, the active form of phytochromes, and are then phosphorylated as a prelude to induce a rapid degradation of these proteins via ubiquitin-proteasome system-mediated proteolysis (Al-Sady et al., 2008; Lorrain et al., 2008; Shen et al., 2008). However, in dark conditions, PIF4, PIF5, and PIF7 dephosphorylated forms accumulate in the nucleus, activating the expression of genes for cell elongation (Lorrain et al., 2008; Li et al., 2012).
The basic domain/leucine zipper (bZIP) transcription factor HY5 functions as a positive regulator of photomorphogenesis downstream of all the photoreceptors and COP1 (Lau and Deng, 2010). HY5 regulates several physiological processes, such as inhibition of hypocotyl elongation, anthocyanin and chlorophyll synthesis, and lateral root formation (Oyama et al., 1997; Holm et al., 2002). HY5 chromatin immunoprecipitation–chip studies have suggested that HY5 preferentially binds to promoter regions throughout the genome (Lee et al., 2007; Zhang et al., 2011), including genes involved in photosynthesis and pigment synthesis, like CAB, RBCS1A, F3H, CHS, and CHI, as well as genes involved in the regulation of the circadian clock (Ang et al., 1998; Chattopadhyay et al., 1998; Lee et al., 2007). However, while HY5 binding is required, it is not sufficient for the transcriptional regulation of the promoters, and HY5 appears to require other cofactors or modifications to regulate the expression of target genes (Lee et al., 2007). For example, HY5 has been reported to act cooperatively with HYH, HFR1, and CAM7 (Holm et al., 2002; Kim et al., 2002; Kushwaha et al., 2008).
B-BOX proteins (BBX), which contain the N-terminal zinc binding B-box motif, function as transcriptional regulators in response to light, circadian cues, and brassinosteroid–light crosstalk signaling. BBX1/CONSTANS (CO) and BBX4/CONSTANS-LIKE3 contain two B-boxes at the N terminus and a CCT domain in the C terminus (Khanna et al., 2009). CO regulates the expression of FT, a flowering time gene, and genetically interacts with COP1. The co-10 allele can suppress the early flowering phenotype of cop1-4 in both long- and short-day conditions, and COP1 targets CO for degradation (Jang et al., 2008; Liu et al., 2008). BBX4, which is a positive regulator of photomorphogenesis in red light, physically interacts with COP1 and partially suppresses the cop1 phenotype in the dark (Datta et al., 2006). Eight of the BBX proteins that belong to structural group IV contain two tandem repeat B-box motifs in the N terminus but lack the CCT domain (Khanna et al., 2009). Among these, BBX21/SALT TOLERANCE HOMOLOG2 (STH2) and BBX22/STH3/LZF1 function as positive regulators (Datta et al., 2007, 2008; Chang et al., 2008, 2011), and BBX18/DBB1a, BBX19/DBB1b, BBX24/STO, and BBX25/STH act as negative regulators of photomorphogenesis (Indorf et al., 2007; Kumagai et al., 2008; Yan et al., 2011; Jiang et al., 2012). BBX21 and BBX22 physically interact with HY5 through their B-box motifs and colocalize with COP1 in nuclear speckles (Datta et al., 2007, 2008; Chang et al., 2008). COP1 ubiquitinates and targets BBX22 protein for 26S proteosome–mediated degradation (Datta et al., 2008; Chang et al., 2011). BBX21 and BBX22 are also involved in transcriptional activation of CAB and CHI in transient protoplast assays (Datta et al., 2007, 2008). Furthermore, BBX21 and BBX22 inhibit elongation, whereas BBX18 and BBX24 play an opposite function by promoting hypocotyl length in shady conditions (Crocco et al., 2010, 2011).
Although the function of BBX24 in light signaling has been investigated in some detail, less is known about the physiological and molecular function of BBX25 in light signaling. Here, we report that BBX25 physically interacts with HY5. BBX25 acts additively with BBX24 during deetiolation and the hypocotyl shade avoidance response. BBX25 enhances COP1 and suppresses HY5 functions through a BBX24-independent genetic pathway. Furthermore, BBX25 and BBX24, both alone and together, regulate the expression of BBX22 via HY5 by interfering with HY5 transcriptional activity. Collectively, we suggest that BBX25 and BBX24 act as transcriptional corepressors of HY5 by downregulating BBX22 expression, probably forming inactive heterodimers with HY5.
RESULTS
BBX25 Physically Interacts with HY5
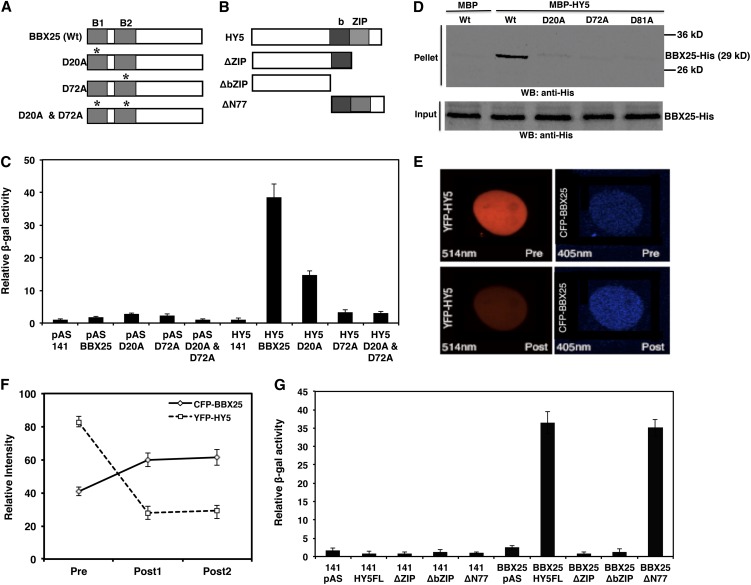
BBX21, BBX22, and BBX24 have been reported to physically and genetically interact with HY5 (Datta et al., 2007, 2008; Chang et al., 2008; Jiang et al., 2012). Since BBX25 shares 69% amino acid sequence similarity with BBX24, we examined whether BBX25 also interacts with HY5 using full-length and truncated versions of BBX25/HY5 (Figures 1A and 1B). Expression of full-length and mutated versions of BBX25 and full-length HY5 were confirmed by immunoblotting (see Supplemental Figures 1A and 1B online). BBX25 expressed together with Gal4-DBD alone did not activate transcription, but, when expressed with Gal4-DBD-HY5, resulted in an ∼28-fold increase in β-galactosidase activity compared with that of the vector control (Figure 1C). Previous reports suggest that the B-boxes in BBX21 and BBX22 are necessary for the interaction with HY5 (Datta et al., 2007, 2008). To see whether the B-boxes in BBX25 were also required for the interaction with HY5, we individually substituted two conserved Asp residues in the B-boxes to Ala. The substituted proteins in BBX25 were named D20A, D72A and D20A, and D72A (Figure 1A). Whereas the substitution in D20A resulted in a drastic reduction in β-galactosidase activity, substitutions in D72A and D20A and D72A resulted in complete abolition of the interaction compared with wild-type levels (Figure 1C). Consistent with the yeast two-hybrid results, in vitro pull-down assays also demonstrated that BBX25 interacts with HY5. These assays showed that HY5 was not able to pull down the mutated versions of BBX25 (D20A, D72A and D20A, and D72A) but that it was able to pull down wild-type BBX25 (Figure 1D). These results suggest that BBX25 interacts with HY5 through its B-boxes.
Figure 1.
BBX25 Physically Interacts with HY5.
(A) and (B) Schematic representation of domain structures of BBX25 (A) and HY5 (B). Asterisk indicates Asp-to-Ala substitutions in the B-boxes at 20 and 72 residues. Wt, the wild type.
(C) Yeast two-hybrid interactions (β-galactosidase enzymatic activity) of mutated versions of BBX25 with HY5. Error bars represent sd (n = 6). The data shown are representative of one of three experiments.
(D) BBX25 interacts with HY5 in vitro pull-down assay. Amylose resin–bound MBP and MBP-HY5 proteins were incubated with BBX25-His protein. BBX25-His was detected by immunoblotting using anti-His antibodies. WB, Western blot.
(E) FRET between CFP-BBX25 and YFP-HY5 as analyzed by acceptor bleaching in nuclei. The top panels show representative prebleach nuclei coexpressing YFP-HY5 and CFP-BBX25 excited with a 514- or a 405-nm laser, resulting in emission from YFP or CFP, respectively. The total nucleus was bleached with the 514-nm laser. The bottom panels show the same nuclei after bleaching excited with a 514- or 405-nm laser.
(F) Relative intensities of both YFP and CFP inside the nucleus measured once before and twice after the bleaching. Error bars represent sd (n = 10). The data shown are representative of one of three experiments.
(G) Yeast two-hybrid interactions (β-galactosidase enzymatic activity) of BBX25 with truncated versions of HY5. Error bars represent sd (n = 6). The data shown are representative of one of three experiments.
[See online article for color version of this figure.]
To determine the subcellular localization of BBX25 and to further characterize its interactions with HY5, we examined whether BBX25 physically interacts with HY5 in the nucleus using fluorescence resonance energy transfer (FRET). The cyan fluorescent protein (CFP) fused to BBX25 and yellow fluorescent protein (YFP) fused to HY5 were coexpressed in onion epidermal cells and checked for the occurrence of FRET. The bleaching of the acceptor (YFP-HY5) resulted in an increased emission from CFP-BBX25 (Figures 1E, bottom panel, and 1F), indicating that FRET had occurred between the two proteins prior to the bleach. However, in control photobleaching experiments using the same microscope settings, we detected no FRET between untagged CFP and YFP (see Supplemental Figure 2 online).
Furthermore, to map which domain of HY5 is responsible for the interaction with BBX25, we used different truncated versions of HY5 (ΔZIP, ΔbZIP, and ΔN77) fused to the DNA binding domain Gal4-DBD (Figure 1B). Expression of the truncated versions of HY5 was confirmed by immunoblot (see Supplemental Figure 1C online). Quantitative measurements of β-galactosidase activity indicated that BBX25 strongly interacts with the full-length version of HY5 and the ΔN77 truncated version of HY5 (Figure 1F). However, deletion of the bZIP domain (either ΔZIP or ΔbZIP) in HY5 resulted in a drastic reduction of β-galactosidase activity (Figure 1G), suggesting that the bZIP domain of HY5 is necessary and sufficient to mediate the interaction with BBX25. Collectively, the results from yeast two-hybrid, pull-down, and FRET experiments demonstrate that BBX25 physically interacts with HY5.
Molecular and Physiological Characterization of bbx25 Mutants
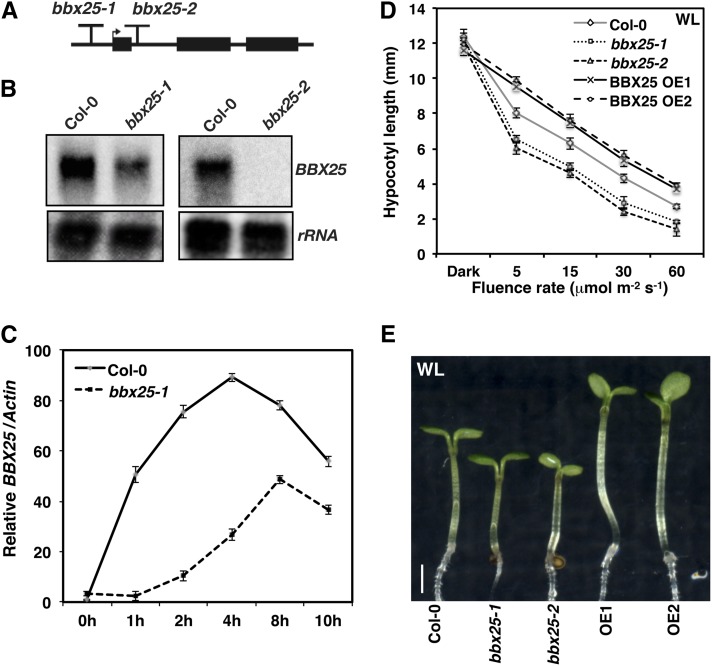
To examine the physiological role of BBX25 in light-mediated seedling development, we obtained two independent T-DNA insertion lines for BBX25 and designated them as bbx25-1 (SAIL_786_F08) and bbx25-2 (SM_3.17575). Genotyping and sequencing of both T-DNA lines revealed that bbx25-1 and bbx25-2 have T-DNA insertions in the promoter and in the first intron of the gene, respectively (Figure 2A). The RNA gel blot results showed that the bbx25-2 mutant carried a null allele of BBX25, whereas the bbx25-1 mutant showed reduced expression of BBX25 transcripts compared with the Columbia-0 (Col-0) wild type (Figure 2B). Since BBX25 expression is light modulated (Kumagai et al., 2008), we monitored the expression of BBX25 in 4-d-old dark-grown wild-type and bbx25-1 seedlings exposed to white light (WL) between 0 and 10 h. In wild-type seedlings, BBX25 transcripts were 50-fold increased at 1 h of light, reached 90-fold maximum expression at 4 h of light, and then gradually decreased (Figure 3C). By contrast, the bbx25-1 mutant showed a reduced fivefold increase at 1 h of light and the 40-fold expression peak was detected after 8 h of light (Figure 3C). In addition, 4-d-old bbx25-1 seedlings grown in constant WL accumulated half of the wild-type transcripts, providing further evidence for a reduced BBX25 activity in the bbx25-1 mutant (see Supplemental Figure 3A online). These results indicate that bbx25-1 is a reduced functional allele of BBX25.
Figure 2.
Molecular and Physiological Characterization of bbx25 Mutants.
(A) Schematic representation of the BBX25 gene. The arrow indicates the position of the start site Met, and the T indicates the T-DNA insertion positions. Black rectangular boxes depict exons, and lines between these boxes represent introns.
(B) RNA gel blots showing BBX25 transcript accumulation just before dawn in Col-0, bbx25-1, and bbx25-2 mutant alleles.
(C) Quantitative RT-PCR analysis of BBX25 expression in wild-type and bbx25-1 in 4-d-old dark-grown seedlings treated with WL (100 µmol m−2 s−1) for the indicated time period before harvesting. Error bars represent se (n = 3).
(D) Fluence response curve of indicated genotypes grown in WL for 6 d. Error bars represent se (n ≥ 28).
(E) Photograph of representative seedlings of indicated genotypes grown in constant WL (30 µmol m−2 s−1) for 6 d. Bar = 1 mm.
[See online article for color version of this figure.]
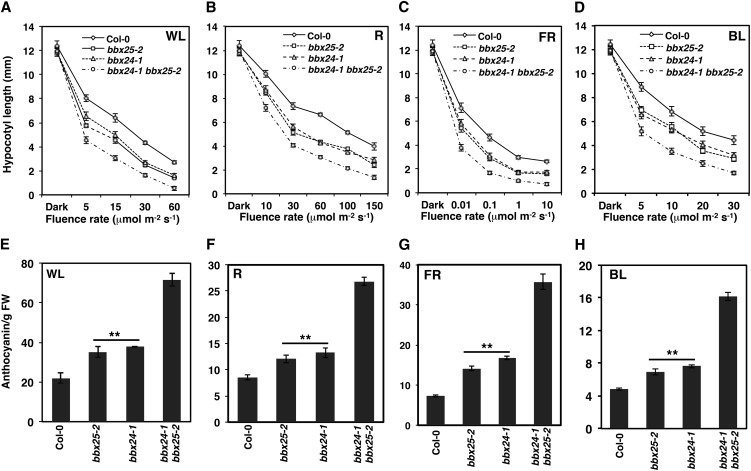
Figure 3.
BBX25 Functions Additively with BBX24 for the Inhibition of Seedling Deetiolation.
(A) to (D) Fluence response curves of 6-d-old Col-0 and single and double mutant seedlings grown in WL, R, FR, and BL. Error bars represent se (n ≥ 28).
(E) to (H) Anthocyanin content of 6-d-old Col-0 and single and double mutant seedlings grown in WL (30 µmol m−2 s−1), R (60 µmol m−2 s−1), FR (0.1 µmol m−2 s−1), and BL (20 µmol m−2 s−1). Error bars represent se (n = 3). Asterisks show genotypes that differ significantly from Col-0 (Student’s t test, *P ≤ 0.01). FW, fresh weight.
To carry out a detailed phenotypic analysis of the role of BBX25 in response to light, we studied the phenotypes of BBX25-overexpressing lines (OE1 and OE2;, see Supplemental Figure 3B online) and mutants (bbx25-1 and bbx25-2). Col-0, bbx25-1, bbx25-2, OE1, and OE2 seedlings were grown in different light conditions for 6 d to examine the hypocotyl length. bbx25-1 and bbx25-2 seedlings displayed significantly shorter hypocotyls, whereas BBX25-overexpressing lines showed larger hypocotyls than Col-0 seedlings in WL (Figures 2D and 2E) and monochromatic light conditions (R, FR, and blue light [BL]; see Supplemental Figure 4 online). We concluded that BBX25 is directly involved in light signaling and functions as a negative regulator of seedling photomorphogenesis irrespective of light quality.
BBX25 Additively Enhances BBX24 Function in Deetiolation
To see whether BBX25 has functions overlapping with those of BBX24, we generated (see Supplemental Figure 5 online) and characterized a bbx25-2 bbx24-1 double mutant. The hypocotyl of bbx25-2 bbx24-1 double mutant seedlings was significantly shorter than that of each of the single mutants, thus suggesting an additive genetic interaction between BBX24 and BBX25 independently of the light treatment (Figures 3A to 3D). In addition, 5-d-old wild-type seedlings showed low levels of anthocyanin in all light conditions, whereas bbx24 and bbx25 single mutant seedlings showed an increase of 30% in their anthocyanin levels and bbx25-2 bbx24-1 double mutant seedlings an increase of >300% compared with wild-type seedlings (Figures 3E to 3H). Again, the stronger enhancement of anthocyanin accumulation in bbx25-2 bbx24-1 compared with single mutants indicates that BBX25 and BBX24 act additively.
BBX25 Suppresses the HY5 Phenotype Together with BBX24
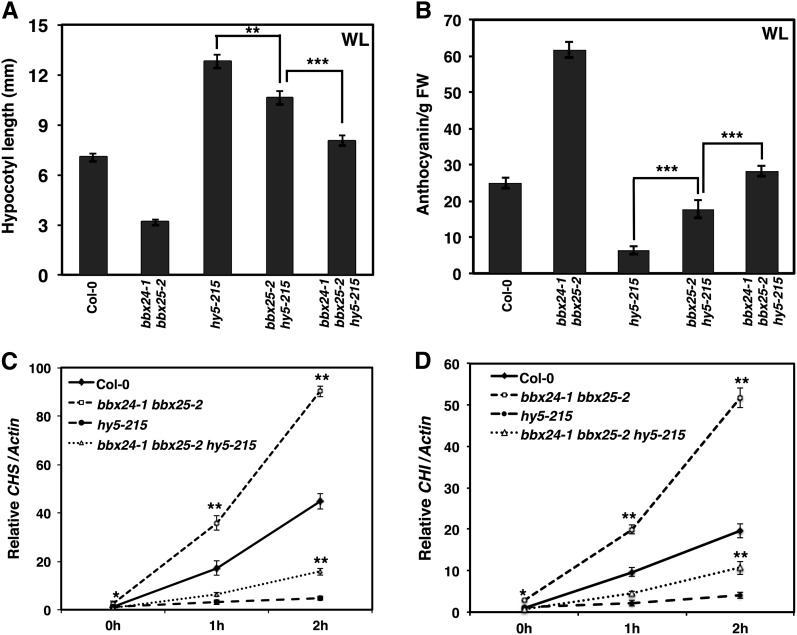
To investigate the genetic relationship between BBX25 and HY5, we generated a homozygous bbx25-2 hy5-215 double mutant. Whereas hy5-215 seedlings showed longer hypocotyls than wild-type seedlings, bbx25-2 hy5-215 double mutant seedlings showed slightly but significantly shorter hypocotyls than hy5-215 in WL and even in R and BL (Figure 4A; see Supplemental Figure 6 online). These data suggest that BBX25 function is partially HY5 dependent. To understand this phenomenon further, we generated a bbx25-2 bbx24-1 hy5-215 triple mutant. The triple mutant had significantly shorter hypocotyls than the bbx25-2 hy5-215 double mutant in WL and different monochromatic light conditions (Figure 4A; see Supplemental Figure 6 online). Taken together, these results suggest that both bbx25-2 and bbx24-1 can partially and additively suppress the hy5-215 hypocotyl phenotype.
Figure 4.
bbx25 and bbx24 Additively Suppress the hy5 Phenotype.
(A) Hypocotyl length phenotype of 5-d-old Col-0 and single, double, and triple mutant seedlings grown in WL (15 µmol m−2 s−1). Error bars represent se (n ≥ 30). Asterisks show pairs of genotypes that differ significantly between them (Student’s t test, **P ≤ 0.01 and ***P ≤ 0.001).
(B) Anthocyanin content of 5-d-old Col-0 and single, double, and triple mutant seedlings grown in WL (30 µmol m−2 s−1). Error bars represent se (n = 3). Asterisks show pair of genotypes that differ significantly between them (Student’s t test, ***P ≤ 0.001). FW, fresh weight.
(C) and (D) Quantitative RT-PCR analysis of CHS and CHI genes, respectively, of 4-d-old Col-0, bbx24-1, bbx25-2, hy5-215, and bbx24-1 bbx25-2 hy5-215 seedlings grown in WL (80 µmol m−2 s−1) for 0, 1, and 2 h. Error bars represent se (n = 3). Asterisks show significant differences between Col-0 and double mutant or hy5-215 and triple mutant at the same time of light exposure (Student’s t test, *P ≤ 0.05 and **P ≤ 0.01).
Since the bbx24-1 bbx25-2 double mutant accumulates significantly more anthocyanin than the single mutants (Figures 3E to 3H), we wanted to know whether enhanced anthocyanin accumulation in the double mutant is HY5 dependent. Although the anthocyanin content in hy5-215 is extremely low in WL, the absence of BBX25 and/or BBX24 in the hy5 background induces the accumulation of anthocyanin, achieving wild-type levels in the bbx24-1 bbx25-2 hy5-215 triple mutant (Figure 4B). By contrast, the anthocyanin content was comparable to that of hy5 in double and triple mutant seedlings grown under monochromatic light conditions (see Supplemental Figure 7 online). Expression of anthocyanin biosynthetic genes, such as CHS and CHI, further confirms the anthocyanin content results. Col-0 seedlings grown in the dark for 4 d and then treated with WL for 1 and 2 h showed increased expression of CHS and CHI (Figures 4C and 4D). In the bbx24-1 bbx25-2 double mutant, the expression of both genes was two- to threefold higher than that of the wild type, and null expression was detected in the hy5-215 single mutant. Moreover, bbx24-1 bbx25-2 hy5-215 triple mutant seedlings showed a slight but significant recovery of CHS and CHI expression compared with hy5-215 but not under R, FR, and BL conditions (Figures 4C and 4D; see Supplemental Figure 8 online). These results suggest that BBX25 and BBX24 regulate anthocyanin gene expression in a HY5-dependent manner.
BBX25 Enhances the COP1 Phenotype Together with BBX24
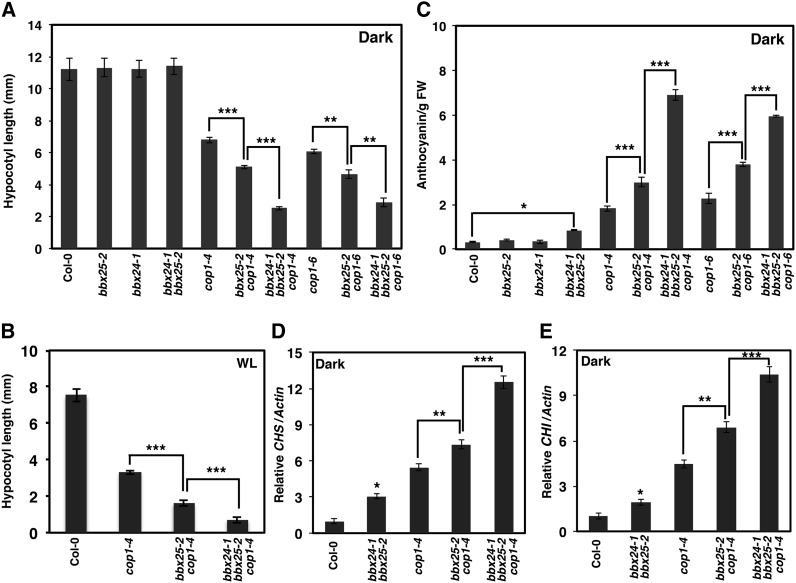
Since COP1 is a central negative regulator of photomorphogenesis, we investigated whether bbx25 alone and together with bbx24 influence the cop1 phenotype. We used two weak cop1 alleles, cop1-4 and cop1-6, to generate bbx25-2 cop1-4 and bbx25-2 cop1-6 double mutants. Five-day-old dark-grown cop1 seedlings displayed a strong photomorphogenic phenotype, whereas the hypocotyls of bbx25-2 bbx24-1 and of their respective single mutants were indistinguishable from the wild-type ones (Figure 5A). By contrast, bbx25-2 cop1-4 and bbx25-2 cop1-6 seedlings showed shorter hypocotyls than cop1-4 and cop1-6 (Figure 5A; see Supplemental Figure 9A online). To further investigate the genetic action of BBX24 and BBX25 on COP1 signaling, we constructed bbx24-1 bbx25-2 cop1-4 and bbx24-1 bbx25-2 cop1-6 triple mutants. Five-day-old dark-grown seedlings of bbx24-1 bbx25-2 cop1-4 and bbx24-1 bbx25-2 cop1-6 showed hypocotyls 61 and 49% shorter, respectively, than their corresponding cop1 single mutants (Figure 5A; see Supplemental Figure 9A online). Similar results were found when seedlings were grown in different light conditions (Figure 5B; see Supplemental Figure 9B online). These results demonstrate that bbx25-2 and bbx24-1 additively enhance the cop1 hypocotyl phenotype in dark and light conditions.
Figure 5.
bbx25 and bbx24 Additively Enhance the cop1 Phenotype.
(A) Hypocotyl length of seedlings from indicated genotypes grown in dark for 5 d. Error bars represent se (n ≥ 25). Asterisks show pairs of genotypes that differ significantly between them (Student’s t test, **P ≤ 0.01 and ***P ≤ 0.001).
(B) Hypocotyl length of seedlings from indicated genotypes grown in WL (5 µmol m−2 s−1) for 5 d. Error bars represent se (n ≥ 35). Asterisks show pairs of genotypes that differ significantly between them (Student’s t test, ***P ≤ 0.001).
(C) Anthocyanin content in seedlings from indicated genotypes grown in dark for 5 d. Error bars represent se (n = 3). Asterisks show pairs of genotypes that differ significantly between them (Student’s t test, *P ≤ 0.05 and ***P ≤ 0.001).
(D) and (E) Quantitative RT-PCR analysis of CHS and CHI genes, respectively, in seedlings from indicated genotypes grown in dark for 5 d. Error bars represent se (n = 3). Asterisks show pairs of genotypes that differ significantly between them (Student’s t test, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
The cop1 mutant accumulates more anthocyanin in both dark and light conditions (Saijo et al., 2003). While neither the bbx24-1 nor the bbx25-2 single mutants showed altered anthocyanin levels, the bbx24-1 bbx25-2 double mutant accumulated slightly but significantly more anthocyanin than the wild type in the dark (Figure 5C). Even more, bbx25-2 cop1-4 and bbx25-2 cop1-6 double mutants showed an increase of 175 and 163% in anthocyanin content, respectively, and the bbx24-1 bbx25-2 cop1-4 and the bbx24-1 bbx25-2 cop1-6 triple mutants showed an increase of 344 and 288% in anthocyanin content over their respective cop1 alleles (Figure 5C). The anthocyanin levels correlated well with the accumulation of CHS and CHI transcripts in 5-d-old dark-grown seedlings (Figures 5D and 5E). These results demonstrate that BBX25 and BBX24 act additively in the COP1 signaling pathway of hypocotyl length, anthocyanin accumulation, and gene expression.
COP1-Mediated Degradation of BBX25 by the 26S Proteasome Pathway
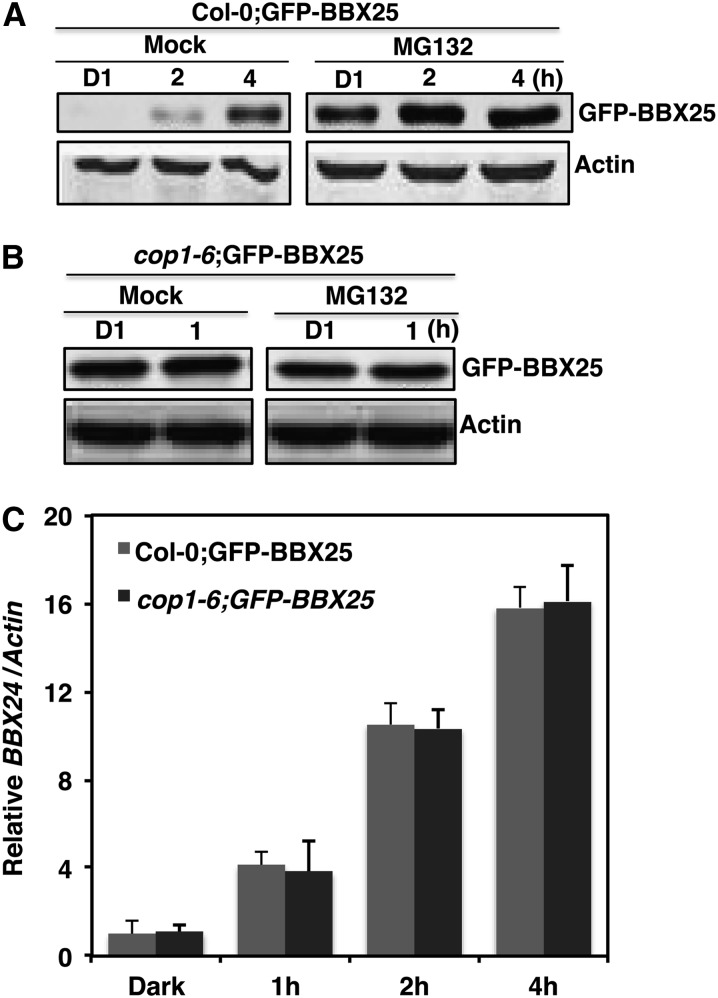
To know how BBX25 protein stability is regulated and whether or not COP1 has any role in the modulation of its stability, we generated transgenic lines overexpressing GFP-BBX25 in Col-0 and cop1-6 mutant background. Overexpression of GFP-BBX25 in the cop1-6 background but not in Col-0 led to longer hypocotyls in the dark (see Supplemental Figure 10 online). Four-day-old dark-grown seedlings were either mock treated with DMSO or treated with the proteasome-specific inhibitor MG132 and then kept in the dark for 1 h more. Immunoblot experiments showed that BBX25 fusion protein was clearly visible in the MG132-treated seedlings but not in mock-treated seedlings. Similarly, when 4-d-old dark-grown seedlings were treated with 2 and 4 h of WL, the BBX25 fusion protein was found to be more stable with MG132 than that of the mock-treated seedlings, suggesting that MG132 can block the degradation of BBX25 even in light conditions (Figure 6A). To know whether BBX25 degradation is mediated by COP1, we examined the BBX25 fusion protein in cop1-6 mutant background. Four-day-old dark-grown seedlings were either mock treated or treated with MG132 and incubated either in the dark or WL for 1 h and subjected to immunoblot analysis. The BBX25 fusion protein was clearly visible in both dark- and WL-grown seedlings either mock treated or treated with MG132 (Figure 6B), indicating that COP1 is mediating BBX25 degradation through the 26S proteasome pathway. Furthermore, the increased stability of BBX25 in cop1 mutant background is not due to an increase in BBX24 transcript levels (Figure 6C).
Figure 6.
BBX25 Undergoes COP1 Mediated Degradation via 26S Proteasome Pathway.
(A) and (B) Immunoblot showing the expression of GFP-BBX25 in 4-d-old seedlings overexpressing GFP-BBX25 in Col-0 or cop1-6, respectively. Seedlings were grown in dark for 4 d and then treated with mock (DMSO) or MG132 and incubated in dark for 1 h (D1) or WL (100 µmol m−2 s−1) for 1 h (in cop1-6 seedlings) or 2 and 4 h (in Col-0 seedlings). The protein blot was probed with GFP antibody. The time point = 0 represents the start of the MG132 that inhibits the degradation of BBX25 protein. Antiactin was used as a loading control.
(C) Quantitative RT-PCR analysis of BBX24 expression in GFP-BBX25 in Col-0 and cop1-6 transgenic seedlings grown in dark for 4 d and then exposed to WL (100 µmol m−2 s−1) for 1, 2, or 4 h. Error bars represent se (n = 3).
BBX25 and BBX24 Negatively Regulate the Expression of BBX22 by Interfering with HY5 Transcriptional Activity
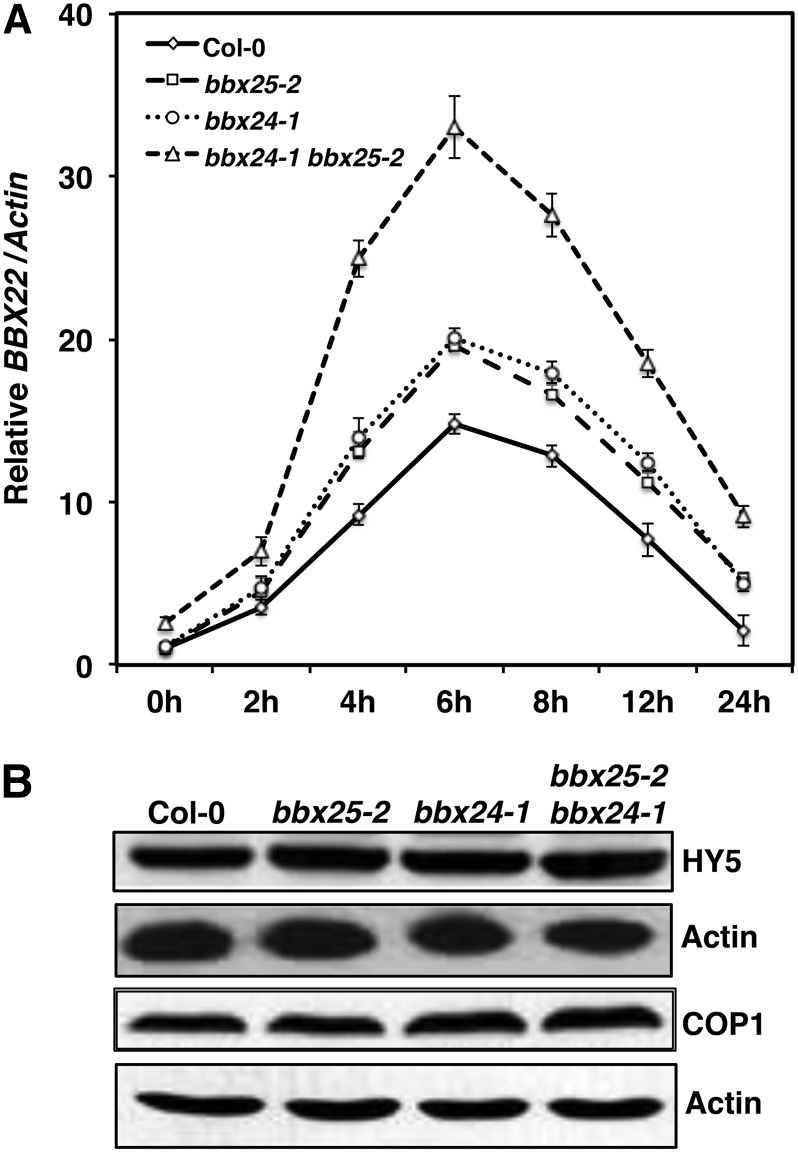
In order to find target genes of BBX24 and BBX25, we analyzed the expression of light-regulated genes like BBX18-BBX23, HY5, and HYH in 4-d dark-grown Col-0 and bbx24-1 bbx25-2 seedlings treated with WL for 2 and 4 h. Quantitative RT-PCR analysis showed that among the genes tested, only BBX22 showed significantly higher expression in bbx25-2 bbx24-1 than in Col-0 seedlings (see Supplemental Figure 11 online). These results suggest that BBX25 and BBX24 together negatively regulate the expression of BBX22. Previously, it has been shown that HY5 promotes the expression of BBX22 by directly binding to its promoter (Chang et al., 2008) and that COP1 ubiquitylates and degrades BBX22 protein in the dark (Datta et al., 2008; Chang et al., 2011). Furthermore, BBX22 physically and genetically interacts with HY5 and enhances the function of HY5 by regulating hypocotyl growth and anthocyanin accumulation (Chang et al., 2008; Datta et al., 2008). To study in detail how the expression of BBX22 could be affected in both bbx25-1 and bbx24-2 single and double mutants, we monitored the expression of BBX22 in seedlings exposed to WL between 0 and 24 h. BBX22 expression increased in a light-dependent manner, with a peak at around 6 h, and then decreased to levels similar to those at 0 h (Figure 7A). Whereas the induction of BBX22 in both bbx24-1 and bbx25-2 single mutants was found to be slightly elevated compared with Col-0, the bbx24-1 bbx25-2 double mutant showed nearly twofold higher BBX22 accumulation than single mutants, suggesting that BBX25 and BBX24 act additively to downregulate the expression of BBX22.
Figure 7.
BBX25 Together with BBX24 Negatively Regulate the Expression of BBX22.
(A) Quantitative RT-PCR analysis of BBX22 expression in Col-0, bbx24-1, bbx25-2, and bbx25-2 bbx24-1. Four-day-old seedlings were irradiated with WL (80 µmol m−2 s−1) for 0 to 24 h. Error bars represent se (n = 3).
(B) Immunoblot of HY5 and COP1 proteins in 4-d-old WL grown seedlings of Col-0, bbx25-2, bbx24-1, and bbx25-2 bbx24-1. Actin was used as loading control. Immunoblot experiments were repeated three times, and similar results were obtained.
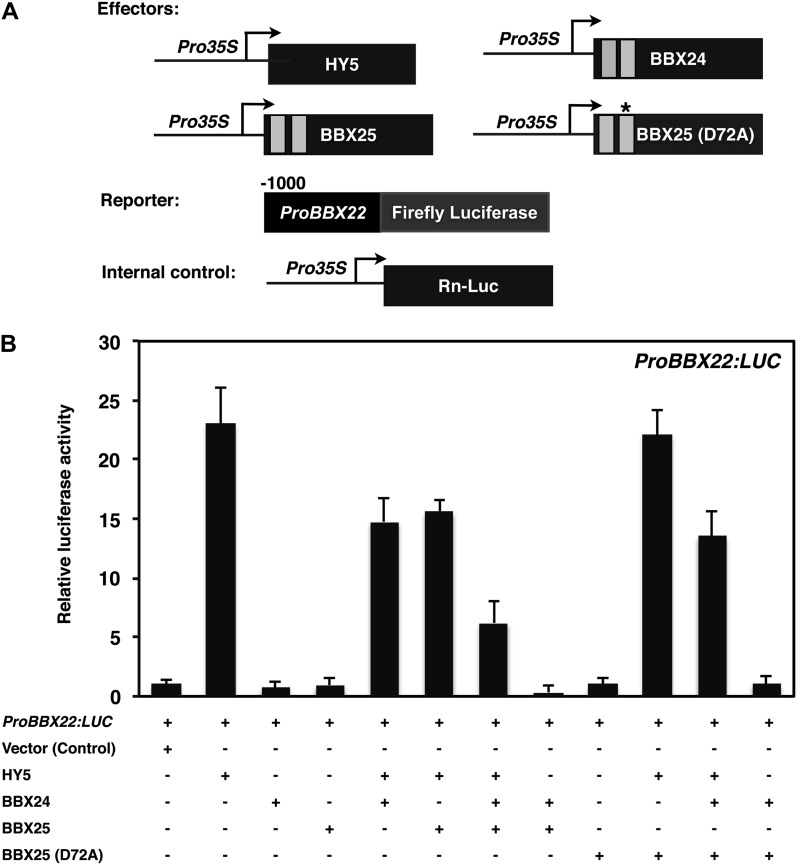
Next, we asked whether the negative regulation of BBX22 by BBX25 and BBX24 is through the altered stability of HY5 and/or COP1. Protein extracts of Col-0, bbx25, bbx24, and bbx24 bbx25 double mutants of 4-d-old WL-grown seedlings were subjected to immunoblot analysis. Neither HY5 nor COP1 was altered in the single and double mutants compared with Col-0 (Figure 7B), suggesting that the increased expression of BBX22 in the bbx24-1 bbx25-2 double mutant is independent of HY5 and COP1 protein levels. Furthermore, we evaluated whether the negative regulation of BBX22 could be due to an altered HY5 action. To this end, we fused 1000 bp of the BBX22 promoter to the luciferase reporter gene. The ProBBX22:LUC reporter was transfected into mesophyll protoplasts along with Pro35S:RnLUC (an internal control of transformation efficiency) and the effectors Pro35S:HY5, Pro35S:BBX25, Pro35S:BBX25(D72A), and Pro35S:BBX24 in different combinations (Figure 8A). The BBX22 promoter activity was null for the vector control, BBX24, BBX25, BBX25(D72A), and BBX24 BBX25 effectors (Figure 8B). However, the cotransfection of HY5 and BBX25(D72A) with ProBBX22:LUC resulted in ∼20-fold activation of the BBX22 promoter, whereas the cotransfection of HY5 with either BBX25 or BXB24 moderately induced the BBX22 promoter activity (Figure 8B). These results suggest BBX25 and BBX24 repress the activation of BBX22 promoter activity by interfering with HY5 action. Since both BBX25 and BBX24 physically interact with HY5 through its bZIP domain, they probably reduce the function of HY5 by forming inactive heterodimers. In fact, the results demonstrate that the mutated version of BBX25 is not able to form heterodimers with HY5.
Figure 8.
BBX25 and BBX24 Regulate BBX22 Expression by Altering HY5 Transcriptional Activity in Vivo.
(A) Constructs used in the transient transfection assay in protoplasts. Arrow after the 35S promoter indicates the transcriptional start site, and -1000 indicates the length of the BBX22 promoter, which was fused to the firefly luciferase to create the reporter construct. The asterisk indicates Asp-to-Ala substitution in the second B-box at position 72.
(B) Activation of ProBBX22:LUC by HY5, BBX24, BBX25, and BBX25 (D72A) either alone or in different combinations as indicated. Error bars represent se (n = 5).
BBX24 and BBX25 Are COP1 Dependent and HY5 Independent under Shade Conditions
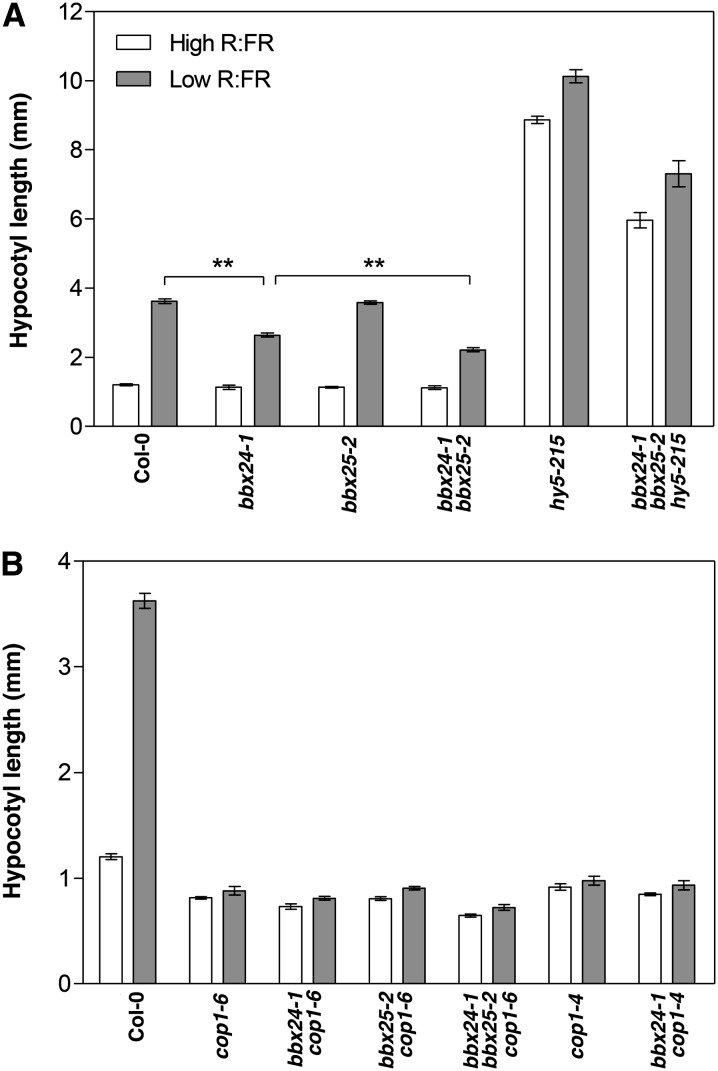
SAS is an adaptive strategy that triggers plant elongation responses in low R:FR light to escape from shade and reach out to the sunlight. BBX21 and BBX22 act as negative factors for hypocotyl growth in dark conditions (Crocco et al., 2010). Furthermore, the bbx21 bbx22 cop1 triple mutant can partially restore the SAS phenotype, suggesting that BBX21 and BBX22 together interact genetically and antagonistically with COP1 under SAS (Crocco et al., 2010). To understand the genetic relationship between BBX25 and BBX24 with HY5 and COP1 in the SAS pathway, we studied the single, double and triple mutant phenotypes under simulated shade conditions. In high R:FR light, bbx24-1 and bbx25-2 displayed hypocotyl elongation similar to that of the wild type. However, bbx24-1 but not bbx25-2 mutants showed significantly shorter hypocotyls than Col-0 seedlings in low R:FR (Figure 9A). Furthermore, the bbx24-1 bbx25-2 double mutant is significantly shorter than bbx24-1 specifically under shade conditions. These results suggest that BBX24 and BBX25 are partially redundant in the promotion of the hypocotyl shade avoidance response. In addition, hy5-215 and bbx24-1 bbx25-2 hy5-215 seedlings had longer hypocotyls than the wild type both in high and low R:FR, suggesting that HY5 is not involved in hypocotyl elongation by shade. We also explored the genetic relationship between COP1, BBX24, and BBX25. cop1-4 and cop1-6 seedlings displayed significantly shorter hypocotyls than wild-type ones both in high and low R:FR, and bbx24 and/or bbx25 mutations in cop1 backgrounds displayed the same phenotypes as cop1, indicating that the functions of BBX24 and BBX25 under shade are completely dependent on COP1 (Figure 9B).
Figure 9.
BBX25 and BBX24 Are HY5 Independent and COP1 Dependent under Shade.
Hypocotyl length of seedlings from Col-0 and single, double, and triple mutants grown in high and low R:FR. Five-day-old seedlings were grown in WL (high R:FR) or simulated shade (low R:FR) with 16-h-light/8-h-dark conditions at 22°C. Error bars represent se (n ≥ 15). Asterisks show pair of genotypes that differ significantly between them (Student’s t test, **P ≤ 0.01).
DISCUSSION
BBX25 acts as a negative regulator of photomorphogenesis and works independently of BBX24. BBX25 and BBX24 enhance COP1 and suppress HY5 functions. Furthermore, BBX25 and BBX24 additively downregulate the expression of BBX22 by interfering with HY5 transcriptional activity to modulate seedling photomorphogenesis in Arabidopsis. This notion is supported by the fact that BBX25 interacts with HY5, as detected by yeast two-hybrid, pull-down, and FRET assays. Furthermore, both B-box domains of BBX25 are necessary to mediate the interaction with the bZIP domain of HY5 (Figure 1). Similar conserved residues in BBX21 and BBX22 have been shown to mediate the interaction with HY5 (Datta et al., 2007, 2008). BBX21, BX22, and BBX24 also interact with HY5 through the bZIP domain (Datta et al., 2007, 2008; Jiang et al., 2012). Considering that BBX24 and BBX25 inhibit and that BBX21 and BBX22 promote seedling photomorphogenesis, our results suggest that B-Box domains and interaction partners of BBX proteins are conserved but that the functions of BBX proteins diverge in the control of plant development.
BBX25 Acts Additively with BBX24 in Seedling Deetiolation
Detailed phenotypic analysis of both mutant alleles of BBX25 and two independent overexpressing lines firmly established that BBX25 is a negative regulator of light signaling in Arabidopsis seedling development (Figure 2; see Supplemental Figure 3 online). Furthermore, epistatic analyses with its closest homolog, BBX24, suggest that BBX25 functions independently of BBX24 (Figure 3). Similar types of additive genetic interactions have been reported between two closely related protein pairs, such as HY5-HYH, BBX21-BBX22, FHY1-FHL, and FHY1-FHY3 (Holm et al., 2002; Kim et al., 2002; Hiltbrunner et al., 2006; Datta et al., 2008). Although they are hypersensitive in the light, neither BBX24 nor BBX25 single or double mutants showed any phenotype in the dark. However, bbx25 individually and together with bbx24 enhanced the hypocotyl phenotypes of cop1 mutants both in the light and in the dark (Figure 5; see Supplemental Figure 9 online). At least two negative regulators, SPA1 and GBF1, are known to be partially redundant with COP1 function in the dark, but independent in light-grown seedlings (Saijo et al., 2003; Mallappa et al., 2008). The genetic interactions of BBX25 and BBX24 together with HY5 and COP1 support the notion that BBX25 and BBX24 exert their effect on COP1 and HY5 signaling. The physical interactions of BBX25 and BBX24 with COP1 further strengthen this notion (Holm et al., 2001). Although BBX25 and BBX24 enhance the function of COP1 in both light and dark conditions, the degradation of BBX24 and BBX25 by COP1 could be part of a fine-tuning mechanism during photomorphogenesis and diurnal dark to light transitions.
Here, we show that BBX25 and BBX24 are involved in anthocyanin accumulation (Figure 3). Recently, BBX24 has been reported to regulate anthocyanin accumulation in UV-B light (Jiang et al., 2012). The enhanced accumulation of anthocyanin in the bbx24 bbx25 double mutant compared with the respective single mutants suggests that BBX25 acts genetically independently of BBX24. The phenotypes of bbx24 and/or bbx25 double and triple mutants in hy5 background indicate that both BBX25 and BBX24 individually can partially suppress HY5 function and that together they are able to completely suppress HY5 action in WL (Figure 4). However, the bbx24 bbx25 mutation in hy5 background cannot restore the wild-type anthocyanin phenotype under monochromatic light conditions, suggesting that R and BL are needed to additively enhance the function of BBX25 and BBX24 downstream of HY5 (Figure 4B; see Supplemental Figure 7 online). Although the molecular mechanism is not known, we hypothesize that WL could promote the coaction of cryptochromes and phytochromes probably by a physical interaction affecting each other’s signaling activities (Casal and Mazzella, 1998; Neff and Chory, 1998). Interestingly, HY5 and HYH have been reported to participate in the synergism between phyB and cry1 (Sellaro et al., 2009), and phytochromes and cryptochromes additively regulate HY5 protein stability (Osterlund et al., 2000).
BBX24 and BBX25 Negatively Regulate the Expression of BBX22 by Interfering with HY5 Transcriptional Activity
Our biochemical, genetic, and physiological studies of BBX25 and BBX24 with HY5 and COP1 suggest a possible functional connection among these proteins. The increased levels of BBX22 transcripts in bbx25 and bbx24 single and double mutants suggest that BBX25 and BBX24 negatively regulate BBX22 expression additively (Figure 7). The fact that BBX22 is a common target of both HY5 and COP1 further supports the function of BBX25 and BBX24 in HY5- and COP1-mediated pathways. HY5 binds to the promoter of BBX22 and regulates its expression, and BBX22 enhances the function of HY5 both individually and together with BBX21 in different developmental processes like hypocotyl growth, anthocyanin accumulation, and gene regulation (Chang et al., 2008; Datta et al., 2008). Furthermore, COP1 ubiquitylates BBX22 and degrades it in dark and light conditions (Datta et al., 2008; Chang et al., 2011). Protoplast assays showed that BBX25 and BBX24 separately suppressed HY5 transcriptional activity moderately, suggesting that both proteins are necessary for effective inactivation of HY5 function (Figure 8). The fact that HY5 protein stability was unaltered in both single and double mutants of bbx25 and bbx24 further supports the observation that BBX25 and BBX24 regulate BBX22 expression by reducing HY5 transcriptional activity without altering its stability (Figure 7).
Moreover, HY5 is known to bind to the CHS and CHI promoters (Ang et al., 1998; Lee et al., 2007; Shin et al., 2007) and regulate their expression. It is therefore likely that BBX25 and BBX24 regulate the expression of CHS and CHI by affecting the ability of HY5 to activate the transcription of these genes. This could be achieved through two possible mechanisms. First, BBX25 and BBX24 could physically interact with HY5, forming inactive heterodimers, and thereby sequester HY5 from the active pool. In fact, HYH, an HY5 homolog, forms heterodimers with HY5 and cooperatively functions in the regulation of anthocyanin pigment accumulation (Holm et al., 2002). Furthermore, the transcription of HFR1, a basic helix-loop-helix factor, is quickly induced by short-term shade but inhibited by long-term shade when its protein directly interacts with PIF4 and PIF5, forming heterodimers that do not bind DNA (Hornitschek et al., 2009). Physical interaction of BBX25 and BBX24 with HY5 at the promoter may recruit corepressors or prevent coactivators from interacting with HY5. Interestingly, two closely related proteins, such as BBX21 and BBX22, enhance the function of HY5 probably by serving as positive coactivators of HY5 (Datta et al., 2007, 2008). Moreover, BBX32 interacts physically with BBX21, but not with HY5, modulating seedling photomorphogenesis in an opposite manner, suggesting that this interaction leads to inactivation of a protein complex containing BBX21 and HY5 (Holtan et al., 2011).
BBX24 and BBX25 Are COP1 Dependent and HY5 Independent under Shade Conditions
Here, we demonstrated that BBX25 and BBX24 play an additive function and that both promote the SAS, as their functions are genetically epistatic to COP1 but HY5 independent (Figure 9). COP1 targets HFR1, but not HY5, for degradation under shade conditions, thus supporting the idea that COP1 is involved in SAS signaling (Crocco et al., 2010; Rolauffs et al., 2012). In addition, it has been demonstrated that HY5 is involved in the light signaling that inhibits the hypocotyl length of Arabidopsis seedlings when the R:FR increases during the formation of sunflecks at the final portion of the photoperiod (Sellaro et al., 2011). How exactly BBX24 and BBX25 collaborate with COP1 to promote hypocotyl growth under shade conditions is unknown. Other BBX proteins can act epistatically with COP1 in low R:FR. In fact, BBX21 and BBX22, which act to enhance HY5 function in light, play an opposite role under long-term shade (Crocco et al., 2010). The fact that different BBX members have antagonistic functions in shade conditions and deetiolation processes allows plants to adjust the expression of hypocotyl elongation genes finely and precisely in different light conditions.
Mode of Action of BBX24 and BBX25 in Arabidopsis Seedling Development
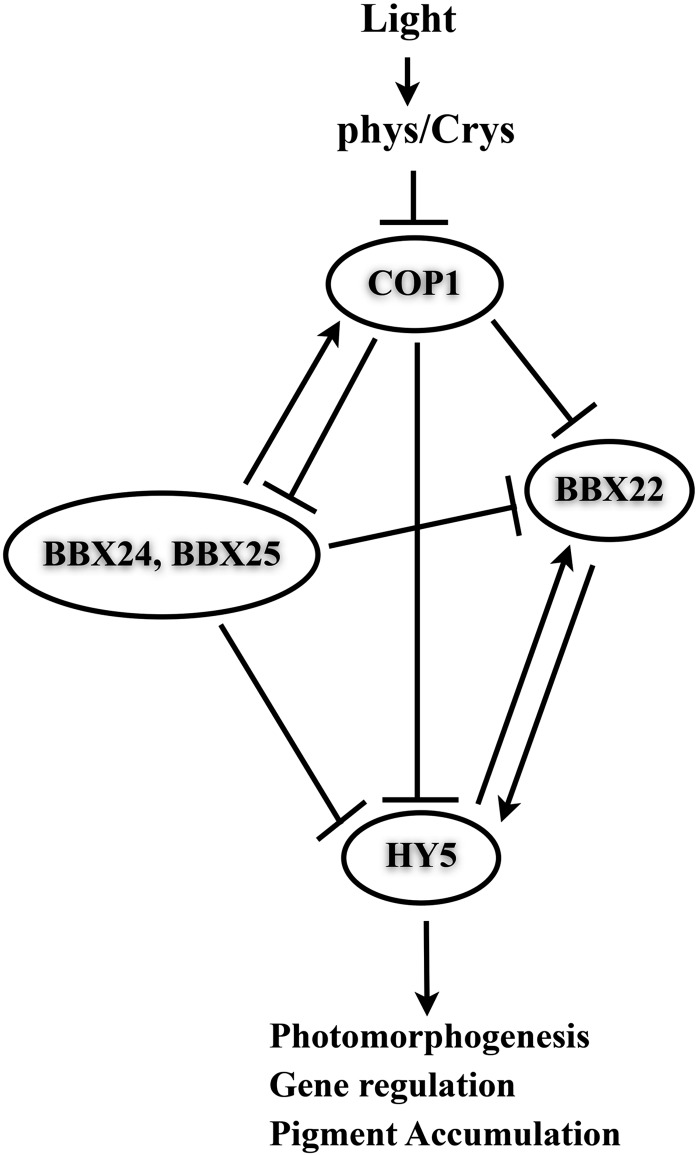
Based on all the results, we propose that BBX25 and BBX24 could serve as transcriptional corepressors of HY5. Since both HY5 and BBX22 are direct targets of COP1 and BBX22 is a direct target of HY5, the increased levels of HY5 protein in cop1 mutants could be indirectly due to increased BBX22 stability. Figure 10 shows a working model for seedling deetiolation. We hypothesize that during dark–light transitions, when a seedling encounters light, HY5 is relieved from COP1, leading to an increase in HY5 stability. HY5 binds to BBX22 and induces the expression of BBX22, a transcriptional coactivator of HY5 that enhances the function of HY5. At the same time, BBX25 and BBX24 accumulate in a light-dependent manner and act to repress the expression of BBX22 by reducing the function of HY5 by interacting through its bZIP domain. Moreover, a fraction of COP1 could be still active in light-grown seedlings degrading HY5 and BBX22 to avoid exaggerated responses mediated by HY5 and cofactors. Furthermore, COP1 negatively regulates BBX24 and BBX25 by degrading them to prevent a feedback regulatory loop. Collectively, we conclude that BBX25 and BBX24 function as corepressors of HY5 as a part of a fine-tuning mechanism for the regulation of seedling deetiolation processes.
Figure 10.
Model of Action of BBX25 and BBX24 in Seedling Deetiolation.
COP1 negatively regulates HY5 and BBX22 proteins by supplying them to 26S proteasome pathway. HY5 induces the expression of BBX22, which in turn enhances the function of HY5. BBX25 and BBX24 negatively regulate the expression of BBX22 by reducing the function of HY5 interacting physically through the bZIP domain. Furthermore, COP1 may attenuate the function of BBX25 and BBX24 by degrading them in the light.
METHODS
Growth Conditions and Plant Materials
The mutants used in this study were bbx25-1 (SAIL_786_F08), bbx25-2 (SM_3.17575), and bbx24-1 (Salk_067473). The bbx24-1, bbx25-1, bbx25-2, hy5-215, cop1-4, and cop1-6 alleles are in the Col-0 accession. Seeds were surface sterilized and plated on Murashige and Skoog medium supplemented with 0.8% Bactoagar (BD Biosciences) and 1% Suc. The plates were then cold treated at 4°C for 3 d and then transferred to light chambers maintained at 22°C with the desired wavelength and intensity of light. For all monochromatic light assays, the plates were transferred to continuous WL for 3 h to induce uniform germination. The plates were then transferred to monochromatic light conditions, incubated at 22°C for the indicated number of days, and measured essentially as previously described (Datta et al., 2007, 2008). Physiological experiments were repeated two or three times, and only a representative one is shown.
For genotyping of homozygous bbx25-1, the T-DNA primer LB3 was used along with gene-specific primers BBX25LP and BBX25RP, whereas for bbx25-2 genotyping, transposon-specific primer Spm1 was used along with gene-specific primers BBX25LP and BBX25RP. For homozygous bbx24-1 genotyping, the T-DNA primer LBb1 was used along with gene-specific primers BBX24LP and BBX24RP (see Supplemental Table 1 online).
For the generation of double and triple mutants, first, bbx25-2 and bbx24-1 were crossed to generate the bbx25-2 bbx24-1 double mutant. The bbx25-2 bbx24-1 double mutant was crossed with cop1-4, cop1-6, and hy5-215 to obtain F1. Through phenotyping and genotyping of F2 plants, we identified different double (bbx25-2 cop1-4, bbx25-2 cop1-6, bbx24-1 cop1-4, bbx24-1 cop1-6, and bbx25-2 hy5-215) and triple (bbx24-1 bbx25-2 cop1-4, bbx24-1 bbx25-2 cop1-6, and bbx24-1 bbx25-2 hy5-215) mutant combinations.
Generation of BBX25-Overexpressing Transgenic Lines
To generate transgenic lines overexpressing BBX25, the full-length BBX25 coding sequence was amplified by PCR from cDNA of 4-d-old light-grown Col-0 seedlings using the primers BBX25OEFP and BBX25OERP (see Supplemental Figure 1 online) and cloned into the pAVA321 vector with NcoI-BglII sites. The expression cassette was subcloned into the pPZP222 binary vector, which was then transformed into the GV3101 Agrobacterium tumefaciens strain and then into Col-0 plants by the floral dip method (Clough and Bent, 1998). In T3 plants, several independent transgenic lines carrying a single-copy transgene were identified, and two of them (OE1 and OE2) were selected for further analysis.
Yeast Two-Hybrid Method and FRET Experiments
For yeast two-hybrid experiments, we cloned full-length and mutated versions of BBX25 (BBX25D20A and BBX25D72A) and BBX25 (D20A and D72A) cDNA into the yeast expression vector pYX141 and full-length HY5 cDNA into the Gal4 DNA binding domain (Gal4-DBD) vector pAS2-1 (see Supplemental Table 1 online for primers). The constructs used for HY5 are described elsewhere (Holm et al., 2002; Datta et al., 2007). We confirmed the expression of different wild-type and mutated versions of BBX25 proteins by immunoblotting, using polyclonal rabbit antibodies raised against full-length BBX25. Meanwhile, full-length and truncated versions of HY5 proteins were examined by immunoblotting using anti-Gal4-DBD antibody. The β-galactosidase assays were performed as described by Holm et al. (2001) in yeast strain Y187.
For the FRET acceptor photobleaching experiments, the pAM- PAT-35SS-YFP-HY5 (Datta et al., 2008) and pAM-PAT-35SS-CFP-BBX25 constructs were introduced into onion epidermal cells by particle bombardment and incubated. Cells were visualized 30 h after particle bombardment using the confocal microscope through a Plan-Neofluor 403/1.3 oil (differential interference contrast) objective. Live-cell images were acquired using an Axiovert 200 microscope equipped with an LSM 510 META laser scanning confocal imaging system (Carl Zeiss). The multitracking mode was used to eliminate spillover between fluorescence channels. The CFP was excited by a diode 405 laser and the YFP by an argon-ion laser (514 nm), both at low intensities. After increasing the intensity of the 514-nm laser, the YFP fluorescence from the acceptor, YFP-HY5, was bleached. The bleaching of the acceptor resulted in an increased emission from CFP-BBX25, indicating that FRET had occurred between the two proteins prior to the bleach. Similarly, in control photobleaching experiments using the microscopic settings, FRET between untagged CFP and YFP was examined. Regions of interest were selected and bleached with 100 iterations using the argon-ion laser at 100%. At least 10 different nuclei were subjected to FRET, and average intensities before and after bleaching were plotted on the graph.
Pull-Down Assay
In vitro protein–protein interaction studies were performed as described by Jang et al. (2007), with some modifications. Wild-type and mutated DNA fragments encoding full-length BBX25 were cloned into the pET20b (+) vector to generate a C-terminal 6×His fusion. The full-length coding sequence of HY5 and BBX24 was cloned into the pMAL vector to generate N-terminal fusion of the maltose binding protein (MBP) tag (MBP-HY5). The proteins were overexpressed and purified from Escherichia coli using a nickel-nitrilotriacetic acid column (Qiagen) or an amylose resin column (GE Healthcare). For in vitro pull-down experiments, 1.5 µg of MBP and MBP-HY5 prey proteins was bound to an amylose resin column by incubating with PBS, pH 7.4, for 2 h at 4°C. Excess unbound protein was washed off and further incubated with 1.5 µg of BBX25-His bait proteins in 250 μL binding buffer for 3 h at 4°C in in vitro pull-down buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM EDTA, pH 8.0, 1 mM PMSF, 0.1% [v/v] protease inhibitor cocktail, and Nonidet P-40) and washed four times with the same buffer. The proteins were eluted from the beads by boiling in equal volume of 2× SDS buffer and loaded onto a 10% SDS- PAGE gel, and the proteins were detected using anti-His antibodies (Sigma-Aldrich; A7058).
Fluorescence Microscopy Analysis of GFP Signal
For the analysis of GFP-BBX25 protein dynamics in hypocotyl cell nuclei, at least 10 to 12 seedlings grown in the respective growth conditions were subjected to fluorescence microscopy analysis and pictures taken with a ×40 objective. Fluorescence signals were detected for GFP (excitation of 488 nm; emission of 505 to 550 nm). In all the experiments, the scans were done with identical microscope settings.
Protein Extraction and Immunoblotting
For the analysis of light-dependent protein accumulation, plants were treated with 100 μM MG132 (Sigma-Aldrich) or DMSO (mock control) for 1 or 4 h as indicated at the time of each experiment. Protein gel blot analysis was performed using the Super Signal West Pico chemiluminescent substrate kit (Fisher Scientific) following the instructions described in the user’s manual. Protein extracts were prepared from wild-type, mutant, or transgenic seedlings. Approximately 100 mg of seedlings was harvested in a microcentrifuge tube, frozen in liquid nitrogen, and ground in 200 mL of grinding buffer (400 mM Suc, 50 mM Tris-Cl, pH 7.5, 10% glycerol, and 2.5 mM EDTA), and PMSF was added (0.5 mL for every 100 mL of grinding buffer). The protein extract was centrifuged at 10,000 rpm for 10 min to pellet the debris. The supernatant was transferred to a fresh tube, and an aliquot of 5 mL was taken out in a separate tube to estimate protein concentration by Bradford assay. Proteins were separated by 10% SDS-PAGE. Prestained protein markers (GE Healthcare) were used for molecular mass determination. The samples were then transferred to Hybond C-Extra (GE Healthcare) at 100 mA for 2 h in trans-blot semidry transfer apparatus following the manufacturer’s protocol (Bio-Rad). The membrane was blocked with 5% milk in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, and 2.7 mM KCl) and probed with GFP (monoclonal), HY5 and COP1 (polyclonal antibodies), or antiactin monoclonal antibodies (Sigma-Aldrich; A0480).
Protoplast Assay
For the protoplast assay, Arabidopsis thaliana mesophyll cell protoplasts were prepared and transfected as described by Yoo et al. (2007). The promoter-reporter used was a 1000-bp fragment of the BBX22 promoter driving firefly luciferase (pPCV814-BBX22-Luc). Full-length HY5, BBX25, BBX25D74A, and BBX24 driven by a cauliflower mosaic virus 35S promoter (pRTL-HY5, pRTL2-BBX25, pRTL2-BBX25, and pRTL2-BBX24) were used as the effectors. For the reporter detection, the Dual Luciferase system was used (Promega Biotech). Renilla luciferase driven by a full-length cauliflower mosaic virus 35S promoter (pRNL) was used as an internal control.
Gene Expression Analysis
To characterize the nature of the mutations in BBX25 and BBX24, we studied the expression of mRNA in bbx25-1, bbx25-2, and bbx24-1 mutant genotypes grown for 15 d under 12 h light/12 h dark and harvested just before dawn when the transcripts are reportedly high (Kumagai et al., 2008). Total RNA was extracted from 15-d-old plants grown in a 12-h-light/12-h-dark cycle, using the RNeasy plant mini kit (Qiagen), and 20 µg was loaded for the RNA gel blot analysis. BBX25 (amplified with BBX25 RNA-FP and BBX25 RNA-RP) and BBX24 (amplified with BBX24 RNA-FP and BBX24 RNA-RP) C-terminal fragments were used as probes to detect transcript levels in the wild type and different bbx24 and bbx25 mutant backgrounds.
For quantitative PCR analysis, RNA was extracted from WL- and red light–treated 4-d-old seedlings using the RNeasy plant mini kit (Qiagen), and RNA was subjected to on-column DNase I digestion following the manufacturer’s instructions (Qiagen). RNA (1 µg) was converted into cDNA using the Revert Aid H minus first-strand cDNA synthesis kit (Fermentas). The cDNA was diluted 1:20 with sterile MQ water. The reaction was set up in 20-μL volume with 2 μL of template using the iTaq Fast SYBR Green PCR Master Mix kit (Applied Biosystems). Quantitative PCR was performed on an optical 96-well plate in an ABI PRISM 7500 real-time PCR system (Applied Biosystems). The thermal cycle used was 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 35 s. Gene-specific primer pairs (see Supplemental Table 1 online) were designed using Primer3 and BLAST software from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/tools/primer-blast/primer3).
Anthocyanin Measurements
Anthocyanin content was measured as described by Gangappa et al. (2010).
Hypocotyl Shade Avoidance Experiments
For shade experiments, we used a light growth chamber with mercury lamps (General Electric HR175/R/DX/FL39 mercury 33026; PAR = 100 µmol m−2 s−1 and R:FR = 2.3) under a long-day photoperiod (16 h light/ 8 h dark) at 22°C. Seedlings were grown for 5 d under WL with a high R:FR or exposed to low R:FR (R:FR = 0.35) obtained by incandescent lamps (Philips R19-100R20/FL/S) covered by Paolini filters (Paolini 2031; La Casa del Acetato) and placed laterally into the growth chamber. Spectral photon fluences of WL and shade conditions were obtained using a Li-Cor Integrating quantum/radiometer/photometer (Li-188B). PAR and R:FR ratios were measured using a SpectroSense2 attached with a SKR-1850SS2 light sensor (Skye Instruments).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BBX25 (At2g31380), BBX24 (At1g06040), HY5 (At5g11260), COP1 (At2g32950), BBX18 (AT2G21320), BBX219 (AT4G38960), BBX20 (AT4G39070), BBX21 (AT1G75540), BBX22 (AT1G78600), BBX23 (AT4G10240), HYH (At3G17609), CHS (At5g13930), CHI (At3g55120), and Actin2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of BBX25 and HY5 Proteins in Yeast.
Supplemental Figure 2. Absence of FRET between Unfused CFP and YFP.
Supplemental Figure 3. BBX25 Transcript Accumulation in Mutant and Overexpressing Lines.
Supplemental Figure 4. Physiological Characterization of bbx25 Mutants and Overexpressing Lines in Monochromatic Lights.
Supplemental Figure 5. Molecular Characterization of BBX24 T-DNA Insertion Mutant Alleles.
Supplemental Figure 6. bbx25-2 and bbx24-1 Additively Suppress hy5 Phenotype in Monochromatic Lights.
Supplemental Figure 7. Elevated Accumulation of Anthocyanin in bbx25-2 and bbx24-1 is HY5 Dependent in Different Monochromatic Lights.
Supplemental Figure 8. Elevated Expression of CHS and CHI Genes in bbx25 bbx24 Mutant Is HY5 Dependent in Monochromatic Lights.
Supplemental Figure 9. bbx25 and bbx24 Additively Enhance the cop1 Phenotype in Monochromatic Lights.
Supplemental Figure 10. Overexpression of GFP-BBX25 in Col-0 and cop1-6 Mutant Backgrounds.
Supplemental Figure 11. Expression Analysis of Double B-Box (DBB), HY5, and HYH Genes.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We acknowledge the Swegene Centre for Cellular Imaging at Gothenburg University for the use of imaging equipment and for the support from the staff. Funding support was received from the Swedish Research Council, The Signhild Engkvist Foundation, The Royal Society of Arts and Science Gothenburg (to M.H.), the Carl Trygger Foundation (to S.N.G.), the University of Buenos Aires (Grant 20020100100774 to J.F.B.), and the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2008-1061 to J.F.B.).
AUTHOR CONTRIBUTIONS
S.N.G., M.H., and J.F.B. designed the research. S.N.G., C.D.C., S.D., and C.H. performed the research. HJ generated essential material for the experiments. S.N.G., M.H., and J.F.B. analyzed data and wrote the article.
Glossary
- R:FR
red:far-red
- SAS
shade avoidance syndrome
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- FRET
fluorescence resonance energy transfer
- Col-0
Columbia-0
- WL
white light
- BL
blue light
References
- Al-Sady B., Kikis E.A., Monte E., Quail P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Casal J.J. (January 25, 2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- Casal J.J., Mazzella M.A. (1998). Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 118: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.S., Li Y.H., Chen L.T., Chen W.C., Hsieh W.P., Shin J., Jane W.N., Chou S.J., Choi G., Hu J.M., Somerville S., Wu S.H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chang C.S., Maloof J.N., Wu S.H. (2011). COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Ang L.H., Puente P., Deng X.W., Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crocco C.D., Holm M., Yanovsky M.J., Botto J.F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64: 551–562 [DOI] [PubMed] [Google Scholar]
- Crocco C.D., Holm M., Yanovsky M.J., Botto J.F. (2011). Function of B-BOX under shade. Plant Signal. Behav. 6: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi C., Johansson H., Holm M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Johansson H., Hettiarachchi C., Irigoyen M.L., Desai M., Rubio V., Holm M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Prasad V.B., Chattopadhyay S. (2010). Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis. Plant Physiol. 154: 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Holm M., Hardtke C.S., Gaudet R., Deng X.W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan H.E., et al. (2011). BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M., Cordero J., Neuhaus G., Rodríguez-Franco M. (2007). Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jang I.C., Yang S.W., Yang J.Y., Chua N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Li Q.F., Björn L.O., He J.X., Li S.S. (2012). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Khanna R., Kronmiller B., Maszle D.R., Coupland G., Holm M., Mizuno T., Wu S.H. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Woo J.C., Song P.S., Soh M.S. (2002). HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 30: 711–719 [DOI] [PubMed] [Google Scholar]
- Kumagai T., Ito S., Nakamichi N., Niwa Y., Murakami M., Yamashino T., Mizuno T. (2008). The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Kushwaha R., Singh A., Chattopadhyay S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-J., Zhang Y.-C., Li Q.-H., Sang Y., Mao J., Lian H.-L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Mallappa C., Singh A., Ram H., Chattopadhyay S. (2008). GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J. Biol. Chem. 283: 35772–35782 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Deng X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolauffs S., Fackendahl P., Sahm J., Fiene G., Hoecker U. (2012). Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160: 2015–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R., Hoecker U., Yanovsky M., Chory J., Casal J.J. (2009). Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol. 19: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R., Yanovsky M.J., Casal J.J. (2011). Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 68: 919–928 [DOI] [PubMed] [Google Scholar]
- Seo H.S., Yang J.-Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.-H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J., Park, E., and Choi, G (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis Plant J. 49: 981–994. [DOI] [PubMed]
- Yan H., Marquardt K., Indorf M., Jutt D., Kircher S., Neuhaus G., Rodríguez-Franco M. (2011). Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 156: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.