This article provides evidence of an interaction between two types of Glu receptor proteins that allow entry of calcium ions into the cytoplasm when activated by an amino acid. They reside in the phloem of Arabidopsis seedling roots, and from there they somehow affect the initial stages of the root branching process.
Abstract
Molecular, genetic, and electrophysiological evidence indicates that at least one of the plant Glu receptor-like molecules, GLR3.4, functions as an amino acid–gated Ca2+ channel at the plasma membrane. The aspect of plant physiology, growth, or development to which GLR3.4 contributes is an open question. Protein localization studies performed here provide important information. In roots, GLR3.4 and the related GLR3.2 protein were present primarily in the phloem, especially in the vicinity of the sieve plates. GLR3.3 was expressed in most cells of the growing primary root but was not enriched in the phloem, including the sieve plate area. GLR3.2 and GLR3.4 physically interacted with each other better than with themselves as evidenced by a biophotonic assay performed in human embryonic kidney cells and Nicotiana benthamiana leaf cells. GLR3.3 interacted poorly with itself or the other two GLRs. Mutations in GLR3.2, GLR3.4, or GLR3.2 and GLR3.4 caused the same and equally severe phenotype, namely, a large overproduction and aberrant placement of lateral root primordia. Loss of GLR3.3 did not affect lateral root primordia. These results support the hypothesis that apoplastic amino acids acting through heteromeric GLR3.2/GLR3.4 channels affect lateral root development via Ca2+ signaling in the phloem.
INTRODUCTION
In the years since GLUTAMATE RECEPTOR-LIKE (GLR) genes were discovered in plants (Lam et al., 1998; Lacombe et al., 2001), research has focused on their phylogeny and evolution (Chiu et al., 1999, 2002; Turano et al., 2001), expression patterns and transcriptional responses (Meyerhoff et al., 2005; Roy et al., 2008), roles in carbon:nitrogen balance (Kang and Turano, 2003), abscisic acid sensing (Kang et al., 2004), and contributions to ionic relations, including Ca2+ signaling (Kim et al., 2001; Qi et al., 2006; Stephens et al., 2008; Cho et al., 2009). The last category relates closely to the presumed molecular function of GLRs because the homologous ionotropic Glu receptors (iGluRs) in animals combine as heterotetramers to form amino acid–gated ion channels with varying permeability to Na+, K+, and Ca2+ (Traynelis et al., 2010). That plant GLRs encode similar ion transporting functions was a logical initial hypothesis, one that was suggested to account for the prevalence of nonselective cation currents across plant cell membranes (Stoeckel and Takeda, 1989; Demidchik et al., 2002, 2004; White et al., 2002) and promised to add some much-needed molecular detail to the mechanism of how cytoplasmic Ca2+ signals are generated (Sanders et al., 2002; Kudla et al., 2010). The case was strengthened by the finding that the large, transient membrane depolarization and rise in cytoplasmic Ca2+ triggered by a defined set of amino acids in wild-type plants depended on GLR genes (Dennison and Spalding, 2000; Qi et al., 2006; Stephens et al., 2008). In another study, the engineering of a chimeric protein in which the pore region of a GLR was transplanted into a mammalian GLR could form a Na+, K+, and Ca+2 conductance when placed in the context of an iGluR protein (Tapken and Hollmann, 2008). These studies genetically linked ionic events, including Ca2+ transport to GLRs, but fell short of demonstrating GLRs to be bona fide Ca2+-permeable channels (Spalding and Harper, 2011). Recently, the evidence of Ca2+-permeable channel activity for GLRs became much more direct. Vincill et al. (2012) transfected human embryonic kidney (HEK) cells with GLR3.4 cDNA. Whole-cell patch clamp analysis showed robust ionic current responses to amino acids in cells transfected with GLR3.4 and green fluorescent protein (GFP) but not in those expressing GFP alone. The GLR3.4 channel functioning in HEK cells was activated by the same three amino acids (Asn, Ser, and Gly) that glr3.4 mutant plants responded poorly to, compared with the wild type, in electrophysiological assays of membrane potential (Stephens et al., 2008). Current voltage analysis of Asn-gated GLR3.4 channels in HEK cells showed that Ca2+ was the predominantly transported ion, being ∼104-fold more permeable than Na+ in the conditions employed (Vincill et al., 2012).
More information about the functions of GLRs at the organismal level in plant physiology, growth, or development is needed to build on the molecular-level functions that have now been established. Phenotypes resulting from mutation or altered expression of GLRs have provided disparate information. Pollen tube growth in glr1.2 mutants (Michard et al., 2011) and root gravitropism in glr3.3 mutants (Miller et al., 2010) are affected in ways that may relate to impaired amino acid–gated Ca2+ signaling. Antisense expression of GLR1.1 affected responses to abscisic acid in ways that may be related to Ca2+ signaling known to occur downstream of this hormone (Kang et al., 2004). Overexpression of GLR3.1 affected stomatal closing behavior without affecting cytoplasmic Ca2+ oscillations (Cho et al., 2009), and overexpression of GLR3.2 had a complicated set of effects on whole-plant ion relations (Kim et al., 2001). These studies undoubtedly provide clues about the biological function of GLRs, but from them it is not yet possible to generate general statements or compelling hypotheses. This work addresses this deficiency with new information about in vivo protein localization, physical and genetic interactions, and developmental phenotypes to provide insight into the biological roles of amino acid–gated Ca2+ channels.
RESULTS
Protein Localization
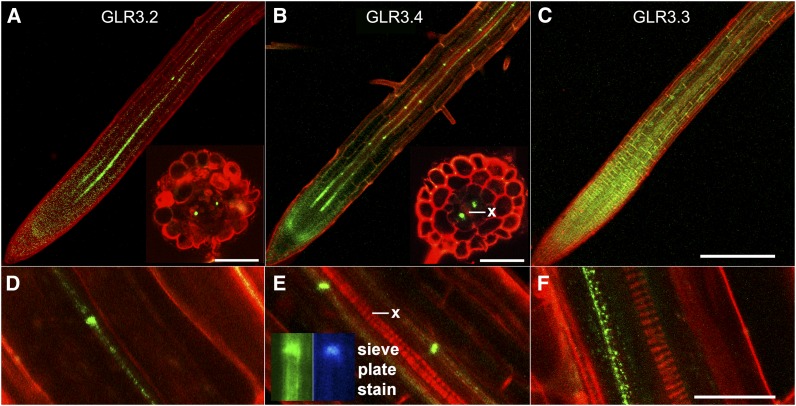
According to promoter strength and mRNA analyses, at least five of the seven genes comprising the clade to which GLR3.4 belongs are expressed at much higher levels in roots than shoots (Chiu et al., 2002; Roy et al., 2008). This localization information was refined by visualizing the localizations of GLR3.2, GLR3.3, and GLR3.4 fused to GFP, each fusion being expressed in the respective glr knockout mutant and under the control of its native promoter. Confocal microscopy confirmed expression of GLR3.2, GLR3.3, and GLR3.4 protein in roots (Figures 1A to 1C). Despite the low fluorescence signal originating from the center of the root, specific expression patterns could be resolved. They compared very favorably with published tissue-specific transcriptome data. The Arabidopsis thaliana eFP browser tool (Winter et al., 2007) indicated GLR3.4 mRNA to be 58-fold more abundant in the phloem than any other tissue or cell type in roots. A transcriptome analysis that distinguished different developmental stages of phloem showed GLR3.4 mRNA to be eightfold higher in the protophloem near the root apex than in mature phloem (Brady et al., 2007). These mRNA patterns closely matched the GLR3.4 protein patterns visualized by confocal microscopy (phloem enrichment and peak expression near the root apex in the protoderm or protophloem). The images shown were difficult to obtain because signal intensity was not high (Ca2+ channels regulating cytoplasmic signals in the micromolar range are not expected to be abundant proteins) and because the majority of expression was restricted to one tissue comprising a minor fraction of the deepest region of the root. Nonetheless, the live-cell visualization results contained an additional, potentially important result. Special concentrations occurred in spots spaced ∼100 µm apart along the mature phloem cell files, readily apparent in the GLR3.4 example shown (Figure 1E). When hand-cut root cross sections contained these special locations of high signal intensity, the spots were observed at positions consistent with phloem (insets in Figures 1A and 1B), and they overlapped with locales of aniline blue staining (Figure 1E, inset), considered diagnostic of the callose that accumulates at specialized cell–cell contact points in sieve tube members known as sieve plates (Knoblauch and Oparka, 2012).
Figure 1.
Localization of GLR3.2, GLR3.3, and GLR3.4 in Roots.
(A) to (C) Confocal microscope images of GFP-tagged GLR3.2, GLR3.4, or GLR3.3 (green) in primary root apices stained with propidium iodide (red) to mark cell boundaries. Insets in (A) and (B) show phloem-localized signal in cross sections cut by hand ∼1 mm from the root tip.
(D) to (F) Higher magnification images of GFP-tagged GLR3.2, GLR3.4, or GLR3.3 signal in mature phloem. Inset in (E) shows that aniline blue, a sieve plate indicator, stains a region of strong GLR3.4-GFP accumulation in the phloem.
Bar in (C) = 200 µm and applies to the main images in (A) to (C); inset bars = 50 µm. Bar in (F) is 20 µm and applies to (D) to (F). x, xylem.
Role in Lateral Root Production
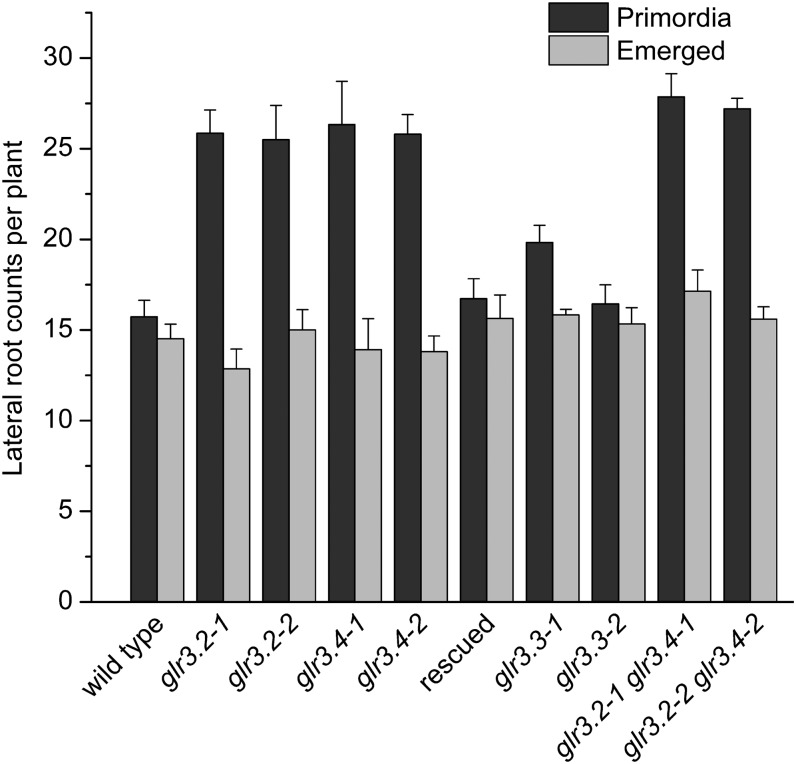
The next clue about GLR3.4 function in roots came from a machine-learning analysis of transcriptome patterns. GLR3.4 was among the genes predicted to play a role in lateral root initiation (Moreno-Risueno et al., 2010). Microscopy inspection of glr3.4 mutants showed they produced twice as many lateral root primordia as the wild type, though the number of emerged lateral roots, and therefore the overt root system architecture, was not affected (Figure 2A). The GFP-tagged GLR3.4 used to visualize the expression pattern rescued this phenotype (Figure 3). The related GLR3.2 gene was included in the same transcript behavior cluster as GLR3.4 (Moreno-Risueno et al., 2010). The Arabidopsis eFP browser tool showed GLR3.2 mRNA to be 418-fold more abundant in the phloem of roots than in the adjacent tissues and cell types and 20-fold enriched in the protophloem compared with mature phloem (Brady et al., 2007; Winter et al., 2007), again consistent with the observed protein localization (Figure 1A). Like GLR3.4, GLR3.2 accumulated at sieve plates (Figure 1D). In agreement with this correlated expression, glr3.2 mutants also hyperproduced lateral root primordia (Figures 2 and 3). The related GLR3.3 should be expressed generally in the root apex according to the Brady et al. (2007) expression atlas. Indeed, the protein was evident in all cell types of the growth zone (Figure 1C), consistent with the root gravitropism phenotype determined by computational image analysis (Miller et al., 2010). GLR3.3 displayed some tendency to accumulate in the phloem in the mature region of the root but it did not accumulate at sieve plates (Figure 1F), and it was not predicted to play a role in lateral root initiation on the basis of its spatiotemporal mRNA patterns (Moreno-Risueno et al., 2010). Accordingly, two independent glr3.3 alleles did not differ from the wild type with respect to production of lateral root primordia (Figure 3).
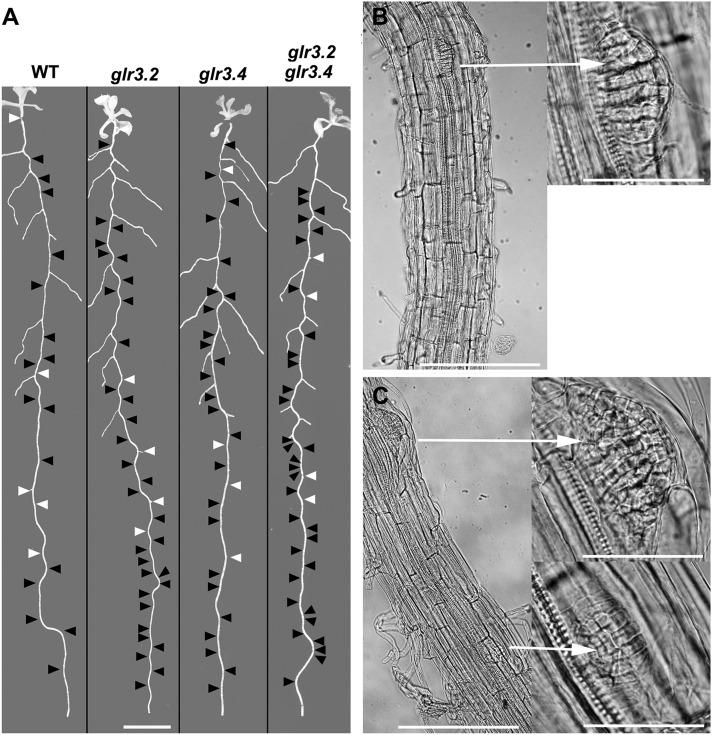
Figure 2.
Spatial Distributions of Lateral Root Primordia and Emerged Lateral Roots in the Wild Type and glr Mutants.
(A) Black arrowheads mark the position of primordia detected by microscopy inspection of the displayed root, and white arrowheads indicate emerged roots not visible in the image. Bar = 8 mm. WT, the wild type.
(B) Segments of primary root showing one lateral root primordium in the wild type and at higher magnification (arrow right).
(C) Two primordia in an equivalent section of a glr3.4 root showing adjacent primordia emerging from the same side of the stele (arrow right).
Bars =200 μm (B) and (C) and 50 μm in the higher magnification images (arrow right).
Figure 3.
Counts of Lateral Root Primordia or Emerged Lateral Roots in Wild-Type and glr Mutant Plants.
The results labeled “rescued” were obtained with glr3.4 mutant plants expressing the ProGLR3.4:GLR3.4-EGFP construct used for visualizing the protein as shown in Figure 1. The values are means ± se (n ≥ 6).
Each primordium and lateral root counted in Figure 3 was also classified as early (stages ii to iv), late (stages v to vii), emerged, or elongated. The results presented in Table 1 show that the glr3.2 and glr3.4 phenotypes were due mostly to extra (∼1.7-fold more) early-stage root primordia, but the higher rate of initiation was offset by a higher rate of subsequent arrest.
Table 1. Developmental Classification of Lateral Roots and Lateral Root Primordia.
| Genotype | Stage ii to iv | Ratio Mutant:Wild Type | Stage v-vii | ratio Mutant:Wild Type | Emerged | Ratio Mutant:Wild Type | Elongated | Ratio Mutant:Wild Type | n |
|---|---|---|---|---|---|---|---|---|---|
| The wild type | 13 ± 1 | NA | 3 ± 1 | NA | 4 ± 0.5 | NA | 10 ± 1 | NA | 19 |
| glr3.4-1 | 22 ± 2 | 1.7 | 5 ± 1 | 1.7 | 4 ± 1 | 1.0 | 10 ± 1 | 1.0 | 12 |
| glr3.4-2 | 23 ± 1 | 1.8 | 3 ± 1 | 1.0 | 4 ± 0.7 | 1.0 | 10 ± 1 | 1.0 | 20 |
| rescued | 13 ± 1 | 1.0 | 3 ± 1 | 1.0 | 6 ± 1 | 1.5 | 9 ± 1 | 0.9 | 11 |
| glr3.2-1 | 22 ± 1 | 1.7 | 4 ± 1 | 1.3 | 4 ± 0.4 | 1.0 | 9 ± 1 | 0.9 | 14 |
| glr3.2-2 | 21 ± 2 | 1.6 | 5 ± 1 | 1.7 | 4 ± 0.6 | 1.0 | 11 ± 1 | 1.1 | 6 |
| glr3.3-1 | 17 ± 1 | 1.3 | 3 ± 1 | 1.0 | 5 ± 1 | 1.2 | 11 ± 1 | 1.1 | 6 |
| glr3.3-2 | 14 ± 1 | 1.1 | 3 ± 1 | 1.0 | 5 ± 0.4 | 1.2 | 11 ± 1 | 1.1 | 9 |
| glr3.2-1 glr2.4-1 | 21 ± 3 | 1.6 | 7 ± 3 | 2.3 | 3 ± 1 | 0.7 | 14 ± 3 | 1.4 | 7 |
| glr3.2-1 glr2.4-1 | 20 ± 4 | 1.5 | 7 ± 4 | 2.3 | 3 ± 1 | 0.65 | 13 ± 2 | 1.3 | 5 |
Each root or primordium was classified according to developmental stage (stage ii-iv, stage v-vii, emerged, or elongated) and enumerated for each genotype. The ratio column shows how the different genotypes compare to the wild type for each stage. The “rescued” genotype refers to a glr3.4 mutant transformed with GFP-GLR3.4 controlled by the GLR3.4 promoter. NA, not applicable.
In addition, glr3.2 and glr3.4 mutations affected primordium positioning. The wild type typically initiated successive primordia on alternate sides of the root but the mutants typically did not. The location of each detected primordium was mapped onto a macroscopic image of the same root as shown by the black arrowheads in Figure 2A. White arrowheads indicate emerged lateral roots that are not visible in the macroscopic image. This display method shows the much higher density of lateral root primordia in representative single and double mutant roots relative to the wild type as well as the high incidence of successive primordia produced on the same side of the root. Light micrographs show wild-type and mutant primordia to be anatomically similar (Figures 2B and 2C). Also shown is an example of successive primordia located on the same side of a glr3.4 root (Figure 2C).
Genetic Evidence of Interaction between GLR3.2 and GLR3.4
The strong similarities in GLR3.2 and GLR3.4 localization patterns, and their similar knockout phenotypes, raised the possibility that these two receptors could function together, possibly as subunits of the same tetrameric channel. If so, the double mutant phenotype would be expected to display a phenotype similar to either single mutant rather than an additive defect. Microscopy inspection of glr3.2 glr3.4 double mutants produced data in support of this GLR3.2/GLR3.4 heteromeric hypothesis. The double mutant hyperproduced lateral root primordia indistinguishably from the glr3.2 and glr3.4 single mutants (Figures 2 and 3). Loss of one subunit from the presumed heterotetramer produced the same phenotype as loss of the other; loss of both was no worse. These genetic results are consistent with GLR3.2 and GLR3.4 subunits forming a heteromeric channel in the phloem that functions in the production of lateral root primordia.
Molecular Evidence of Interaction between GLR3.2 and GLR3.4
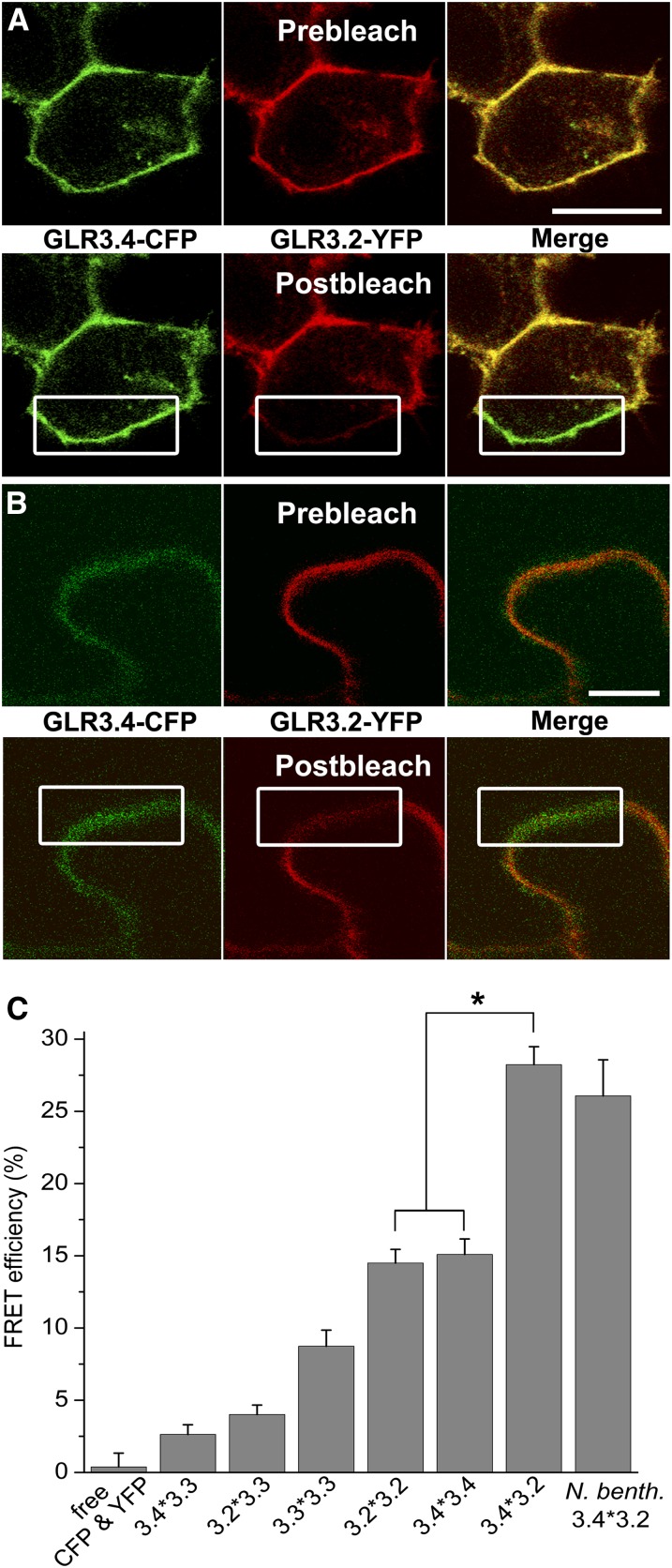
A phenomenon known as Förster resonance energy transfer (FRET) can occur between excited cyan fluorescent protein (CFP; donor) and yellow fluorescent protein (YFP; acceptor) if the two are not more than 10 nm apart, a distance consistent with a physical interaction (Truong and Ikura, 2001; Piston and Kremers, 2007). CFP or YFP was fused to the C terminus of GLR3.2, GLR3.3, or GLR3.4 and then expressed in HEK cells, where fluorescence was observed at the cell periphery with a confocal laser scanning microscope (Figure 4). To test for FRET between one GLR fused to CFP and the same or a different GLR fused to YFP, fluorescence from the donor within a region of interest including the plasma membrane was measured before and after photobleaching of the acceptor in coexpressing cells (Figure 5A). The relative increase in CFP fluorescence after photobleaching was quantified to determine the FRET efficiency values shown in Figure 5C. FRET did not occur between free CFP and free YFP, as expected. Substantial amounts of FRET occurred between identical GLRs, as expected at least for GLR3.4 given the previously published evidence of channel activity from GLR3.4 homomers (Vincill et al., 2012). GLR3.3 showed very little or no FRET with GLR3.2 or GLR3.4. By contrast, FRET efficiency between GLR3.2 and GLR3.4 was high, ∼25%, similar to that observed between aquaporin subunits that interact to form water channels (Zelazny et al., 2007). The GLR3.2/GLR3.4 signal was greater than either homomeric signal to a degree that is highly significant (P < 0.001), indicating a preference for heteromerization.
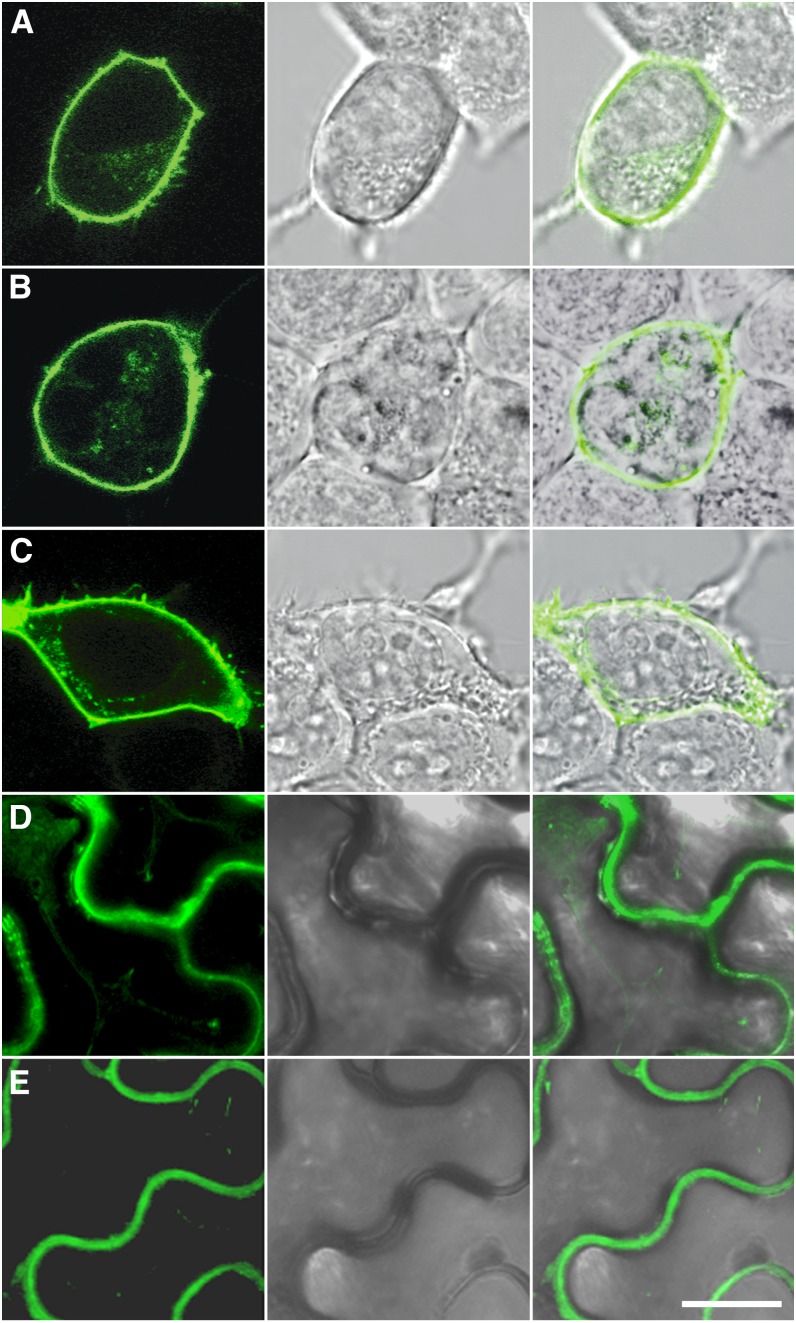
Figure 4.
Subcellular Localization of GLR3.2, GLR3.3, and GLR3.4 in Animal and Plant Cells.
Confocal laser scanning microscope images of HEK293T cells expressing GLR3.2-YFP (A), GLR3.3-YFP (B), and GLR3.4-YFP (C) or N. benthamiana leaf epidermal cells expressing GLR3.2-YFP (D) and GLR3.4-YFP (E) show YFP fluorescence at the plasma membrane. Middle panels show bright-field light micrographs corresponding to the confocal images. Rightmost panels show overlay of the florescence signal with the light micrograph. Bar = 10 μm.
Figure 5.
Close Interaction between GLRs Evidenced by FRET in Animal and Plant Cells.
(A) and (B) GLR3.4-CFP fluorescence shown in green and GLR3.2-YFP fluorescence shown in red were acquired simultaneously in transfected HEK cells (A) or N. benthamiana leaves (B). The percentage of increase in CFP fluorescence after photobleaching YFP (FRET efficiency) was determined in a region including plasma membrane (white rectangle) in HEK (A) or N. benthamiana (B) cells to test for spacing small enough to allow FRET between GLR subunits. Bars = 10 µm.
(C) FRET efficiencies quantified for different pairs of GLRs (denoted as x*y) in acceptor photobleaching experiments performed as shown in (A) and (B). N. benth, N. benthamiana. Number of replicates for each FRET pair represented: free CFP and YFP (n = 6), 3.4*3.3 (n = 28), 3.2*3.3 (n = 30), 3.3*3.3 (n = 35), 3.2*3.2 (n = 33), 3.4*3.4 (n = 22), 3.4*3.2 (n = 29), and N. benth 3.4*3.2 (n = 8).
Transient expression of GLR3.2 and GLR3.4 in tobacco (Nicotiana benthamiana) leaf cells produced essentially the same degree of FRET as observed in HEK cells (Figures 5B and 5C), greatly reducing concerns about physiological relevance that sometimes accompany results obtained with heterologous systems. The fact that two subunits displaying a high FRET efficiency (Figure 5C) show a similar subcellular localization pattern in phloem (Figure 1), along with their equivalent and nonadditive mutant phenotypes, leads to the conclusion that GLR3.2 and GLR3.4 are components of heteromeric amino acid–gated channels in the phloem.
Channel Activity
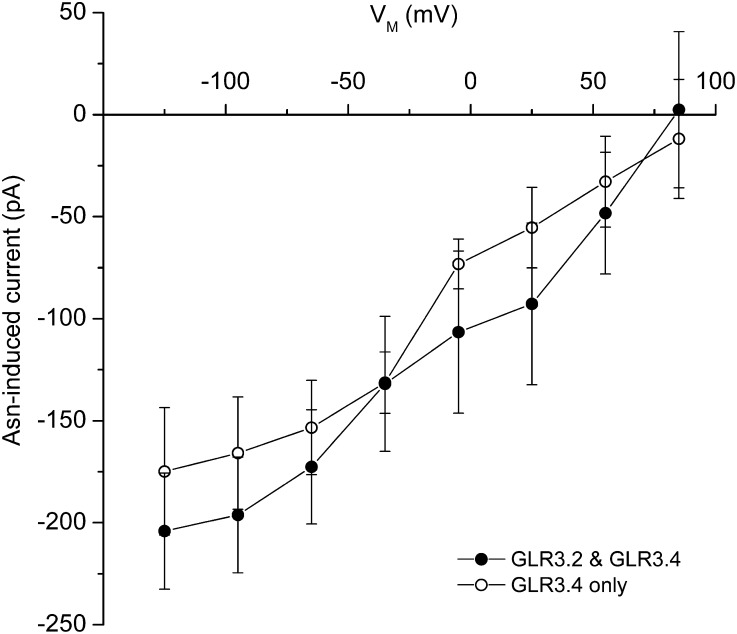
Patch clamp electrophysiology assays for GLR3.2 channel function in transfected HEK cells did not produce positive results. GLR3.2 homomers may not form functional channels in the HEK cell due to the lack of some important factor, or the amino acids tested (Asn and Gly) are not effective agonists for GLR3.2. Lack of information about homomeric GLR3.2 channel activity left open the question of what activity to expect from GLR3.2/GLR3.4 heteromeric channels predicted to form, based on the FRET results in Figure 5, when coexpressed in HEK cells. Current voltage (I-V) analysis of Asn-gated currents in GLR3.2/GLR3.4-coexpressing cells were performed with the patch clamp technique (Figure 6) as previously described (Vincill et al., 2012). The expectation was that any such currents would reflect the activity of GLR3.2/GLR3.4 heteromers because the FRET studies and genetics indicated that these two subunits interacted. The slopes of the difference I-V curves and their zero-current voltages (reversal potentials), which indicate large selectivity for Ca2+ of the Asn-gated current, were essentially the same as those previously obtained for GLR3.4 alone (Figure 6). The small differences between the two curves could be due to different levels of expression; heteromerization with GLR3.2 did not have a large effect on the Asn-induced activity of GLR3.4 in these conditions. However, differences in properties not yet tested, such as a comprehensive agonist profile or kinetic properties, remain possible. Another possible explanation of why coexpressing GLR3.2 with GLR3.4 did not create a substantially new activity is that the only functional channels produced in the coexpressing cells were GLR3.4 homomers formed from subunits that failed to partner with GLR3.2 subunits.
Figure 6.
Current Voltage Analysis of Asn-Activated GLR3.2/GLR3.4 Channels Expressed in HEK Cells Compared with Asn-Activated GLR3.4-Only Channels.
Steady state currents after treatment with 1 mM Asn minus currents recorded before Asn are plotted as a function of membrane potential. The similar results obtained with GLR3.4 alone (dashed line) are replotted from Vincill et al. (2012). The conditions of this and previous experiments were identical except this one used 1 mM extracellular CaCl2 and the previous used 2 mM extracellular CaCl2. The plotted data are means ± se (n ≥ 3).
An interesting possibility not supported by the coexpression experiment is that the phenomenon of GLR desensitization so apparent in planta (Stephens et al., 2008) but apparently absent in GLR3.4-expressing HEK cells is an emergent property of native heteromers. Asn-activated GLR3.2/GLR3.4 currents were sustained, maintaining steady state during agonist presentation as in cells expressing only GLR3.4. Otherwise, the analysis shown in Figure 6 would not have been possible because the currents in Figure 6 are the difference between steady state currents recorded before and after Asn. Some factor in the plant is apparently required for desensitization, possibly a third GLR to join GLR3.2 and GLR3.4 in the tetramer.
DISCUSSION
The results presented in this article establish a role for phloem-localized GLR3.2/GLR3.4 heteromeric, amino acid–gated Ca2+ channels in regulating the production of lateral root primordia. The consequence of mutating this control mechanism is ectopic hyperproduction of lateral root primordia. These results could be interpreted to mean that GLR3.2/GLR3.4 is a negative regulator, restricting primordia numbers and position along the root axis by a signaling process originating in the phloem. A less direct but plausible interpretation is that GLR3.2/GLR3.4 channels act downstream of initiation to promote lateral root emergence; hyperproduction of primordia in the mutants would, in this scenario, compensate for the lower percentage that successfully develop into roots, a feedback explanation similar to that invoked to explain the phenotype of lax3 mutants (Swarup et al., 2008). However, abcb19 (formerly mdr1) mutants are defective in lateral root elongation due to an auxin transport deficiency and do not display a compensatory increase in primordia production (Wu et al., 2007). If GLR3.2/GLR3.4 were necessary for the proper growth of primordia after their production and the hyperproduction of primordia in the mutants served to compensate for low rates of emergence, expression of the channels in the expanding primordia would be expected. However, this was not observed. In any case, an intriguing possibility is that GLR3.2/GLR3.4 informs the root branching mechanism via Ca2+ signaling about the apoplastic concentration of amino acids in the adjacent xylem (Lalonde et al., 2004), which is expected to contain activating amounts of all GLR3.4 agonists, especially Asn (Lea et al., 2007).
If GLR-mediated Ca2+ signals are generated in the phloem as they are in other cell types (Meyerhoff et al., 2005; Qi et al., 2006; Michard et al., 2011; Vincill et al., 2012), which is a potentially testable question given improved fluorescent Ca2+ reporters (Swanson et al., 2011), and if those Ca2+ signals affect phloem transport rates by altering sieve plate properties (Knoblauch et al., 2001; Furch et al., 2009), novel mechanisms for appropriately balancing carbon and nitrogen levels at root sinks can be envisioned. Carbohydrate transport rates could be adjusted by GLR3.2/GLR3.4-mediated Ca2+ signaling according to nitrogen status, which is known to affect lateral root production (Tester, 1990; Forde and Walch-Liu, 2009). It may be significant that Asn, a primary agonist of GLR3.4 (Stephens et al., 2008; Vincill et al., 2012), is also a major carrier of nitrogen and indicator of plant carbon:nitrogen status (Lea et al., 2007). Conceptually different from a role in modifying phloem sap transport rates by affecting conductance through the sieve plate is a role for GLR3.2/GLR3.4 in passing systemic signals through the phloem (Thompson and Holbrook, 2004), perhaps even an ionic or electrical signal (Fromm and Lautner, 2007; Furch et al., 2009), that ultimately affects the production of lateral root primordia. The role for Ca2+ in this scenario is consistent with the finding that primordium production induced by mechanical bending of the primary root requires a Ca2+ signaling step (Richter et al., 2009). Future work may find a connection between GLR signaling and a recently discovered phloem protein of unknown function that affects lateral root production (Ingram et al., 2011).
A role for GLR1.1 in regulating carbon:nitrogen ratios was suggested in one of the earliest publications on this gene family, which provided immunohistochemical evidence of phloem localization in the Arabidopsis inflorescence stem (Kang and Turano, 2003). Therefore, the hypothesis that GLRs form a signaling bridge between xylem and phloem can be widened to more than the root system and merits further investigation.
Expression patterns provided important clues that led to the identification of GLR3.2 and GLR3.4 as potentially interacting subunits, but expression patterns alone probably do not determine the subunit composition of the tetrameric channels in planta. Instead, structural features of the proteins influence the strength of interactions between subunits to determine functional channel compositions. The results of the FRET studies (Figure 5) indicate that interaction between GLR3.2, GLR3.3, and GLR3.4 may not be equal despite a close phylogenetic relationship (Chiu et al., 2002). The strongest FRET signal was obtained between GLR3.2 and GLR3.4, while very little was detected between GLR3.3 and GLR3.4. Such results are often interpreted as a measure of interaction strength, but a weak or absent FRET signal can also occur despite a bona fide interaction between the proteins if the fluorophores are for some structural reason prevented from achieving the close separation necessary for the energy transfer. Future structural studies may identify the amino acid differences that account for the lack of potential for interaction displayed by GLR3.3 with the other subunits, which was an unexpected result given the electrophysiological evidence that GLR3.3 was a subunit of all GLR heteromers (Stephens et al., 2008). It may be that GLR3.3 interacts well with GLR3.2/GLR3.4 pairs better than with itself or either single molecular species. No new channel properties were obvious in cells expected to form GLR3.2/GLR3.4 heteromers compared with GLR3.4 homomers (Figure 6), but perhaps incorporation of a third or fourth species in the tetramer creates the type of functional diversity attributable to heteromerization in animal iGluRs (Monyer et al., 1992; Furukawa et al., 2005; Ferre et al., 2007; Gielen et al., 2009; Karakas et al., 2011; Kumar et al., 2011).
While much about GLR functions in plants remains to be elucidated, it now seems clear that heteromeric, amino acid–gated, Ca2+-permeable channels in the phloem play a regulatory role in the development of the root system. Future work should focus on elucidating the mechanism that creates this role. Useful would be a transmission electron microscopy study of GLR3.2-GFP or GLR3.4-GFP root samples decorated with gold-labeled anti-GFP antibodies designed to determine where the protein is localized with respect to sieve plate structures, companion cells, plasmodesmata, or adjacent xylem. Localizing the proteins with light microscopy, despite the technical problems encountered with a weak signal restricted to the phloem in the center of the root, generated ideas useful to this study, and so increasing the resolution ∼100-fold with electron microscopy may stimulate the next generation of mechanistic hypotheses. Examining different positions along the root axis (i.e., different stages of phloem development) may address the interesting question of how a membrane protein becomes concentrated at the ends of mature phloem cells over time. Precise localization of the GLR molecules and live-cell imaging of Ca2+ signals in response to amino acids using new optical probes controlled by tissue-specific promoters may provide critical clues about how phloem channels influence developmental decisions in the pericycle (Swanson et al., 2011).
METHODS
Plant Growth and Root Primordia Analysis
Seeds of Arabidopsis thaliana were surface sterilized and sown on Petri plates containing B5 medium diluted 50-fold and supplemented with 0.5% Suc (w/v) and 23 mM MES, adjusted to pH 5.7 with NaOH, and gelled with 1% agarose. The plates with seeds were maintained at 4°C for 2 to 4 d before being placed vertically in a growth chamber under constant light. After 4 d of growth, seedlings were transferred to 150 × 15-mm Petri plates containing the same medium and spaced such that their lateral roots would not overlap during an additional 8 d of growth under constant light. The primordia measurements were performed on the resulting 12-d-old plants as described previously (Zhang and Forde, 1998; Wu et al., 2007).
DNA Cloning
Full-length GLR3.2, GLR3.3, and GLR3.4 cDNA was isolated from total RNA by RT-PCR as previously described (Vincill et al., 2012) and amplified using AccuPrime Pfx DNA polymerase (Invitrogen) using the following amplification primers: GLR3.2, forward, 5′-CACCATGTTTTGGGTTTTGGTTCTGTTGAGC-3′, and reverse, 5′-TATTGGTCTAGAAGGCTTTAAAGAAAGATCATCG-3′; GLR3.3, forward, 5′-CACCATGAAGCAACTCTGGACTTTCTTC-3′, and reverse, 5′-GTCTAATGGATTTACCGAATTGAAGCTTC-3′; GLR3.4, forward, 5′-CACCATGGGATTTTTGGTGATGATAAGAGAAGTTTC-3′, and reverse, 5′-AGTAATTTCGCCATGTTGTGATTG-3′. The Gateway directional cloning modification (CACC) in italics was added at the 5′ end of the forward primers. The resulting PCR fragment was cloned into the pENTR-D entry vector (Invitrogen). For FRET experiments in Nicotiana benthamiana, the pENTR-D vectors containing GLR3.2 and GLR3.4 cDNA were shuttled into the Gateway destination vectors pEARLEYGATE 101(CFP) and pEARLEYGATE 102(YFP), which fuse the indicated fluorescent tag to the C terminus of the translated gene product (Earley et al., 2006) to generate Pro35S:GLR3.2-YFP and Pro35S:GLR3.4-CFP constructs. To generate the GLR native promoter constructs for plant expression, the GLR3.2, GLR3.3, and GLR3.4 full-length cDNAs in the respective pENTR-D entry vectors were shuttled into the modified pEARLEYGATE 102 plant expression vector in which the 35S promoter was replaced with the genomic DNA sequence upstream from the transcriptional start site (native promoter) of GLR (2006 bp for GLR3.2, 3500 bp for GLR3.3, and 1902 bp for GL3.4). The fluorescent CFP fragment was replaced with enhanced GFP for enhanced fluorescence to produce the respective ProGLR3.2:GLR3.2-EGFP, ProGLR3.3:GLR3.3-EGFP, and ProGLR3.4:GLR3.4-EGFP constructs. The tobacco leaf infiltration method was used to transiently express the Pro35S:GLR3.2-YFP and Pro35S:GLR3.4-CFP constructs (Sparkes et al., 2006). The floral dip method (Clough and Bent, 1998) was used to transform the null mutant background lines with these constructs.
To generate GLR receptor subunits fused to either CFP or YFP, the pENTR-D vectors described above containing the respective full-length GLR3.2, GLR3.3, or GLR3.4 cDNAs were shuttled into the pDS_EF1-XB-CFP and pDS_EF1-XB-YFP mammalian expression vectors from American Type Culture Collection using the Gateway recombination reaction (Invitrogen) to generate a library of vectors that C-terminally tag the translated GLR3.2, GLR3.3, and GLR3.4 receptor subunits with either CFP or YFP (see Supplemental Table 1 online). All constructs were confirmed by DNA sequencing.
HEK Cell Culture and Transfection
HEK293T cells from the American Type Culture Collection were cultured in Dulbecco’s modified Eagle’s medium-GlutaMAX (Invitrogen) with 10% fetal bovine serum, 100 IU mL−1 penicillin, and 100 µg mL−1 streptomycin in a 37°C incubator with 95% air and 5% CO2. Trypsin-treated HEK293T cells (5 × 105 cells per well) were plated into six-well tissue culture plates containing collagen-coated glass cover slips 8 to 12 h before being transfected with 1 µg of the indicated plasmid DNA using FuGENE 6 transfection reagent (Roche Scientific) following the manufacturer’s protocol. In the case of cotransfections, a plasmid ratio of 1:1 (0.5 µg + 0.5 µg) was used. Vectors employed are listed in Supplemental Table 1 online. All experiments were performed 12 to 48 h after transfection and imaged live at room temperature in Hank’s balanced buffer solution without phenol (Invitrogen).
FRET
A Zeiss LSM 510 Meta confocal imaging system with a 30-mW argon laser and a ×63 1.4–numerical aperture oil immersion Plan-Apochromat objective was used to visualize live HEK392T cells or N. benthamiana epidermal cells coexpressing subunit pairs consisting of GLR3.2, GLR3.3, or GLR 3.4 C-terminally tagged with CFP or YFP as indicated in Figure 5. FRET was measured by acceptor photobleaching (Herrick-Davis et al., 2006), with the following modifications. Prebleach CFP and YFP images were collected simultaneously following excitation at 458 nm (11% laser intensity). A selected region of interest was irradiated with the 514-nm laser line (100% intensity) using a 458-nm/514-nm dual dichroic mirror for 5 to 10 s to photobleach YFP. Postbleach CFP and YFP images were collected simultaneously immediately following photobleaching. CFP and YFP fluorescence were separated using online fingerprinting and linear unmixing with the Zeiss Meta detector and Zeiss Aim Software, as previously described (Herrick-Davis et al., 2006). FRET was measured as an increase in CFP fluorescence intensity following YFP photobleaching. FRET efficiency was calculated as 100× [(CFP postbleach – CFP prebleach)/CFP postbleach]. The FRET ratios at all the pixels in a region of interest within the cell were averaged to quantify the interactions of the GLR receptor subunits.
Electrophysiology
Whole-cell patch clamping and current voltage analysis was performed as previously described. Briefly, HEK293T cells were transfected with both GLR3.2-CFP and GLR3.4-YFP constructs. Cells expressing both constructs were selected on an Olympus BX51Wi upright fixed-stage fluorescence microscope equipped with a ×40 dipping lens with an emission filter setup that allows simultaneous separation and imaging of CFP and YFP fluorescence. Whole-cell patch clamp and current voltage analysis protocol was essentially the same as described by Vincill et al. (2012), including both bath and pipette solutions, except the bath solution contained 1 mM CaCl2 instead of 2 mM CaCl2. The methods for analyzing the steady state currents before and after ligand to generate the difference I-V curve shown in Figure 6 were exactly as described by Vincill et al. (2012).
Root Microscopy
The Zeiss 510 confocal laser scanning microscope was used to visualize GLR3.4-EGFP in roots stained with propidium iodide (50 µg mL−1) for 5 min. Optics employed were a plan-Apochromat ×20 lens or a C-Apochromat ×40 water immersion lens. The samples were excited with the 488-nm laser line (11% power), and channel mode detection was used to record the emission of enhanced GFP (500 to 530 nm) and propidium iodide (560 nm). For sieve plate imaging, root tissue was stripped of epidermal and cortical layers with fine tweezers to expose the central stele region and stained with aniline blue (0.1% [w/v]) for 5 min and visualized by epifluorescence microscopy using a short-wavelength emission filter set (330- to 385-nm excitation; 420-nm emission) or enhanced GFP (470- to 490-nm excitation; 515-nm emission). To count and map lateral root primordia, plants grown as described above were first digitally imaged with an Epson Perfection V700 scanner at 1200-dpi resolution to obtain a macroscopic image. Plants were then water mounted on a glass slide with cover slip for examination at ×20 using an Olympus BX60 microscope equipped with Nomarksi optics. Primordia were mapped to the macroscopy image of the same plant, which was annotated using Photoshop CS4 (Adobe). For high-magnification primordia imaging, plants were fixed and cleared before mounting as previously described (Ivanchenko et al., 2006).
Mutant Lines
The T-DNA insertion lines used in this study were used to generate the double glr mutants; glr3.2-1 was crossed with glr3.4-1, and glr3.2-2 was crossed with glr3.4-2 by cross pollination. The glr3.2-1/glr3.4-1 and glr3.2-2/glr3.4-2 double knockout T-DNA insertion lines were identified in the F2 generation and verified by PCR genotyping (see Supplemental Figure 1 online). Primers for genotyping all alleles in this study are presented in Supplemental Table 1 online.
Electrophysiology experiments with mutant plants gave evidence that the glr3.3 and glr3.4 mutant alleles used here lacked detectable function (Qi et al., 2006; Stephens et al., 2008). For this study it was particularly important to assess the mRNA levels in the previously unstudied glr3.2 alleles and the glr3.4 alleles with which they were combined in the genetic analyses. Total RNA was isolated from the roots of 7-d-old glr3.2-1, glr3.2-2, glr3.4-1, glr3.4-2, and wild-type seedlings and then treated with DNase as previously described (Vincill et al., 2012). Five hundred nanograms of total RNA from the indicated preparations was resolved by electrophoresis on an ethidium bromide agarose gel to verify RNA integrity (see Supplemental Figure 2 online). Reverse transcriptase reactions employing 4 μg of total RNA and random hexamer primers produced cDNA that was subsequently amplified with AccuPrime high fidelity Taq DNA Polymerase (Invitrogen) using the gene-specific primer sets listed in Supplemental Table 2 online. The results in Supplemental Figure 2 online show that the glr3.2 and glr3.4 alleles do not contain detectable mRNA for the disrupted genes and may be considered nulls.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for genes described in this article are as follows: At4g35290, GLR3.2, formerly known as At-GluR2 (Kim et al., 2001); At1g42540 (GLR3.3); and At1g05200, GLR3.4, formerly known as At-GLR4 (Chiu et al., 1999). T-DNA insertion lines glr3.2-1 (SALK_150710), glr3.2-2 (SALK_125952), glr3.3-1 (Salk_040458), glr3.3-2 (Salk_066009), glr3.4-1 (Salk_079842), and glr3.4-2 (Salk_016904) were obtained from the Salk Institute (http://signal.salk.edu/cgi-bin/tdnaexpress).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of Homozygous glr3.2, glr3.4, and glr3.2 glr3.4 Plants by PCR-Based Genotyping.
Supplemental Figure 2. Analysis of GLR3.2 and GLR3.4 Expression in Respective Mutant Alleles.
Supplemental Table 1. Plant and Mammalian Expression Vectors Used in This Study.
Supplemental Table 2. Primers Used for Genotyping and Expression Analysis.
Supplementary Material
Acknowledgments
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (Grant DE-FG02-10ER15527 to E.P.S.).
AUTHOR CONTRIBUTIONS
E.D.V. designed the research, performed research, analyzed data, and wrote the article. A.E.C. and J.N.M. performed research. E.P.S. designed the research and wrote the article.
Glossary
- iGluR
ionotropic Glu receptor
- HEK
human embryonic kidney
- GFP
green fluorescent protein
- FRET
Förster resonance energy transfer
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
References
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Chiu J., DeSalle R., Lam H.M., Meisel L., Coruzzi G. (1999). Molecular evolution of glutamate receptors: A primitive signaling mechanism that existed before plants and animals diverged. Mol. Biol. Evol. 16: 826–838 [DOI] [PubMed] [Google Scholar]
- Chiu J.C., Brenner E.D., DeSalle R., Nitabach M.N., Holmes T.C., Coruzzi G.M. (2002). Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol. Biol. Evol. 19: 1066–1082 [DOI] [PubMed] [Google Scholar]
- Cho D., Kim S.A., Murata Y., Lee S., Jae S.K., Nam H.G., Kwak J.M. (2009). De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J. 58: 437–449 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Davenport R.J., Tester M. (2002). Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Essah P.A., Tester M. (2004). Glutamate activates cation currents in the plasma membrane of Arabidopsis root cells. Planta 219: 167–175 [DOI] [PubMed] [Google Scholar]
- Dennison K.L., Spalding E.P. (2000). Glutamate-gated calcium fluxes in Arabidopsis. Plant Physiol. 124: 1511–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ferre S., Agnati L.F., Ciruela F., Lluis C., Woods A.S., Fuxe K., Franco R. (2007). Neurotransmitter receptor heteromers and their integrative role in 'local modules': The striatal spine module. Brain Res. Brain Res. Rev. 55: 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B.G., Walch-Liu P. (2009). Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ. 32: 682–693 [DOI] [PubMed] [Google Scholar]
- Fromm J., Lautner S. (2007). Electrical signals and their physiological significance in plants. Plant Cell Environ. 30: 249–257 [DOI] [PubMed] [Google Scholar]
- Furch A.C., van Bel A.J., Fricker M.D., Felle H.H., Fuchs M., Hafke J.B. (2009). Sieve element Ca2+ channels as relay stations between remote stimuli and sieve tube occlusion in Vicia faba. Plant Cell 21: 2118–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H., Singh S.K., Mancusso R., Gouaux E. (2005). Subunit arrangement and function in NMDA receptors. Nature 438: 185–192 [DOI] [PubMed] [Google Scholar]
- Gielen M., Siegler Retchless B., Mony L., Johnson J.W., Paoletti P. (2009). Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick-Davis K., Weaver B.A., Grinde E., Mazurkiewicz J.E. (2006). Serotonin 5-HT2C receptor homodimer biogenesis in the endoplasmic reticulum: Real-time visualization with confocal fluorescence resonance energy transfer. J. Biol. Chem. 281: 27109–27116 [DOI] [PubMed] [Google Scholar]
- Ingram P., Dettmer J., Helariutta Y., Malamy J.E. (2011). Arabidopsis Lateral Root Development 3 is essential for early phloem development and function, and hence for normal root system development. Plant J. 68: 455–467 [DOI] [PubMed] [Google Scholar]
- Ivanchenko M.G., Coffeen W.C., Lomax T.L., Dubrovsky J.G. (2006). Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J. 46: 436–447 [DOI] [PubMed] [Google Scholar]
- Kang J., Turano F.J. (2003). The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.M., Mehta S., Turano F.J. (2004). The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 45: 1380–1389 [DOI] [PubMed] [Google Scholar]
- Karakas E., Simorowski N., Furukawa H. (2011). Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Kwak J.M., Jae S.K., Wang M.H., Nam H.G. (2001). Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 42: 74–84 [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Oparka K. (2012). The structure of the phloem—Still more questions than answers. Plant J. 70: 147–156 [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Peters W.S., Ehlers K., van Bel A.J.E. (2001). Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell 13: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Batistič O., Hashimoto K. (2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Schuck P., Mayer M.L. (2011). Structure and assembly mechanism for heteromeric kainate receptors. Neuron 71: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B., et al. (2001). The identity of plant glutamate receptors. Science 292: 1486–1487 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Wipf D., Frommer W.B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lam H.M., Chiu J., Hsieh M.H., Meisel L., Oliveira I.C., Shin M., Coruzzi G. (1998). Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Lea P.J., Sodek L., Parry M.A.J., Shewry P.R., Halford N.G. (2007). Asparagine in plants. Ann. Appl. Biol. 150: 1–26 [Google Scholar]
- Meyerhoff O., Müller K., Roelfsema M.R., Latz A., Lacombe B., Hedrich R., Dietrich P., Becker D. (2005). AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 222: 418–427 [DOI] [PubMed] [Google Scholar]
- Michard E., Lima P.T., Borges F., Silva A.C., Portes M.T., Carvalho J.E., Gilliham M., Liu L.H., Obermeyer G., Feijó J.A. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Miller N.D., Durham Brooks T.L., Assadi A.H., Spalding E.P. (2010). Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 186: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H., Sprengel R., Schoepfer R., Herb A., Higuchi M., Lomeli H., Burnashev N., Sakmann B., Seeburg P.H. (1992). Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Van Norman J.M., Moreno A., Zhang J., Ahnert S.E., Benfey P.N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston D.W., Kremers G.-J. (2007). Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem. Sci. 32: 407–414 [DOI] [PubMed] [Google Scholar]
- Qi Z., Stephens N.R., Spalding E.P. (2006). Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 142: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter G.L., Monshausen G.B., Krol A., Gilroy S. (2009). Mechanical stimuli modulate lateral root organogenesis. Plant Physiol. 151: 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.J., et al. (2008). Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 31: 861–871 [DOI] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (suppl.): S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P., Harper J.F. (2011). The ins and outs of cellular Ca2+ transport. Curr. Opin. Plant Biol. 14: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stephens N.R., Qi Z., Spalding E.P. (2008). Glutamate receptor subtypes evidenced by differences in desensitization and dependence on the GLR3.3 and GLR3.4 genes. Plant Physiol. 146: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel H., Takeda K. (1989). Calcium-activated, voltage-dependent, non-selective cation currents in endosperm plasma membrane from higher plants. Proc. R. Soc. Lond. B Biol. Sci. 237: 213–231 [Google Scholar]
- Swanson S.J., Choi W.G., Chanoca A., Gilroy S. (2011). In vivo imaging of Ca2+, pH, and reactive oxygen species using fluorescent probes in plants. Annu. Rev. Plant Biol. 62: 273–297 [DOI] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tapken D., Hollmann M. (2008). Arabidopsis thaliana glutamate receptor ion channel function demonstrated by ion pore transplantation. J. Mol. Biol. 383: 36–48 [DOI] [PubMed] [Google Scholar]
- Tester M. (1990). Plant ion channels: Whole-cell and single-channel studies. New Phytol. 114: 305–340 [DOI] [PubMed] [Google Scholar]
- Thompson M.V., Holbrook N.M. (2004). Scaling phloem transport: Information transmission. Plant Cell Environ. 27: 509–519 [Google Scholar]
- Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. (2010). Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 62: 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong K., Ikura M. (2001). The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 11: 573–578 [DOI] [PubMed] [Google Scholar]
- Turano F.J., Panta G.R., Allard M.W., van Berkum P. (2001). The putative glutamate receptors from plants are related to two superfamilies of animal neurotransmitter receptors via distinct evolutionary mechanisms. Mol. Biol. Evol. 18: 1417–1420 [DOI] [PubMed] [Google Scholar]
- Vincill E.D., Bieck A.M., Spalding E.P. (2012). Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol. 159: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.J., Bowen H.C., Demidchik V., Nichols C., Davies J.M. (2002). Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochim. Biophys. Acta 1564: 299–309 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lewis D.R., Spalding E.P. (2007). Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Borst J.W., Muylaert M., Batoko H., Hemminga M.A., Chaumont F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 104: 12359–12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.