This work shows that, similarly to green algae, vascular plants can acclimate to singlet oxygen (1O2). The Arabidopsis ch1 mutant is highly photosensitive due to increased release of 1O2 by photosystem II, but preexposure of ch1 plants to moderately elevated light intensities eliminated oxidative damage in high light without suppressing 1O2 formation. Regulation of the jasmonate biosynthesis pathway is a key factor in this acclimation process.
Abstract
Singlet oxygen (1O2) is a reactive oxygen species that can function as a stress signal in plant leaves leading to programmed cell death. In microalgae, 1O2-induced transcriptomic changes result in acclimation to 1O2. Here, using a chlorophyll b–less Arabidopsis thaliana mutant (chlorina1 [ch1]), we show that this phenomenon can also occur in vascular plants. The ch1 mutant is highly photosensitive due to a selective increase in the release of 1O2 by photosystem II. Under photooxidative stress conditions, the gene expression profile of ch1 mutant leaves very much resembled the gene responses to 1O2 reported in the Arabidopsis mutant flu. Preexposure of ch1 plants to moderately elevated light intensities eliminated photooxidative damage without suppressing 1O2 formation, indicating acclimation to 1O2. Substantial differences in gene expression were observed between acclimation and high-light stress: A number of transcription factors were selectively induced by acclimation, and contrasting effects were observed for the jasmonate pathway. Jasmonate biosynthesis was strongly induced in ch1 mutant plants under high-light stress and was noticeably repressed under acclimation conditions, suggesting the involvement of this hormone in 1O2-induced cell death. This was confirmed by the decreased tolerance to photooxidative damage of jasmonate-treated ch1 plants and by the increased tolerance of the jasmonate-deficient mutant delayed-dehiscence2.
INTRODUCTION
Photosynthetic organisms have to cope with the toxicity of reactive oxygen species (ROS), which are continually produced by various cellular functions, especially in the chloroplasts (Apel and Hirt, 2004; Li et al., 2009). A variety of sophisticated systems participate in the protection of cells against ROS toxicity, including low molecular weight antioxidants (ascorbate, carotenoids, glutathione, and tocopherols) and a panoply of enzymatic proteins distributed in all cellular compartments. When ROS production overwhelms the cellular antioxidant capacity, ROS can induce oxidative damage by reacting with a large range of biomolecules, including lipids, proteins, and nucleic acids, essential for the activity and integrity of the cells. Light-induced ROS formation is exacerbated by various environmental stress conditions that inhibit the photosynthetic processes, resulting in overreduction of the electron transport chain and accumulation of excited chlorophylls unable to transfer rapidly their energy to the photosynthetic reaction centers. Triplet chlorophylls are thus formed, which can interact with triplet state molecular oxygen to generate singlet oxygen (1O2) (Krieger-Liszkay et al., 2008; Triantaphylidès and Havaux, 2009). 1O2 has been proposed to be the major ROS produced in plant cells exposed to excess light (González-Pérez et al., 2011) and to be responsible for photooxidative damage to plant leaves (Triantaphylidès et al., 2008). Its high reactivity and short lifetime limit its capacity of diffusion in cells (Moan, 1990) and explain its reaction in close proximity to its production site (Triantaphylidès and Havaux, 2009).
In addition to its cytotoxicity, 1O2 is known to be a key signal molecule involved in cellular responses (Mittler et al., 2004; Suzuki et al., 2012). Indeed, ROS have several important physiological functions, such as regulation of plant development (Foreman et al., 2003) or defense against pathogens (O’Brien et al., 2012). However, the study of the specific biological activity of 1O2 is difficult due to the fact that various chemically distinct ROS can be generated simultaneously in cells under high-light stress. In addition to ROS coproduction, 1O2 signaling can communicate with other ROS signals, as shown by the antagonizing effects of O2−/hydrogen peroxide (H2O2) on 1O2-induced gene expression (Laloi et al., 2007; Kim et al., 2008). Other signaling molecules, including hormones, such as auxins, ethylene, and abscisic acid, and defense-related signals, such as salicylate and jasmonate, are also interconnected, making the signaling network complex (Reymond and Farmer, 1998; Reinbothe et al., 2009; Ballaré, 2011) but allowing the metabolism and physiology of the plants to be modulated and to adapt to changes in the environmental conditions.
The conditional flu mutant of Arabidopsis thaliana enabled a breakthrough in the elucidation of 1O2 effects on plants. The flu mutant allows selective accumulation of 1O2 within plastids in a noninvasive and controlled manner, providing the opportunity to link plant cell responses to a particular ROS. This mutant accumulates photosensitizing chlorophyll precursors in the dark and therefore generates massive amounts of 1O2 in the stroma of the chloroplasts during dark-to-light transitions (Meskauskiene et al., 2001; op den Camp et al., 2003; Wagner et al., 2004). Studies performed on the flu mutant have identified distinct sets of genes specifically activated by 1O2 (op den Camp et al., 2003; Wagner et al., 2004; Gadjev et al., 2006). 1O2 also promotes programmed cell death by activating two nucleus-encoded chloroplast-localized proteins denoted EXECUTER1 (EX1) and EX2, leading to growth inhibition and death (Lee et al., 2007). The current view is that 1O2-mediated cell death program does not operate as a linear pathway but rather forms a complex signaling network interconnected with other signaling routes.
Specific acclimation to 1O2 was found in the unicellular alga Chlamydomonas reinhardtii in response to a pretreatment with sublethal doses of 1O2 generated by the photosensitizer dye rose bengal (Ledford et al., 2007). This pretreatment was sufficient to induce changes in 1O2-responsive gene expression, such as GLUTATHIONE PEROXIDASE H and GLUTATHIONE S TRANSFERASE S1 (Leisinger et al., 2001; Fischer et al., 2005). No cross-acclimation to other ROS generators could be observed; however, it was discovered that high light induces acclimation to 1O2 stress, demonstrating an overlap between 1O2 responses and high-light exposure (Ledford et al., 2007). Even if several plastid and cytosolic proteins or signaling molecules are involved in chloroplast-to-nucleus retrograde signaling (Estavillo et al., 2011, 2012; Inaba et al., 2011; Leister, 2012), the molecules transmitting the plastid 1O2 signal out of the chloroplast are still largely unknown. However, one of them seems to have been recently identified in Arabidopsis: β-Cyclocitral, a volatile derivative of β-carotene that accumulates in Arabidopsis leaves under high-light stress, was found to induce changes in the expression of a large set of 1O2-specific genes (Ramel et al., 2012a). Treatments of Arabidopsis plants with this volatile molecule were also associated with increased tolerance to photooxidative stress, suggesting that β-cyclocitral is an intermediate in the 1O2 signaling pathway leading to acclimation.
The ability of Arabidopsis to acclimate to 1O2 is investigated in this work using the chlorina1 (ch1) mutant. This pale-green mutant is characterized by a deficiency in chlorophyll b (Espineda et al., 1999), and, as a consequence, it is completely devoid of photosystem II (PSII) chlorophyll-protein antenna complexes (Havaux et al., 2007). Loss of the normal structural architecture around the PSII reaction centers in chlorophyll b–less plant mutants has significant effects on the functionality of the PSII core reaction center complexes, including loss of nonphotochemical energy quenching (NPQ), impaired oxidizing side of PSII, and reduced grana stacking, which lead in fine to enhanced sensitivity to PSII photoinactivation (Leverenz et al., 1992; Havaux and Tardy, 1997; Kim et al., 2009). Accordingly, the ch1 Arabidopsis mutant displayed an extreme sensitivity to photooxidative stress in high light (Havaux et al., 2007), and this was attributed to an enhanced release of 1O2 from the naked PSII centers (Triantaphylidès et al., 2008; Dall’Osto et al., 2010). Like in the flu mutant, this mutant allows large amounts of 1O2 to be produced without significant coproduction of other ROS. However, in contrast with the flu mutant, 1O2 formation is localized in the natural site of production (i.e., PSII). Moreover, the ch1 mutant does not require any special growth conditions and thus appears to be a more appropriate model for studying 1O2 acclimation mechanisms in plants. Using this 1O2-producing mutant, we show here that vascular plants do acclimate to 1O2 when exposed to mild light stress conditions that produce low, sublethal internal 1O2 concentrations, and we provide data emphasizing the regulation of jasmonate synthesis as a key factor in this acclimation.
RESULTS
Photosensitivity of the Arabidopsis ch1 Mutant
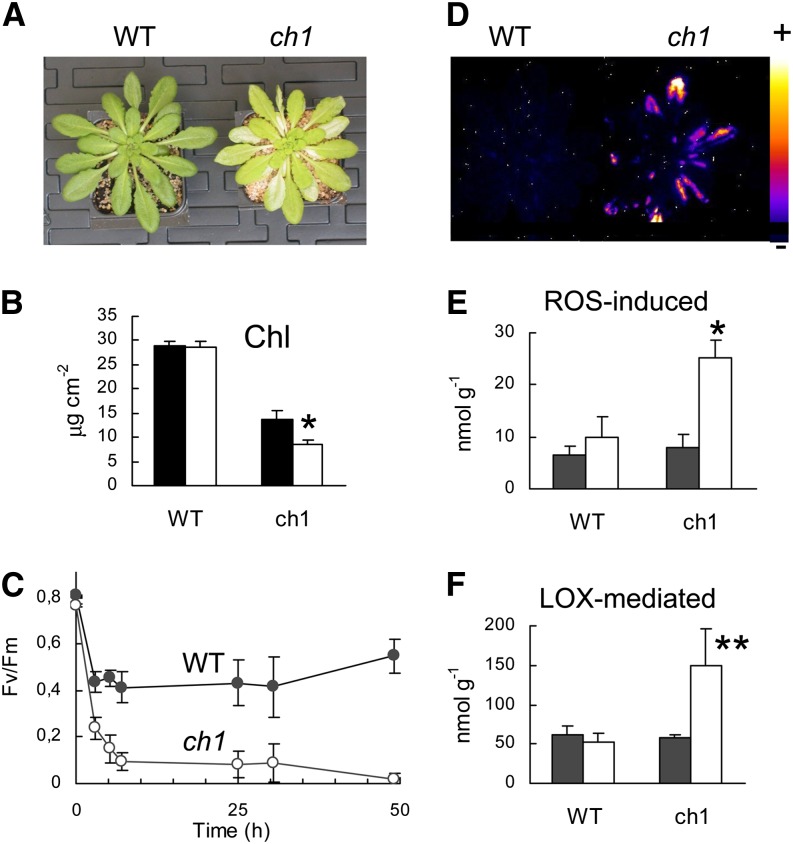
Wild-type and ch1 Arabidopsis plants were transferred from the growth light conditions (180 μmol photons m−2 s−1) to high-light stress (1000 μmol photons m−2 s−1; Figure 1). To avoid leaf heating during the high-light treatment, the air temperature was decreased to 10°C, leading to leaf temperature (approximately 18°C) close to the temperature of control leaves exposed to 180 μmol photons m−2 s−1. Most leaves of ch1 plants bleached within 2 d (Figure 1A), indicating extensive photooxidation of the pigmented photosynthetic complexes. This was confirmed by HPLC analysis of photosynthetic pigments, showing a strong reduction (∼40%) of the chlorophyll content in ch1 mutant leaves after light stress (Figure 1B), and by chlorophyll fluorescence measurements revealing a drastic inhibition of PSII photochemistry (Figure 1C). No such effects were observed in the wild type: No leaf bleaching was visible after the high-light treatment, the chlorophyll concentration remained unchanged, and PSII photoinhibition was much less pronounced than in ch1. Moreover, lipid peroxidation was induced in ch1 mutant plants as shown by autoluminescence imaging of lipid peroxides (Figure 1D). We also measured lipid peroxidation by HPLC quantification of products derived from the oxidation of linolenic acid (hydroxy-octadecatrienoic acids [HOTEs]) after reduction of the extract with triphenyl phosphine (i.e., after conversion of the peroxides into alcohols) (Figures 1E and 1F). Both ROS-induced lipid peroxidation (Figure 1E) and lipoxygenase (LOX)–dependent lipid peroxidation (Figure 1F) were stimulated by high light in the mutant. By contrast, no induction of lipid peroxidation was found in high-light-treated wild-type plants. The results presented in Figure 1 illustrate the high photosensitivity of the ch1 mutant compared with its wild type, thus confirming previous observations (Havaux et al., 2007; Dall’Osto et al., 2010).
Figure 1.
Photosensitivity of the ch1 Arabidopsis Mutant.
Plants were exposed for 2 d to high-light stress (1000 μmol photons m−2 s−1 at 10°C).
(A) Wild-type (WT) and ch1 mutant plants after high-light stress, showing leaf bleaching in the mutant.
(B) Effect of high-light stress on the total chlorophyll content (Chl) of wild-type and ch1 leaves. Black, before stress; white, after the stress treatment. n = 3.
(C) PSII inhibition, as measured by the decrease in the chlorophyll fluorescence ratio Fv/Fm, during exposure of wild-type and ch1 plants to high-light stress. n = 9.
(D) Autoluminescence imaging of lipid peroxidation in wild-type and ch1 mutant plants after light stress. No luminescence was detected in wild-type and ch1 plants before light stress. The color palette on the right indicates the intensity of the luminescence signal from low (dark blue) to high (white).
(E) and (F) Effects of high-light stress on ROS-induced lipid peroxidation ([E]; HOTE levels, in nmol g−1 fresh weight) and LOX-mediated lipid peroxidation ([F]; HOTE levels, nmol g−1 fresh weight) in leaves of wild-type and ch1 plants. Black, before stress; white, after stress. n = 3. Data are mean values + sd. Asterisks indicate significant difference from the control (unstressed) at *P < 0.001 and **P < 0.05 (Student’s t test).
The ch1 Mutant Is a 1O2 Overproducer
As a major consequence of the lack of chlorophyll b, the ch1 mutant completely lacks assembled pigment-protein antenna complexes of PSII (light-harvesting complex II) with relatively little effect on photosynthetic electron flow at high photon flux densities (PFDs) (Havaux et al., 2007). The photosynthetic capacity of ch1 leaves was previously reported to reach ∼80% of the wild-type level (Kim et al., 2009), and this was confirmed in our study by measuring in vitro the photosynthetic electron flow rate in isolated thylakoids exposed to a high PFD (107 ± 11 versus 91 ± 6 μmol O2 mg−1 chlorophyll h−1 for wild-type and ch1 thylakoids, respectively, corresponding to a 15% decrease in the electron flow capacity in the mutant). Using the 1O2-specific fluorescent probe singlet oxygen sensor green (SOSG), Dall’Osto et al. (2010) previously showed that 1O2 production during illumination was substantially enhanced in ch1 leaves compared with wild-type leaves, whereas no difference was observed with fluorescent probes indicative of superoxide or hydroxyl radical. The high sensitivity of the ch1 mutant to high light stress was thus attributed to 1O2 accumulation and toxicity. Accordingly, the HOTE signature of lipid peroxidation in ch1 leaves was found to be typical of 1O2-induced lipid peroxidation (Triantaphylidès et al., 2008).
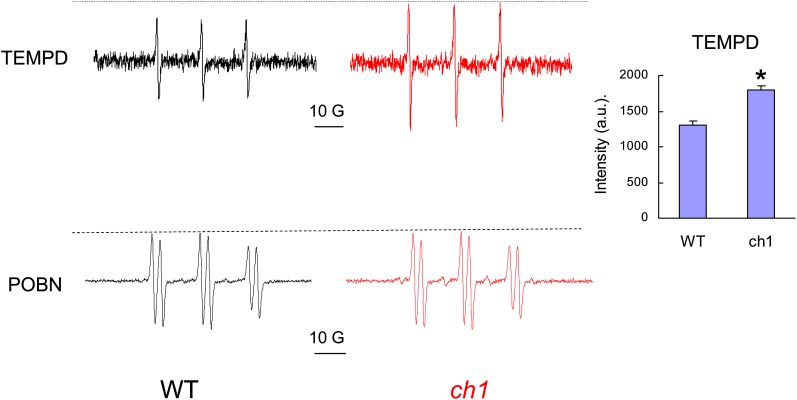
To confirm the increased 1O2 production in ch1 compared with the wild type, we prepared thylakoid membranes from wild-type and ch1 mutant leaves and measured light-induced ROS production using spin trapping electron paramagnetic resonance (EPR) spectroscopy. The spin trap 2,2,6,6-tetramethyl-4-piperidone chloride (TEMPD) was used to detect 1O2 (Lion et al., 1980). As shown from the EPR traces presented in Figure 2, the formation of 1O2 by ch1 thylakoids exposed for 3 min to high light was enhanced by ∼20% compared with wild-type thylakoids. By contrast, the EPR trace of 4-pyridyl-1-oxide-N-tert-butylnitrone (POBN), a hydroxyl radical probe, did not reveal any difference between the wild type and ch1. This confirms the selective accumulation of 1O2 in ch1 under high-light stress.
Figure 2.
1O2 Production in the ch1 Mutant.
EPR signals of TEMPD and POBN spin traps in wild-type (WT) and ch1 thylakoids exposed to high light (1500 μmol m−2 s−1, 3 and 1 min, respectively). Thylakoid suspensions were adjusted for the same chlorophyll concentration (40 μg mL−1). Inset: intensity of the TEMPD EPR signal. Data are mean values of three experiments + sd. Asterisk indicates significant difference from the wild type at P < 0.001 (Student’s t test). a.u., arbitrary units
[See online article for color version of this figure.]
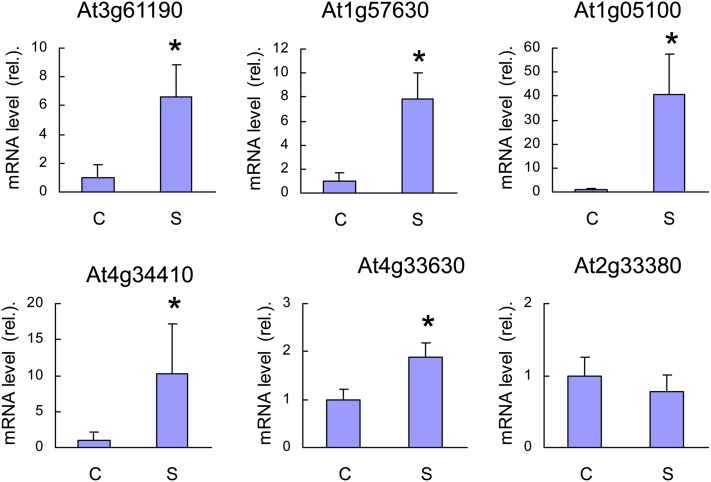
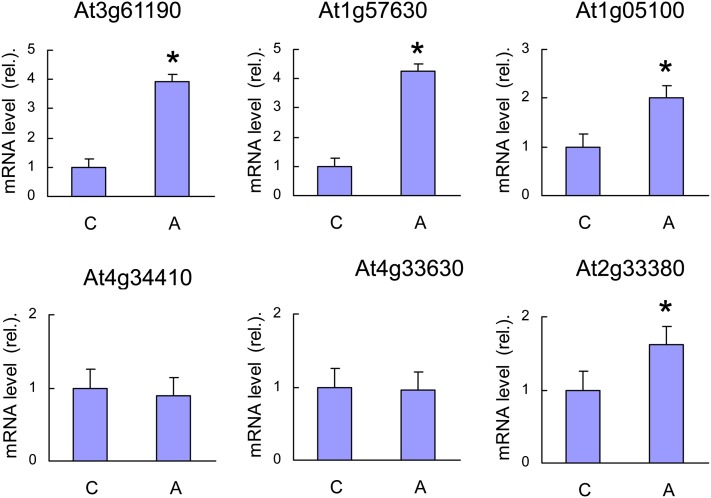
As another way of monitoring 1O2 production in ch1 leaves, we measured the expression levels of different 1O2-responsive genes in ch1 before and after high-light stress (1000 μmol m−2 s−1, 10°C, 2 d) (Figure 3): At3g61190 (BAP1), At1g57630 (disease resistance protein [TIR class] putative), At1g05100 (MAPKKK18), At4g34410 (AP2 domain-containing transcription factor), At4g33630 (EX1), and At2g33380 (RD20). All genes except At2g33380 were noticeably induced by the light treatment, indicating that ch1 leaves do produce 1O2 during high-light stress. Taken together with previously published results, the data presented in Figures 2 and 3 clearly show that ch1 is a 1O2 producer in the light. Additional evidence for this phenomenon will be provided below using fluorescent and biochemical 1O2 markers (see below).
Figure 3.
qRT-PCR Analysis of the Expression Levels of 1O2-Responsive Genes in ch1 Leaves.
C, control conditions; S, stress conditions (1000 μmol photons m−2 s−1, 2 d). At3g61190 = BAP1, At1g57630 = disease resistance protein (TIR class) putative, At1g05100 = MAPKKK18, At4g34410 = AP2 domain-containing transcription factor, At4g33630 = EX1, and At2g33380 = RD20. Data are mean values of five measurements from independent RNA extractions + sd. Asterisk indicates significant difference from control at P < 0.05 (Student’s t test).
[See online article for color version of this figure.]
Transcriptomic Analysis of the ch1 Mutant under Photooxidative Stress Conditions
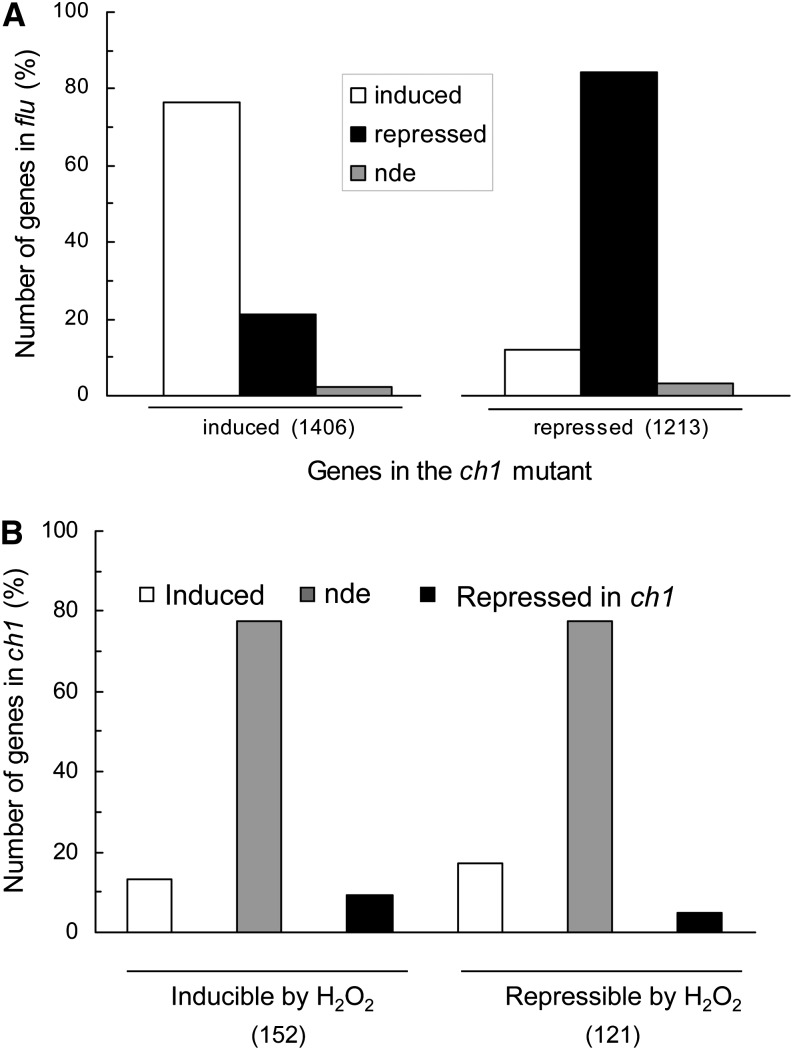
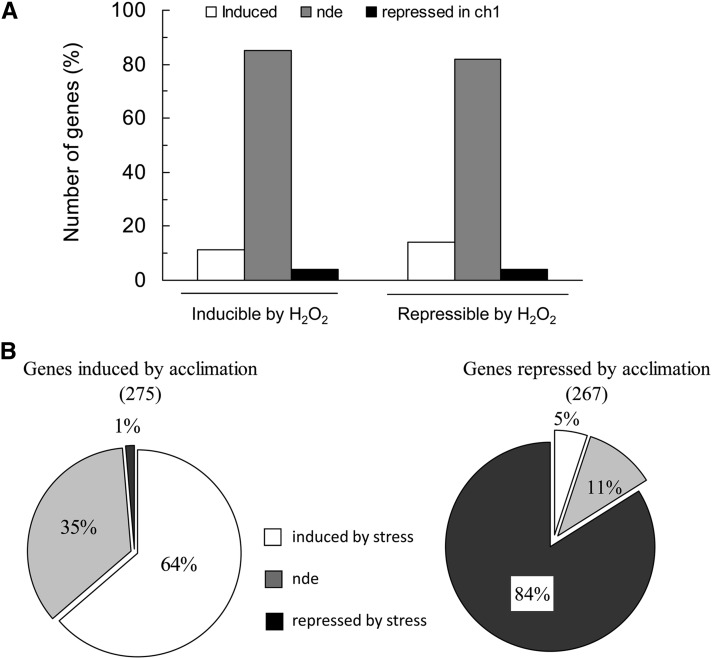
Transcriptomic analysis of ch1 mutant plants exposed for 2 d to high-light stress was performed using CATMAv5 microarrays (Crowe et al., 2003). Compared with control conditions, 1406 genes were induced by high-light stress conditions, while 1213 genes were repressed (number of genes that showed statistically significant and greater than twofold change in expression; see Methods and Supplemental Data Set 1 online; more than twofold changes in expression were used to limit the number of genes and to identify interesting genes exhibiting relatively strong responses to stress conditions). A comparison of the gene expression profile of ch1 leaves exposed to high-light stress with the expression profile induced by 1O2 in flu leaves (op den Camp et al., 2003) shows a strong similarity between the two mutants: Of the 1406 genes induced by high-light stress in ch1, ∼80% were also induced in flu, while the rest were either repressed or not differentially expressed in flu (Figure 4A). Similarly, of the 1213 genes repressed in the ch1 mutant, ∼80% were also repressed in the flu mutant. This high similarity in gene expression between the two mutants is consistent with the finding that, similarly to flu, ch1 is a 1O2-producing mutant. Moreover, Supplemental Table 1 online shows that many 1O2-specific genes (which do not respond to other ROS, such as H2O2, according to Gadjev et al., 2006) were induced in ch1 plants exposed to high-light stress, providing additional evidence of 1O2 formation in the mutant exposed to high-light stress. A similar induction of 1O2-specific genes was not observed in wild-type leaves exposed to the same changes in the light conditions (see Supplemental Table 1 online). We also analyzed genes that were previously identified as H2O2 inducible or repressible (Gadjev et al., 2006). The expression of most of those genes (∼75%) was not modified by high light in ch1: Less than 15% of the H2O2-inducible genes were induced by high light in ch1 and ∼5% of the H2O2-repressible genes were repressed by high light (Figure 4B). Thus, the expression of most H2O2-responsive genes was not affected by high-light stress in ch1. Clearly, the changes in gene expression in ch1 mutant plants exposed to high-light stress do not correspond to a response to H2O2 stress.
Figure 4.
Microarray-Based Analysis of Gene Expression Changes in ch1 Mutant Leaves during High-Light Stress (1000 μmol Photons m−2 s−1, 2 d).
(A) Comparison of the gene expression profile in ch1 and flu leaves. The plot shows the percentage of genes induced or repressed in ch1 leaves during high-light stress (more than twofold change) that were induced, repressed, or not differentially expressed (compared with control conditions) in the flu Arabidopsis mutant after a dark-to-light transfer. In parentheses: number of genes induced or repressed in the ch1 mutant. This comparison is based on the microarray study of the flu mutant performed by op den Camp et al. (2003) (2-h illumination after a dark period). The latter study used the Affymetrix Gene Chip Arabidopsis ATHI Genome Array containing >22,500 probe sets representing around 24,000 genes.
(B) Percentage of genes inducible or repressible by H2O2 that were induced, repressed, or not differentially expressed (compared with control conditions) in the ch1 mutant during high-light stress. This comparison is based on Gadjev et al. (2006). The latter study used the Affymetrix Gene Chip Arabidopsis ATHI genome array. In parentheses: number of genes from the Gadjev study used for the comparison with ch1. nde, not differentially expressed.
Although the ch1 and flu mutants showed strong similarities in terms of gene expression changes in response to 1O2 stress, photooxidative damage of ch1 plants was found to be independent of the EX1 protein that regulates cell death in the flu mutant (Lee et al., 2007). We could not distinguish the double mutant ch1 ex1 from the single mutant ch1 during photooxidative stress: Lipid peroxidation and PSII photoinhibition after high-light stress were both induced to a similar extent in ch1 and ch1 ex1 plants (see Supplemental Figure 1 online). Consequently, we can conclude that 1O2-induced damage and cell death are mediated by different mechanisms in ch1 and flu. Possibly, differences in the intensity of the 1O2 stress and of the associated gene responses as well as in the way 1O2 is produced between the two mutants could explain the involvement of distinct signaling pathways.
The ch1 Mutant Is Able to Acclimate to 1O2
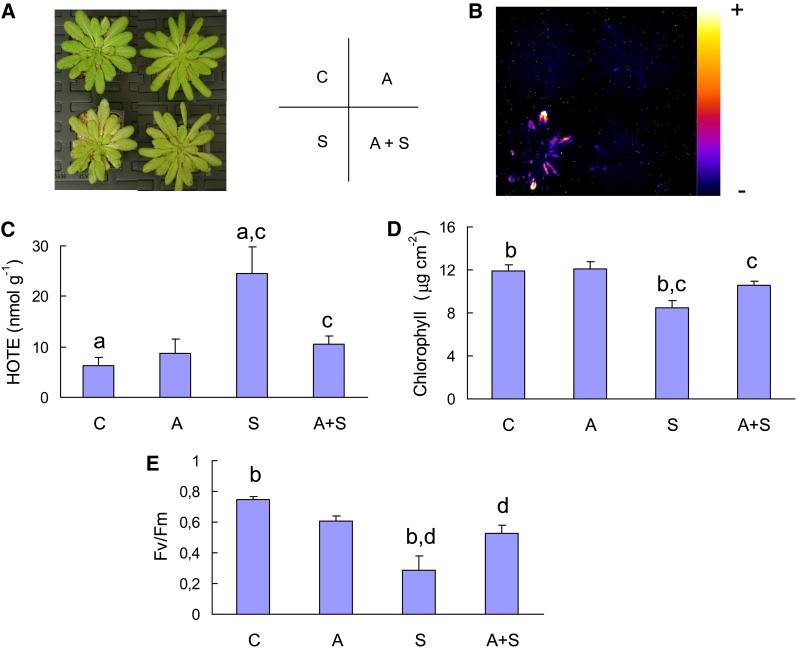
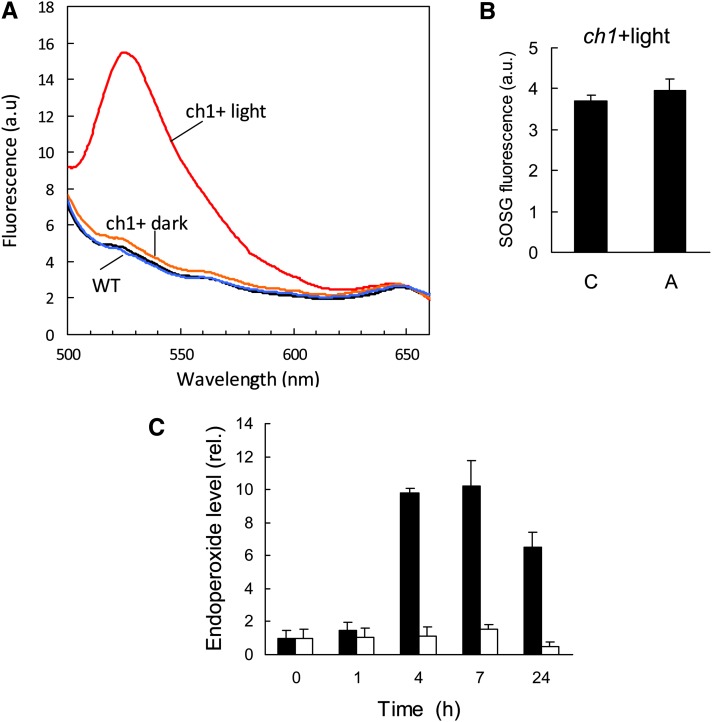
Although the ch1 mutant is highly photosensitive, it is able to acclimate to high-light stress. Indeed, transfer of ch1 plants for 2 d to a moderately elevated PFD of 450 μmol m−2 s−1 (at 20°C) eliminated lipid peroxidation (Figures 5B and 5C) and leaf bleaching (Figures 5A and 5D) during subsequent high-light stress. Also, the light pretreatment protected PSII from photoinhibition (Figure 5E). Nevertheless, some of the 1O2 gene markers that were induced during high-light stress were also induced during the acclimatory treatment (Figure 6), but the increases in the expression of 1O2-responsive genes during acclimation (Figure 6) were low compared with the gene expression changes observed during high-light stress (Figure 3B), suggesting lower production of 1O2 during acclimation. Formation of 1O2 in ch1 leaves during photoacclimation is also supported by the fluorescence of SOSG that was induced by illumination of ch1 leaves with light of PFD 450 μmol m−2 s−1 (Figure 7A). This phenomenon was not observed in the wild type exposed to the same PFD. Interestingly, 1O2 production at 450 μmol m−2 s−1 was still observed in ch1 plants after 2 d of exposure to the acclimation conditions (Figure 7B). In other words, acclimation was not associated with a suppression of 1O2 production by leaves.
Figure 5.
Acclimation of the ch1 Mutant to High-Light Stress.
Plants were exposed to acclimatory conditions (A = 450 μmol m−2 s−1 for 2 d at 20°C) and/or exposed to stress conditions (S = 1000 μmol m−2 s−1 for 2 d).
(A) Plants after the different treatments (A, S, C = control; A+S = acclimation followed by stress treatment). The position of the different plants (C, A, S, A+S) is indicated by the letters on the right.
(B) Autoluminescence imaging of lipid peroxidation.
(C) ROS-induced lipid peroxidation (HOTE levels, nmol g−1 fresh weight). n = 3.
(D) Chlorophyll concentration. n = 5.
(E) PSII inhibition (Fv/Fm chlorophyll fluorescence parameter). n = 8. Data are mean values + sd. Experimental values indicated by the same letter are significantly different. a and c, significantly different at P < 0.05 and 0.015, respectively (Student’s t test). b or d, significantly different at P < 0.001.
[See online article for color version of this figure.]
Figure 6.
Expression Levels of 1O2-Responsive Genes in ch1 Leaves during Acclimation at a PFD of 450 μmol m−2 s−1.
The same 1O2-responsive genes as those shown in Figure 3 were analyzed by qRT-PCR. See legend of Figure 3 for the identification of the genes. C = control; A = acclimated (450 μmol m−2 s−1 for 2 d). Data are mean values of five measurements from independent RNA extractions + sd. Asterisk indicates significant difference from control (C) at P < 0.05 (Student’s t test).
[See online article for color version of this figure.]
Figure 7.
1O2 Production in ch1 Leaves under the Acclimatory Conditions (450 μmol m−2 s−1 for 2 d).
(A) SOSG fluorescence in wild-type and ch1 leaves in the dark or after 20 min illumination at 450 μmol m−2 s−1.
(B) SOSG fluorescence (as measured by the ratio between leaf fluorescence measured at 525 nm and leaf fluorescence at 649 nm) in control (C) and acclimated (A) leaves of the ch1 mutant. Fluorescence was measured after 30-min illumination at 450 μmol photons m−2 s−1.
(C) β-Carotene endoperoxide levels in wild-type (white) and ch1 plants (black) exposed to 450 μmol photons m−2 s−1 and 20°C. n = 4. Data are mean values + sd.
[See online article for color version of this figure.]
We also measured β-carotene endoperoxide, a product derived from the oxidation of β-carotene in the reaction center of PSII (Ramel et al., 2012b). This oxidized carotenoid derivative is a good biochemical marker of 1O2 since it cannot be formed by free radicals or by enzymatic catalysis. Figure 7C shows that β-carotene endoperoxide accumulated very rapidly during the acclimation treatment in ch1, confirming the production of 1O2 in the PSII reaction centers. Again, this phenomenon was not observed in the wild type, indicating no or very little production of 1O2 under this condition. The data in Figure 7 clearly confirm that the ch1 mutant is a 1O2 overproducer compared with the wild type.
The volatile carotenoid derivative β-cyclocitral has been identified as a signal molecule that can induce gene expression changes leading to resistance to photooxidative stress (Ramel et al., 2012a). Consistent with this, the concentration of β-cyclocitral was found to increase during photoacclimation of ch1 plants (see Supplemental Figure 2 online).
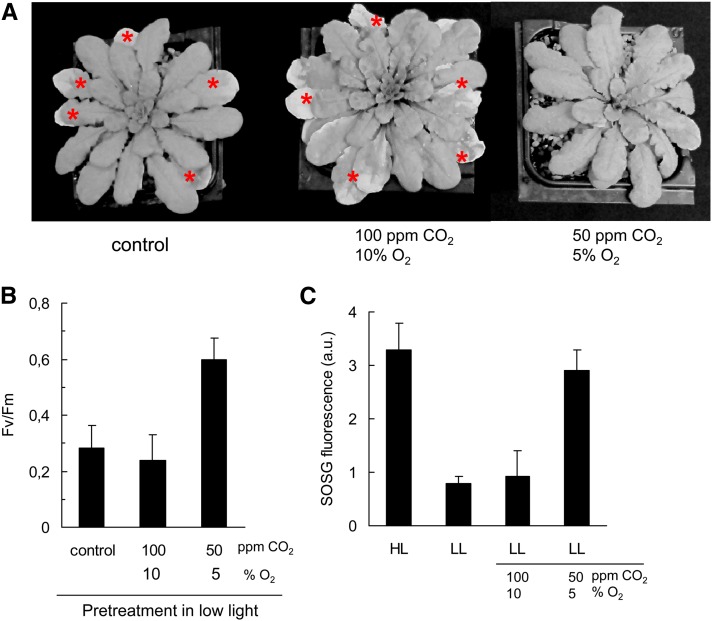
To rule out the possibility that the acclimation response of the ch1 mutant reported in Figure 5 results from the increase in PFD (from 180 to 450 μmol m−2 s−1) (e.g., through the action of photoreceptors, rather than an increase in 1O2 production), we exposed plants to an atmosphere with reduced levels of CO2 and O2 (Figure 8). By reducing the final electron acceptors, it is possible to increase the excitation pressure in PSII (and hence to increase 1O2 production in the PSII centers) at constant PFD (Montané et al., 1998). CO2 and O2 concentrations in the atmosphere were reduced to 50 ppm and 5%, respectively, while PFD was maintained at 180 μmol m−2 s−1. Exposing plants to this low CO2 and O2 atmosphere noticeably increased 1O2 production (as measured by SOSG fluorescence) compared with the control conditions (180 μmol m−2 s−1 in the air) (Figure 8C). The 1O2 levels reached in leaves exposed to 50 ppm CO2 and 5% O2 were close to the levels measured in high light (450 μmol m−2 s−1) in air. Another condition was also tested, 100 ppm CO2 and 10% O2, which did not increase substantially 1O2 production. As shown in Figures 8A and 8B, 2 d of exposure of ch1 plants to 50 ppm CO2 and 5% O2 in low light enhanced the tolerance to a subsequent high-light stress (1000 μmol m−2 s−1 at 10°C in normal air). No leaf bleaching was observed after high-light stress, and the PSII activity was much higher than in stressed control plants (maximal PSII photochemical efficiency [Fv/Fm], 0.6 versus 0.28). Thus, the plants appeared to be as tolerant to photooxidative stress as the plants pretreated at 450 μmol photons m−2 s−1 (Figure 5). The CO2/O2 concentrations (100 ppm/10%) that did not increase 1O2 production had no protective effect against photooxidative stress. We can conclude from the experiments of Figure 8 that the acclimation of ch1 mutant plants to 1O2 was not due to the change in PFD per se, but it was closely correlated with the levels of 1O2 released from PSII.
Figure 8.
Induction of the Acclimation Response to Photooxidative Stress in ch1 Mutant Plants in Low Light by Decreasing CO2 and O2 Availability.
Plants were preexposed for 2 d to an atmosphere with reduced levels of CO2 and O2: 100 ppm CO2 and 10% O2 or 50 ppm CO2 and 5% O2.
(A) Plants exposed for 2 d to high-light stress (1000 μmol photons m−2 s−1 and 10°C, in air). The asterisks indicate leaves with symptoms of chlorophyll bleaching.
(B) PSII photochemical efficiency (Fv/Fm) after high-light stress. Data are mean values +sd. n = 7
(C) 1O2 production, as measured by the increase in SOSG fluorescence, under different conditions: HL, 450 μmol photons m−2 s−1 in air; LL, 180 μmol photons m−2 s−1 in air or in atmosphere with reduced CO2 and O2 levels. Data are mean values +sd. n = 4 to 9.
[See online article for color version of this figure.]
Transcriptomic Analysis of the ch1 Mutant under Photoacclimatory Conditions
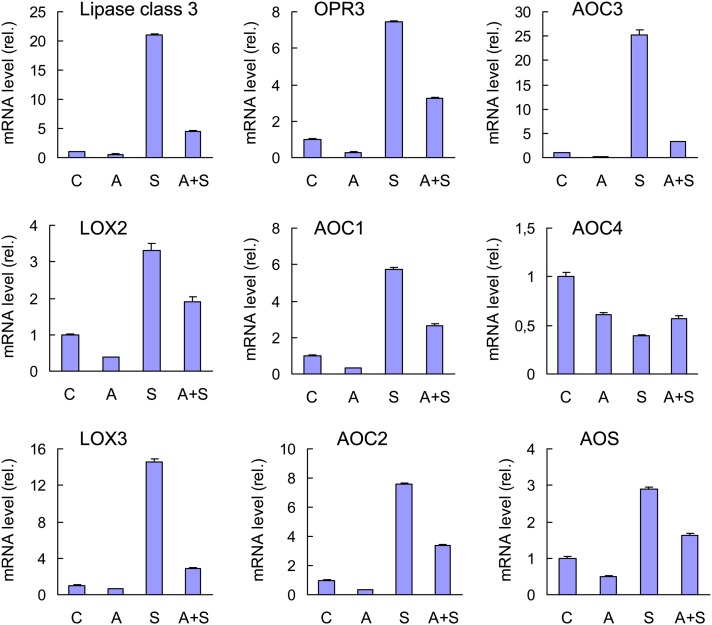
Microarray-based transcriptomic analysis of ch1 was performed during acclimation (see Supplemental Data Set 1 online), and the data were compared with the transcriptomic data obtained during high-light stress. A total of 275 genes were induced during acclimation and 267 genes were repressed. Among the genes induced by acclimation to photooxidative stress, we found a number of 1O2-specific genes (see Supplemental Table 2 online), confirming that 1O2 is produced during the acclimatory treatment. The number of induced genes and the intensity of expression were much lower than the changes observed for high-light stress conditions (see Supplemental Table 1 online), confirming that acclimation induced lower 1O2 production than did stress. Similarly to high-light stress, acclimation did not induce marked changes in the expression of H2O2-responsive genes (Figure 9A). Moreover, 64% of the 275 induced genes were also induced during stress, while the expression of 35% of the genes was not induced by stress (Figure 9B). Among the 267 genes repressed by acclimation, 16% were either not differentially expressed (compared with the control condition) or were induced by high-light stress. Thus, although many genes displayed the same expression responses in stress and acclimation conditions, significant differences exist, like some genes presenting specifically induction or repression by acclimation. In particular, we found that many genes coding for proteins involved in the biosynthesis of jasmonate were induced during stress, whereas most of them were repressed during acclimation (see Supplemental Table 3 online). This differential behavior was checked by quantitative RT-PCR (qRT-PCR). Figure 10 shows that high-light stress brought about a marked induction of all tested genes except AOC4, which was repressed in any conditions. In particular, the induction of the lipoxygenases LOX2 and LOX3 is in line with the accumulation of LOX-induced lipid metabolites observed in stressed ch1 plants (Figure 1F). 1O2 induction of the jasmonate pathway in ch1 plants is also in agreement with previous work by Przybyla et al. (2008), who found induction of enzymatic lipid peroxidation and accumulation of the LOX oxidation product 13-HOTE during 1O2 stress in the flu Arabidopsis mutant.
Figure 9.
Microarray-Based Analysis of Gene Expression Changes in ch1 Leaves during Acclimation to Light Stress (450 μmol m−2 s−1 for 2 d).
(A) Percentage of genes inducible or repressible by H2O2 that were induced, repressed, or not differentially expressed (compared with control conditions) in the ch1 mutant during acclimation.
(B) Percentage of genes induced or repressed in the ch1 mutant during acclimation that were induced, repressed, or not differentially expressed during high-light stress (1000 μmol m−2 s−1 for 2 d). nde, not differentially expressed.
Figure 10.
The Jasmonate Pathway in the ch1 Arabidopsis Mutants Exposed to Stress and/or Acclimation Conditions.
qRT-PCR analysis of the expression levels of various genes of the jasmonate pathway during stress (S), acclimation (A), and acclimation followed by stress (A+S). C = control conditions. AOC, ALLENE OXIDE CYCLASE; AOS, ALLENE OXIDE SYNTHASE; OPR3, 12-OXOPHYTODIENOATE REDUCTASE3. Data are mean values +sd. n = 4.
[See online article for color version of this figure.]
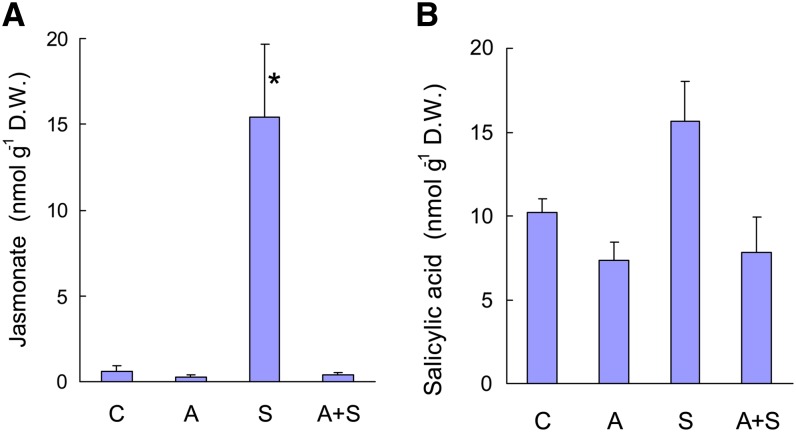
Conversely, acclimation of ch1 plants to high-light stress was found to repress all the jasmonate pathway genes that were induced during the stress treatment (Figure 10). When acclimated plants were exposed to high-light stress, gene induction was very modest compared with the expression changes observed when unacclimated plants were exposed to high-light stress. Thus, the acclimation process repressed the jasmonate pathway and counteracted the induction of this pathway during high-light stress. Moreover, those changes in gene expression were correlated with changes in the jasmonate concentration in the leaves (Figure 11A). Jasmonate levels were strongly increased (×15) after stress, while acclimated plants (before and after high-light stress) contained less jasmonate that control plants. In comparison, there were relatively small changes in salicylic acid levels during high-light stress and/or acclimation (Figure 11B), and the concentration of the jasmonate precursor 12-oxo phytodienoic acid (OPDA) was not affected by the treatments (on average, 12 nmol g−1 dry weight).
Figure 11.
Jasmonate and Salicylic Acid Levels in ch1 Leaves under Various Conditions.
Jasmonate (A); salicylic acid (B). C = control, A = acclimation, S = stress, and A+S = acclimation followed by stress. D.W., dry weight. Data are mean values +sd. n = 4. Asterisk indicates significant difference from control (C) at P < 0.001 (Student’s t test).
[See online article for color version of this figure.]
Jasmonate Biosynthesis and Photooxidative Stress Tolerance
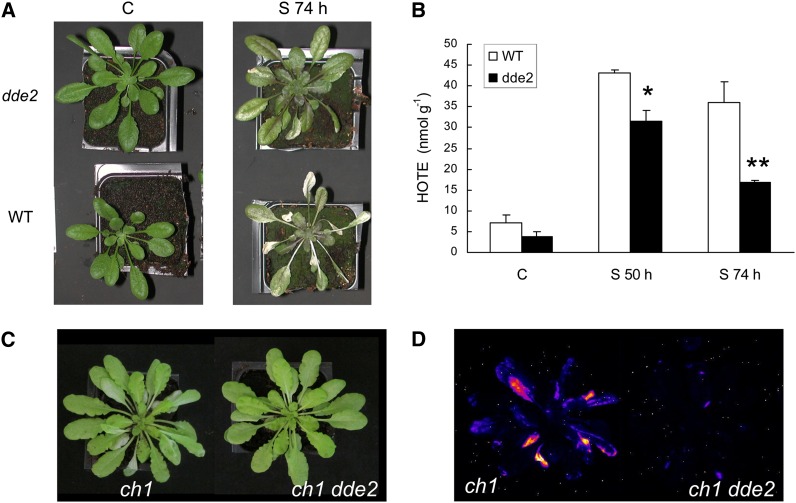
In the experiments described above, induction of the jasmonate biosynthesis pathway was correlated with cell death, whereas repression of this pathway during acclimation was associated with an increased tolerance to photooxidative stress. To determine if there is a causal relationship between repression of jasmonate biosynthesis and tolerance to 1O2, we examined the light response of the jasmonate mutant delayed-dehiscence2 (dde2). Because of a deficiency in the gene encoding allene oxide synthase (von Malek et al., 2002), dde2 plants cannot metabolize 13-hydroperoxy linolenic acid; therefore, they cannot synthesize jasmonate. Wild-type and dde2 mutant plants were exposed to severe light stress treatment (1500 μmol photons m−2 s−1 at 7°C air temperature) that induced photooxidative damage to wild-type plants, as shown by leaf bleaching and tissue necrosis (Figure 12A). This treatment brought about accumulation of ROS-generated HOTEs in wild-type leaves, indicating the occurrence of nonenzymatic lipid peroxidation. HPLC–mass spectrometry analysis of the HOTE isomer distribution in high-light-stressed wild-type leaves revealed the presence of 10-HOTE and 15-HOTE (Triantaphylidès et al., 2008), indicating the occurrence of 1O2-induced lipid peroxidation. By striking contrast, jasmonate-deficient dde2 plants were much more tolerant to this treatment: Leaf bleaching was much less pronounced (Figure 12A), and the intensity of ROS-induced lipid peroxidation was substantially reduced compared with the wild type (Figure 12B).
Figure 12.
Effects of Jasmonate Deficiency on the Photoresistance of Arabidopsis.
(A) and (B) Wild-type and dde2 mutant plants were exposed to high-light stress (1500 μmol m−2 s−1 at 7°C) for 50 or 74 h.
(A) Plants before (C) and after 74-h exposure to the light stress (S 74 h).
(B) ROS-induced lipid peroxidation (HOTE levels) before (C) and after high-light stress (S 50 h and S 74 h, exposure to the light stress for 50 and 74 h, respectively). Data are mean values + sd. n = 3. Asterisks indicate significant difference from the wild type at *P < 0.005 and **P < 0.010, respectively (Student’s t test).
(C) and (D) Arabidopsis ch1 and ch1 dde2 mutant plants were exposed to high-light stress (1000 μmol m−2 s−1 at 10°C) for 26 h.
(C) Plants after high-light stress.
(D) Autoluminescence imaging of lipid peroxidation after high-light stress.
[See online article for color version of this figure.]
We crossed the dde2 mutant with the ch1 mutant to generate a 1O2-producing double mutant (ch1 dde2) that is unable to produce jasmonate. As shown in Figures 12C and 12D, the ch1 dde2 double mutant was resistant to high-light stress compared with the ch1 single mutant: Leaf bleaching (Figure 12C) and lipid peroxidation (Figure 12D) were much more marked in ch1 plants compared with ch1 dde2 plants. Also, PSII photochemical efficiency (Fv/Fm) was higher in ch1 dde2 relative to ch1 (0.49 ± 0.11 versus 0.22 ± 0.09). These data confirm that 1O2-induced cell death is associated with jasmonate biosynthesis.
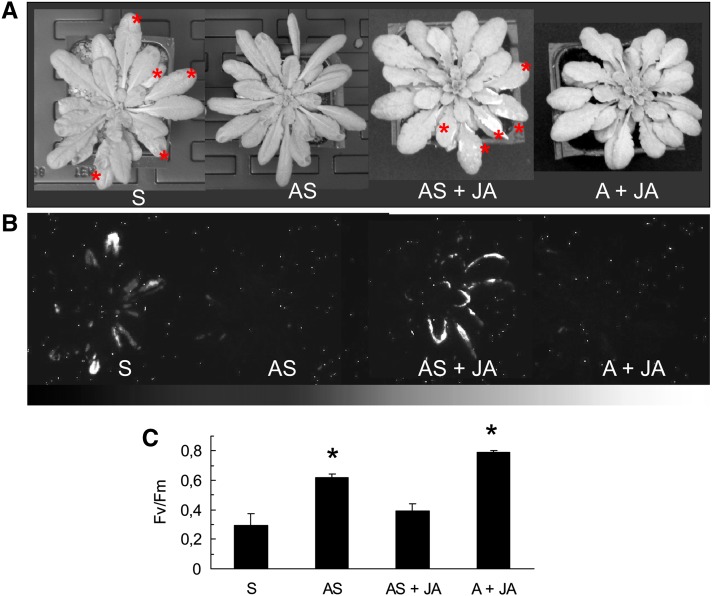
Light-acclimated ch1 plants were sprayed with a solution of methyl jasmonate (1 mM) during the high-light treatment. As shown in Figure 13, exogenously applied jasmonate cancelled the acclimation process: Pigment bleaching (Figure 13A) and lipid peroxidation (Figure 13B) were observed in many leaves of the jasmonate-sprayed plants, and PSII photochemistry was severely affected (falling from 0.6 in acclimated plants to 0.4 in sprayed acclimated plants; Figure 13C). This clearly indicates that jasmonate accumulation during high-light stress can induce cell death. It should be noted that jasmonate itself was not toxic, as shown by the absence of stress symptoms in plants sprayed with methyl jasmonate and kept in low light. From the experiments in Figures 12 and 13, one can conclude that jasmonate is associated with high-light-induced cell death. A block of jasmonate synthesis has beneficial effects on plants exposed to photooxidative stress conditions by enhancing tolerance against 1O2-induced damage.
Figure 13.
Effects of Methyl Jasmonate Applications on the Acclimation Response to Photooxidative Stress in the Arabidopsis ch1 Mutant.
(A) Plants after 2-d exposure to high-light stress (1000 μmol m−2 s−1 at 10°C). S, control plants exposed to high-light stress; AS, plants acclimated for 2 d at 450 μmol m−2 s−1 and then exposed to high-light stress; AS + JA, plants acclimated to 450 μmol m−2 s−1 were sprayed with 1 mM methyl jasmonate (400 μL, twice a day) during high-light stress; A + JA, plants acclimated to 450 μmol m−2 s−1 and then sprayed with 1 mM methyl jasmonate in low light (180 μmol photons m−2 s−1). The asterisks indicate leaves showing symptoms of bleaching.
(B) Autoluminescence imaging of lipid peroxidation in the S, AS, AS + JA, and A + JA.
(C) PSII photochemical efficiency (Fv/Fm) in the S, AS, AS + JA, and A + JA plants. Data are mean values +sd. n = 7. Asterisk indicates significant difference from S at P < 0.05 (Student’s t test).
[See online article for color version of this figure.]
DISCUSSION
The Arabidopsis Mutant ch1 as a Physiological Model for 1O2 Studies
As mentioned in the Introduction, so far most of the information on the 1O2 signaling pathway in plants comes from studying the conditional flu mutant of Arabidopsis. This study provides another 1O2-producing physiological model that is complementary to the flu mutant. The latter mutant is characterized by the accumulation of free protochlorophyllide in plastids when kept in the dark (Meskauskiene et al., 2001). In the light, protochlorophyllide acts as a photosensitizer and generates 1O2, activating a cell death program mediated by the two chloroplast-located proteins EX1 and EX2 (op den Camp et al., 2003; Wagner et al., 2004; Lee et al., 2007). However, the physiological significance of the 1O2-dependent cell death pathway identified in the flu mutant is still elusive, and questions about how well this mutant represents the physiological situation in wild-type plants have been raised (Mullineaux and Baker, 2010; Gutiérrez et al., 2011; Suzuki et al., 2012). Indeed, in wild-type plants, 1O2 is produced by triplet chlorophylls within PSII, particularly when the PSII reaction center is submitted to excess excitation (Krieger-Liszkay et al., 2008; Triantaphylidès and Havaux, 2009). This differs markedly from the way 1O2 is produced in the flu mutant, namely, from a membrane-soluble chlorophyll precursor in the absence of PSII overexcitation. In wild-type Arabidopsis plants, photooxidative damage to leaves is associated with direct oxidation of membrane lipids by 1O2 (Triantaphylidès et al., 2008). Nevertheless, the 1O2-mediated signaling pathway can be activated in the wild type under light stress, but this seems to occur without decreasing the viability of the seedlings (Kim et al., 2012). It should also be kept in mind that 1O2 oxidation of protochlorophyllide could generate oxidized molecules that are not normally present in the wild type, and we cannot exclude an effect of those metabolites on the gene responses in flu. In this context, it is important to mention that porphyrins are able to operate as cell death factors, even in their unexcited state (Hirashima et al., 2009).
Elucidation of the relevance of the 1O2 effects observed in the Arabidopsis flu mutant to other mutants or to other plant species would be very useful, in particular to validate the flu model. The chlorophyll b–less Arabidopsis ch1 mutant provides an interesting alternative model. In this mutant, PSII is restricted to its reaction center (Havaux et al., 2007) since PSII light-harvesting antennae cannot assemble in the absence of chlorophyll b (Plumley and Schmidt, 1987). Because LHCII-deficient PSII reaction centers do not benefit from the photoprotective mechanisms associated with the chlorophyll antennae, such as NPQ, PSII overexcitation can be readily reached with increasing PFD, promoting 1O2 formation (Dall’Osto et al., 2010). Consistent with this, inhibition of NPQ in the Arabidopsis npq4 mutant brought about an increased oxidation of β-carotene by 1O2 in the PSII reaction centers (Ramel et al., 2012b). As confirmed in this study, the ch1 mutant is characterized by increased production of 1O2 resulting in high photosensitivity. Additional perturbations of the PSII functionality previously reported in chlorophyll b–less plants, such as alteration of the PSII donor side (Havaux and Tardy, 1997; Kim et al., 2009), can also contribute to this increased production of 1O2 by PSII.
A good correlation was found between the transcription profiles of the ch1 and flu mutants, confirming the induction of a 1O2 signaling pathway in the ch1 leaves in high light. However, important differences were observed between the two mutants. First, cell death in ch1 was not dependent on EX1. Possibly, photooxidative damage in ch1 was due to direct cytotoxicity of 1O2 which masked the EX1-dependent signaling cascade, as previously suggested for wild-type Arabidopsis (Kim et al., 2012). Consistent with this, high-light stress induced LOX genes in ch1, and leaf bleaching was associated with both LOX-induced and 1O2-induced HOTE accumulation, suggesting a combination of genetically regulated lipid oxidation and direct oxidation by 1O2. It is worth mentioning that photooxidative damage appeared in ch1 leaves after 2 d of stress treatment, while cell death was reported to occur much faster in flu leaves, with visible necrotic lesions appearing within a few hours after the onset of illumination (op den Camp et al., 2003). Second, unlike the flu mutant (Ochsenbein et al., 2006), the ch1 mutant exposed to high-light stress did not exhibit increased expression of EDS1 encoding the ENHANCED DISEASE SUSCEPTIBILITY1 protein required for the modulation of the 1O2-mediated cell death response. This is consistent with the observation that the concentration of salicylic acid did not change substantially in ch1 leaves during high-light stress since this compound relies on EDS1 expression for its accumulation (Wiermer et al., 2005). Similarly, Arabidopsis cell suspensions were reported to respond to high light in a manner that resembles the defense responses of flu except that it was independent of EDS1 and salicylic acid (Gutiérrez et al., 2011).
The flu mutant must be grown in continuous light to avoid accumulation of protochlorophyllide. On the contrary, ch1 does not require any particular growth conditions; hence, it is a more convenient model for physiological studies. By altering the light environment, we were able to find conditions that lead to a marked increase in the tolerance to high-light stress and to suppression of 1O2 toxicity, providing an interesting model for studying acclimation to 1O2. The acclimatory conditions were found to induce 1O2 production in ch1 plants, which was not eliminated in the acclimated state. Consequently, we can speculate that acclimation to 1O2 stress was induced in ch1 chloroplasts by 1O2 itself. This suggests also that acclimation involves mainly increased resistance to 1O2, rather than suppression of 1O2 production in the PSII centers. It must be kept in mind that, similarly to the flu mutant, the ch1 mutant is characterized by a specific increase in 1O2 production in the light. The situation might be more complex in the wild type, where several ROS can be produced simultaneously, possibly leading to more complex responses to high-light stress.
Acclimation of Arabidopsis to 1O2
It is long known that plants are able to acclimate to changes in their light environment in order to optimize their photosynthetic performance and to avoid imbalance between energy absorption and utilization, which can promote ROS production in the chloroplasts. The acclimatory response to light can involve a variety of mechanisms occurring in different time ranges (Walters, 2005; Eberhard et al., 2008; Li et al., 2009; Murchie and Niyogi, 2011). In the mid and long terms, light acclimation involves changes in gene expression and reorganization of the photosynthetic apparatus, with the ROS themselves as possible triggers. Indeed, besides their toxic effects, ROS can also act as inducers of signaling cascades leading to changes in gene expression (Suzuki et al., 2012). This role has long been recognized for the long-lived hydrogen peroxide (Petrov and Van Breusegem, 2012), and a similar role as a signal messenger has been identified more recently for the short-lived 1O2 in plants (op den Camp et al., 2003; Wagner et al., 2004). Gene expression reprogramming by 1O2 has been reported in a variety of other organisms including bacteria (Ziegelhoffer and Donohue, 2009) and algae (Ledford et al., 2004). In the microalga C. reinhardtii, growth in the presence of low 1O2 concentrations enhances the resistance of the cells to high 1O2 concentrations (Ledford et al., 2007). This study demonstrates that vascular plants can also acclimate to 1O2, resulting in a marked enhancement of their tolerance to photooxidative stress. Preexposure of ch1 mutant plants to a moderately elevated PFD that induced a low production of 1O2 rendered them much more resistant to damage by 1O2 produced in higher amounts at higher PFDs. In accordance with this, a product derived from 1O2 oxidation of the carotenoid β-carotene can concomitantly induce the expression or repression of 1O2-responsive genes and enhance tolerance to high-light stress in Arabidopsis (Ramel et al., 2012a), suggesting the existence of an acclimatory response to 1O2. Similarly, gene responses to 1O2 that do not lead to cell death were previously reported in the lut2 npq1 double mutant of Arabidopsis deficient in two 1O2 quenchers (Alboresi et al., 2011). Arabidopsis cell suspensions exposed to high light display gene responses that resemble very much the gene expression profile induced in the flu mutant by 1O2, but again this occurs in the absence of programmed cell death (González-Pérez et al., 2011; Gutiérrez et al., 2011). The orientation of the signaling pathway toward cell death or acclimation may be determined by the intensity of the 1O2 stress, with low concentrations leading to acclimation and high concentrations inducing direct toxicity or programmed cell death (Kim et al., 2008). The acclimation treatment used in this study was associated with 1O2 production in ch1 leaves, but the resulting photooxidative stress was moderate, as judged by the rather modest inhibition of PSII (Fv/Fm), suggesting that it belongs to the former category of stress conditions.
Acclimation of the ch1 mutant to high-light stress was associated with the induction of genes related to different metabolic processes (see Supplemental Data Set 1 online): plastoquinone and ubiquinone synthesis (SOLANESYL DIPHOSPHATE 1 and 2 [SPS1 and SPS2]), anthocyanin and flavonoid synthesis (PRODUCTION OF ANTHOCYANIN PIGMENT1 and 2, [PAP1, PAP2] CHALCONE SYNTHASE [CHS], FLAVONONE SYNTHASE [FLS], CINNAMYL ALCOHOL DEHYDROGENASE4 [CAD4], and TRANSPARENT TESTA5 [TT5]), pyridoxine synthesis (PDX2), and methionine synthesis (At-MS2). Interestingly, all those processes concern molecules that are known to have high reactivity with 1O2 (Triantaphylidès and Havaux, 2009). In general, genes induced during acclimation were related to metabolism, in particular, secondary metabolism, such as metabolism of phenylpropanoids and flavonoids, to oxidative stress responses and to systemic interaction with the environment such as ethylene response (see Supplemental Table 4 online). In particular, we observed induction of several genes involved in the response to stress (the ethylene responsive gene AT2G38210, the glutathione peroxidases 6 and 7, and EIN3-BINDING F BOX PROTEIN2 [EBF2]) or in transport processes (AT1G78410, AT1G80300, and MATE protein). In addition, many genes related to the regulation of transcription were also induced by the acclimation treatment, including MYB DOMAIN PROTEIN 13 (ATMYB13), MYB DOMAIN PROTEIN114 (MYB114), zinc finger (B-box type) family proteins, HIGH MOBILITY GROUP B2 (HMGB2), RELATED TO AP 2.3 (ATEBP/RAP2.3), and dof-type zinc finger domain-containing protein At1G21340, PAP1, PAP2, and HIGH CHLOROPHYLL FLUORESCENCE152 (HCF152). Some of those genes were not significantly induced by high-light stress (ATMYB13, ATEBP/RAP2.3, and At1G21340), indicating a specific response to acclimation conditions.
As far as gene repression is concerned, acclimation affected many genes coding for proteins of the jasmonate pathway or proteins related to the wound response (AT3G10985 and AT4G28240). Genes encoding transcription factors (HOMEOBOX PROTEIN16 [ATHB16], ARABINOGALACTAN PROTEIN8 [FLA8], PHYTOCHROME INTERACTING FACTOR4 [PIF4], INDOACETIC ACID-INDUCED PROTEIN8 [IAA8], AT1G68520, and AT1G25440) and proteins related to photosynthesis PSII LIGHT HARVESTING COMPLEX GENE2.3 (LHCB2.3) and PSII LIGHT HARVESTING COMPLEX GENE3 (LHCB3) were also downregulated during the acclimation treatment. The acclimation-induced downregulation of genes involved in jasmonate biosynthesis is of particular interest because it contrasts very much with the strong induction of this pathway in ch1 leaves exposed to high-light stress.
Jasmonate Biosynthesis and the Response to 1O2
Jasmonates are essential plant hormones that regulate defense responses and developmental processes. In particular, these signal molecules are important regulators of plant responses to environmental stresses, such as ozone, wounding, or water deficit, and attack by pathogens and pests (Reymond and Farmer, 1998; Wasternack, 2007; Balbi and Devoto, 2008; Reinbothe et al., 2009; Avanci et al., 2010). A wealth of molecular genetic studies have provided evidence for the involvement of jasmonates in the regulation of the complex and highly regulated cell death program. Moreover, sustained exposure of leaves and suspension-cultured cells to exogenous methyl jasmonate induces cell death (Repka et al., 2004; Zhang and Xing, 2008). Jasmonates also increase the fungal toxin fumonisin B1-induced apoptosis-like programmed cell death in Arabidopsis protoplasts (Asai et al., 2000; Mur et al., 2006). In cultured animal cells, jasmonates and some of their synthetic derivatives were found to induce cell death in cancer cell lines (Cohen and Flescher, 2009). Actually, the role played by jasmonates in plant cell death regulation is complex, involving interactions with other low molecular mass signal molecules, such as salicylic acid and ethylene (Reymond and Farmer, 1998; Reinbothe et al., 2009; Ballaré, 2011). For instance, salicylic acid is known to repress jasmonate signaling (Pena Cortes et al., 1993).
The work done on flu has shown that jasmonate and its precursor OPDA are synthesized in response to 1O2 (Ochsenbein et al., 2006; Przybyla et al., 2008). Genes encoding enzymes involved in the biosynthesis of jasmonic acid were upregulated in flu seedlings in response to nonpermissive dark-to-light shifts (op den Camp et al., 2003), and this was followed by the rapid synthesis of the corresponding proteins (Samol et al., 2011). It was suggested that jasmonic acid may be required for cell death manifestation, whereas OPDA would counteract the establishment of the cell death phenotype (Przybyla et al., 2008). Our results are in line with this suggestion: 1O2-induced cell death and leaf necrosis in the ch1 mutant were associated with jasmonate accumulation, while the OPDA level remained unchanged. In fact, the whole jasmonate biosynthesis pathway was upregulated in ch1 plants during high-light stress. On the contrary, the EX1/EX2- and EDS1-dependent signaling pathway appeared to remain inactive in ch1 under high-light stress conditions. This constitutes a major difference with the response of the flu mutant, which was characterized by a strong accumulation of EDS1 transcripts and salicylic acid (Ochsenbein et al., 2006). In other words, an oxylipin-dependent pathway was specifically activated in ch1 under high-light stress where jasmonate had a prominent role in the activation of transcripts associated with oxidative stress. Moreover, acclimation and increased resistance to photooxidative damage were accompanied by strong downregulation of the jasmonate biosynthesis pathway, reinforcing the idea that jasmonate is involved in the modulation of 1O2-induced cell death. This was confirmed in the dde2 and ch1 dde2 mutants, which are unable to synthesize jasmonate. Leaf bleaching and lipid peroxidation were notably attenuated in the jasmonate-deficient mutants compared with their jasmonate-producing counterparts (the wild type or ch1) when exposed to photooxidative stress conditions. Importantly, exogenous applications of methyl jasmonate were observed to cancel the acclimation response.
In sum, this study has established that, similarly to green algae (Ledford et al., 2007), vascular plants have the capacity to acclimate to 1O2 stress. This acclimation process is accompanied by gene transcription changes. In particular, this study emphasizes the role of the jasmonate pathway in the sensitivity to 1O2. Acclimation of Arabidopsis to photooxidative stress was associated with a downregulation of jasmonate synthesis, and mutational suppression of jasmonate synthesis enhanced phototolerance. Conversely, photodamage in nonacclimated plants was correlated with jasmonate accumulation. This close correlation between jasmonate levels and sensitivity to 1O2 was not observed for other hormones (salicylic acid) or other oxylipins (OPDA), which exhibited little change during photo stress in ch1 mutant plants. It would be interesting to analyze other jasmonate mutants, such as those with reduced sensitivity to jasmonate or with constitutively active jasmonate signaling (Berger, 2002), to characterize further the link between jasmonate and 1O2-induced cell death.
1O2 is a particularly important ROS in photosynthetic organisms because chlorophyll molecules are potential photosensitizers present in high amounts in their photosynthetic apparatus. When light energy is absorbed by chlorophylls in excess to what can be used by the photosynthetic processes, overexcitation of the photosynthetic system can increase the lifetime of singlet excited chlorophylls favoring their transition to the triplet excited state, the main source of 1O2 (Krieger-Liszkay et al., 2008; Triantaphylidès and Havaux, 2009). Also, any stress conditions that perturb the assembly of the chlorophyll binding proteins can favor 1O2 formation from uncoupled chlorophyll molecules (Santabarbara and Jennings, 2005). For instance, upon infection with Pseudomonas syringae, Arabidopsis leaves accumulate 1O2-specific hydroxy fatty acids (10- and 15-HOTEs) (Grun et al., 2007), reflecting the generation of 1O2 during pathogen attack (Vellosillo et al., 2010). Interestingly, the hypersensitive response to pathogenic challenge has been associated with disruption of the light-harvesting pigment-protein complexes and the formation of chlorophyll catabolites that act as 1O2 generators (Mur et al., 2010). Moreover, 1O2 was shown to be the major ROS involved in photooxidative damage to plant leaves (Triantaphylidès et al., 2008). Therefore, the capacity to enhance tolerance toward 1O2 toxicity, as reported here for Arabidopsis, can be of prime importance for the survival of plants under natural, stressful environments. Challenges for future research will be to establish in wild-type plants the occurrence of the acclimation response reported here and to reveal the genes that are crucial to this response.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown for 5 to 8 weeks in a phytotron under controlled conditions of temperature (20°C/18°C, day/night), PFD (180 μmol photons m−2 s−1, 8-h photoperiod), and relative air humidity (65%). The experiments were done on wild-type Arabidopsis (ecotype Columbia-0), a chlorophyll b–deficient mutant ch1 (ch1-1) (Havaux et al., 2007), and dde2, a jasmonate mutant deficient in allene oxide synthase (von Malek et al., 2002). A detailed description of the phenotype of the Arabidopsis ch1 mutant can be found in a previous publication (Havaux et al., 2007). The ch1 mutant was crossed with the dde2 mutant and the EX1-deficient mutant (ex1) (Wagner et al., 2004; Lee et al., 2007). Photooxidative stress was induced in the ch1 mutant by transferring the plants to a PFD of 1000 μmol photons m−2 s−1 (photoperiod, 8 h) and an air temperature of 10°C. For acclimation to photooxidative stress, plants were exposed for 2 d to a PFD of 450 μmol m−2 s−1 (photoperiod, 8 h) at 20°C. PFDs were measured with a Li-Cor quantum meter. The CO2 and O2 concentrations of the atmosphere were controlled in a hermetically closed chamber as described by Montané et al. (1998). In some experiments, 400 μL of methyl jasmonate (1 mM in 25 mM phosphate buffer) was sprayed on plants twice a day.
Chlorophyll Fluorometry
The maximal quantum yield of PSII photochemistry (Fv/Fm) was measured in dark-adapted attached leaves by chlorophyll fluorometry using a PAM-2000 Walz system, as previously described (Havaux et al., 2007). Fv/Fm was calculated as (Fm − Fo)/Fm, where Fo is the initial level of chlorophyll fluorescence and Fm is the maximal level (induced by an 800-ms flash of intense white light). A 2-s pulse of far-red light was used to oxidize QA (the primary quinone electron acceptor of PSII) and to measure the true Fo level.
Chlorophyll Quantification
Chlorophyll pigments were extracted from leaf discs in methanol and were quantified by HPLC as previously described (Havaux et al., 2007), using authentic standards of chlorophyll a and b (Sigma-Aldrich).
Lipid Peroxidation Analysis and Imaging
Lipids were extracted from 0.5 g frozen plant material by grinding with 2 × 1 mL CHCl3 containing 1 mg/mL triphenyl phosphine and 0.05% (w/v) butylated hydroxytoluene, with 15-hydroxy-11,13(Z,E)-eicosadienoic acid as internal standard. The organic phase was evaporated under a stream of N2. The residue was recovered in 1.25 mL ethanol and 1.25 mL 3.5 M NaOH and hydrolyzed at 80°C for 15 min. After addition of 2.2 mL 1 m citric acid, hydroxy fatty acids were extracted with 2 × 1 mL hexane/ether (50/50). The organic phase was used for the straight phase HPLC-UV analysis of HOTE isomers derived from the oxidation of linolenic acid, as previously described (Montillet et al., 2004). ROS-induced HOTEs were separated from LOX-dependent HOTEs according to the method described by Montillet et al. (2004).
Lipid peroxides were visualized in whole plants by autoluminescence imaging. Imaged autoluminescence signal is attributed to the spontaneous decomposition of lipid peroxides (Havaux et al., 2006; Birtic et al., 2011). Spontaneous photon emission from whole Arabidopsis plants was measured after 2 h dark adaptation with a liquid N2 cooled charge-coupled device camera, as detailed by Birtic et al. (2011). Integration time was 20 min.
Detection of ROS Production
1O2 production was measured in attached leaves with the SOSG fluorescent probe (Invitrogen; see Flors et al., 2006), as previously described (Ramel et al., 2012b). Leaves were infiltrated with 100 μM SOSG under pressure. A 1-mL syringe, without needle and filled with the solution to be infiltrated, was pushed against the lower surface of the leaf, and the solution (100 μL) was forced to enter inside the leaf under pressure. Plants with SOSG infiltrated leaves were exposed for 30 min to a PFD of 450 μmol m−2 s−1 at 20°C. SOSG fluorescence was then measured using a Perkin-Elmer spectrofluorometer (LS 50B) with a 475-nm exciting light.
ROS production by thylakoid membranes was measured in vitro by spin trapping EPR spectroscopy. Thylakoid membranes were isolated from fresh leaves as described by Takahashi and Asada (1982) and suspended in a HEPES buffer (20 mM, pH 7.5) containing 0.4 M Suc, 15 mM NaCl, and 3 mM MgCI2. The suspensions were adjusted to the same chlorophyll concentration (40 µg mL−1) and were illuminated for 1 or 3 min with white light (1500 µmol m−2 s−1) in the presence a spin trap (100 mM TEMPD or 50 mM 4-pyridyl-1-oxide-N-tert-butylnitrone, 4% ethanol, and 50 µM FeEDTA). EPR spectra were recorded at room temperature in a standard quartz flat cell using an ESP-300 X-band (9.73 GHz) spectrometer (Bruker). The following parameters were used: microwave frequency of 9.73 GHz, modulation frequency of 100 kHz, modulation amplitude of 1G, microwave power of 6.3 mW, receiver gain of 2 × 104, time constant of 40.96 ms, and four scans.
β-Carotene-5,8-endoperoxide, as specific 1O2 marker, was extracted from 500 mg leaves in 5 mL acetone and quantified by HPLC–tandem mass spectrometry (MS/MS) as previously described in detail (Ramel et al., 2012b). The co-called multiple reaction monitoring mode using transition 659 to 109 (with a collision energy of 45 eV) was employed to specifically detect and quantify the β-carotene endoperoxide.
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted using the NucleoSpin RNA plant kit (Macherey-Nagel) and treated with the Turbo DNA-free (Ambion) according to the manufacturers’ instructions. Each extraction from leaves of three different plants was performed at least five times.
Quantitative RT-PCR (qRT-PCR) experiments were performed with cDNA synthesized with the SuperScript III reverse transcriptase (Invitrogen) from 500 ng of total RNA. Specific primers for each gene selected for analysis were designed using Primer3plus software (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Gene-specific primers and oligonucleotide sequences are listed in Supplemental Table 5 online. qRT-PCR was performed using the LightCycler 480 SYBR Green I Master (Roche) in the quantitative PCR thermal cycler (LightCycler 480 real-time PCR system; Roche). Each reaction was prepared using 2 μL of cDNA diluted 20-fold, 2 μL of SYBR Green I Master, and 1 μM forward and reverse primers, in a total volume of 5 μL. The amplification profile consisted of 95°C for 10 min and 45 cycles (95°C for 15 s, 62°C for 15 s, and 72°C for 15 s). All reactions were performed in triplicates. The profilin PRF1 was taken as housekeeping gene to normalize the expression of genes of interest.
Microarray Transcriptomic Analyses
Microarray analysis was performed on CATMAv5 (Complete Arabidopsis Transcriptome MicroArray) arrays containing 31,987 gene-specific tags from Arabidopsis corresponding to 31,599 genes (Hilson et al., 2004). The transcriptome analysis compared RNA of wild-type and ch1 plants grown for 5 to 8 weeks, respectively, under control conditions (C = temperature [20°C/18°C, day/night], PFD [180 μmol photons m−2 s−1, 8 h photoperiod]), after 2 d of acclimation treatment (A = 450 µmol m−2 s−1, 8-h photoperiod, 20°C), after 2 d of stress treatment (S = 1000 µmol m−2 s−1, 8-h photoperiod, 10°C) and after acclimation and stress treatment (AS). Each treatment was repeated three times in independent experiments. In each experiment, five leaves of five different plants corresponding to a given treatment were harvested, frozen in liquid nitrogen, and extracted for RNA. cDNA synthesis, amplification, labeling, as well as hybridizations and scanning of the slides were performed as described elsewhere (Lurin et al., 2004). For each comparison, two technical replicates with fluorochrome reversal were performed for each pool of RNA (i.e., one dye swap per comparison). A global intensity-dependent normalization using the loess procedure (Yang et al., 2002) was performed to correct for the dye bias. Then, differential analysis was based on the log ratios averaged on the dye-swap, and these values were used to perform a paired Student’s t test. A trimmed variance is calculated from spots that do not display extreme variance (Gagnot et al., 2008). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate to keep a strong control of the false positives in a multiple comparison context (Ge et al., 2003). The probes with a Bonferroni P value < 0.05 were considered as being differentially expressed. The microarray data are available at http://urgv.evry.inra.fr/CATdb (project: CEA10-02 Light).
Jasmonate and Salicylate Analysis
Jasmonate, its precursor OPDA, and salicylate were quantified by UPLC-MS/MS, as described elsewhere (Stingl et al., 2013). Extraction from frozen leaves was performed in ethylacetate:formic acid (99:1), with addition of an internal standard solution, in a bead mill for 3 min at 20 Hz. After centrifugation, the supernatant was evaporated and the residue was resuspended in acetonitrile:water (1:1). An aliquot of the solution was injected in the liquid chromatography–MS/MS system constituted by a Waters Quattro Premier triple-quadrupole mass spectrometer with an electrospray interface coupled to a Waters Acquity ultraperformance liquid chromatograph. Analysis and quantification were done as explained in detail by Stingl et al. (2013).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CH1 (At1g44446), DDE2 (At5g42650), BAP1 (At3g61190, disease resistance protein (TIR class) putative (At1g57630), MAPKKK18 (At1g05100), AP2 domain-containing transcription factor (At4g34410), EX1 (At4g33630), and RD20 (At2g33380).The microarray data are available at http://urgv.evry.inra.fr/CATdb (project: CEA10-02 Light).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Responses to High-Light Stress of the ch1 Single Mutant and the ch1 ex1 Double Mutant.
Supplemental Figure 2. Levels of Different Volatile Derivatives of β-Carotene (β-Cyclocitral, β-Ionone, and Dihydroactinidiolide) in ch1 Mutant Leaves Exposed to Acclimatory Conditions.
Supplemental Table 1. List of 1O2-Specific Genes Induced in ch1 Mutant Leaves during High-Light Stress.
Supplemental Table 2. List of 1O2-Specific Genes Induced in ch1 Mutant Leaves during Acclimation.
Supplemental Table 3. Changes in the Expression of Genes Involved in the Jasmonate Pathway in Wild-Type and ch1 Leaves during Acclimation, Stress, and Acclimation followed by Stress as Measured by Microarray-Based Transcriptomic Analyses.
Supplemental Table 4. Functional Categories Significantly Enriched in Genes Induced or Repressed in ch1 Leaves during Acclimation at 450 μmol Photons m−2 s−1 in Comparison with Their Relative Abundance in the Genome.
Supplemental Table 5. Gene-Specific Primers and Oligonucleotide Sequences Used for the qRT-PCR Analyses.
Supplemental Data Set 1. List of the Genes Induced or Repressed in ch1 Mutant Leaves Exposed for 2 d to High-Light Stress or to Acclimation Relative to the Control.
Supplementary Material
Acknowledgments
We thank the Agence Nationale de la Recherche for financial support (Program ‘blanc’, Photox project). We thank the ‘Groupe de Recherches Appliquées and Phytotechnologie’ (Commissariat à l’Energie Atomique et aux Energies Alternatives/Cadarache) for help growing Arabidopsis plants, Christophe Laloi (Aix-Marseille University) for sharing transcriptomic data of the flu mutant, and Christian Triantaphylidès (Commissariat à l’Energie Atomique et aux Energies Alternatives/Cadarache) and Bilal Camara (University of Strasbourg) for useful discussions.
AUTHOR CONTRIBUTIONS
F.R. and M.H. designed the experiments. F.R., B.K., E.A., A.S.M., A.K.-L., F.M., F.B., and J.-L.R. performed the experiments. F.R., M.J.M., F.B., and M.H. analyzed data. F.R. and M.H. wrote the article.
Glossary
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- 1O2
singlet oxygen
- NPQ
nonphotochemical energy quenching
- PSII
photosystem II
- HOTE
hydroxy-octadecatrienoic acid
- LOX
lipoxygenase
- PFD
photon flux density
- SOSG
singlet oxygen sensor green
- EPR
electron paramagnetic resonance
- TEMPD
2,2,6,6-tetramethyl-4-piperidone chloride
- Fv/Fm
maximum quantum yield of PSII photochemistry = ratio between the amplitude of the variable chlorophyll fluorescence and the maximal fluorescence level
- qRT-PCR
quantitative RT-PCR
- OPDA
12-oxo phytodienoic acid
- MS/MS
tandem mass spectrometry
References
- Alboresi A., Dall’osto L., Aprile A., Carillo P., Roncaglia E., Cattivelli L., Bassi R. (2011). Reactive oxygen species and transcript analysis upon excess light treatment in wild-type Arabidopsis thaliana vs a photosensitive mutant lacking zeaxanthin and lutein. BMC Plant Biol. 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asai T., Stone J.M., Heard J.E., Kovtun Y., Yorgey P., Sheen J., Ausubel F.M. (2000). Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanci N.C., Luche D.D., Goldman G.H., Goldman M.H.S. (2010). Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet. Mol. Res. 9: 484–505 [DOI] [PubMed] [Google Scholar]
- Balbi V., Devoto A. (2008). Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (2011). Jasmonate-induced defenses: A tale of intelligence, collaborators and rascals. Trends Plant Sci. 16: 249–257 [DOI] [PubMed] [Google Scholar]
- Berger S. (2002). Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214: 497–504 [DOI] [PubMed] [Google Scholar]
- Birtic S., Ksas B., Genty B., Mueller M.J., Triantaphylidès C., Havaux M. (2011). Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. 67: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Cohen S., Flescher E. (2009). Methyl jasmonate: A plant stress hormone as an anti-cancer drug. Phytochemistry 70: 1600–1609 [DOI] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., Havaux M., Bassi R. (2010). Enhanced photoprotection by protein-bound vs free xanthophyll pools: A comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant 3: 576–593 [DOI] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G., Wollman F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Espineda C.E., Linford A.S., Devine D., Brusslan J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 10507–10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo G.M., Chan K.X., Phua S.Y., Pogson B.J. (2012). Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front. Plant Sci. 3: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo G.M., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B.B., Krieger-Liszkay A., Eggen R.I. (2005). Oxidative stress induced by the photosensitizers neutral red (type I) or rose bengal (type II) in the light causes different molecular responses in Chlamydomonas reinhardtii. Plant Sci. 168: 747–759 [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H.F., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D.G., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36 (Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Dudoit S., Speed T.P. (2003). Resampling-based multiple testing for microarray data analysis. TEST 12: 1–77 [Google Scholar]
- González-Pérez S., Gutiérrez J., García-García F., Osuna D., Dopazo J., Lorenzo Ó., Revuelta J.L., Arellano J.B. (2011). Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol. 156: 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun C., Berger S., Matthes D., Mueller M.J. (2007). Early accumulation of non-enzymatically synthesised oxylipins in Arabidopsis thaliana after infection with Pseudomonas syringae. Funct. Plant Biol. 34: 65–71 [DOI] [PubMed] [Google Scholar]
- Gutiérrez J., González-Pérez S., García-García F., Lorenzo O., Arellano J.B. (2011). Does singlet oxygen activate cell death in Arabidopsis cell suspension cultures?: Analysis of the early transcriptional defense responses to high light stress. Plant Signal. Behav. 6: 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Dall'Osto L., Bassi R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Tardy F. (1997). Thermostability and photostability of photosystem II in leaves of the Chlorina-f2 barley mutant deficient in light-harvesting chlorophyll a/b protein complexes. Plant Physiol. 113: 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Triantaphylidès C., Genty B. (2006). Autoluminescence imaging: A non-invasive tool for mapping oxidative stress. Trends Plant Sci. 11: 480–484 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima M., Tanaka R., Tanaka A. (2009). Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 50: 719–729 [DOI] [PubMed] [Google Scholar]
- Inaba T., Yazu F., Ito-Inaba Y., Kakizaki T., Nakayama K. (2011). Retrograde signaling pathway from plastid to nucleus. Int. Rev. Cell Mol. Biol. 290: 167–204 [DOI] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Apel K., Laloi C. (2008). No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Zhang S., Lee K.P., Lakshmanan Ashok M., Blajecka K., Herrfurth C., Feussner I., Apel K. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-H., Li X.-P., Razeghifard R., Anderson J.M., Niyogi K.K., Pogson B.J., Chow W.S. (2009). The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: A study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 1787: 973–984 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Fufezan C., Trebst A. (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H.K., Baroli I., Shin J.W., Fischer B.B., Eggen R.I., Niyogi K.K. (2004). Comparative profiling of lipid-soluble antioxidants and transcripts reveals two phases of photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas reinhardtii. Mol. Genet. Genomics 272: 470–479 [DOI] [PubMed] [Google Scholar]
- Ledford H.K., Chin B.L., Niyogi K.K. (2007). Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot. Cell 6: 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisinger U., Rüfenacht K., Fischer B., Pesaro M., Spengler A., Zehnder A.J., Eggen R.I. (2001). The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Mol. Biol. 46: 395–408 [DOI] [PubMed] [Google Scholar]
- Leister D. (2012). Retrograde signaling in plants: From simple to complex scenarios. Front. Plant Sci. 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz J.W., Öquist G., Wingsle G. (1992). Photosynthesis and photoinhibition in leaves of chlorophyll b-less barley in relation to absorbed light. Physiol. Plant. 85: 495–502 [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lion Y., Gandin E., van de Vorst A. (1980). On the production of nitroxide radicals by singlet oxygen reaction: An EPR study. Photochem. Photobiol. 31: 305–309 [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Moan J. (1990). On the diffusion length of singlet oxygen in cells and tissues. J. Photochem. Photobiol. B Biol. 6: 343–347 [Google Scholar]
- Montané M.-H., Tardy F., Kloppstech K., Havaux M. (1998). Differential control of xanthophylls and light-induced stress proteins, as opposed to light-harvesting chlorophyll a/b proteins, during photosynthetic acclimation of barley leaves to light irradiance. Plant Physiol. 118: 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet J.L., Cacas J.L., Garnier L., Montané M.H., Douki T., Bessoule J.J., Polkowska-Kowalczyk L., Maciejewska U., Agnel J.P., Vial A., Triantaphylidès C. (2004). The upstream oxylipin profile of Arabidopsis thaliana: A tool to scan for oxidative stresses. Plant J. 40: 439–451 [DOI] [PubMed] [Google Scholar]
- Mullineaux P.M., Baker N.R. (2010). Oxidative stress: Antagonistic signaling for acclimation or cell death? Plant Physiol. 154: 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A., Aubry S., Mondhe M., Kingston-Smith A., Gallagher J., Timms-Taravella E., James C., Papp I., Hörtensteiner S., Thomas H., Ougham H. (2010). Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 188: 161–174 [DOI] [PubMed] [Google Scholar]
- Mur L.A., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H., Niyogi K.K. (2011). Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J.A., Daudi A., Butt V.S., Bolwell G.P. (2012). Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236: 765–779 [DOI] [PubMed] [Google Scholar]
- Ochsenbein C., Przybyla D., Danon A., Landgraf F., Göbel C., Imboden A., Feussner I., Apel K. (2006). The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 47: 445–456 [DOI] [PubMed] [Google Scholar]
- op den Camp, R.G.L., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Göbel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed]
- Pena-Cortes H., Albrecht T., Prat S., Weiler E.W., Willmitzer L. (1993). Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191: 123–128 [Google Scholar]
- Petrov, V.D., and Van Breusegem, F. (2012). Hydrogen peroxide - A central hub for information flow in plant cells. AoB Plants 2012: pls014. [DOI] [PMC free article] [PubMed]
- Plumley F.G., Schmidt G.W. (1987). Reconstitution of chlorophyll a/b light-harvesting complexes: Xanthophyll-dependent assembly and energy transfer. Proc. Natl. Acad. Sci. USA 84: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla D., Göbel C., Imboden A., Hamberg M., Feussner I., Apel K. (2008). Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J. 54: 236–248 [DOI] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Cuiné S., Triantaphylidès C., Ravanat J.L., Havaux M. (2012b). Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158: 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012a). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C., Springer A., Samol I., Reinbothe S. (2009). Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276: 4666–4681 [DOI] [PubMed] [Google Scholar]
- Repka V., Fischerova I., Silharova K. (2004). Methyl jasmonate is a potent elicitor of multiple defense responses in garpevine leaves and cell-suspension cultures. Biol. Plant. 48: 273–283 [Google Scholar]
- Reymond P., Farmer E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Samol I., Buhr F., Springer A., Pollmann S., Lahroussi A., Rossig C., von Wettstein D., Reinbothe C., Reinbothe S. (2011). Implication of the oep16-1 mutation in a flu-independent, singlet oxygen-regulated cell death pathway in Arabidopsis thaliana. Plant Cell Physiol. 52: 84–95 [DOI] [PubMed] [Google Scholar]
- Santabarbara S., Jennings R.C. (2005). The size of the population of weakly coupled chlorophyll pigments involved in thylakoid photoinhibition determined by steady-state fluorescence spectroscopy. Biochim. Biophys. Acta 1709: 138–149 [DOI] [PubMed] [Google Scholar]
- Stingl N., Krischke M., Fekete A., Mueller M.J. (2013). Analysis of defense signals in Arabidopsis thaliana leaves by ultra performance liquid chromatography/tandem mass spectrometry: Jasmonates, salicylic acid, abscisic acid. Methods Mol. Biol., in press [DOI] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Asada K. (1982). Dependence of oxygen affinity for Mehler reaction on photochemical activity of chloroplast thylakoids. Plant Cell Physiol. 23: 1457–1461 [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C., Krischke M., Hoeberichts F.A., Ksas B., Gresser G., Havaux M., Van Breusegem F., Mueller M.J. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T., Vicente J., Kulasekaran S., Hamberg M., Castresana C. (2010). Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol. 154: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Walters R.G. (2005). Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 56: 435–447 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xing D. (2008). Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant Cell Physiol. 49: 1092–1111 [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer E.C., Donohue T.J. (2009). Bacterial responses to photo-oxidative stress. Nat. Rev. Microbiol. 7: 856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.