This work characterizes three genes that control pollen maturation in rice, finding that they are closely related to Arabidopsis genes involved in the same functions, indicating strong conservation of the developmental control of pollen maturation for about 150 million years. However, the rice genes show less functional redundancy than the Arabidopsis genes.
Abstract
There are two groups of MADS intervening keratin-like and C-terminal (MIKC)-type MADS box genes, MIKCC type and MIKC* type. In seed plants, the MIKCC type shows considerable diversity, but the MIKC* type has only two subgroups, P- and S-clade, which show conserved expression in the gametophyte. To examine the functional conservation of MIKC*-type genes, we characterized all three rice (Oryza sativa) MIKC*-type genes. All three genes are specifically expressed late in pollen development. The single knockdown or knockout lines, respectively, of the S-clade MADS62 and MADS63 did not show a mutant phenotype, but lines in which both S-clade genes were affected showed severe defects in pollen maturation and germination, as did knockdown lines of MADS68, the only P-clade gene in rice. The rice MIKC*-type proteins form strong heterodimeric complexes solely with partners from the other subclade; these complexes specifically bind to N10-type C-A-rich-G-boxes in vitro and regulate downstream gene expression by binding to N10-type promoter motifs. The rice MIKC* genes have a much lower degree of functional redundancy than the Arabidopsis thaliana MIKC* genes. Nevertheless, our data indicate that the function of heterodimeric MIKC*-type protein complexes in pollen development has been conserved since the divergence of monocots and eudicots, roughly 150 million years ago.
INTRODUCTION
Development of the male gametophyte (pollen) is highly conserved among angiosperms and results in one vegetative and two sperm cells that act together in double fertilization of the female gametophyte. Monocots and eudicots, despite their evolutionary differences, express a huge number of homologous transcripts during pollen development (Wei et al., 2010). However, hard evidence that this is due to conserved developmental processes remains scarce. Since MADS box genes play crucial roles in plant development, they are promising candidates for comparative analyses of key regulatory networks.
In plants, MADS box genes, encoding MADS domain transcription factors, have acquired considerable phylogenetic diversity (Becker and Theissen, 2003; Nam et al., 2004). Type II MADS domain proteins in plants all share the same domain structure, comprising a MADS (M), intervening (I), keratin-like (K), and C-terminal (C) domain; therefore, they are also known as MIKC-type MADS domain proteins (Ma et al., 1991; Fan et al., 1997; Riechmann and Meyerowitz, 1997; Alvarez-Buylla et al., 2000). Two different types of MIKC-type genes have been defined: the classic MIKC-type (MIKCC-type) and the MIKC*-type genes (Henschel et al., 2002). MIKC*-type genes probably originated more than 450 million years ago in the lineage that led to extant land plants, by duplication of an ancestral MIKCC-type gene, followed by an elongation of the I-region or a duplication of parts of the K-box (Henschel et al., 2002; Kwantes et al., 2012). MIKCC-type MADS box genes have been shown to regulate diverse aspects in development of the sporophyte of higher plants (Gramzow and Theissen, 2010; Smaczniak et al., 2012), and MIKC*-type genes are crucial for development of the male gametophyte in the model plant Arabidopsis thaliana (Verelst et al., 2007a, 2007b; Adamczyk and Fernandez, 2009). The Arabidopsis genome encodes six MIKC*-type genes (AGAMOUS-LIKE30 [AGL30], AGL65, AGL66, AGL67, AGL94, and AGL104), all of which, except AGL67, are almost exclusively expressed during pollen development (Kofuji et al., 2003; Honys and Twell, 2004; Verelst et al., 2007a). They activate late pollen developmental genes and repress early genes at late stages of pollen development and thus are crucial for pollen maturation (Verelst et al., 2007b). In Arabidopsis, MIKC*-type genes act in a highly redundant manner, with no aberrant phenotype being described for any single loss-of-function mutant (Verelst et al., 2007a; Adamczyk and Fernandez, 2009).
MIKC*-type genes are not only present in higher eudicots but have also been identified in representatives of all major groups of land plants, including bryophytes, lycophytes, ferns, gymnosperms, basal angiosperms, eudicots, and monocots (Henschel et al., 2002; Kofuji et al., 2003; Riese et al., 2005; Rensing et al., 2008; Zobell et al., 2010; Gramzow et al., 2012; Kwantes et al., 2012). In ferns and seed plants, two different monophyletic lineages of MIKC*-type genes could be identified, the P- and S-clades, which are not present in bryophytes or lycophytes (Nam et al., 2004; Gramzow et al., 2012; Kwantes et al., 2012). This high degree of conservation of different subgroups is not observed for MIKCC-type genes (Kwantes et al., 2012). Also, MIKC*-type genes were shown to be expressed predominantly during the gametophytic stage in all land plants that have been studied so far, suggesting an ancient function during the haploid phase of the life cycle which was progressively restricted to the male gametophyte in the lineage leading to higher eudicots (Kwantes et al., 2012). However, when and in which common ancestors that happened is unknown.
Protein–protein and protein–DNA interactions of MIKC*-type proteins have been studied in Arabidopsis and in the liverwort (bryophyte) Marchantia polymorpha, the lycophytes Selaginella moellendorffii and Selaginella pallescens, the fern Ceratopteris richardii, and the basal eudicot Eschscholzia californica (Verelst et al., 2007a; Zobell et al., 2010; Kwantes et al., 2012). Generally, MADS domain proteins bind to DNA sequences called CArG-boxes (for C-A-rich-G). While most MADS domain proteins prefer the so-called serum response element (SRE)-type CArG-box, 5′-CC(A/T)6GG-3′, for binding (Hayes et al., 1988; Riechmann et al., 1996; de Folter and Angenent, 2006), some others, such as the mammalian MYOCYTE ENHANCER FACTOR2A (MEF2A) and the MIKC*-type proteins of all species investigated so far, preferentially bind to the N10-type CArG-box or MEF2 consensus binding site. This motif has the consensus sequence 5′-CTA(A/T)4TAG-3′ but might be more loosely defined as 5′-C(A/T)8G-3′ (Pollock and Treisman, 1991; Shore and Sharrocks, 1995; Verelst et al., 2007a; Zobell et al., 2010; Wu et al., 2011).
Arabidopsis MIKC*-type proteins form obligate heterodimers exclusively with one interaction partner from the P-clade and one from the S-clade (Verelst et al., 2007a). Interclade heterodimerization was also observed for MIKC*-type proteins from more basal species, such as the fern C. richardii and the basal eudicot E. californica. However, MIKC*-type proteins isolated from these species also showed homodimerization and intraclade heterodimerization (Kwantes et al., 2012).
In contrast with their orthologs from Arabidopsis, MIKC*-type genes from monocotyledonous plants have been little investigated so far. Hence, it remains unclear whether the high sequence conservation between monocot and eudicot MIKC*-type genes is also reflected in conserved gene expression and protein interaction patterns and function in pollen maturation.
Here, we provide a comprehensive investigation of MIKC*-type MADS box genes from a monocot species. We characterize all three MIKC*-type genes from rice (Oryza sativa): the S-clade genes MADS62 and MADS63 and the P-clade gene MADS68. Expression of all three MIKC*-type genes was detected in late developmental stages of pollen and proteins of different subclades form heterodimers, features highly similar to the situation in Arabidopsis. Furthermore, we report loss-of-function phenotypes of the rice MIKC*-type genes, including the single gene knockdown of a MIKC*-type gene that resulted in a mutant phenotype (i.e., severe defects in pollen maturation and germination). Our results suggest a highly conserved role of MIKC*-type genes in monocots and eudicots.
RESULTS
Phylogenetic Analysis of MIKC*-Type Genes
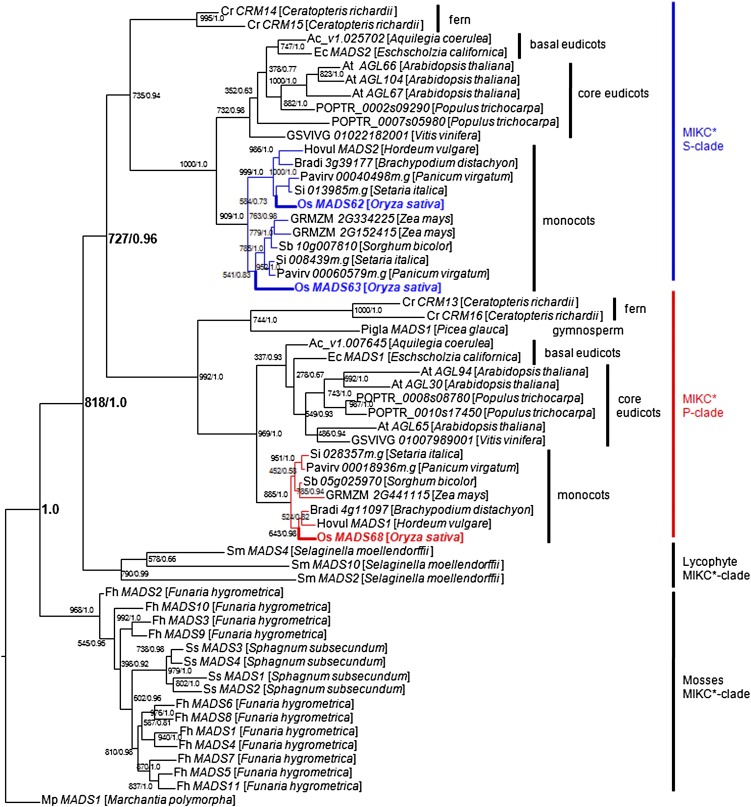
To gain insight into the evolutionary relationships of MIKC*-type genes within land plants and especially monocots, a comprehensive search was performed in the National Center for Biotechnology Information, Phytozome, and PlantGDB databases. Homologous sequences were retrieved from a broad variety of land plants, including bryophytes, lycophytes, ferns, gymnosperms, monocots, basal eudicots, and core eudicots (see Supplemental Table 1 online). In particular, the MIKC*-type genes of different Poaceae (grass) species were obtained through BLAST for the available genome sequences, including those of barley (Hordeum vulgare), sorghum (Sorghum bicolor), maize (Zea mays), purple false brome (Brachypodium distachyon), switchgrass (Panicum virgatum), and foxtail millet (Setaria italica). In rice, even though six genes grouped with MIKC*-type genes of Arabidopsis in some previous analyses (Arora et al., 2007), only three of them, MADS62, MADS63, and MADS68, were identified as genuine MIKC*-type genes (Gramzow et al., 2012; Kwantes et al., 2012). All the obtained sequences were aligned (see Supplemental Data Set 1 online) and then phylogenetic trees were constructed using maximum likelihood and Bayesian methods (Figure 1). Four highly supported clades can be observed within the superclade of MIKC*-type genes. One contains the MIKC*-type genes from mosses and one consists of sequences from lycophytes, while the other two monophyletic subgroups, the S- and P-clades, include sequences from both ferns and seed plants. In grasses, MIKC*-type genes of the S-clade constitute two subclades, MADS62-like and MADS63-like genes. These subclades comprise MIKC*-type genes from rice together with putative orthologs from other grass species. Only a single P-clade MIKC*-type gene, MADS68 and its orthologs, could be found in each of the grass species (Figure 1).
Figure 1.
Phylogenetic Tree of MIKC*-Type Genes.
Maximum likelihood and Bayesian phylogenies were conducted from an alignment of 58 cDNAs of MIKC*-type genes from a broad variety of land plants (an overview of all sequences can be found in Supplemental Table 1 online). Mp MADS1, the MIKC*-type gene from the liverwort M. polymorpha, representing the most early branching bryophyte lineage, was used as the outgroup. Maximum likelihood bootstrap/posterior probability values are shown at the branching points. Terminal nodes are labeled with a gene name followed by the species name. The S-clade genes are highlighted in blue and include the two rice genes MADS62 and MADS63. The P-clade genes are highlighted in red and include the only rice P-clade gene MADS68.
Rice MIKC*-Type Genes Are Specifically Expressed in Pollen
To determine the temporal and spatial expression patterns of rice MIKC*-type genes, diverse tissues at different developmental stages were analyzed by RT-PCR, quantitative RT-PCR (qRT-PCR), promoter-β-glucuronidase (GUS) fusion, and in situ hybridization.
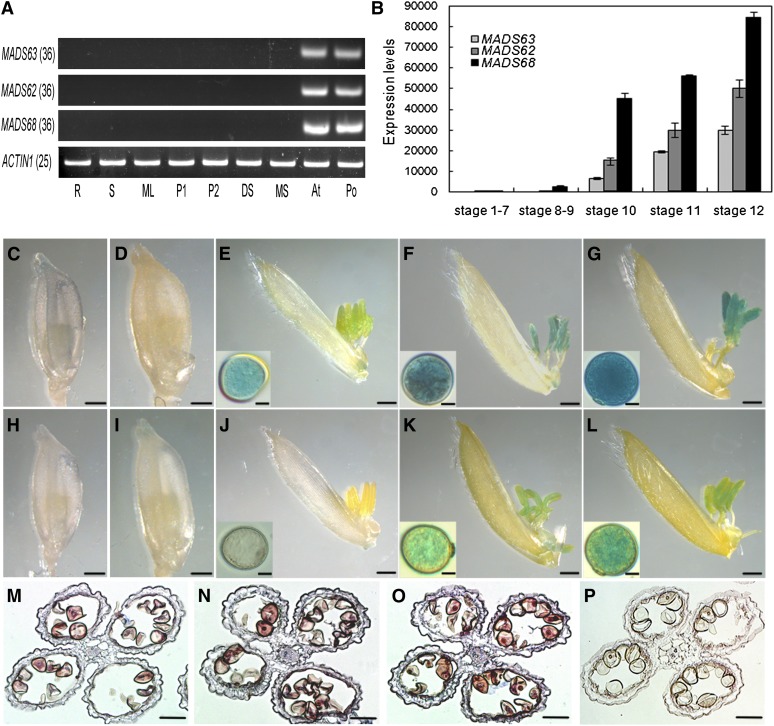
RT-PCR analysis revealed that transcripts of MADS62, MADS63, and MADS68 are specifically present in large quantities in the anthers and pollen at stage 13, when pollen achieves maturation, as classified previously (Zhang and Wilson, 2009). No expression was detectable in other tissues, including roots, stems, leaves, a mixture of rice panicles at stages P1 (1 to ∼3 cm in length) and P2 (3 to ∼6 cm in length), developing seeds, and the other three whorls of mature rice floral organs (lemma/palea, lodicule, and pistil) (Figure 2A).
Figure 2.
Expression Pattern of Rice MIKC*-Type Genes.
(A) Expression analyses of three rice MIKC*-type genes in various tissues of rice by RT-PCR. Total RNA was isolated from of 7-d-old roots (R), 7-d-old stems (S), mature leaves (ML), 1- to 3-cm panicles (P1), 3- to 6-cm panicles (P2), a mixture of developing seeds (DS), a mixture of lemmas/paleas, lodicules, and pistils of mature spikelets (MS), anthers at stage 13 (An), and mature pollen (Po). ACTIN1 was used as control. The cycle number of PCR is shown in parentheses for each gene.
(B) Expression levels of three rice MIKC*-type genes in the anther at different developmental stages as revealed by qRT-PCR analysis. Total RNA was pooled from anthers of stage 1 to 7, stage 8 and 9, stage 10, stage 11, and stage 12. Data show the mean ± sd (n = 3).
(C) to (G) GUS staining analyses of florets and pollen grains at different developmental stages in MADS68pro:GUS transgenic plants. A stained floret with anther before stage 8 (C), at stage 9 (D), at stage 10 after the removal of lemma (E), at stage 11 after the removal of lemma (F), and at stage 12 after the removal of lemma (G). Insets ([E] to [G]) show a magnified pollen grain at the corresponding stage.
(H) to (L) GUS staining analyses of florets and pollen grains at different developmental stages in MADS63pro:GUS transgenic plants. A stained floret with anther before stage 8 (H), at stage 9 (I), at stage 10 after the removal of lemma (J), at stage 11 after the removal of lemma and insets (K), and at stage 12 after the removal of lemma (L). Insets ([J] to [L]) show a magnified pollen grain at the corresponding stage.
(M) to (P) In situ hybridization of MADS63 transcripts in anther at stage 11 (M), MADS62 transcripts in anther at stage 11 (N), MADS68 transcripts in anther at stage 11 (O), and negative control with sense probes of MADS68 (P).
Bars = 1 mm in (C) to (L), 10 µm in insets, and 50 µm in (M) to (P).
To further analyze the expression profiles of rice MIKC*-type genes in anthers, qRT-PCR analyses were performed. Results showed that the three genes are only expressed at very low levels or not expressed at all in a mixture of rice anthers of stages 1 to 7 and stages 8 and 9. Expression of the three MIKC*-type genes was first detected in anthers at stage 10, when vacuolated microspores form. Later during pollen development, the expression increased continuously through stage 11, when bicellular pollen forms, and reached the highest levels in anthers at stage 12 (Figure 2B). However, the expression levels of the three genes differ, with MADS68 showing the highest and MADS63 the lowest level of expression (Figure 2B).
Additionally, we constructed reporter constructs by fusing the gusA (also known as uidA) gene encoding GUS to 3-kb sequences upstream of the translation start sites, which we assumed to contain the promoters of the different MIKC*-type genes, and transformed these constructs into rice plants. GUS activity from the MADS68pro:GUS fusion was first detectable in anthers at stage 10, increasing gradually from stage 11 to 12 (Figures 2C to 2G), consistent with the RT-PCR and the qRT-PCR results. For the MADS63pro:GUS transgenic lines, GUS expression was hardly observed in anthers before stage 10 (Figures 2H to 2J) but became obvious in anthers at stages 11 and 12 (Figures 2K and 2L). Similar results were observed for the MADS62pro:GUS constructs (see Supplemental Figure 1 online).
To determine whether rice MIKC*-type genes are also expressed in the anther walls, RNA in situ hybridization analyses were performed. The results showed that transcripts of MADS62, MADS63, and MADS68 are specifically localized in pollen but not in anther walls (Figures 2M to 2P). Taken together, these results reveal that rice MIKC*-type genes are exclusively expressed in pollen at late developmental stages, suggesting that they may be involved in pollen maturation.
Dimerization and DNA Binding of Rice MIKC*-Type Proteins
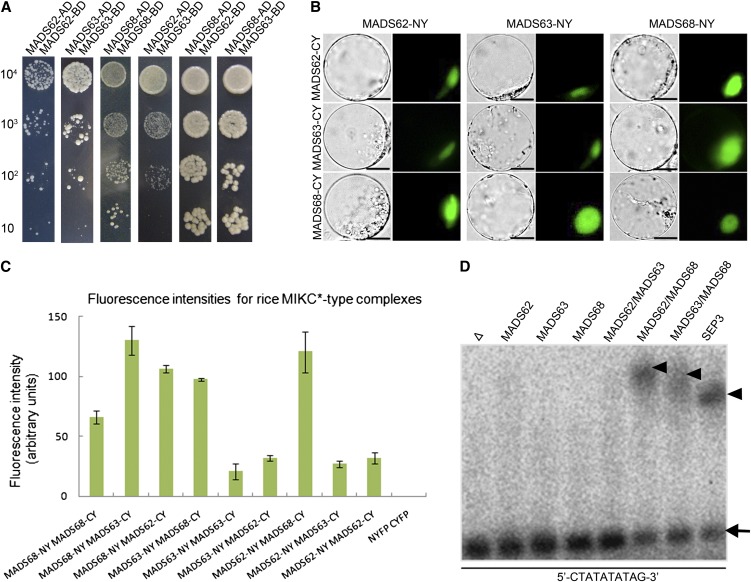
Given that MADS domain transcription factors need to form dimeric complexes to bind DNA (Riechmann et al., 1996), the interaction patterns between the three MIKC*-type proteins in rice were examined by yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays. In the Y2H system, strong interactions between MADS62 and MADS68 as well as between MADS63 and MADS68 were observed. Interactions between MADS62 and MADS63 as well as MADS68 homodimerization were of medium strength, while MADS62 and MADS63 each were observed to homodimerize only weakly (Figure 3A; see Supplemental Figure 2 online).
Figure 3.
Interaction Patterns and DNA Binding Ability of the Rice MIKC*-Type Proteins.
(A) Rice MIKC*-type protein interactions as revealed by Y2H. Each MIKC*-type protein was fused to the activation domain (AD) as a prey and the DNA binding domain (BD) as a bait. Serial dilutions of 104 to 101 AH109 cells containing different constructs combinations indicated were grown on the selective medium.
(B) BiFC yellow fluorescent protein fluorescence and bright-field images of rice protoplasts cotransfected with constructs encoding the indicated fusion proteins. Each MIKC*-type protein fused with the N-fragment or C-fragment of yellow fluorescent protein is labeled with its gene name followed by -CY and -NY, respectively. Bars = 10 µm
(C) Quantification of BiFC fluorescence intensities in transiently transfected rice protoplasts. Fluorescence intensity (arbitrary units) for each combination was determined to assess the strength of protein interactions. The mean and sd of six independent measurements are shown. Empty NYFP (N-terminal fragment of yellow fluorescent protein) and CYFP (C-terminal fragment of yellow fluorescent protein) were used as negative control.
(D) EMSA assay for rice MIKC*-type protein complexes binding N10-type CArG-box DNA. A probe containing an N10-type CArG-box (5′-CTATATATTAG-3′) was incubated with in vitro–translated MADS62, MADS63, MADS68, and combinations of these proteins. Free DNA is indicated by an arrow and shifted complexes by arrowheads. In vitro translation with SEPALLATA3 (SEP3) and an empty vector (Δ) served as positive and negative control, respectively.
[See online article for color version of this figure.]
To determine rice MIKC*-type protein interactions in planta, BiFC was performed using the full-length coding sequences. The fluorescence intensity and percentage of BiFC signal-emitting protoplasts, which reflect the strength of interactions, were measured and determined. The strongest interaction could be detected in rice protoplasts transiently transformed with MADS62/MADS68 and MADS63/MADS68 combinations (Figures 3B and 3C; see Supplemental Figure 3 online), which is in agreement with the results of the Y2H assays. However, considerably weaker interactions were observed when MADS62 together with MADS63 was transformed as well as MADS62, MADS63, and MADS68 alone, thereby reflecting the ability of weak heterodimerization of MADS62/MADS63 and homodimerization of all the rice MIKC*-type proteins in vivo.
To determine their ability to bind DNA, the rice MIKC*-type proteins were synthesized in vitro in a cell-free system and tested in electrophoretic mobility shift assay (EMSA) experiments. In vitro, only MADS63/MADS68 and MADS62/MADS68 heterodimeric complexes were found to bind to DNA probes containing CArG-boxes of the N10 or SRE type (Figure 3D; see Supplemental Figure 4 online). However, MADS62/MADS63, MADS62/MADS62, MADS63/MADS63, and MADS68/MADS68 complexes neither bound to N10-type nor to SRE-type CArG-boxes (Figure 3D; see Supplemental Figure 4 online). Competitive EMSA experiments were further performed to test the binding specificity of these complexes. The binding of MADS62/MADS68 heterodimer to a labeled probe containing the N10-type motif 5′-CTATATATAG-3′ was strongly competed by an excess of a probe containing the same motif or the N10-type motif CTATTTTTAG. However, an excess of a competitor probe containing the SRE-type motif CCATATATGG showed very little competition for binding (see Supplemental Figure 5 online). Thus, rice MIKC*-type heterodimers have a strong preference for the N10-type compared with the SRE-type motifs. Our findings suggest that even though rice MIKC*-type proteins are able to form homodimers and heterodimers among S-clade proteins, these might not function as transcription factors, since these dimers are not able to bind to DNA (Figures 3A to 3D; see Supplemental Figures 2 to 4 online). Thus, the heterodimers formed between members of the P- and S-clades may be the only dimers of rice MIKC*-type proteins that participate in directly controlling target genes and pollen development.
Loss of Function of Single Rice S-clade Genes Does Not Cause a Visible Mutant Phenotype
To explore the functions of the rice S-clade MIKC*-type genes, for MADS63, a mutant (PFG_ 2D-10691) that harbors a T-DNA insertion in the seventh intron (see Supplemental Figure 6A online) was obtained from the Postech collection. For MADS62, RNA interference (RNAi) transgenic lines were generated. Homozygous mads63 mutant plants were confirmed by PCR of genomic DNA (see Supplemental Figure 6B online) and used for further analysis. Transcripts of MADS63 could not be detected by RT-PCR analysis in mutant plants, suggesting that mads63 is a null mutant (see Supplemental Figure 6C online).
Genetic analyses were performed to determine whether the mads63 mutant showed gametophytic defects. When heterozygous lines were selfed, plants with wild-type, heterozygous, and homozygous genotypes were produced with a ratio of nearly 1:2:1, thus indicating Mendelian segregation. When heterozygous lines were test-crossed with wild-type pollen, two types of offspring resulted: plants with homozygous wild-type and heterozygous genotypes with a ratio of ∼1:1. Also, homozygous mutants pollinated with wild-type pollen all generated heterozygous progeny (see Supplemental Table 2 online). These results demonstrate that the male and female gametophytes, as well as the surrounding sporophytic tissues, are properly functioning. Moreover, I2-KI staining, Alexander staining, transmission electron microscopy (TEM), and pollen germination assays in vitro all showed that mads63 pollen is well developed and viable (see Supplemental Figures 6D to 6G online).
Likewise, no aberrant morphologies were found in the MADS62_RNAi transgenic lines.
Downregulation of the P-Clade Gene MADS68 Affects Pollen Maturation and Germination
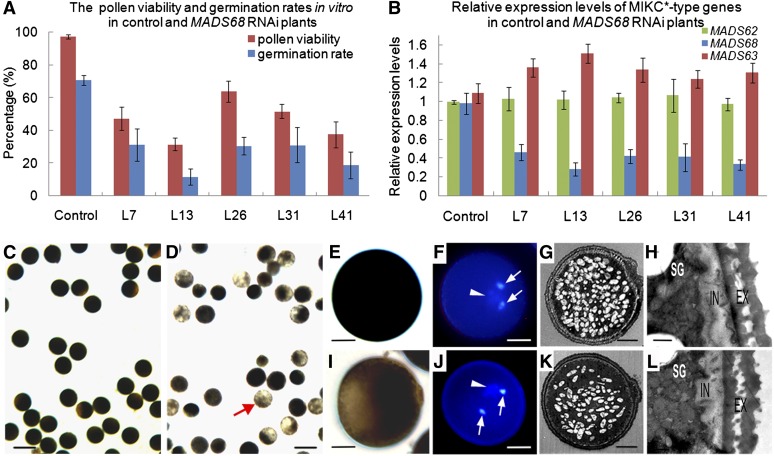
To elucidate the role of the P-clade gene MADS68 during rice pollen development, transgenic RNAi knockdown lines of MADS68 were generated. Forty-nine independent transgenic lines were obtained. No differences in vegetative growth and floral organ development were observed between knockdown plants and control plants transformed with empty vector. However, 26 RNAi lines had obviously reduced pollen fertility compared with control plants. Out of these, we selected five RNAi lines (lines 7, 13, 26, 31, and 41) for further phenotype analysis and measurement of MADS68 expression levels by qRT-PCR. Viability of pollen, tested with I2-KI staining and Alexander staining, was lower in MADS68 RNAi plants (31 to 64%) compared with control plants (97%; Figure 4A), and the frequency of in vitro pollen germination (12 to 30%) was much lower than that of control plants (71%; Figure 4A). Moreover, the reduced expression level of MADS68 (Figure 4B) was overall consistent with the degree of pollen viability and germination rate in the MADS68 RNAi transgenic plants. No significant decrease in MADS62 or MADS63 expression was observed in the MADS68 RNAi plants (Figure 4B). These results indicate that the observed phenotype of abnormal pollen in the transgenic plants was caused by specific silencing of MADS68.
Figure 4.
Molecular Detection and Pollen Phenotypes of MADS68 RNAi Lines.
(A) The percentage of viable pollen grains and the germination rates in vitro of control plants and different MADS68 RNAi lines. The data show mean ± sd of three independent experiments.
(B) Relative expression levels of MADS68, MADS62, and MADS63 in control plants and five selected MADS68 RNAi lines (L7, L13, L26, L31, and L41). Total RNAs were pooled from anthers at stage 13. The samples were quantified by qRT-PCR using UBIQUITIN as a reference gene. The data are presented as mean ± sd (n = 3).
(C) to (L) Pollen phenotypes at the maturation stage. I2-KI staining showing pollen viability in control plant (C) and MADS68 RNAi L7 (D). The higher magnification of I2-KI staining pollen of control plant (E) and abnormal pollen of line MADS68 RNAi L7 (I). DAPI staining showing three nuclei of pollen grain in the control plant (F) and line MADS68 RNAi L7 (J). TEM showing starch accumulation in pollen of control plant (G) and abnormal pollen of line MADS68 RNAi L7 (K); wall structure in pollen of control plant (H) and abnormal pollen of line MADS68 RNAi L7 (L). Arrowheads indicate the vegetative nucleus, and arrows indicate sperm-cell nuclei in pollen of control plant (F) and abnormal pollen of line MADS68 RNAi L7 (J).
EX, extine; IN, intine; SG, starch granules. Bars = 100 µm in (C) and (D), 12.5 µm in (E), (F), (I), and (J), 5 µm in (G) and (K), and 0.5 µm in (H) and (L).
Compared with the pollen grains of control plants, which were deeply stained with I2-KI and enriched with starch (Figures 4C and 4E), we observed only weak staining and defects of starch accumulation in the abnormal pollen grains of RNAi plants (Figures 4D and 4I). To further determine whether mitotic cell division was affected, 4′,6-diamidino-2-phenylindole (DAPI) staining and fluorescence microscopy of RNAi pollen grains were performed. Nuclear staining showed two brightly stained small organelles, the sperm nuclei (Figure 4J, arrow), and a larger and diffusely stained structure, the vegetative cell nucleus (Figure 4J, arrowhead) in the abnormal pollen grains of the RNAi plants, similar to those of pollen grains of control plants (Figure 4F). Pollen morphology was investigated in detail by TEM to check for structural mutant phenotypes. A decrease of starch grains in the mutant pollen compared with pollen of the control plants was observed (Figures 4G and 4K). Beyond that, no obvious morphological differences in exine and intine structures of the abnormal pollen were observed in comparison to those of control plants (Figures 4H and 4L). These analyses demonstrate that knockdown of MADS68 causes defects in pollen maturation and germination but did not affect pollen mitosis or pollen wall development.
Knockdown of MADS68 and MADS62 in the mads63 Background Generates More Severely Affected Pollen
Because the mads63 mutant and MADS62 single RNAi plants displayed no obvious morphological alteration, and the three rice MIKC*-type genes exhibited similar expression patterns, the possible functional redundancy among the three genes was investigated.
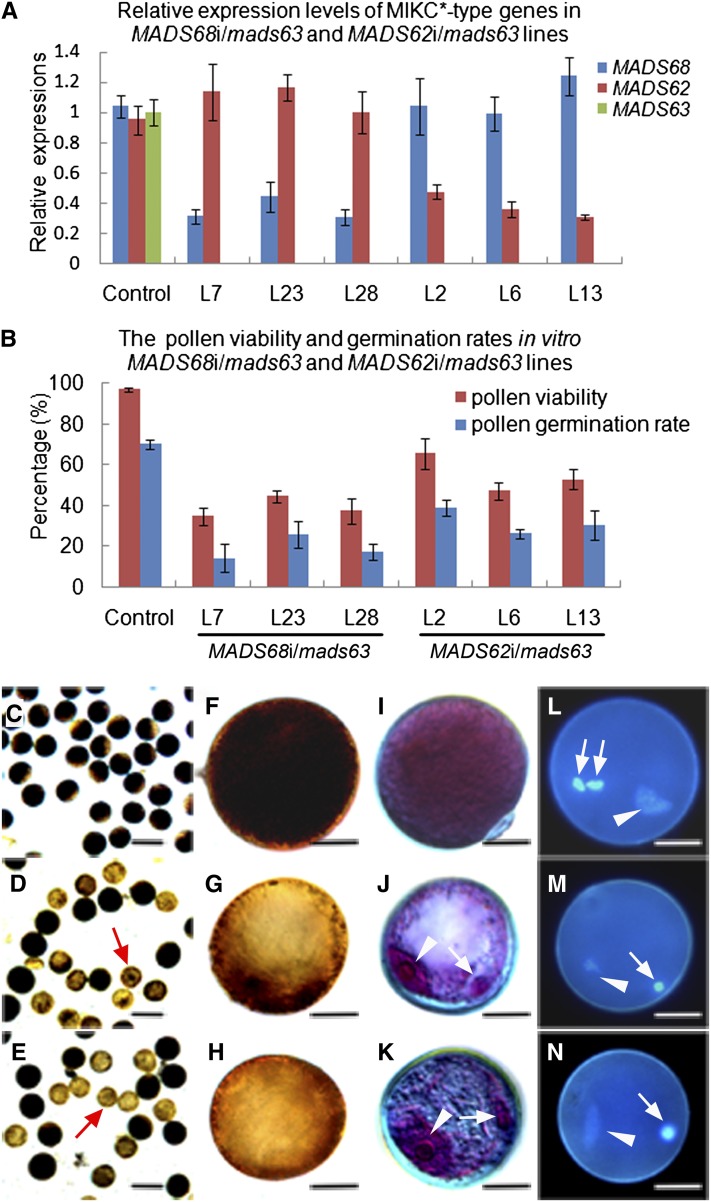
Given that the mads63 mutant investigated here was hygromycin resistant, pCam23A-Ubi, a vector conferring G418 resistance, was used for silencing MADS62 and MADS68 in the mads63 mutant background (see Supplemental Figure 7A online). The double-transgenic lines were confirmed by PCR of genomic DNA (see Supplemental Figures 7B and 7C online). RNAi knockdown of either MADS68 or MADS62 in the mads63 background exhibited aberrant pollen phenotypes in 22 of 28 transgenic lines and in nine of 16 transgenic lines, respectively. The mutant phenotypes were similar to those observed in the MADS68 RNAi lines, but to a more severe extent. Six RNAi lines (lines 7, 23, and 28 for MADS68i/mads63; lines 2, 6, and 13 for MADS62i/mads63) were selected for more detailed phenotypic analyses and quantitative analysis of gene expression levels (Figure 5). Expression of the genes was diminished as expected from single knockdown and knockout lines (Figure 5A). Using I2-KI staining and Alexander staining, we observed that a certain proportion of pollen grains (56 to 66% for MADS68i/mads63; 35 to 53% for MADS62i/mads63) were small and very weakly stained, indicating little starch accumulation at the mature stage compared with control plants (Figures 5B to 5K). Furthermore, the mutant pollen displayed an in vitro germination rate (14 to 25% for MADS68i/mads63; 26 to 39% for MADS62i/mads63; Figure 5B) that was much lower than in the case of control plants (71%). In contrast with control plants, only two nuclei, a vegetative-like nucleus and a generative-like nucleus (Figures 5J to 5N), could be identified in the abnormal pollen of MADS68i/mads63 and MADS62i/mads63 lines by Alexander and DAPI staining. These closely resembled the vegetative and generative nucleus, respectively, of wild-type binucleate pollen, thus suggesting the abnormal pollen of both MADS68i/mads63 and MADS62i/mads63 plants is unable to complete the second mitosis to generate three nuclei.
Figure 5.
Severe Defects in the Pollen Maturation and Germination for MADS68i/mads63 Plants and MADS62i/mads63 Plants.
(A) Relative expression levels of MADS68, MADS62, and MADS63 in control plants, three selected MADS68i/mads63 lines (L7, L23, and L28), and three selected MADS62i/mads63 lines (L2, L6, and L13). Total RNAs were pooled from anthers at stage 13. The samples were quantified by qRT-PCR using UBIQUITIN as a reference gene. Data show mean ± sd (n = 3).
(B) The percentage of viable pollen grains and the germination rates in vitro of control plants and different MADS68i/mads63 lines (L7, L23, and L28) and MADS62i/mads63 lines (L2, L6, and L13). The data show mean ± sd of three independent experiments.
(C) to (E) I2-KI staining showing starch accumulation of pollen from a control plant (C), line MADS68i/mads63 L23 (D), and line MADS62i/mads63 L6 (E).
(F) to (H) A higher magnification of I2-KI stained pollen from a control pollen (F) and pollen of line MADS68i/mads63 L23 (G) and line MADS62i/mads63 L6 (H).
(I) to (K) Alexander staining of pollen from the control plant (I) and lines MADS68i/mads63 L23 (J) and MADS62i/mads63 L6 (K).
(L) to (N) DAPI staining showing three nuclei in pollen of control plants (L), but only two nuclei in abnormal pollen of lines MADS68i/mads63 L23 (M) and MADS62i/mads63 L6 (N).
Arrowheads indicate the vegetative nucleus in pollen of control plants (L) and in abnormal pollen of MADS68i/mads63 L23 ([J] and [M]) and MADS62i/mads63 L6 ([K] and [N]); arrows indicate sperm-cell nuclei in pollen of a control plant (L) and the single generative-like cell nucleus in an abnormal pollen of MADS68i/mads63 L23 ([J] and [M]) and MADS62i/mads63 L6 ([K] and [N]). Bars = 100 µm in (C) to (E) and 12.5 µm in (F) to (N).
Arrest in Pollen Development Occurs during the Bicellular Microspore Stage When MIKC*-Type Genes Are Affected
To determine exactly when pollen development is aborted in RNAi plants, pollen collected at different developmental stages was examined by light microscopy.
For single knockdown MADS68 RNAi lines, no obvious phenotypic alterations were observed in the pollen of RNAi plants until stage 11 (see Supplemental Figure 8D online), compared with control plants (see Supplemental Figure 8A online). At stage 12, bicellular pollen of control plants was enriched with starch granules (see Supplemental Figure 8B online), whereas the development of nearly half of the pollen grains of MADS68 RNAi plants did not have as many inclusions as pollen of control plants (see Supplemental Figure 8E online). Eventually, a significant number of the abnormal pollen grains failed to reach the mature stage (see Supplemental Figure 8F online).
For MADS68i/mads63 and MADS62i/mads63 plants, microscopy showed that microspores of RNAi and control plants before stage 10 were similar in appearance. However, alterations became apparent at late stage 11, when starch accumulation started in bicellular pollen of control plants. Two types of pollen grains could be found in the anthers of MADS68i/mads63 and MADS62i/mads63 plants. Whereas some of the grains (34.8% for MADS68i/mads63; 52.7% for MADS62i/mads63) developed as normally as those in the wild type, the rest displayed apparent abnormalities: smaller pollen and delayed formation of starch granules (see Supplemental Figures 8G and 8J online), compared with control plants (see Supplemental Figure 8A online). Also, the delayed and aberrant pollen was evident at stages 12 and 13, and pollen development eventually arrested or aborted at the bicellular stage (Figure 5; see Supplemental Figures 8H, 8I, 8K, and 8L online).
Late-Stage Pollen Genes with N10-Type CArG-Boxes in Their Putative Regulatory Regions Are Potential Targets of Rice MIKC*-Type Genes
We reanalyzed the rice pollen transcriptome data set from Wei et al. (2010), containing the expression levels of the rice genes in the five stages of pollen development represented on the Affymetrix microarray chip. Excluding transposon and retrotransposon elements, 437 transcripts were identified that were enriched in late stage pollen, including 113 in bicellular pollen, 140 in immature tricellular pollen, 68 in mature pollen grains, and 116 in germinated pollen grains. Subsequently, we screened the 3-kb upstream regions of these genes for the presence of N10-type CArG-boxes using the PLACE tool (Higo et al., 1999). In total, 62 genes possessed N10-type motifs in their putative regulatory regions: 18 bicellular pollen–enriched, 21 tricellular pollen–enriched, four mature pollen grain–enriched, and 19 germinated pollen grain–enriched transcripts (listed in Supplemental Data Set 2 online). These genes are hence potential targets of rice MIKC*-type transcription factor complexes.
RT-PCR and qRT-PCR were further performed in the wild-type plants, mads63, MADS68 single RNAi, MADS62i/mads63, and MADS68i/mads63 plants to test the alteration in expression of a selected nine of these in silico predicted target genes. Results indicate that four genes (Os05g45370, Os02g02460, Os05g11790, and Os06g49860) are substantially downregulated, and two (Os01g47050 and Os10g26470) are upregulated in MADS68i/mads63 and/or MADS62i/mads63 plants (see Supplemental Figures 9 and 10 online).
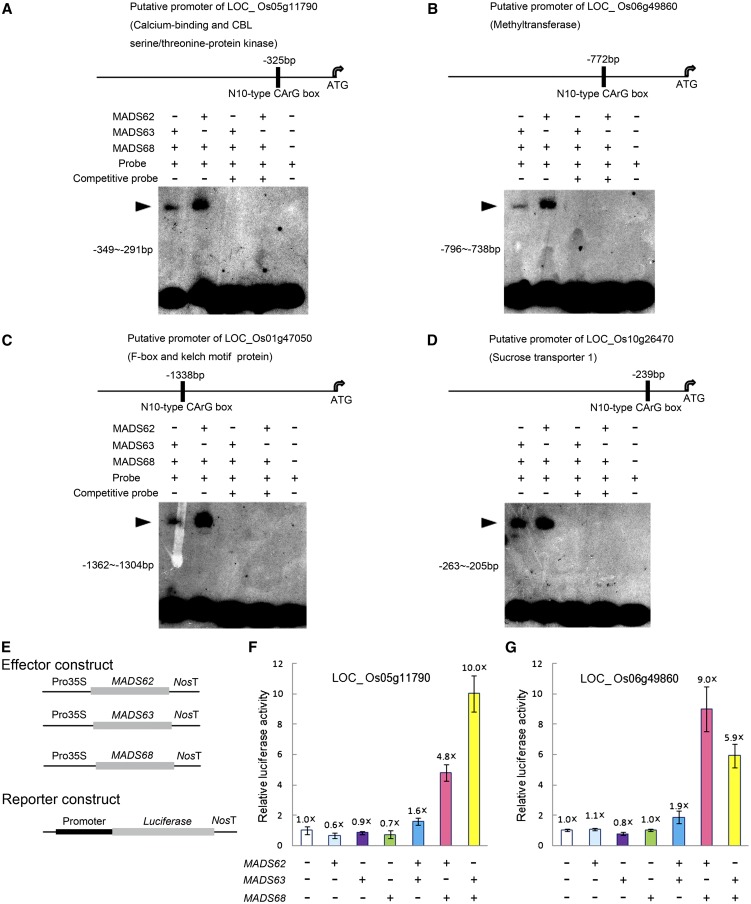
To further test whether these genes are targets of rice MIKC*-type genes, EMSA experiments and protoplast transient transfection assays were performed. The EMSA experiments showed that MADS62/MADS68 and MADS63/MADS68 heterodimeric complexes directly bind to the putative promoter regions of Os10g26470, Os05g11790, Os01g47050, and Os06g49860 and that these interactions are outcompeted by the addition of excess unlabeled fragments (Figures 6A to 6D). Transient transfection assays confirmed that MADS62/MADS68 and MADS63/MADS68 combinations can activate Os05g11790pro:LUC and Os06g49860pro:LUC expression in vivo. By contrast, MADS62/MADS63 combinations and any of the three proteins alone could not activate Os05g11790pro:LUC nor Os06g49860pro:LUC expression (Figures 6E to 6G). Therefore, these genes may be primary targets of MADS62/MADS68 and MADS63/MADS68 complexes during pollen development.
Figure 6.
EMSA and Transient Transfection Analyses for Heterodimeric Complexes of Rice MIKC*-Type Proteins.
(A) to (D) EMSA assays show that MADS62/MADS68 and MADS63/MADS68 dimers directly bind to fragments of putative promoter regions of Os05g11790 (A), Os06g49860 (B), Os01g47050 (C), and Os10g26470 (D). The positions of the N10-type CArG-boxes site in the promoter regions are shown (top panels). DNA fragments (each shown in left of bottom panel) were labeled as probes and unlabeled DNA fragments in 20-fold excess were added as competition probes. The shifted bands are indicated with arrowheads.
(E) to (G) Transient transfection assays indicate that heterodimers of MIKC*-type proteins activate LUC reporter gene expression under the control of putative promoters of Os05g11790 (F) and Os06g49860 (G) in Arabidopsis protoplasts. The luciferase activity of protoplasts transfected with empty effector and reporter vectors (E) was set to 1×, and other values for the different constructs combinations were normalized relative to this value. Data show mean ± sd (n = 3).
[See online article for color version of this figure.]
DISCUSSION
The Function of MIKC*-Type Genes Is Conserved between Arabidopsis and Rice
Phylogenetic studies revealed that there are four well-supported clades within the superclade of MIKC*-type genes, comprising genes of mosses, lycophytes, and the P-clade and S-clade genes of ferns and seed plants, respectively. The latter ones probably originated by a duplication event in the lineage that led to the ferns and seed plants after the lineage that led to mosses and lycophytes had branched off (Figure 1; Gramzow et al., 2012; Kwantes et al., 2012). Within the S- and the P-clades, genes from ferns are basal to angiosperm MIKC*-type genes, which further show a split into a monocot and a eudicot subgroup, thus resembling species phylogeny. In rice and other grass species, two different subclades, MADS62-like and MADS63-like genes can be found within the S-clade. In combination with the colinearity found between the two rice genes and their corresponding orthologs (Tang et al., 2008a, 2008b), this finding suggests that the duplication event that led to the two subclades had already occurred before the diversification of grass species. The existence of S-clade genes only from either the MADS62 or the MADS63 subclade in barley, sorghum, maize, and B. distachyon is probably due to gene loss after duplication or lack of comprehensive genome data. Only a single P-clade MIKC*-type gene could be found in each of the grass species, which is in contrast with eudicots, where several recent duplications could be observed within the P- and S-clades (Figure 1).
Expression studies in several representatives of the major land plant lineages revealed that most MIKC*-type genes are highly expressed in the male gametophyte (Verelst et al., 2007a; Zobell et al., 2010; Kwantes et al., 2012). Five of the six Arabidopsis MIKC*-type genes (AGL30, AGL65, AGL94, AGL66, and AGL104) are predominantly expressed in pollen, while the other one (AGL67) is expressed in embryonic tissue (Kofuji et al., 2003; Honys and Twell, 2004; Lehti-Shiu et al., 2005; Verelst et al., 2007a). Within the P-clade genes, different temporal expression profiles were observed between AGL30 and AGL65, which represent a pair of paralogs that originated from a recent gene duplication event (Veron et al., 2007). Like the two S-clade genes AGL66 and AGL104, AGL30 is expressed already at the unicellular microspore stage, whereas AGL65 is only activated during the tricellular stage (Honys and Twell, 2004; Verelst et al., 2007a, 2007b). In this study, the three rice MIKC*-type genes MADS62, MADS63, and MADS68 were observed to be expressed exclusively during pollen development, from the unicellular microspore stage to pollen maturation (Figure 2), which resembles the expression of AGL66, AGL104, and AGL30 in Arabidopsis. This suggests that a pollen-specific expression pattern was already established in the most recent common ancestor of monocots and eudicots.
It has been proposed that MIKC*-type genes may provide conserved and essential functions to the male gametophytes of all vascular land plants, including lycophytes, ferns, and seed plants (Kwantes et al., 2012). Mutant analyses also confirmed that in the eudicot Arabidopsis, the MIKC*-type genes are required for pollen maturity and tube growth (Verelst et al., 2007a, 2007b; Adamczyk and Fernandez, 2009). The mads63 mutant and the MADS62 RNAi transgenic plants display no aberrant pollen morphologies, but in the MADS62i/mads63 double mutants, pollen maturation and germination is impaired (Figure 5; see Supplemental Figures 6 and 8 online). This indicates that the two S-clade genes of MADS62 and MADS63 act largely redundantly in pollen development, as their homologs AGL66 and AGL104 in Arabidopsis do (Verelst et al., 2007a; Adamczyk and Fernandez, 2009). Previously, neither in Arabidopsis nor in any other species was a complete knockout or a knockdown of P-clade genes achieved. However, the downregulation of the sole rice P-clade gene MADS68 in RNAi transgenic lines resulted in defects in pollen maturation and germination (Figure 4; see Supplemental Figure 8 online), similar to that of MADS62i/mads63 double mutants. These data illustrate that P- and S-clade genes are not functionally redundant and that both clades are indispensable for pollen development.
However, in the double loss-of-function line MADS62i/mads63, the extent of pollen abnormality is more severe than in the MADS68 single RNAi lines. This could possibly be due to incomplete downregulation of MADS68 via RNAi (Figure 4), meaning that a certain degree of MADS68 function may have remained intact even in the knockdown lines. Furthermore, the MADS68i/mads63 mutant lines show a more severe phenotype than the single knockdown MADS68 transgenic lines do. This could hint at an incomplete redundancy of MADS62 and MADS63, since a complete redundancy of both genes should enable the intact MADS62 to replace MADS63 function completely in this double mutant, even though neither the mads63 mutant nor the MADS62 RNAi transgenic plants showed a mutant phenotype. However, the more severe mutant phenotype of MADS68i/mads63 lines compared with MADS68 RNAi lines may also suggest that there is some redundancy between the S- and P-clade genes that escaped previous analyses.
In MADS68 RNAi, MADS68i/mads63, and MADS62i/mads63 plants, starch accumulation during pollen development is affected (Figures 4 and 5; see Supplemental Figure 8 online). Starch filling is a prominent metabolic process during pollen maturation. Starch can serve as an energy resource for subsequent germination, and starch levels hence can be considered as a checkpoint for pollen maturity (Datta et al., 2002). The degree of starch accumulation below a certain threshold can result in premature termination of pollen development and failure of pollen to complete second mitosis (Wen and Chase, 1999; Zhang et al., 2001; Datta et al., 2002; Han et al., 2006). Accordingly, the fact that pollen development of MADS68i/mads63 and MADS62i/mads63 plants is delayed and eventually arrested at the bicellular pollen stage while MADS68 single RNAi can finish the second mitosis to produce three nuclei is assumed to be related to the apparently different degrees of starch deficiency, which are much more severe in the double loss-of-function plants than in MADS68 single RNAi lines (Figures 4 and 5; see Supplemental Figure 8 online). By contrast, many of agl66 agl104-2 double mutant pollen grains showed signs of structural damage and ∼40% of the double mutant pollen grains had at least one visible nucleus compared with 70% of wild-type pollen in Arabidopsis (Adamczyk and Fernandez, 2009). However, it is unknown as to precisely when the structural abnormalities begin to appear and whether pollen second mitosis occurs in agl66 agl104-2 double mutant pollen. Therefore, our findings provide clues at the cellular level for causes of pollen defects in loss-of-function mutants of MIKC*-type genes, at least in rice.
Taken together, our data underline the fact that despite independent duplications in both P- and S-clade genes, and sequence divergence after the separation of monocots and dicots more than 150 million years ago (Wikström et al., 2001), MIKC*-type genes confer an indispensable and highly conserved function in pollen maturation and germination in the monocot rice as well as in the higher eudicot Arabidopsis. This strongly suggests that P- and S-clade genes already had such a role in pollen development in the most recent common ancestor of monocots and eudicots. Further investigation of MIKC*-type genes in gymnosperms and basal angiosperms may reveal whether functional conservation of MIKC*-type genes for male gametophytes could be extended to all angiosperms or seed plants.
The Formation of Heterodimeric Complexes between S- and P-Clade Proteins Is Crucial for the Function of MIKC*-Type Proteins
It was hypothesized that homodimerization might be an ancestral state of MIKC*-type complexes, and obligatory heterodimerization probably evolved from homodimerization after gene duplication and subsequent sequence divergence (Zobell et al., 2010). Shortly after the divergence of the S- and P-clade genes, MIKC*-type homodimers and heterodimers between members of the same clade might still have been functional, as one still finds in ferns. Based on expression and interaction data, it is suggested that the MIKC*-type protein complexes might function not only in male and female gametophytes but also in sporophytic tissues (Kwantes et al., 2012). However, in basal eudicots, the functional MIKC*-type protein complexes in the form of S- and P-clade heterodimers might only exist in male gametophytes (Kwantes et al., 2012). In the higher eudicot Arabidopsis, the MIKC*-type proteins form obligate heterodimeric complexes between S- and P-clade members and function exclusively in pollen (Verelst et al., 2007a, 2007b; Adamczyk and Fernandez, 2009). Our protein–protein interaction analyses revealed that the MIKC*-type proteins in rice are able to form both homodimeric and heterodimeric complexes, but our EMSA analyses demonstrated that only heterodimeric complexes between S- and P-clade proteins are able to bind to DNA (Figure 3). Therefore, our study suggests that only heterodimeric complexes of S- and P-clade members are of functional relevance, even though a regulatory role of protein dimerization that does not lead to DNA binding complexes cannot be completely ruled out at the moment.
Obligate heterodimerization of proteins from the S- and P-clades is in line with the severe phenotypes that were observed when all genes from one phylogenetic clade were knocked down, as in MADS62i/mads63 and MADS68i lines. In these cases, putatively heterodimeric complexes cannot form, thus leading to aberrant pollen development. By contrast, the normal phenotype of the single mutants of S-clade genes MADS62 and MADS63 might be due to redundancy of the MADS62/MADS68 and the MADS63/MADS68 complexes.
Furthermore, we showed that rice MIKC*-type protein heterodimers are very strict with respect to their binding motif, N10-type CArG-boxes, which is very similar to the behavior of MIKC*-type proteins heterodimers of Arabidopsis (Verelst et al., 2007a). In contrast with SEP3, which is a MIKCC-type protein, MIKC*-type heterodimers did not bind to SRE-type CArG-boxes in competition assays. This suggests a very high target specificity for MIKC*-type genes, not only in rice but very likely also in Arabidopsis, which might be essential for their exclusive and crucial role in pollen development.
Taken together, our data suggest that MIKC*-type protein complexes have confined their function to pollen development in the most recent common ancestor of monocots and eudicots.
The Heterodimeric MIKC*-Type Protein Complexes Are Involved in Pollen Maturation and Germination via Binding N10-Type CArG-Box Motifs of Target Genes
A considerable number of genes have been demonstrated to be associated with the process of pollen maturation and subsequent pollen germination (Honys and Twell, 2003, 2004). Gene expression analyses employing microarrays revealed that in Arabidopsis heterodimeric complexes of MIKC*-type proteins regulate pollen maturation and germination by activating or repressing many pollen-specific genes (Verelst et al., 2007b; Adamczyk and Fernandez, 2009). However, no direct biochemical or molecular genetic experiments were performed to verify that these genes are direct targets of heterodimeric complexes of MIKC*-type proteins. In rice, binding sites typical for MIKC*-type protein complexes are enriched in putative promoters of genes expressed during late pollen development (see Supplemental Data Set 2 online). These genes may well be involved in pollen maturation and germination and are potential targets involved in the pollen regulatory network of MIKC*-type protein complexes. Several of these genes have been demonstrated to be substantially up- or downregulated in the double loss-of-function lines (see Supplemental Figures 9 and 10 online), and EMSA assays confirmed that MADS62/MADS68 and MADS63/MADS68 complexes can directly bind to N10-type CArG-boxes that are present in the putative promoter regions of these genes (Figures 6A to 6D). Furthermore, two of these genes, Os05g11790 and Os06g49860, can be activated by MADS62/MADS68 and MADS63/MADS68 combinations in vivo in transient transfection assays (Figures 6E to 6G). Os05g11790 (CIPK20) is annotated as encoding a calcium binding and CBL-interacting protein kinase (CIPK), which has not yet been functionally characterized. However, RNAi suppression of CIPK23, another member of the rice CIPK family, results in partial male sterility due to a proportion of pollen without starch accumulation (Yang et al., 2008), which is similar to the pollen phenotype of rice plants with MIKC*-type gene loss of function. Intriguingly, seven members of the Arabidopsis CIPK family are significantly downregulated in agl66 agl104-2 double, agl65 agl66 agl104-1 triple, and agl65 agl66 agl94 agl104-1 quadruple mutants (Adamczyk and Fernandez, 2009). For AT45820 (CIPK20), which is the putative ortholog of rice CIPK20, a N10-type CArG-box has been found within its 1-kb putative promoter region (see Supplemental Data File S1 in Adamczyk and Fernandez, 2009). Given that MIKC*-type heterodimeric complexes preferentially bind N10-type motifs both in rice and Arabidopsis (Verelst et al., 2007a; this study), it is conceivable that they regulate orthologous targets (for example, rice and Arabidopsis CIPK20). Hence, the gene regulatory network involving MIKC*-type protein complexes is likely conserved between rice and Arabidopsis.
Additionally, Os06g49860 is a member of a methyltransferase family. These enzymes play crucial roles in many development processes, and some of them have been reported to be associated with pollen development (Mou et al., 2002; Grini et al., 2009). These data thus provide some clues concerning the molecular mechanisms of pollen development, in which a large number of genes are regulated by complexes of MIKC*-type proteins.
Taken together, complexes of MIKC*-type proteins are essential components of the gene regulatory network for pollen maturation and germination in rice, in which they activate or repress the expression of many downstream target genes via binding to N10-type CArG-boxes in regulatory regions of their DNA. In combination with previous studies in Arabidopsis, we conclude that the function and underlying regulatory network of MIKC*-type protein complexes in pollen developmental processes was already established in the most recent common ancestor of monocots and eudicots and hence has been strongly conserved for at least ∼150 million years.
METHODS
Plant Materials and Growth Conditions
The rice cultivars ‘Zhonghua 11’ and ‘Hwayoung’ (Oryza sativa ssp japonica) were grown under natural conditions in an experimental field of the Institute of Botany, Chinese Academy of Sciences, Beijing, China.
Mutant Identification and Cultivation
The T-DNA mutant line PFG_ 2D-10691 for MADS63, derived from cultivar Hwayoung, was obtained from the Postech collection (Jeong et al., 2002; An et al., 2003). Homozygous lines were identified by PCR screening with a pair of MADS63 specific primers and a T-DNA border primer (see Supplemental Table 3 online). Then, the homozygous lines were backcrossed to wild-type plants to generate F1 seeds to make the genetic background uniform. F1 plants were cultivated for two generations, and homozygous descendants (F3 generation) were isolated as described above for further analysis. Total RNAs from the anthers of F3 homozygous mutants were extracted and RT-PCR was performed to analyze the expression of MADS63.
Generation of Knockdown Plants in Wild-Type and mads63 Backgrounds
To construct the MADS62 and MADS68 knockdown vectors, gene-specific DNA fragments of 307 bp (MADS62; nucleotides 745 to 1051 counted from ATG) or 402 bp (MADS68; nucleotides 179 to 581 from ATG) were amplified as RNAi fragments. The intron and cauliflower mosaic virus 35S terminator cassette of pJawohl3-RNAi (GenBank accession number AF404854) was cloned into the HindIII-NotI site of the pBluescript SK vector (Stratagene), thus yielding pBIntron as an intermediate vector. The RNAi fragments of MADS62 and MADS68 were subcloned into the pBIntron vector in both sense (EcoRI-SmaI) and antisense (HindIII-XhoI) orientations. Finally, the RNAi cassettes were mobilized with restriction enzymes KpnI and SacI and introduced into the vector pCAMBIA1301-Ubi (Cui et al., 2010), thus yielding the final binary vectors. These constructs or an empty vector were introduced into cultivar Zhonghua 11 by Agrobacterium tumefaciens–mediated transformation, as described previously (Hiei et al., 1994).
To generate the MADS68 and MADS62 RNAi binary vectors for transformation of the mads63 mutant, the RNAi cassettes described above were digested with restriction enzymes KpnI and SacI and finally ligated into the vector pCam23A-Ubi (a derivative of pCAMBIA 2300 carrying the maize [Zea mays] UBIQUITIN promoter). A schematic diagram of the resulting RNAi constructs can be found in Supplemental Figure 7 online. Then, F3 homozygous mads63 mutants were used for subsequent transformation with MADS68 and MADS62 RNAi constructs under selection with 120 mg/mL G418, as described previously (Huang et al., 2001). The empty vector pCam23A-Ubi was used in parallel transformations as control for vector-dependent effects.
Promoter-GUS Fusion Studies and Histochemical Analysis of GUS Activity
The sequences ∼3.0 kb upstream of the translation start sites of MADS62, MADS63, and MADS68 were amplified with specific pairs of primers (see Supplemental Table 3 online) and recombined into the vector pCAMBIA1301 upstream of the GUS (uidA) gene. The resultant constructs were transformed into rice by the method described above. GUS staining was performed as previously described (Jefferson et al., 1987).
Phylogenetic Analyses
All sequences for phylogenetic analyses were retrieved from the databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), Phytozome v7.0 (http://www.phytozome.net/), and PlantGDB (http://www.plantgdb.org/) using the BLAST program (Altschul et al., 1990). Fifty-eight full-length amino acid sequences of MIKC*-type proteins were aligned with the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/) and manually optimized using Genedoc software (Nicholas and Nicholas, 1997). Then, the alignment was reverse translated using aa2dna (http://www.personal.psu.edu/nxm2/software.htm) into the corresponding nucleotide alignment, which is provided in Supplemental Data Set 1 online. Phylogenetic analyses were performed based on the alignment of DNA sequences that encode MIK regions using maximum likelihood and Bayesian methods. Maximum likelihood analysis was performed using PHYML version 3.0 with GTR + I + Γ model and 1000 bootstrap replicates (Guindon and Gascuel, 2003). Bayesian phylogeny was determined using MrBayes version 3.2.1 (Ronquist et al., 2012), sampling every 500 generations for 1,000,000 generations and discarding 500 of the sampled trees; the posterior probability was used to estimate nodal robustness.
In Situ Hybridization Analyses
MADS62-, MADS63-, and MADS68-specific regions were amplified using corresponding primer pairs (see Supplemental Table 3 online) and then transcribed in vitro as probes using the Digoxigenin RNA labeling kit (Roche). Fresh wild-type flowers from different developmental stages were fixed immediately, embedded in paraffin (Sigma-Aldrich), and sectioned at 8-µm thickness. Hybridization and immunological detection were performed according to the previous description (Cui et al., 2010).
RT-PCR and qRT-PCR Analyses
Total RNAs were isolated from various wild-type rice tissues and anthers at different developmental stages of wild-type, mutant, and transgenic lines using TRIzol reagent (Invitrogen) digested with DNase I and reverse transcribed using Superscript-III reverse transcriptase (Invitrogen) into cDNA samples as templates for subsequent RT-PCR and qRT-PCR. For RT-PCR, the internal standard ACTIN1 was used. qRT-PCR analyses were performed using SYBR Premix Ex Taq Mix (Takara) on a Rotor-Gene 3000 (Corbett Research) according to the manufacturer’s instructions. For analysis of the expression of MADS62, MADS63, and MADS68 in wild-type anthers at different stages, a dilution series and standard curve were created to determine the initial quantity of each gene and then their expressions levels were normalized to the geometric average the expression of internal genes UBIQUITIN and ACTIN1 (Vandesompele et al., 2002). For other qRT-PCR analyses, relative expression levels were calculated using UBIQUITIN as reference gene. The primer sequences are listed in Supplemental Table 3 online.
Y2H and BiFC assays
For Y2H assays, the full-length coding sequence of each rice MIKC*-type gene was amplified and fused into the GAL4 activation domain vector pGADT7 or the GAL4 binding domain vector pGBKT7. Yeast cells containing different construct combinations were grown on the selective medium SD-LTHA + 12.5 mM 3-amino-1,2,4-triazole (L, Leu; T, Trp; H, His; A, adenine). Yeast transformations and interaction analyses were conducted as previously described (Cui et al., 2010). For BiFC assays, full-length coding sequences of three rice MIKC*-type genes were amplified and fused into the pUC-SPYNE and the pUC-SPYCE vectors (Walter et al., 2004). Rice protoplast isolation and transfection was performed essentially as described (Chen et al., 2006). The experiments were repeated six times. Fluorescence intensity (arbitrary units) was determined using Image J software. More than 450 rice protoplasts for each complex were counted to determine the percentage of BiFC fluorescence signal-emitting protoplasts after transient transfection.
EMSA Analyses
For cell-free expression, the coding sequences of MADS62, MADS63, and MADS68 were subcloned into the pTNT vector via digestion and ligation. In vitro translation was done with a SP6 TNT QuickCoupled Transcription/Translation mix (Promega) according to the manufacturer’s instructions. DNA fragments were labeled using polynucleotide kinase (Fermentas) and then used as probes (see Supplemental Table 4 online); unlabeled DNA fragments were used as competition probes. The protein-DNA binding and competition reactions, electrophoretic separation, and visualization of bands were conducted as described (Verelst et al., 2007a). The experiments were performed three times independently.
I2-KI, Alexander, and DAPI Staining and Pollen Germination in Vitro
To assess pollen viability, mature anthers were incubated with 1% I2-KI staining and Alexander staining solution (Alexander, 1969). For pollen germination assays, mature pollen grains were gently applied to plates in a medium consisting of 20% Suc, 40 mg/L boric acid, 3 mM/L Ca(NO3)2, 5 mg vitamin B1, and 10% polyethylene glycol 4000 and cultured at room temperature for 10 min before observation. The percentage of pollen viability and pollen germination rate for each line was calculated using three independent biological samples, and at least 500 pollen grains for each sample were scored. For DAPI staining, pollen grains were fixed in an ethanol:acetic acid (3:1) solution, rehydrated, and then incubated in 1 µg/mL DAPI solution for 1 h at room temperature. Photography was performed using a Leica microscope (DM4500B) under UV light.
TEM for Pollen Ultrastructural Analyses
Anthers, sampled from wild-type, mads63 mutant, and MADS68 RNAi transgenic plants at developmental stage 13, were fixed with 3% glutaraldehyde and 2% osmic acid, dehydrated, and embedded in Spurr’s resin (Sigma-Aldrich). Then, ultrathin sections (50-nm thick) were mounted on a grid, stained with 4% uranylacetate, and observed under a transmission electron microscope (JEM-1230; JEOL).
Protoplast Transient Transfection Assays
To generate effector constructs, full-length sequences of MADS62, MADS63, and MADS68 were amplified and inserted into the p2GW7,0 vector (Karimi et al., 2002). About 2-kb (upstream of ATG) putative promoter regions of Os05g11790 and Os06g49860 were amplified and inserted into pUC-35sLUC (Tang et al., 2012) to produce the corresponding promoter-LUC reporter vectors. The primers used are listed in Supplemental Table 3 online.
Arabidopsis thaliana protoplasts were isolated and transfected as described (Yoo et al., 2007). The reporter and effector constructs as well as 35S:GUS vector (internal control) were cotransformed into protoplasts. After cotransformation, samples were incubated for 12 h in darkness and then the luciferase and GUS enzymatic assays were performed as previously described (Tang et al., 2012). The reporter gene expression levels were determined as the relative ratio of luciferase to GUS activity.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MADS62 (Os08g38590), MADS63 (Os06g11970), MADS68 (Os11g43740), UBIQUITIN (NM_001056014, Os03g13170), and ACTIN1 (AK100267, Os03g50890); tested predicted target genes (Os05g45370, Os02g02460, Os05g11790, Os06g49860, Os01g47050, Os10g26470, Os03g25460, Os03g27900, and Os12g37690) from rice; and SEP3 (NM_102272, AT1G24260) from Arabidopsis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GUS Staining Analyses of Florets and Pollen Grains in MADS62pro:GUS Transgenic Plants.
Supplemental Figure 2. Schematic Depiction of All Rice MIKC*-Type Protein Interactions as Revealed by Y2H Analyses.
Supplemental Figure 3. The Percentage of BiFC Fluorescence Signal-Emitting Protoplasts in Rice Protoplasts after Transient Transfection.
Supplemental Figure 4. EMSA Assay for Rice MIKC*-Type Heterodimeric Complexes Binding SRE-Type CArG-Box DNA.
Supplemental Figure 5. Competitive EMSA for MIKC*-Type Heterodimers and SEP3.
Supplemental Figure 6. The Identification and Microscopy Analyses of Pollen of MADS63 T-DNA Insertion Mutants.
Supplemental Figure 7. Schematic Diagram of RNAi Constructs and Identification of MADS68i/mads63 and MADS68i/mads63 Transgenic Plants.
Supplemental Figure 8. Delay and Arrest of Pollen Development Occurs from the Bicellular Stage.
Supplemental Figure 9. RT-PCR Analyses for a Selection of Nine in Silico Predicted Target Genes of the Rice MIKC*-Type Protein Complexes.
Supplemental Figure 10. qRT-PCR Analyses of the Expression of Putative Target Genes of the Rice MIKC*-Type Protein Complexes.
Supplemental Table 1. Detailed Description of MIKC*-Type Genes Used for Alignment and Phylogenetic Analyses.
Supplemental Table 2. Genetic Analyses of the mads63 Mutant.
Supplemental Table 3. Gene-Specific Primers Used in This Study.
Supplemental Table 4. EMSA Probes Used in This Study.
Supplemental Data Set 1. Text File of Alignment of Nucleotide Sequences of cDNAs of 58 MIKC*-Type Genes Used for the Phylogenetic Analyses Shown in Figure 1.
Supplemental Data Set 2. List of Rice Late Pollen-Enriched Genes with Their Expression Levels and Spatial Distribution of N10-Type CArG-Box Motifs in Their Putative Promoters.
Supplementary Material
Acknowledgments
We thank Lihuang Zhu for providing the pCam23A vector and Fengqin Dong for providing ultrathin sections and TEM observations. S.S. and G.T. thank Friedrich Schiller University Jena for general support. We thank three anonymous reviewers for their encouraging comments and helpful suggestions for improvement on a previous version of our article. This work was supported by grants from the National Nature Science Foundation of China (Grants 31270280 and 31100867) and the Ministry of Science and Technology of China (Grants 2011CB100405 and 2011ZX08009-004).
AUTHOR CONTRIBUTIONS
Y.L. and Z.M. designed the research. Y.L., S.C., S.S., and S.Y. performed the research. Y.L. and S.S. analyzed the data. X.L., F.W., and X.D. contributed reagents/materials/analysis tools. Y.L., Z.M., S.S., and G.T. wrote the article. F.W. and K.C. gave suggestions for the draft article.
Glossary
- SRE
serum response element
- qRT-PCR
quantitative RT-PCR
- GUS
β-glucuronidase
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- EMSA
electrophoretic mobility shift assay
- RNAi
RNA interference
- TEM
transmission electron microscopy
- DAPI
4′,6-diamidino-2-phenylindole
- CIPK
CBL-interacting protein kinase
References
- Adamczyk B.J., Fernandez D.E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., et al. (2003). Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133: 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., Xu Q., Chong K., Theissen G., Meng Z. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Datta R., Chamusco K.C., Chourey P.S. (2002). Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Angenent G.C. (2006). trans meets cis in MADS science. Trends Plant Sci. 11: 224–231 [DOI] [PubMed] [Google Scholar]
- Fan H.Y., Hu Y., Tudor M., Ma H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12: 999–1010 [DOI] [PubMed] [Google Scholar]
- Gramzow L., Barker E., Schulz C., Ambrose B., Ashton N., Theissen G., Litt A. (2012). Selaginella genome analysis - Entering the “homoplasy heaven” of the MADS world. Front. Plant Sci. 3: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L., Theissen G. (2010). A hitchhiker’s guide to the MADS world of plants. Genome Biol. 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini P.E., Thorstensen T., Alm V., Vizcay-Barrena G., Windju S.S., Jørstad T.S., Wilson Z.A., Aalen R.B. (2009). The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 4: e7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Han M.J., Jung K.H., Yi G., Lee D.Y., An G. (2006). Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. 47: 1457–1472 [DOI] [PubMed] [Google Scholar]
- Hayes T.E., Sengupta P., Cochran B.H. (1988). The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 2 (12B): 1713–1722 [DOI] [PubMed] [Google Scholar]
- Henschel K., Kofuji R., Hasebe M., Saedler H., Münster T., Theissen G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 19: 801–814 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D., Twell D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.Q., Wel Z.M., An H.L., Zhu Y.X. (2001). Agrobacterium tumefaciens-mediated transformation of rice with the spider insecticidal gene conferring resistance to leaffolder and striped stem borer. Cell Res. 11: 149–155 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kofuji R., Sumikawa N., Yamasaki M., Kondo K., Ueda K., Ito M., Hasebe M. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20: 1963–1977 [DOI] [PubMed] [Google Scholar]
- Kwantes M., Liebsch D., Verelst W. (2012). How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29: 293–302 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Adamczyk B.J., Fernandez D.E. (2005). Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol. Biol. 58: 89–107 [DOI] [PubMed] [Google Scholar]
- Ma H., Yanofsky M.F., Meyerowitz E.M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Mou Z., Wang X., Fu Z., Dai Y., Han C., Ouyang J., Bao F., Hu Y., Li J. (2002). Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Kim J., Lee S., An G., Ma H., Nei M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B.J. (1997). GeneDoc: A tool for editing and annotating multiple sequence alignments. EMBNEW. NEWS 4: 14 [Google Scholar]
- Pollock R., Treisman R. (1991). Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 5 (12A): 2327–2341 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378: 1079–1101 [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese M., Faigl W., Quodt V., Verelst W., Matthes A., Saedler H., Münster T. (2005). Isolation and characterization of new MIKC*-type MADS-box genes from the moss Physcomitrella patens. Plant Biol. (Stuttg.) 7: 307–314 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G., Angenent G.C., Kaufmann K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. (2008a). Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Tang H., Wang X., Bowers J.E., Ming R., Alam M., Paterson A.H. (2008b). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed]
- Verelst W., Saedler H., Münster T. (2007a). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W., Twell D., de Folter S., Immink R., Saedler H., Münster T. (2007b). MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 8: R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron A.S., Kaufmann K., Bornberg-Bauer E. (2007). Evidence of interaction network evolution by whole-genome duplications: a case study in MADS-box proteins. Mol. Biol. Evol. 24: 670–678 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wei L.Q., Xu W.Y., Deng Z.Y., Su Z., Xue Y., Wang T. (2010). Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L.Y., Chase C.D. (1999). Mitochondrial gene expression in developing male gametophytes of male-fertile and S male-sterile maize. Sex. Plant Reprod. 11: 323–330 [Google Scholar]
- Wikström N., Savolainen V., Chase M.W. (2001). Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 268: 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Huang X., Cheng J., Li Z., de Folter S., Huang Z., Jiang X., Pang H., Tao S. (2011). Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Mol. Biol. Evol. 28: 501–511 [DOI] [PubMed] [Google Scholar]
- Yang W., Kong Z., Omo-Ikerodah E., Xu W., Li Q., Xue Y. (2008). Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J. Genet. Genomics 35: 531–543, S1–S2 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang D., Wilson Z. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 2342–2353 [Google Scholar]
- Zhang Y., Shewry P.R., Jones H., Barcelo P., Lazzeri P.A., Halford N.G. (2001). Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28: 431–441 [DOI] [PubMed] [Google Scholar]
- Zobell O., Faigl W., Saedler H., Münster T. (2010). MIKC* MADS-box proteins: Conserved regulators of the gametophytic generation of land plants. Mol. Biol. Evol. 27: 1201–1211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.