This work identifies a mucilage extrusion factor, PER36, which is transiently secreted into the periclinal cell wall at the early stage of seed development. The polarized and temporal secretion of PER36 functions in cell wall modification followed by degradation for efficient mucilage extrusion.
Abstract
The epidermal cells of the Arabidopsis thaliana seed coat, which correspond to the second layer of the outer integument (oi2), contain large quantities of a pectic polysaccharide called mucilage within the apoplastic space beneath the outer periclinal cell wall. Immediately after seed imbibition, the mucilage is extruded and completely envelops the seed in a gel-like capsule. We found that a class III peroxidase family protein, PEROXIDASE36 (PER36), functions as a mucilage extrusion factor. Expression of PER36 occurred only in oi2 cells for a few days around the torpedo stage. A PER36–green fluorescent protein fusion was secreted into the outer cell wall in a polarized manner. per36 mutants were defective in mucilage extrusion after seed imbibition due to the failure of outer cell wall rupture, although the mutants exhibited normal monosaccharide composition of the mucilage. This abnormal phenotype of per36 was rescued by pectin solubilization, which promoted cell wall loosening. These results suggest that PER36 regulates the degradation of the outer cell wall. Taken together, this work indicates that polarized secretion of PER36 in a developmental stage-dependent manner plays a role in cell wall modification of oi2 cells.
INTRODUCTION
The seed coat of angiosperm seeds surrounds the next-generation tissues (embryo and endosperm), protecting them from environmental stresses and mediating the supply of nutrients to the embryo during seed formation (Weber et al., 1995; King et al., 1997; Sheen et al., 1999; Wobus and Weber, 1999; Debeaujon et al., 2000). The angiosperm seed coat is differentiated from the outer integument (oi) and inner integument (ii). In Arabidopsis thaliana, the outer integument consists of two cell layers (oi1 and oi2), and the inner integument consists of three cell layers (ii1, ii2, and ii3). These cell layers play important roles during early seed development in the formation of specialized structures of the seed coat (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000; Debeaujon et al., 2003; Haughn and Chaudhury, 2005). The innermost cell layer ii1 produces flavonoids, which are responsible for the characteristic brown color of Arabidopsis seeds (Beeckman et al., 2000; Debeaujon et al., 2003). The cell layers ii2 and ii3 transiently express δ-type VACUOLAR PROCESSING ENZYME (δVPE), which regulates programmed cell death at the torpedo-shaped embryo stage (Nakaune et al., 2005). The oi1 cell layer forms a thickened primary cell wall attached to the inner integument (Beeckman et al., 2000; Windsor et al., 2000).
The outer cell layer of the outer integument (oi2) deposits large quantities of a pectic polysaccharide, mucilage, around a volcano-shaped structure of secondary cell wall called the columella (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). The oi2 cells of developing seeds actively synthesize mucilage in the Golgi apparatus and secrete it into the apoplastic space underneath the primary cell wall, where it is directly exposed to the external environment (Young et al., 2008). Several mucilage biosynthesis-related factors have been identified, including transcription factors and cell wall modification enzymes (Western, 2012). TRANSPARENT TESTA GLABRA1 (TTG1) is a WD-repeat transcription factor that regulates seed coat development (Walker et al., 1999). Seeds of the ttg1 mutant show aberrant mucilage accumulation in oi2 cells and seed pigmentation in ii1 cells (Koornneef, 1981; Western et al., 2001). TTG1, together with other transcriptional regulators, such as APETALA2 and GLABRA2, regulates the expression of a putative NDP-l-rhamnose synthase, MUCILAGE-MODIFIED4 (MUM4) (Western et al., 2004).
Mucilage exists in a dehydrated form in the dry seed and becomes rehydrated during seed imbibition (Western et al., 2000). The rehydrated mucilage then envelops the entire seed, forming a gel-like capsule. Mucilage is proposed to function to protect the embryo from dehydration during germination among other roles (Penfield et al., 2001; Arsovski et al., 2009; Western, 2012). The rehydrated mucilage mechanically ruptures the primary cell wall and starts to be extruded from the seed coat within a minute of imbibition (Western et al., 2000; Arsovski et al., 2009). The mechanism that mediates the rapid extrusion of mucilage from the seed coat after imbibition is not fully understood. MUM1/LEUNING_HOMOLOG (LUH; Bui et al., 2011; Huang et al., 2011; Walker et al., 2011), MUM2 (Dean et al., 2007; Macquet et al., 2007), SUBTILASE1.7 (SBT1.7; Rautengarten et al., 2008), β-XYLOSIDASE1 (BXL1; Arsovski et al., 2009), and PECTIN METHYLESTERASE INHIBITOR6 (PMEI6; Saez-Aguayo et al., 2013) are thought to be involved in extrusion, although they are also expressed in tissues other than the outer integument.
In this article, we show that PEROXIDASE (PER36), a member of the class III peroxidase family (Tognolli et al., 2002), acts as a mucilage extrusion factor in Arabidopsis. PER36 showed spatiotemporal expression in oi2 cells and polarized localization in the outer periclinal cell wall. PER36 appears to be required for degrading and loosening the cell wall to extrude mucilage from the seed coat. Taken together, the findings of this study provide a model of the role of PER36 in modifying the cell wall of the seed coat.
RESULTS
PER36 Is Specifically Expressed in the Outermost Integument Cells (oi2 Cells)
Previously, we reported that the NAC transcription factors NAC-REGULATED SEED MOLPHOLOGY1 (NARS1) and NARS2 regulate seed coat formation (Kunieda et al., 2008). Staining of seeds with ruthenium red to visualize pectic polysaccharides revealed that the nars1 nars2 double mutant fails to extrude mucilage (see Supplemental Figure 1A online), suggesting that NARS1 and NARS2 may regulate the expression of mucilage-related factors. The At3g50990 gene encoding PER36 was remarkably downregulated in the nars1 nars2 silique, which contains the developing seeds (see Supplemental Figure 1B online). These results suggested that PER36 is a mucilage-related factor.
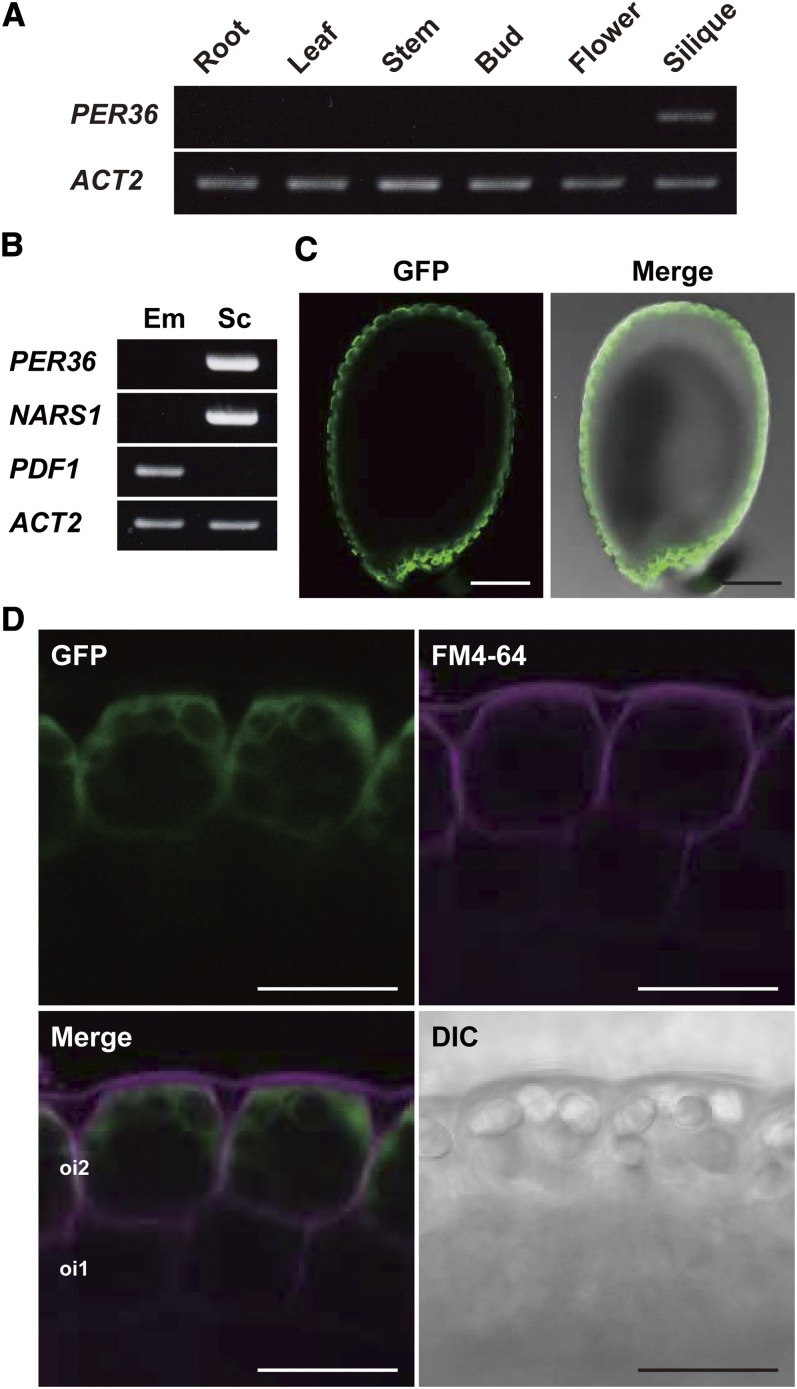
Expression of PER36 was restricted to the young silique containing the developing seeds (Figure 1A). AtGenExpress expression profile analysis indicated that PER36 expression occurred in developing seeds isolated from the silique (Schmid et al., 2005) (see Supplemental Figure 2 online). To determine where PER36 was expressed in the developing seed, RT-PCR was performed using RNA extracted from the embryo and from the remaining tissue consisting of endosperm and integument. The PER36 transcript was not detected in the embryo but was detected in the tissue comprising endosperm and integument (Figure 1B). The distribution pattern of transcripts of NARS1 and PROTODERMAL FACTOR1 (PDF1), which are markers of the outer integument and embryo, respectively (Abe et al., 1999; Kunieda et al., 2008), confirmed that the tissue had been prepared correctly (Figure 1B). These results indicate that PER36 is expressed in the endosperm and/or integument.
Figure 1.
Specific Expression of PER36 in the Outermost Integument (oi2) of Developing Arabidopsis Seeds.
(A) RT-PCR showing the expression of PER36 mRNA in the silique. ACT2 was used as an internal control.
(B) RT-PCR showing the expression of PER36 mRNA in the seed coat of developing seeds. Developing seeds were separated into two parts, the embryo (Em) and the seed coat plus endosperm (Sc). PDF1 and NARS1 are tissue-specific markers of the embryo and seed coat, respectively. ACT2 was used as an internal control.
(C) Fluorescence images showing the PER36 promoter activity in the seed coat of developing seeds of ProPER36:sGFP-GUS transgenic plants. Developing seeds at the torpedo-shaped embryo stage were inspected with confocal laser scanning microscopy and differential interference contrast microscopy. Bars = 100 μm.
(D) Fluorescence images of the outer integuments of developing ProPER36:sGFP-GUS seeds show specific expression driven by the PER36 promoter in the outermost integument (oi2) of the seed coat. FM4-64 was used to visualize the plasma membrane. DIC, differential interference contrast image. Bars = 20 μm.
Transgenic Arabidopsis plants expressing ProPER36:sGFP-GUS, the synthetic green fluorescent protein–β-glucuronidase fusion under the control of the PER36 promoter, were generated to examine the expression pattern of PER36 in tissues of developing seeds. The GFP fluorescence signal was detected in the integument but not in the endosperm during the torpedo stage (Figure 1C). Higher magnification of the outer integument revealed that GFP fluorescence was restricted to oi2 cells, whereas the fluorescent dye FM4-64, a marker of the plasma membrane, was present in the inner cell layer of the integument including oi1 and oi2 cells (Figure 1D). The observed PER36 expression pattern indicates that PER36 is specifically localized to oi2 cells.
Transient Accumulation of PER36 in oi2 Cells around the Torpedo Stage
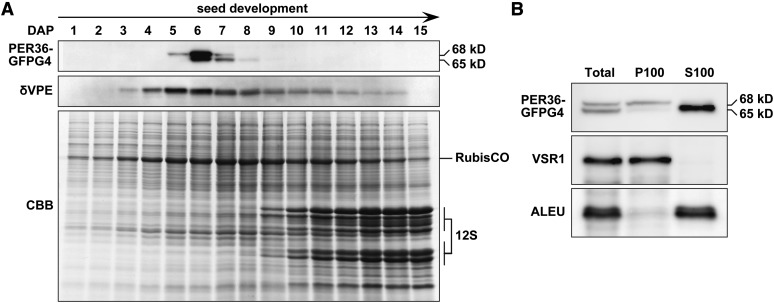
The protein accumulation levels of PER36 during seed formation were investigated using ProPER36:PER36-GFPg4 transgenic plants (Figure 2A), which were generated to express the PER36-GFPg4 fusion protein under the control of the PER36 promoter in the per36-1 genetic background (described below). GFPg4 is a modified GFP in which four consecutive Gly residues are added to the C terminus to avoid mislocalization of the fusion protein to vacuoles (Nishizawa et al., 2003). The siliques of ProPER36:PER36-GFPg4 were harvested at different stages ranging from 1 d after pollination (DAP) to 15 DAP and subjected to immunoblot analysis. PER36-GFPg4 emerged at 5 DAP, and its accumulation reached a maximum at 6 DAP, which corresponded to the torpedo stage, followed by rapid degradation. Thus, PER36-GFPg4 was detected only for a few days during early seed formation, which is different from the accumulation pattern of δVPE. In previous work, we showed that δVPE accumulates until the middle stages of seed formation, despite programmed cell death of the inner integument at the end of early seed formation (Nakaune et al., 2005). These results suggest that PER36 functions transiently in living oi2 cells around the torpedo stage.
Figure 2.
The Rapid Emergence and Degradation of PER36 Results in the Transient Detection of PER36 in Developing Seeds at the Torpedo-Shaped Embryo Stage.
(A) Siliques at sequential stages (from 1 to 15 DAP) of ProPER36:PER36-GFPg4 transgenic plants were subjected to SDS-PAGE followed by either immunoblot analysis with anti-GFP and anti-δVPE antibodies or Coomassie blue (CBB) staining. δVPE is a marker of the inner integument during early seed development. 12S, the major seed storage protein 12S globulin. RubisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Differences in the subcellular distribution of the 68- and 65-kD molecular species of PER36-GFPg4. The microsomal (P100), soluble (S100), and total fractions from developing seeds at the torpedo-shaped embryo stage were subjected to immunoblot analysis with an anti-GFP antibody and the following specific antibodies for organelle markers: anti-VSR1 (a marker for the microsomal fraction) and anti-ALEU (a marker for the soluble fraction).
The molecular mass of PER36-GFPg4 shifted from 68 to 65 kD during seed development (Figure 2A), suggesting that the 68-kD form is processed to yield the 65-kD form. Suborganellar fractionation followed by immunoblot analyses revealed that the 68-kD form was present in the microsomal fraction (P100), whereas the 65-kD form was present in the soluble fraction (S100) (Figure 2B). The 65-kD form might be generated after removal of the C-terminal sequence of PER36-GFPg4. Similar processing of the GFP fusions on the secretory pathway was reported previously (Tamura et al., 2003).
PER36-GFPg4 Is Secreted into the Outer Periclinal Cell Wall of oi2 Cells in a Polarized Manner
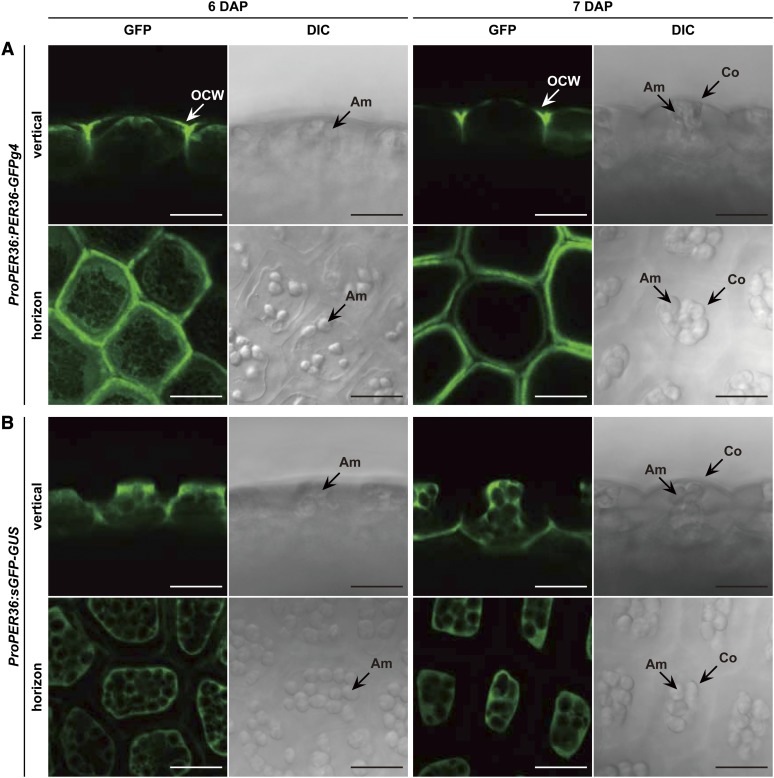
Because the high accumulation of PER36-GFPg4 is restricted to the torpedo stage (Figure 2A), its subcellular localization was examined in the oi2 cells of developing seeds at 6 and 7 DAP using a confocal laser scanning microscope (Figure 3A). sGFP-GUS, which is localized specifically to the cytosol, was used as a control (Figure 3B). At 6 DAP, PER36-GFPg4 fluorescence was detected in both the intra- and extracellular spaces of oi2 cells. Intracellular fluorescence was distributed throughout the endoplasmic reticulum network-like structures (Figure 3A; 6 DAP, horizon). Extracellular fluorescence was primarily localized to the outer periclinal cell wall, which is exposed to the external environment. No fluorescence was observed at the inner periclinal cell wall. These results suggest that PER36-GFPg4 secretion is highly regulated in a polarized manner.
Figure 3.
Polarized Cell Wall Localization of PER36 on the External Side of the Outermost Integument Cells.
The images represent vertical and horizontal sections of the outer integument. Am, amyloplast; Co, columella; OCW, outer cell wall. Bars = 20 μm.
(A) The outermost integument of 6- and 7-DAP seeds of ProPER36:PER36-GFPg4 transgenic plants was inspected with confocal laser scanning microscopy (GFP) and differential interference contrast microscopy (DIC).
(B) ProPER36:sGFP-GUS plants, which produce cytosolic fluorescence in the outermost integument, were used for visualization of tissue-specific cell morphology.
Accumulating extracellular fluorescence was still observed at 7 DAP; however, intracellular fluorescence was no longer detected (Figure 3A; 7 DAP). Considering the developmental shift in the molecular mass of PER36-GFPg4 (Figure 2A), the extra- and intracellular fluorescent signals likely correspond to the 65- and 68-kD forms, respectively. Together, these results suggest that the endoplasmic reticulum–localized 68-kD protein is secreted into the outer cell wall of oi2 cells, where the processed 65-kD form accumulates.
Seed Coat Structures in per36 Mutants
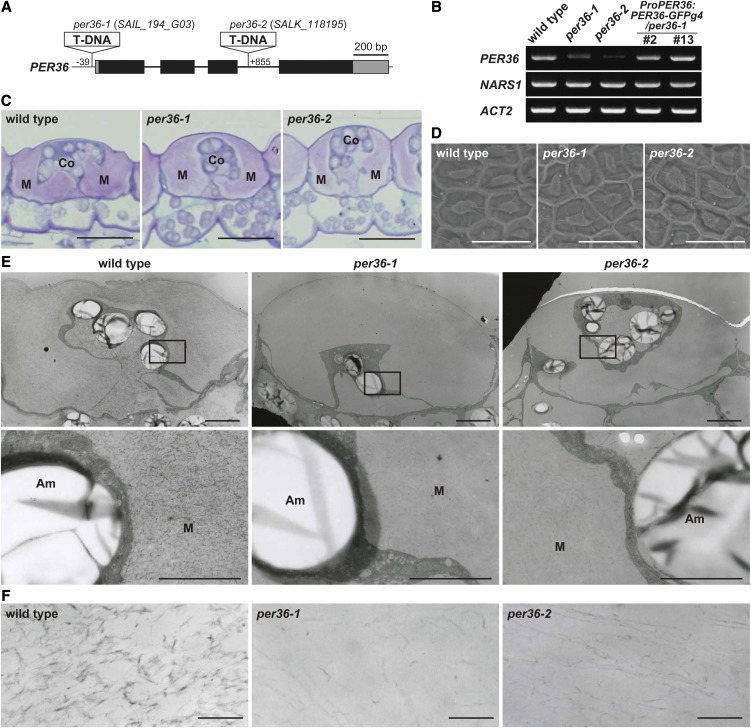
Next, two T-DNA insertion mutants of PER36, per36-1 and per36-2, were isolated (Figure 4A). Analysis of gene expression confirmed that per36-1 and per36-2 were knockdown mutants of PER36 (Figure 4B; see Supplemental Figure 3 online). Toluidine blue staining of transverse sections cut from developing seeds showed normal accumulation of mucilage around the columella in the oi2 cells of per36 mutants, similar to that observed in wild-type seeds (Figure 4C). Scanning electron microscopy revealed no obvious differences in the surfaces of dry wild-type and per36 mutant seeds (Figure 4D). These results indicate that oi2 cells develop normally in per36 mutants.
Figure 4.
Seed Coat Structure of the Wild Type and per36 Mutants.
(A) Schematic representation of the PER36 gene, which consists of four exons (filled boxes) and three introns (thick lines). The coding and untranslated regions are shown in black and gray boxes, respectively. The T-DNAs of per36-1 (SAIL_194_G03) and per36-2 (SALK_118195) mutants are inserted at nucleotide base positions −39 and +855, respectively, from the adenine of the start codon of the PER36 gene.
(B) Results of RT-PCR with 35 cycles showing that PER36 was downregulated in developing seeds of both per36-1 and per36-2 mutants, whereas the PER36 transcript was similarly expressed in wild-type plants and two independent transgenic lines (#2 and #13) that express ProPER36:PER36-GFPg4 in the background of per36-1 (ProPER36:PER36-GFPg4/per36-1). NARS1 is a seed coat marker and ACT2 is an internal control.
(C) Toluidine blue–stained sections of seed coats of 9-DAP developing seeds. No obvious differences between the wild type and two per36 mutants were observed. Co, columella; M, mucilage. Bars = 20 μm.
(D) Scanning electron microscopy of the surface of dry seeds reveals no obvious differences between the wild type and two per36 mutants. Bars = 50 μm.
(E) Ultrastructural analysis of the outermost integument cells of 9-DAP developing seeds of wild-type plants and two per36 mutants, showing that the densities of filamentous structures are reduced in per36 mutants. The bottom panels are higher magnifications of the boxes shown in the top panels. Am, amyloplast; M, mucilage. Bars = 50 μm (top panels) and 2 μm (bottom panels).
(F) Enlarged views of the filamentous structure in the mucilage region of wild-type and two per36 mutants seeds. Bars = 200 nm.
Transmission electron micrographs of transverse sections of developing seeds at 9 DAP showed no differences in the outer cell wall of the oi2 cells of wild-type and per36 mutant seeds (Figure 4E). Higher magnification images of wild-type mucilage revealed the presence of a large number of highly electron-dense filaments that formed a meshwork within the mucilage (Figure 4F; the wild type). However, the per36 mutants showed a marked reduction in the number of filamentous structures, giving the meshwork a rough appearance in contrast with the fine appearance in the wild type (Figure 4F; per36-1 and per36-2). This abnormal phenotype of the per36-1 mutant was restored by expressing PER36-GFPg4 in oi2 cells (see Supplemental Figure 4 online). These results suggest that mucilage components are affected by downregulation of PER36. Because peroxidases are known to catalyze the cross-linking of cell wall components (Passardi et al., 2004), the filamentous structures might be formed by a cross-linking reaction between the mucilage components in a peroxidase-dependent manner (the peroxidative cycle; see Discussion).
PER36 Is Involved in Mucilage Extrusion from the Seed Coat
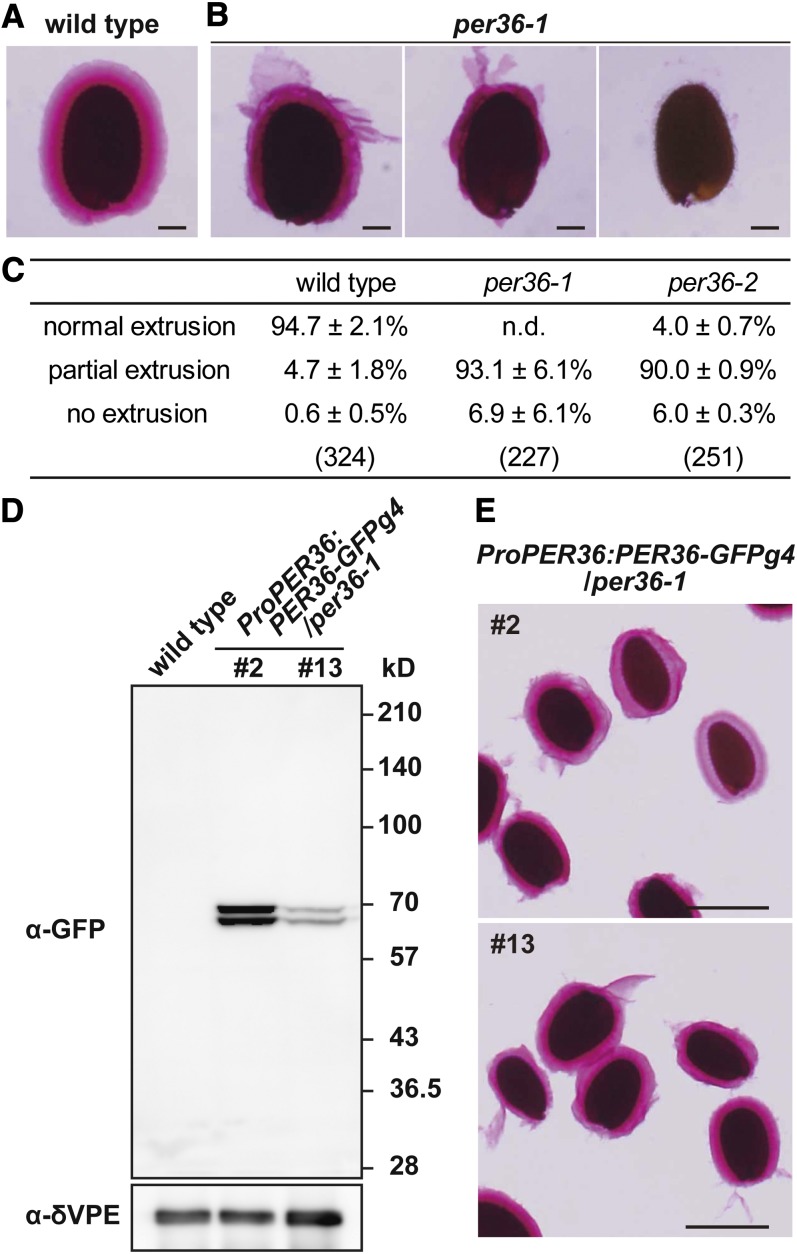
To examine mucilage extrusion from seed coat, dry seeds of the wild type and per36 mutants were directly shaken in ruthenium red solution, which stains pectic polysaccharides. Approximately 94.7% ± 2.1% of wild-type seeds were completely enrobed in mucilage gel matrix, whereas 4.7% ± 1.8% showed partial extrusion of mucilage from the seed coat (Figures 5A and 5C). By contrast, in per36 mutants, complete mucilage extrusion was rarely observed (<10%) and partial mucilage extrusion occurred in ∼90% of the seeds (Figures 5B and 5C).
Figure 5.
Knockdown Mutants of per36 Exhibited Abnormal Mucilage Extrusion.
(A) Visualization of mucilage extrusion from wild-type dry seeds. Mucilage extrusion was induced by shaking seeds in ruthenium red solution for 2 h, after which it completely surrounds the seeds. Bar = 100 μm.
(B) Three patterns of mucilage extrusion from per36-1 dry seeds after shaking for 2 h in ruthenium red solution: normal extrusion (left panel), partial extrusion (center panel), and no extrusion (right panel). Bars = 100 μm.
(C) Quantification of mucilage extrusion classified into three patterns in the wild type and per36 mutants. Dry seeds of the wild type or per36 mutants were shaken in ruthenium red solution for 1 h. The stained seeds were classified into three patterns depending on mucilage extrusion. The data were calculated from three independent experiments and are shown ±sd. The values in parentheses show the total number of seeds examined. n.d., not detected.
(D) Immunoblot analysis with an anti-GFP antibody showing the different accumulation of PER36-GFPg4 in developing seeds of transgenic plants at the torpedo-shaped embryo stage (#2 and #13; ProPER36:PER36-GFPg4/per36-1; see Figure 4B). δVPE was used as a loading control.
(E) Ruthenium red staining of dry seeds of transgenic plants (#2 and #13; ProPER36:PER36-GFPg4/per36-1) treated by shaking for 2 h. Normal mucilage extrusion occurred in these seeds. Bars = 500 μm.
To determine whether the abnormal mucilage extrusion of per36 mutants correlated with PER36 downregulation, molecular complementation studies were conducted using two independent lines of ProPER36:PER36-GFPg4 transgenic plants. Immunoblotting using an anti-GFP antibody showed that the transgenic plants expressed PER36-GFPg4 in developing seeds at the torpedo stage, as observed by the detection of the 65- and 68-kD bands (Figure 5D). Expression of PER36-GFPg4 rescued the mucilage extrusion deficiency of per36-1 mutant seeds, and the mucilage gel matrix of the transgenic plants was indistinguishable from that of wild-type seeds (Figure 5E). Quantification of mucilage extrusion in seeds of the wild type, per36 mutants, and ProPER36:PER36-GFPg4 transgenic plants was achieved by staining seeds with ruthenium red solution without shaking (see Supplemental Figure 5 online). In this case, ∼30% of per36 mutant seeds showed complete or partial mucilage extrusion, whereas ∼60 to 80% of seeds of wild-type and transgenic plants showed normal or partial mucilage extrusion. Taken together, these results indicate that PER36 is involved in mucilage extrusion.
PER36 Regulates Degradation of the Outer Cell Wall of oi2 Cells
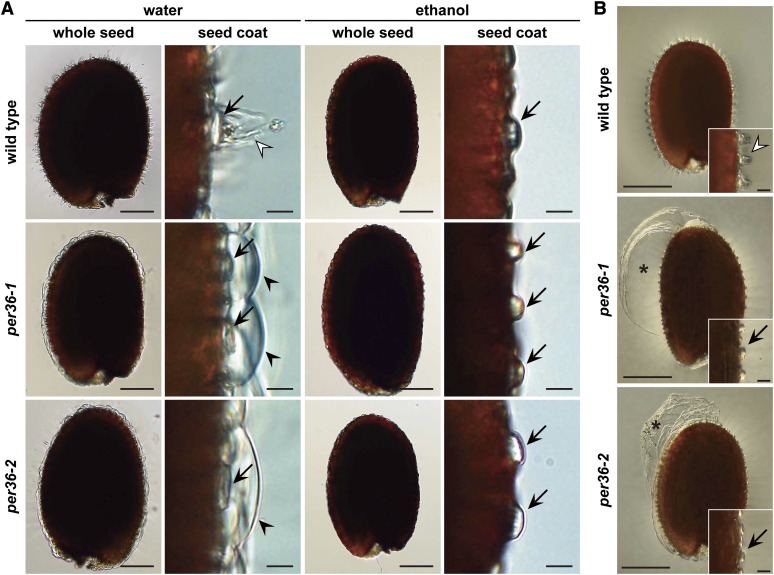
The outer cell walls of wild-type and per36 mutant seeds were inspected under a light microscope after imbibition. Seed imbibition resulted in mucilage-mediated rupture of the outer cell wall in wild-type seeds (Figure 6A). By contrast, the outer cell wall of per36 mutant seeds formed dome-shaped structures that were detached from the columella; these structures appeared to be caused by mucilage expansion. No obvious differences were observed between the cell walls of wild-type and per36 mutant seeds incubated in ethanol, indicating that the dome-shaped walls observed on the per36 mutant seeds were formed due to imbibition. These dome-shaped outer cell walls were frequently peeled away as a large sheet by shaking the seeds (Figure 6B), possibly due to the mechanically induced collisions between the seeds. This peeling resulted in the partial extrusion of mucilage (Figures 5B and 5C). These results suggest that per36 mutants have rigid outer cell wall, compared with that of the wild type.
Figure 6.
An Intact Outer Cell Wall in Imbibed Seeds of per36 Mutants Prevents Mucilage Extrusion and Frequently Peels off as a Large Sheet from the Seed Coat after Shaking.
(A) Dry seeds of the wild type and two per36 mutants were shaken in water for 2 h (left panels) or ethanol (right panels; mock treatment). The outer cell wall of wild-type seeds ruptured (open arrowheads). By contrast, imbibed seeds of per36 mutants typically maintained an intact outer cell wall (filled arrowheads), although the mucilage was expanded. Arrows indicate columellae. Bars = 100 μm (whole seed) and 10 μm (seed coat).
(B) Dry seeds of the wild type and per36 mutants were observed after shaking in water for 2 h. The per36 mutants showed a large sheet of the outer cell wall (asterisks) and columellae without the fragmented residue of the outer cell wall (arrows). By contrast, wild-type seeds had the residue of the fragmented outer cell wall on the columella (open arrowhead). The insets are magnified views of the seed coat. Bars = 200 μm (whole seed) and 20 μm (inset).
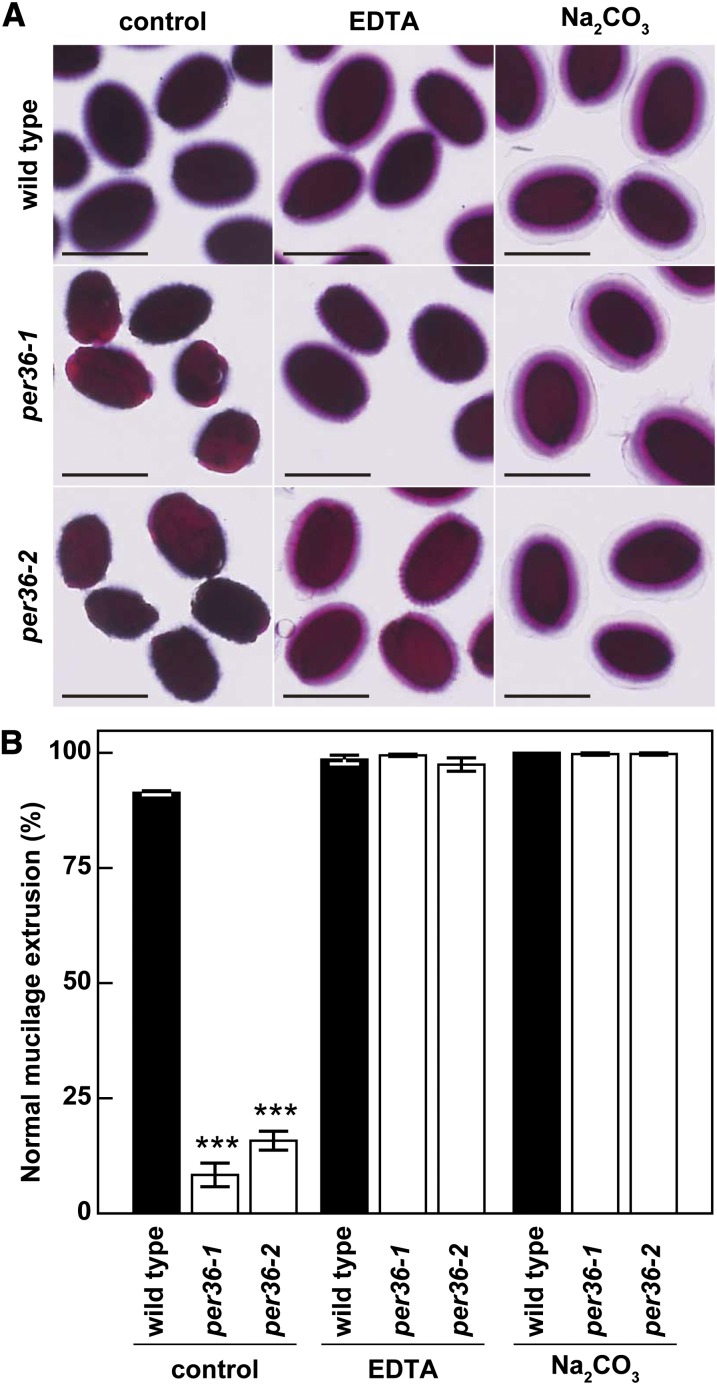
Finally, to examine the influence of pectin solubilization on mucilage extrusion in the per36 mutants, wild-type and per36 mutant seeds were shaken in a solution containing the chelating agent EDTA or the weak alkali Na2CO3 and then stained with ruthenium red. per36 mutant seeds showed normal mucilage extrusion, similar to wild-type seeds (Figure 7A). Quantification of ruthenium red staining demonstrated that treatment with EDTA and Na2CO3 significantly increased mucilage extrusion in per36 mutant seeds (Figure 7B). A similar result was reported for some mucilage extrusion-deficient mutants (Dean et al., 2007; Rautengarten et al., 2008; Arsovski et al., 2009). Taken together, these results suggest that PER36 plays a role in mucilage extrusion by regulating the degradation of the outer cell wall of oi2 cells.
Figure 7.
Pectin Solubilization Enhanced Mucilage Extrusion in per36 Mutants.
(A) Ruthenium red staining of wild-type and per36 mutant seeds treated with shaking for 1 h after pectin solubilization with either 50 mM EDTA or 1 M Na2CO3 for 30 min. Bars = 500 μm.
(B) Comparison of the normal mucilage extrusion patterns of wild-type and per36 mutant seeds with or without EDTA or Na2CO3 treatments, which cause the solubilization of pectin. Error bars indicate ±sd of three biological replicates; more than 80 seeds were examined in each experiment. Asterisks indicate significant differences between the wild type and per36 mutants according to Student’s t test (***P < 0.001).
PER36 Has Peroxidase Activity
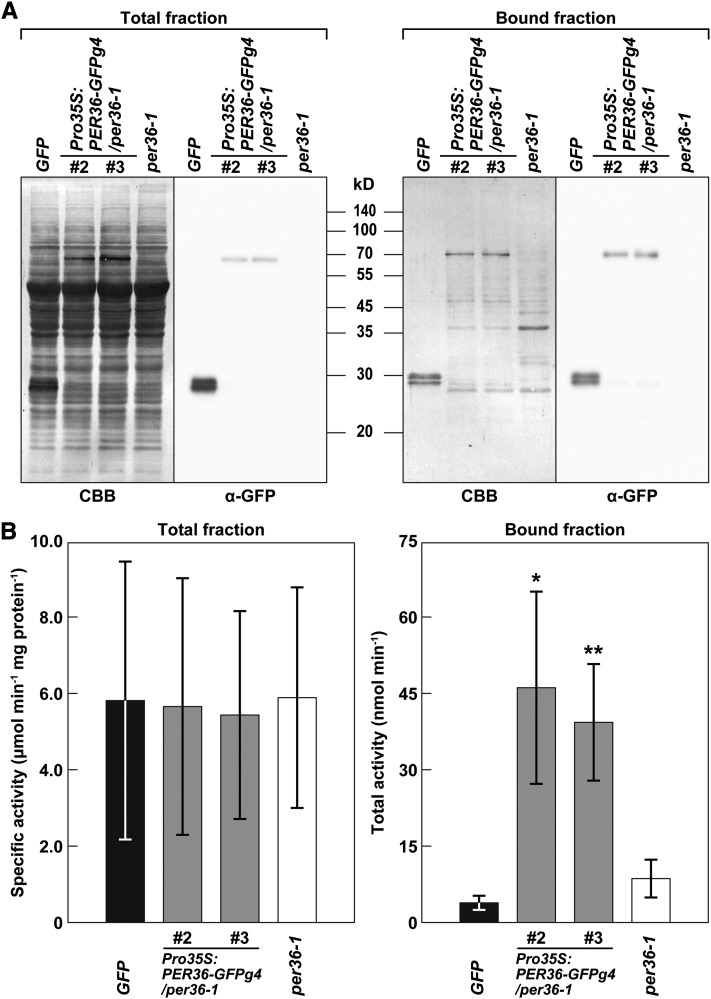
The PER36 sequence has both a catalytic and distal heme binding domain and a proximal heme binding domain that are conserved in peroxidase family proteins (see Supplemental Figure 6 online). This raised the question of whether PER36 actually functions as a peroxidase. To address it, we generated transgenic plants that overexpressed the PER36-GFPg4 protein under the control of the cauliflower mosaic virus 35S promoter in the per36-1 background and isolated the PER36-GFPg4 protein from two independent transgenic lines by affinity chromatography with anti-GFP antibody (Figure 8A). The PER36-GFPg4 fraction showed peroxidase activity toward the typical peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (Figure 8B). The activities of the PER36-GFPg4 fractions were significantly higher than those of the control fractions that were obtained from the transgenic line expressing free GFP and the per36-1 mutant line. It should be noted that similar levels of activity were found in the total extract fractions from the lines examined (Figure 8B). These results indicate that the PER36-GFPg4 protein had a peroxidase activity, suggesting that PER36 functions as a peroxidase in oi2 cells.
Figure 8.
PER36-GFPg4 Has Peroxidase Activity.
(A) Protein profiles (Coomassie blue [CBB]) and immunoblot with anti-GFP antibody (α-GFP) of total extract (total fraction) and the affinity-purified proteins (bound fraction) from 3-week-old plants of each of transgenic plants expressing free GFP (GFP), two independent lines of Pro35S:PER36-GFPg4/per36-1 (#2 and #3), and per36-1.
(B) Analysis of peroxidase activity of PER36-GFPg4. Peroxidase activities in both total and eluted fractions were measured using TMB as a substrate. Error bars indicate ±sd of three biological replicates. Asterisks indicate significant differences between GFP and others according to Student’s t test (*P < 0.05; **P < 0.01).
DISCUSSION
PER36-Dependent Mucilage Extrusion Machinery
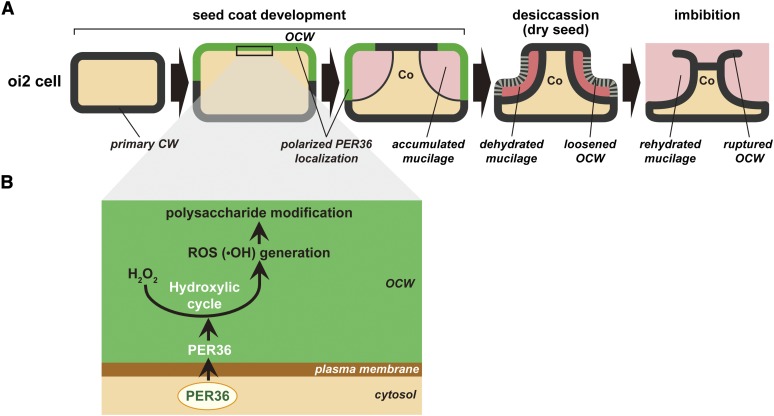
Seventy-three class III peroxidase family members have been identified in Arabidopsis (Tognolli et al., 2002). Comprehensive transcriptome and proteome analyses suggest that these members are involved in various physiological processes, such as biotic and abiotic stress responses, and cell wall modification (Cosio and Dunand, 2009). This study suggests that PER36 plays a role in oi2 cell wall degradation. This role of PER36 may be supported by its peroxidase activity (Figure 8). Class III peroxidases are involved in two cycles: the hydroxylic cycle, which generates various reactive oxygen species (ROS), and the peroxidative cycle, which oxidizes various substrates (Passardi et al., 2004; Cosio and Dunand, 2009). The hydroxylic cycle is thought to play a role in cell wall loosening. Peroxidases catalyze the formation of hydroxyl radicals (•OH) through the hydroxylic cycle (Passardi et al., 2004), and polysaccharides such as pectin and xyloglucan are broken down by •OH in vitro (Fry, 1998; Schweikert et al., 2000). The predominant accumulation of PER36 in the outer cell wall (Figures 2A and 3A) would generate large amounts of •OH, resulting in cell wall loosening (i.e., degradation), suggesting that the rapid extrusion of mucilage is most likely regulated by the hydroxylic cycle (Figure 9).
Figure 9.
Schematic Representation of PER36 Function in Cell Wall Modification of oi2 Cells.
(A) Morphological changes of oi2 cells during seed coat development, desiccation, and imbibition. PER36 is temporarily secreted into the outer periclinal cell wall in a polarized manner. See text for details.
(B) A hypothetical model of PER36 function in cell wall modification: ROS-mediated degradation of polysaccharides of the outer cell wall through the hydroxylic cycle. See text for details.
Co, columella; CW, cell wall; OCW, outer cell wall.
The question is: Which cell wall components are targeted by PER36 in the cell wall of oi2 cells? No significant differences were found in monosaccharide composition of the mucilage between the wild type and per36 mutants (see Supplemental Table 1 online), indicating that PER36 is not required for polysaccharide chain turnover. PER36 might be involved in structurally modifying cell wall polysaccharides via ROS-mediated polysaccharide scission. The partial mucilage extrusion phenotype shown by the per36 mutants is similar to that of BXL1-deficient mutants (Arsovski et al., 2009). BXL1 degrades (1→5)-linked arabinans in the outer cell wall at the early stage of seed formation (Arsovski et al., 2009), which is when PER36 specifically accumulates in the outer cell wall (Figures 2A and 3A). (1→5)-Linked arabinans form one of the side-chain polysaccharides of pectin rhamnogalacturonan I (Mohnen, 2008). Rhamnogalacturonan I within the cell wall might be a target for PER36-dependent polysaccharide modification. Furthermore, a mutant deficient in PMEI6, a modulator of the degree of homogalacturonan methylesterification, exhibits a large sheet of the outer cell wall after water imbibition (Saez-Aguayo et al., 2013), and this phenotype is similar to that of per36 mutants (Figure 6B), suggesting that another possible target for PER36-dependent modification is pectin homogalacturonan. PER36 might modify rhamnogalacturonan I and/or homogalacturonan of the outer cell wall of oi2 cells to promote degradation of the cell wall for efficient mucilage extrusion.
Temporal and Polarized Secretion of PER36 for Cell Wall Modification
PER36 begins to accumulate at 5 DAP and reaches the maximum level at 6 DAP (Figure 2A). At the early stage, PER36 is temporarily secreted into the outer periclinal cell wall (Figure 3), where it likely functions to loosen the cell wall, as depicted in Figure 9. At the later stage, large amounts of mucilage begin to accumulate in the space between the plasma membrane and the outer cell wall (Western et al., 2000). This mucilage accumulation necessarily inhibits newly secreted proteins from accessing the outer cell walls. Hence, PER36 secretion before mucilage accumulation makes sense. Significant features of PER36 include its polarized secretion into the outer periclinal cell wall and its transient accumulation (Figures 2A and 3A). In other words, PER36 is a short-lived protein on the outer cell wall. Considering the fact that ROS widely damages living cells, the polarized secretion and transient accumulation of the ROS-generating enzyme PER36 might contribute to minimize the effects of ROS in oi2 cells.
This study demonstrates polarized secretion of PER36-GFPg4 (Figure 3A), although the intracellular transport of mucilage-related factors that function in the apoplastic space is still poorly understood. Polarized secretion of soluble proteins has been reported during cuticle development. The cuticle formation-related factor, BODYGUARD (BDG), which is a member of the α/β-hydrolase fold protein family, specifically accumulates in the outermost cell wall of the epidermis (Kurdyukov et al., 2006). The polarized secretion of PER36 may be regulated by a mechanism similar to that involved in BDG secretion because oi2 cells also possess a cuticular layer (Watanabe et al., 2004).
PER36 Is Rapidly Degraded
PER36 is a short-lived protein (Figure 2A). Its manner of degradation is unknown but might be different from that of other seed coat proteins such as δVPE and SBT1.7. δVPE remains in the extracellular space of ii2 and ii3 cells, even after cell death (Nakaune et al., 2005) (Figure 2A). SBT1.7 is present in oi2 cells at the mature embryo stage, although it is also highly expressed during early seed formation (Rautengarten et al., 2008). The rapid degradation of PER36 may prevent excess modification or degradation of the cell wall.
The localization of secreted PER36-GFPg4 to the outer cell wall and away from the plasma membrane (Figure 3A) suggests that its rapid degradation must take place at the cell wall but not within the cell via the endocytosis pathway. Two possible pathways for PER36 degradation are proposed. The first pathway is ROS-dependent protein fragmentation. The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is rapidly fragmented by ROS (such as •OH) in wheat (Triticum aestivum) chloroplast lysates (Ishida et al., 1997). Therefore, the •OH generated by PER36 could cause fragmentation of polysaccharides and outer cell wall proteins, including PER36 itself, in oi2 cells. The second pathway is protease-dependent degradation. Various proteases belonging to several families (including subtilisin-like Ser proteases) have been identified in apoplastic and/or cell wall fractions by proteome analysis (Jamet et al., 2006; Irshad et al., 2008) and are thought to be involved in proteolysis at the extracellular space. A subtilisin-like Ser protease, SBT1.7, is necessary for mucilage extrusion from the Arabidopsis seed coat (Rautengarten et al., 2008). The enzymatic activity of pectin methylesterase, which functions in modification of cell wall, is higher in sbt1.7 mutant seeds than in wild-type seeds, suggesting that SBT1.7 is involved in the degradation or inhibition of pectin methylesterase (Rautengarten et al., 2008). It is possible that PER36 may also be degraded by SBT1.7. Alternatively, considering the fact that exposure to •OH, together with O2•−, alters primary, secondary, and tertiary protein structures and increases susceptibility to proteolysis (Davies, 1987; Davies and Delsignore, 1987; Davies et al., 1987a, 1987b), both ROS and apoplastic proteases may regulate the degradation of PER36 within a coordinated pathway.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used as the wild type. The T-DNA insertion mutants SAIL_194_G03 (per36-1) and SALK_118195 (per36-2) were obtained from the ABRC at Ohio State University, and the T-DNA insertion sites were confirmed by DNA sequencing. The nars1, nars2, and nars1 nars2 double mutants were obtained as reported previously (Kunieda et al., 2008). The transgenic Arabidopsis plant expressing sGFP-BE under control of the cauliflower mosaic virus 35S promoter were described previously (Mano et al., 1999). Surface-sterilized seeds were sown onto 0.8% agar in Murashige and Skoog medium containing B5 vitamins and 1% Suc and grown at 22°C under continuous light. Three-week-old plants were transferred onto soil and grown at 22°C under 16-h-light conditions.
Generation of ProPER36:sGFP-GUS, ProPER36:PER36-GFPg4, and Pro35S:PER36-GFPg4 Transgenic Plants
For generation of transgenic plants expressing sGFP-GUS or PER36-GFPg4 under the control of the PER36 promoter or the cauliflower mosaic virus 35S promoter, R4pGWB533/ProPER36:sGFP-GUS, R4pGWB501/ProPER36:PER36GFPg4, and pH2GW7/Pro35S:PER36GFPg4 vectors were constructed. This cloning information is shown in Supplemental Table 2 online. The vectors were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformed bacteria were then infiltrated into Arabidopsis plants (Columbia-0 or per36-1) by the floral dip method (Clough and Bent, 1998). The sequences of the primers used are shown in Supplemental Table 3 online.
Staining with FM4-64 and Ruthenium Red
Developing seeds were stained with 50 μM FM4-64 (Invitrogen) for 0.5 h. Seed coat mucilage was stained with 0.01% ruthenium red under appropriate conditions for each experiment. For details, see the corresponding figure legends.
Confocal Laser Scanning Microscopy, Light Microscopy, and Electron Microscopy
Fluorescent samples were observed with a confocal laser scanning microscope (model LSM 510 META; Carl Zeiss) using the 488-nm line of a 40-mW Ar/Kr laser for GFP or the 544-nm line of a 1-mW He/Ne laser for FM4-64. The exported data were processed using Adobe Photoshop CS3 (Adobe Systems).
For observation of extruded mucilage, the seeds were examined using a stereoscopic microscope (model MVX10; Olympus) or a light microscope (Axioskop 2 plus system; Carl Zeiss) equipped with a CCD camera (model VB-7010; Keyence).
Developing seeds were fixed, dehydrated, and embedded as described previously (Kunieda et al., 2008). For histological studies, thin sections were stained with toluidine blue and examined with a light microscope (Axioskop 2 plus system). For ultrastructural studies, the ultrathin sections were stained with 0.4% uranyl acetate and lead citrate and examined using a transmission electron microscope (model H-7600; Hitachi High-Technologies) operated at 80 kV.
The surface of dry seeds was observed with a scanning electron microscope (model Miniscope TM-1000; Hitachi High-Technologies) without fixation.
Isolation of RNA, RT-PCR, and Quantitative RT-PCR
Total RNA was isolated from Arabidopsis tissues with the RNeasy Plant Mini Kit (Qiagen). Total RNA was used for the synthesis of cDNA with Ready-to-Go RT-PCR beads (GE Healthcare Bio-Sciences). PCR was performed with the cDNAs and ExTaq polymerase (Takara). The primer sets used are shown in Supplemental Table 3 online. Quantitative RT-PCR was performed essentially as described previously (Nakano et al., 2012) using the following gene-specific primer sets (Applied Biosystems): At02297801_g1 for PER36 and At02335270_gH for ACT2. The relative quantity of target mRNA was calculated using ACT2 as a standard.
SDS-PAGE and Immunoblot Analysis
Developing seeds or siliques of ProPER36:PER36-GFPg4/per36-1 transgenic plants were harvested at the proper developmental stages for each experiment. Protein extracts were subjected to SDS-PAGE followed by either Coomassie Brilliant Blue staining or immunoblot analysis. The following antibodies were used as primary and secondary antibodies: anti-GFP (JL-8) (Clontech; diluted 1:1000), anti-δVPE (1:5000) (Nakaune et al., 2005), anti-VSR1 (1:5000) (Yamada et al., 2005), anti-ALEU (1:5000) (Ueda et al., 2006), horseradish peroxidase–conjugated sheep anti-mouse IgG (GE Healthcare Bio-Sciences; diluted 1:2000), and horseradish peroxidase–conjugated goat anti-rabbit IgG (Pierce; diluted 1:2000). The antibodies were detected using the ECL or ECL Plus Western Blotting Detection Systems (GE Healthcare Bio-Sciences). The proteins were stained with Coomassie Brilliant Blue R 250.
Suborganellar Fractionation
Siliques with developing seeds at the torpedo-shaped embryo stage were homogenized in homogenizing buffer containing 50 mM HEPES-KOH, pH 7.5, 5 mM EDTA, 0.4 M Suc, and a protease inhibitor cocktail (Roche Diagnostics). The homogenate was filtered through a cell strainer (100-μm nylon; BD Biosciences) to remove cellular debris. The filtrate was centrifuged at 100,000g for 1 h at 4°C and separated into pellet (microsomal fraction) and supernatant (soluble fraction).
Enzyme Assay
Three-week-old plants (0.5 g) of sGFP-BE, Pro35:PER36-GFPg4/per36-1, and per36-1 were homogenized in three volumes of cold homogenizing buffer containing 20 mM HEPES-NaOH, pH 7.0, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (Roche Diagnostics). Homogenates were centrifuged at 20,000g for 10 min at 4°C, and the supernatants were collected as total fractions. The total fractions were incubated with magnetic beads conjugated to an anti-GFP antibody (Miltenyi Biotec) on ice for 30 min and then applied to μColumns (Miltenyi Biotec) in a magnetic field. After extensive washing with the buffer, proteins bound to magnetic beads were collected. The total and bound fractions were subjected to enzyme assay and immunoblot analysis.
Peroxidase activity was determined spectrophotometrically using 3,3′,5,5′-tetrametylbenzidine (TMB) as the substrate. The reaction mixture contained 50 mM sodium acetate buffer, pH 5.5, 0.5 mM TMB, 0.00525% (v/v) hydrogen peroxide, and the enzyme in a total volume of 200 μL. The reaction was initiated by the addition of the enzyme, and the change in absorbance at 650 nm was monitored for 5 min with a spectrophotometer (model Infinite 200 PRO; TECAN). The extinction coefficient for TMB was 39.0 mM−1 cm−1 at 653 nm.
Monosaccharide Composition Analysis
Mucilage for high-performance anion exchange chromatography was prepared essentially as described by Dean et al. (2007). Dry seeds (30 mg) were pretreated with 50 μL 1 M sodium carbonate for 30 min. After adding 1.2 mL of water, the samples were shaken for 2 h and then the extract was collected as a sodium carbonate fraction. The seeds were further shaken with 300 μL 4 M KOH containing 0.2% sodium borohydride for 1 h and then the extract was collected and neutralized with 75 μL of acetic acid. We repeated this treatment twice and collected the extract (total 1125 µL) as a KOH fraction. Either the sodium carbonate fraction or the KOH fraction was mixed with four volumes ethanol and then centrifuged. The precipitate mucilage was hydrolyzed with 2 n trifluoroacetic acid for 90 min at 120°C. The neutral and acidic monosaccharide sugars derived from the mucilage were analyzed by high-performance anion exchange chromatography (model ICS-5000; Dionex) with a CarboPac PA1 column (Dionex).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PER36 (At3g50990), ACT2 (At3g18780), ALEU (At5g60360), BXL1 (At5g49360), HRPC1 (M37156), MUM1/LUH (At2g32700), MUM2 (At5g63800), MUM4 (At1g53500), SBT1.7 (At5g67360), PMEI6 (At2g47670), NARS1 (At3g15510), NARS2 (At1g52880), PDF1 (At2g42840), δVPE (At3g20210), and VSR1 (At3g52850).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. NAC Transcription Factors NARS1 and NARS2 Regulate PER36 Expression.
Supplemental Figure 2. Analysis of PER36 Gene Expression during Early Seed Development.
Supplemental Figure 3. Quantitative RT-PCR Showing the Expression Levels of PER36 mRNA in Developing per36-1 and per36-2 Seeds at the Torpedo-Shaped Embryo Stage.
Supplemental Figure 4. Transmission Electron Micrographs Showing Filamentous Structures in the Mucilage of the Outermost Integument Cells.
Supplemental Figure 5. Comparison of Mucilage Extrusion in Dry Seeds of the Wild Type, Two per36 Mutant Alleles, and Two Independent Lines of ProPER36:PER36-GFPg4/per36-1.
Supplemental Figure 6. Comparison of the Amino Acid Sequences between PER36 and HORSERADISH PEROXIDASE C1.
Supplemental Table 1. Monosaccharide Composition of Mucilage in Dry Seeds of the Wild Type and Two per36 Mutant Alleles.
Supplemental Table 2. DNA Constructs Used in This Study.
Supplemental Table 3. Primer Sets Used in This Study.
Supplementary Material
Acknowledgments
We thank Tsuyoshi Nakagawa (Shimane University) for his kind donation of the Gateway vectors (R4pGWB501 and R4pGWB533), the Flanders Institute for Biotechnology (VIB) for providing the pH2GW7 vector, and the ABRC for providing seeds of Arabidopsis T-DNA insertion mutants (SAIL_194_G03 and SALK_118195). We thank Kazuyuki Wakabayashi (Osaka City University), Arata Yoshinaga (Kyoto University), Keiji Takabe (Kyoto University), and Ryusuke Yokoyama (Tohoku University) for valuable discussion and Mitsue Fukazawa (National Institute for Basic Biology) for her technical support. This work was supported by Grants-in-Aid for Specially Promoted Research to I.H-.N. (no. 22000014), by Kyoto University Start-Up Grant-in-Aid for young scientists to T.K., by a research fellowship to T.K. (nos. 19004891 and 22003439) from the Japan Society for the Promotion of Science, and by the Global Center of Excellence Program “Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
AUTHOR CONTRIBUTIONS
T.K., T.S., and I.H-.N. designed the research. T.K. performed all experiments except transmission electron microscopy. M.K. and M.N. performed light microscopy of cross sections and transmission electron microscopy. K.N. performed monosaccharide composition analysis. T.K., T.S., and I.H-.N. wrote the article.
Glossary
- GFP
green fluorescent protein
- DAP
days after pollination
- ROS
reactive oxygen species
- TMB
3,3′,5,5′-tetrametylbenzidine
References
- Abe M., Takahashi T., Komeda Y. (1999). Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 40: 571–580 [DOI] [PubMed] [Google Scholar]
- Arsovski A.A., Popma T.M., Haughn G.W., Carpita N.C., McCann M.C., Western T.L. (2009). AtBXL1 encodes a bifunctional β-D-xylosidase/α-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T., Rycke R.D., Viane R., Inze D. (2000). Histological Study of Seed Coat Development in Arabidopsis thaliana. J. Plant Res. 113: 139–148 [Google Scholar]
- Bui M., Lim N., Sijacic P., Liu Z. (2011). LEUNIG_HOMOLOG and LEUNIG regulate seed mucilage extrusion in Arabidopsis. J. Integr. Plant Biol. 53: 399–408 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosio C., Dunand C. (2009). Specific functions of individual class III peroxidase genes. J. Exp. Bot. 60: 391–408 [DOI] [PubMed] [Google Scholar]
- Davies K.J. (1987). Protein damage and degradation by oxygen radicals. I. General aspects. J. Biol. Chem. 262: 9895–9901 [PubMed] [Google Scholar]
- Davies K.J., Delsignore M.E. (1987). Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J. Biol. Chem. 262: 9908–9913 [PubMed] [Google Scholar]
- Davies K.J., Delsignore M.E., Lin S.W. (1987a). Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J. Biol. Chem. 262: 9902–9907 [PubMed] [Google Scholar]
- Davies K.J., Lin S.W., Pacifici R.E. (1987b). Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J. Biol. Chem. 262: 9914–9920 [PubMed] [Google Scholar]
- Dean G.H., Zheng H., Tewari J., Huang J., Young D.S., Hwang Y.T., Western T.L., Carpita N.C., McCann M.C., Mansfield S.D., Haughn G.W. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Léon-Kloosterziel K.M., Koornneef M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C. (1998). Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochem. J. 332: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G., Chaudhury A. (2005). Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Huang J., DeBowles D., Esfandiari E., Dean G., Carpita N.C., Haughn G.W. (2011). The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol. 156: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. (2008). A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Nishimori Y., Sugisawa M., Makino A., Mae T. (1997). The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol. 38: 471–479 [DOI] [PubMed] [Google Scholar]
- Jamet E., Canut H., Boudart G., Pont-Lezica R.F. (2006). Cell wall proteins: A new insight through proteomics. Trends Plant Sci. 11: 33–39 [DOI] [PubMed] [Google Scholar]
- King S.P., Lunn J.E., Furbank R.T. (1997). Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol. 114: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M. (1981). The complex syndrome of ttg mutants. Arabid. Inf. Serv. 18: 45–51 [Google Scholar]
- Kunieda T., Mitsuda N., Ohme-Takagi M., Takeda S., Aida M., Tasaka M., Kondo M., Nishimura M., Hara-Nishimura I. (2008). NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 20: 2631–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Nawrath C., Bär S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Métraux J.P., Yephremov A. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A., Ralet M.C., Loudet O., Kronenberger J., Mouille G., Marion-Poll A., North H.M. (2007). A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Hayashi M., Nishimura M. (1999). Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17: 309–320 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Nakano R.T., Matsushima R., Nagano A.J., Fukao Y., Fujiwara M., Kondo M., Nishimura M., Hara-Nishimura I. (2012). ERMO3/MVP1/GOLD36 is involved in a cell type-specific mechanism for maintaining ER morphology in Arabidopsis thaliana. PLoS ONE 7: e49103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaune S., Yamada K., Kondo M., Kato T., Tabata S., Nishimura M., Hara-Nishimura I. (2005). A vacuolar processing enzyme, deltaVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 17: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa K., Maruyama N., Satoh R., Fuchikami Y., Higasa T., Utsumi S. (2003). A C-terminal sequence of soybean β-conglycinin α’ subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 34: 647–659 [DOI] [PubMed] [Google Scholar]
- Passardi F., Penel C., Dunand C. (2004). Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Penfield S., Meissner R.C., Shoue D.A., Carpita N.C., Bevan M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Usadel B., Neumetzler L., Hartmann J., Büssis D., Altmann T. (2008). A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S., Ralet M.C., Berger A., Botran L., Ropartz D., Marion-Poll A., North H.M. (2013). PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schweikert C., Liszkay A., Schopfer P. (2000). Scission of polysaccharides by peroxidase-generated hydroxyl radicals. Phytochemistry 53: 565–570 [DOI] [PubMed] [Google Scholar]
- Sheen J., Zhou L., Jang J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2: 410–418 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., Takahashi T., Ohtomo I., Nishimura M., Hara-Nishimura I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47: 164–175 [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M., Tehseen M., Doblin M.S., Pettolino F.A., Wilson S.M., Bacic A., Golz J.F. (2011). The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol. 156: 46–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Tanaka H., Watanabe D., Machida C., Machida Y. (2004). The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39: 298–308 [DOI] [PubMed] [Google Scholar]
- Weber H., Borisjuk L., Heim U., Buchner P., Wobus U. (1995). Seed coat-associated invertases of fava bean control both unloading and storage functions: Cloning of cDNAs and cell type-specific expression. Plant Cell 7: 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L. (2012). The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 22: 1–25 [Google Scholar]
- Western T.L., Burn J., Tan W.L., Skinner D.J., Martin-McCaffrey L., Moffatt B.A., Haughn G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Young D.S., Dean G.H., Tan W.L., Samuels A.L., Haughn G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor J.B., Symonds V.V., Mendenhall J., Lloyd A.M. (2000). Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Wobus U., Weber H. (1999). Seed maturation: Genetic programmes and control signals. Curr. Opin. Plant Biol. 2: 33–38 [DOI] [PubMed] [Google Scholar]
- Yamada K., Fuji K., Shimada T., Nishimura M., Hara-Nishimura I. (2005). Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 41: 888–898 [DOI] [PubMed] [Google Scholar]
- Young R.E., McFarlane H.E., Hahn M.G., Western T.L., Haughn G.W., Samuels A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.