Abstract
We have developed a computational design strategy based on the α-helical coiled-coil to generate modular peptide motifs capable of assembling into metalloporphyrin arrays of varying lengths. The current study highlights the extension of a two-metalloporphyrin array to a four-metalloporphyrin array through the incorporation of a coiled-coil repeat unit. Molecular dynamics simulations demonstrate that the initial design evolves rapidly to a stable structure with a small r.m.s.d. compared to the original model. Biophysical characterization reveals elongated proteins of the desired length, correct cofactor stoichiometry, and cofactor specificity. The successful extension of the two-porphyrin array demonstrates how this methodology serves as a foundation to create linear assemblies of organized electrically and optically responsive cofactors.
Keywords: protein design, nanowires, metalloproteins, porphyrins, coiled-coil
INTRODUCTION
De novo protein design allows the design of proteins and nanostructured arrays that bind a variety of cofactors.1,2 Because the proteins are designed from scratch, one is not limited to naturally occurring cofactors, and it becomes possible to incorporate a variety of non-natural optically and electrically active small molecules. This approach, therefore, has the potential to bring the versatility and programmability of the biological world to bear on the design of nanostructured materials. As a first step in this direction, we have developed a computational strategy for designing α-helical coiled-coils that assemble into metalloporphyrin arrays.3 The coiled-coil was chosen as the structural scaffolding, because its periodic structure enables one to control the length and assembly properties of the proteins. Our initial work focused on porphyrin-containing proteins similar in size and shape to cytochromes;3 here we extend the approach to design longer helical bundles encapsulating linear arrays of electrically and optically responsive cofactors. The parameters defining the coiled-coil geometry afford control over the distribution and alignment of cofactors, while the protein environment around the cofactors modulates the chemical properties of the system.
Our original design consisted of a D2-symmetric α-helical coiled-coil that binds two non-biological iron porphyrin (DPP-FeII/III) cofactors (Figure 1a).3 The two-porphyrin array (2PA) displayed significant cofactor specificity and functional redox properties.3 We have extended the initial coiled-coil design by inserting a three-heptad repeat enabling the protein to bind four iron porphyrins (Figure 1b and e). The modularity of a three-heptad repeat permits the placement of cofactors at periodic intervals, demonstrating the potential for using this approach to generate metalloporphyrin arrays.
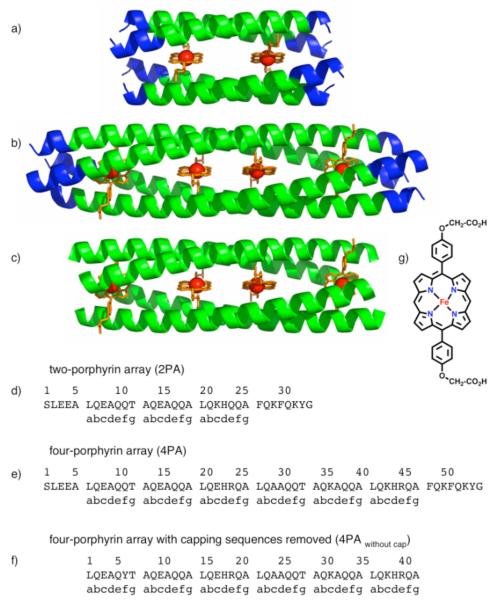
Figure 1.
(a) (b) (c) Models of designed D2-symmetric α-helical coiled-coil proteins containing two (a) and four (b), (c) iron porphyrin cofactors. Blue helices represent capping sequences. Green helices represent the metalloporphyrin-binding region, (d) Sequence of the two-porphyrin array (2PA) (e) Sequence of the four-porphyrin array (4PA). (f) Sequence of the four-porphyrin array with capping sequences removed (4PAwithout cap). Sequences (d), (e), and (f) correspond to models (a), (b), and (c), respectively. (g) Chemical structure of the iron porphyrin cofactor.
EXPERIMENTAL SECTION
Computational Design
An idealized D2-symmetric α-helical coiled-coil backbone4 was generated with the following parameters: 3.5 residues/turn, α-helix radius (r1) = 2.23 Å, the angular frequency of the minor helix (ω1) = 102.76°, rise per residue (h) = 1.59 Å, phase of the minor helix (φ1) = −58.98°, superhelical radius (r0) = 8.00 Å, superhelical pitch (P) = 191.7 Å, the phase of the superhelix, (φ0) = 31.96°, and a z-axis translation (ztrans) = 0.57 Å.3 The superhelical radius defines the radius of the coiled-coil.4 The z-axis translation defines the offset displacement between antiparallel helix pairs obtained by the translation of the supercoiled helix along the superhelical axis.4 The phase of the superhelix describes the rotation applied around the z-axis that determines the squareness of the bundle.4
The sequence was derived by inserting three heptads between positions A26 and F27 of the 34 amino acid sequence previously designed to bind two iron porphyrins (Figure 1d and e).3 The three-heptad repeat matched the region of the 34-mer spanning positions L6 to L26 except charge patterning mutations at K22E, E29A, and E36K were performed to enforce anti-parallel alignment, and mutations at Q24R and Q45R were selected to improve the electrostatic interactions between the cofactor carboxylates. This extended 55 amino acid sequence contains two histidines allowing for axial ligation of four iron porphyrins per monomer within the interior of the D2-symmetric tetramer (Figure 1e).
Within this four porphyrin array (4PA), the iron porphyrins and the histidine and threonine residues were aligned according to the procedure previously reported.3 Along the z-axis, the resulting metal-to-metal distances between the four porphyrins were 19 Å between the first and second porphyrins, 13.3 Å between the second and third porphyrins, and 19 Å between the third and fourth porphyrins.
Molecular Dynamics Simulations
The designed cofactor-bound four porphyrin array was taken as the starting structure for the simulations. The protein was solvated within a 50 × 50 × 108 Å3 water box, and periodic boundary conditions were employed in all cell dimensions. Simulations were performed using the NAMD 2.5 package5 and the CHARMM22 all-atom force field.6 CHARMM226 was also implemented to derive the force fields of the six coordinate planar DPP-FeIII macrocycle and the corresponding macrocycle substituents. Newton’s equation was integrated with a time step of 2 fs, and a cutoff distance of 10 Å was maintained to calculate non-bonded interactions. Bonded and nonbonded forces were evaluated at every time step, and full electrostatic forces were evaluated at every other time step through application of the particle-mesh Ewald (PME) method.7 All bonds between hydrogen atoms and heavy atoms were constrained using the SHAKE algorithm.8 In the production period, the system was simulated in the constant temperature and pressure (NPT) ensemble at 300 K for over 4 ns.
Simulation structures were fit to idealized D2-symmetric backbone conformations according to a previously published modified Crick parameterization.4 The method generates ideal Cα coordinates using the following parameters to define D2-symmetric α-helical coiled-coils: α-helix radius (r1), the angular frequency of the minor helix (ω1), rise per residue (h), phase of the minor helix (φ1) superhelical radius (r0), superhelical pitch (P), the phase of the superhelix (φ0), and a z-axis translation (ztrans).3 During the procedure, r1 = 2.23 Å, ω1 = 102.86°, and h = 1.51Å were held fixed while parameter fitting was performed initially by an exhaustive grid search, followed by a minimization of the Cα coordinates r.m.s.d. using the Simplex algorithm.9
Peptide Synthesis
Automated solid phase synthesis was performed on an ABI 433A Peptide Synthesizer using standard Fmoc-chemistry and Rink Amide resin. All sequences were acetylated and amidated at their respective N- and C- termini. Cleavage of the peptidyl-resin and removal of protecting groups was achieved by stirring the resin at ambient temperature in a mixture containing 95% TFA/2.5% TIS/2.5% H2O for two hours. Peptides were purified by reversed phase HPLC using a Varian ProStar High Performance Liquid Chromatography System equipped with a Vydac C4 22 mm × 250 mm column. The product eluted from a linear gradient of acetonitrile in water containing 0.1% TFA. Product identity was confirmed by MALDI-TOF MS using a PerSeptive Biosystems Voyager-DE RP Biospectrometry Workstation. Purity was further assessed by reversed phase HPLC at 220 nm using a Agilent 1100 Series Liquid Chromatograph equipped with a Vydac C4 4.6 mm × 250 mm column. All peptides were purified to levels exceeding 95% purity (Supporting Info S6 – S9).
Synthesis of DPP-FeIII
5,15-Di[4-carboxymethylene-oxy)phenyl]porphinato iron(III)–chloride (DPP-FeIII-Cl) was synthesized according to previously published procedures.3 Cofactor stock solutions were prepared in dimethylsulfoxide or in 0.1 M NaOH. Stock solutions were used within 24 hours of preparation. ε in DMSO at 522 nm = 9121 M−1cm−1. ε in 0.1 M NaOH at 580 nm = 5995 M−1cm−1.
Size Exclusion Chromatography (SEC)
50 μM peptide solutions were prepared in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 in the presence and absence of 50 μM DPP-FeIII. Samples were heated at 75 °C for 15 minutes, equilibrated at room temperature for 3 hours, and subsequently filtered through 0.1 μm PVDF centrifuge filter membranes (Millipore). 100 μL of each solution was injected onto a GE Healthcare FPLC system equipped with a Superdex 75 10/300GL column. The column was equilibrated in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 with a mobile phase flow rate of 0.5 mL/min, and the absorbance at 280 nm was recorded. Calibration curves were obtained using the molecular weight standard kit, MW-GF-70 (Sigma). Blue dextran was used to determine the void volume.
Binding Stoichiometry (Titration Method)
Peptide solutions ranging from 1.25 μM to 80 μM and containing 10 μM DPP-FeIII were prepared in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 and were equilibrated at ambient temperature for 24 hours. UV-vis spectra were subsequently collected in 1 cm path length quartz cuvettes using a Agilent HP 8453 UV-vis spectrophotometer. The absorbance measured at 409 nm was subtracted from the absorbance of 10 μM DPP-FeIII prepared in the absence of peptide, and the resulting absorbance values were plotted as a function of [4PA peptide] [DPP-FeIII] ratio; data collected from two separate titrations were averaged and used to calculate the binding stoichiometry.
Binding Stoichiometry (Hemochrome Method)10
A solution containing 100 μM peptide and 100 μM DPP-FeIII was prepared and chromatographed according to the procedure described in the SEC section. The fraction containing the cofactor-bound protein was collected and subsequently concentrated using an Amicon Ultra 4 10,000 MWCO centrifuge filter membrane (Millipore).
An aliquot of the concentrate was used to determine to amount of DPP-FeIII present in the SEC fraction. 25 μL of 50 mM potassium ferricyanide, 115 μL buffer, and 300 μL of 40% v/v pyridine in 200 mM NaOH were added to a 160 μL aliquot of the SEC concentrate ensuring that the protein was denatured, and the DPP-Fe cofactor was fully oxidized and unbound from the denatured protein. The sample was subsequently reduced upon addition of 4 mg of sodium dithionite. The absorbances of the ferric and ferrous forms of DPP-Fe were measured in a 1 cm path length quartz cuvette using an Agilent HP 8453 UV-vis spectrophotometer, and the concentration of DPP-FeIII was determined by subtracting the oxidized cofactor spectrum from that of the corresponding ferrous state at 520 nm. A standard curve was generated from samples containing 40% v/v pyridine, 200mM NaOH, and varying concentration of DPP-FeIII. All samples were allowed to equilibrate for 10 minutes before spectra were acquired. λmax Q-band oxidized = 565 nm, λmax Q-band reduced = 520 nm. Δε (εreduced − εoxidized at 520 nm) = 1.84 10−4 M−1cm−1.
A second aliquot of the concentrate was used to determine the amount of peptide present in the SEC fraction. In order to dissociate the DPP-FeIII from the protein, the aliquot was diluted with acetonitrile and TFA to a final concentration of 18% acetonitrile and 2% TFA. The peptide concentration was measured by reversed phase HPLC at 220 nm using an Agilent 1100 Series Liquid Chromatograph equipped with a Vydac C4 4.6 mm × 250 mm column. The peptide eluted from a linear gradient of acetonitrile in water containing 0.1% TFA. A standard curve was generated from samples containing 20 to 200 μM peptide.
Binding Selectivity
Solutions containing 10 μM DPP-FeIII or 10 μM FeIII protoporphyrin IX were prepared in 20 mM sodium phosphate pH 7.5 and were equilibrated in the presence and absence of 10 μM peptide at ambient temperature for 4 hours. UV-vis spectra were subsequently collected in 1 cm path length quartz cuvettes using an Agilent HP 8453 UV-vis spectrophotometer.
Electron Paramagnetic Resonance
Samples were prepared in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 with 250 μM peptide and 225 μM DPP-FeIII Samples were heated to 75 °C for 15 minutes and equilibrated at room temperature for three hours. Samples were then frozen in liquid nitrogen, and EPR spectra were acquired at 7 K on a Bruker Model ESP300E Spectrometer equipped with an Oxford ESR Model 900 Continuous Flow Cryostat, an Oxford Model ITC4 Temperature Controller, and a Hewlett-Packard Model 5350B Frequency Counter using the following parameters: microwave frequency, 9.448 GHz; microwave power, 20 mW; modulation amplitude, 20 gauss; receiver gain, 105; conversion time, 81.9 milliseconds; time constant, 81.9 milliseconds. Blank spectra were acquired in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 and subtracted from the sample spectra.
Circular Dichroism
Spectra were acquired on a Jasco 810 Spectropolarimeter. Samples containing 20 mM sodium phosphate, pH 7.5 and 50 μM peptide were prepared in the presence and absence of stoichiometric amounts of DPP-FeIII. Samples were heated to 75 °C for 15 minutes and allowed to equilibrate at room temperature for at least three hours prior to measurement. Measurements were obtained in 0.1 cm path length quartz cells. Isothermal scans were collected at 25 °C. For thermal unfolding experiments, the ellipticity at 222 nm was monitored as a function of temperature; data were collected every 2 °C with a heating rate of 1 °C/min followed by a 4 minute incubation time.
Analytical Ultracentrifugation
Peptide solutions were prepared in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5. Samples were heated to 75 °C for 15 minutes and equilibrated at room temperature for at least three hours prior to analysis. Sedimentation velocity and sedimentation equilibrium experiments were performed at 25 °C using a Beckman XL-I analytical ultracentrifuge.
For equilibrium experiments, samples were centrifuged at 25,000 and 35,000 rpm, and consecutive scans were acquired ensuring equilibrium conditions were attained. The absorbance was monitored at 409 nm and at 230 nm for peptide samples prepared in the presence and absence of DPP-FeIII, respectively. The data were analyzed using a modified global fitting routine in IgorPro (Wavemetricsjnc,).11 Partial specific volumes and buffer densities were calculated by the program Sedentrp.12
For sedimentation velocity experiments, samples were centrifuged at 45,000 rpm. The absorbance was monitored at 409 nm and at 230 nm for peptide samples prepared in the presence and absence of DPP-FeIII, respectively. Sedimentation coefficients (s values) were determined by fitting sedimentation data using the direct boundary method implemented in the program “SEDFIT”.13 Prolate ellipsoid axis ratios were estimated using the Teller method implemented in the program “Sedentrp”.12
RESULTS AND DISCUSSSION
Design
The original two-porphyrin array (2PA) (Figure 1a) consists of an anti-parallel tetramer with each iron porphyrin (Figure 1g) coordinated to the s-nitrogens of two axial histidine ligands, located at “d” heptad positions on diagonally opposed parallel helices.3 Second-shell hydrogen bonds between the δ-histidine nitrogens and the γ-threonine oxygens stabilize the orientations of the His residues in a perpendicular manner. The functional structural unit of 2PA is a homo-tetrameric 21-residue sequence that binds two iron porphyrin cofactors. Because the length of this sequence is an integral multiple of the 7-residue repeat of the α-helical coiled-coil, its repetition should lead to elongated, multiporphyrin structures. Thus, as described in the Experimental Section, an elongated four-porphyrin array (4PA) was designed by inserting this geometric motif into the original 34 amino acid structure3 (Figure 1d and e). The extended 55 amino acid sequence (Figure 1e) now contains two histidines per monomer, allowing for axial ligation of four DPP-FeII/III cofactors within the interior of the D2-symmetric tetramer (Figure 1b).
Both the previous two-porphyrin and the newly designed four-porphyrin arrays incorporate helical capping sequences to stabilize the proteins (Figure 1a and 1b blue helices). In order to probe the specific contribution of these capping sequences on the four-porphyrin array, residues 1-5 and residues 48-55 were removed. Since the removal of these capping resides caused the loss of a tyrosine, the glutamine at position 11 of the 55 residue 4PA was mutated to a tyrosine to aid in spectroscopic quantitation. The resulting 42-residue peptide (4PAwithout cap) contains only the two modular units of the three-heptad porphyrin binding motif (Figure 1c and 1f).
Molecular Dynamics Calculations
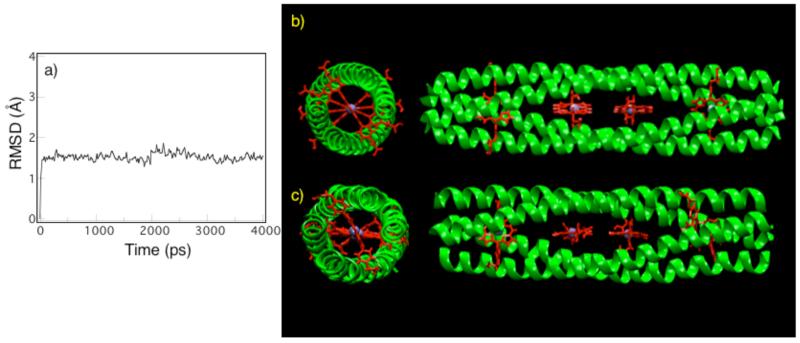
Molecular dynamics calculations test the viability of a given design. When examined by molecular dynamics, a poorly designed sequence may undergo large structural fluctuations on the nanosecond timescale never evolving to a stable structure, whereas a well-designed sequence equilibrates quickly, attaining a stable structure that persists over the remainder of the simulation. Molecular dynamics calculations6,14 of the 4PA solvated in a 50 × 50 × 108 Å3 water box were performed to validate the retention of key design elements during an all-atom relaxation of the molecular structure. The initial model was generated using a Monte Carlo coarse-grained ridged-body optimization of the helical bundle (Figure 2b). The protein equilibrated within 100 ps and remained stable during the final 4 ns of the trajectory (Figure 2a). Upon equilibration, the average r.m.s.d. of the central superhelical structure (residues 7-48) with respect to the starting structure was 1.50±0.14 Å. The rapid equilibration time and the small fluctuations of the r.m.s.d. from the initial model highlight the ability of the design methodology to produce a stable structure in a deep energy minimum. In contrast, previous “maquette” designs have tended to drift from their starting configurations.1 Fitting the simulation structure (Figure 2c) to an idealized D2-symmetric backbone conformation,4 the superhelical pitch decreased from 191 Å to 179 Å, while the superhelical radius increased from 8.00 Å to 8.41 Å. This caused a contraction along the superhelical axis and an expansion between the helices that simultaneously retained the D2 symmetry (0.4 r.m.s.d. from an idealized structure). The resulting relaxation of the helical bundle maintained the porphyrin edge-to-edge distance but slightly altered the relative orientation between adjacent cofactors (Figure 2c). The rapid equilibration time, the small fluctuations of the r.m.s.d., the preservation of D2 symmetry, and the small changes between the simulated structure and the initial model demonstrate a highly stable designed configuration and strongly support the potential of these scaffolds to control the local environment and assembly of the bound metalloporphyrins.
Figure 2.
Evolution of the root-mean-squared deviation (r.m.s.d.) for the backbone atoms of the central superhelical structure (residues 7-48) of the 4PA with respect to the model structure calculated from the molecular dynamics simulation. (b) Model of designed D2-symmetric α-helical coiled-coil 4PA protein containing four DPP-FeIII cofactors. (c) Configuration of the model after 4 ns of molecular dynamics simulations.
UV-Visible Spectroscopy
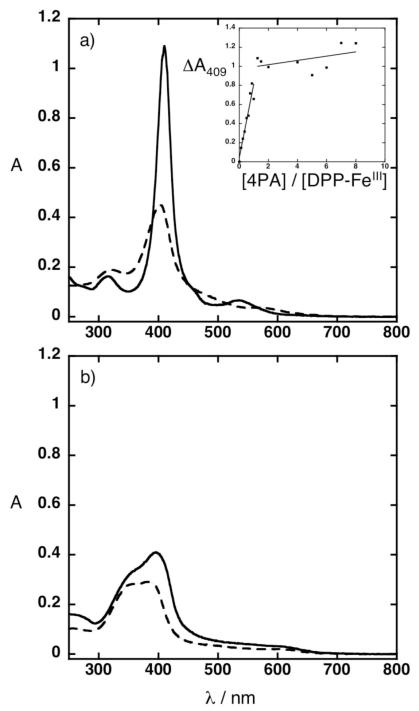
Inserting the three-heptad repeat enabled the protein to bind four DPP-FeIII cofactors as determined by titrating the 4PA peptide into a fixed concentration of cofactor while monitoring the increase in the Soret absorbance (Figure 3a inset). The 4:4 binding stoichiometry was further confirmed by the hemochrome method (Supporting Info S1)10 and by molecular weight determination of the cofactor-bound protein. UV-vis spectra of the 4PA peptide in the presence of stoichiometric amounts of DPP-FeIII cofactor exhibit an increase, sharpening, and shift in the Soret and Q bands to 409 and 530 nm, respectively when compared to free DPP-FeIII cofactor (Figure 3a).
Figure 3.
UV-vis spectra of solutions prepared in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5. (a) (inset) Titration of the 4PA peptide into 10 μM DPP-FeIII. (a) Solid line represents 10 μM 4PA peptide and 10 μM DPP-FeIII Dashed line represents 10 μM DPP-FeIII (b) Solid line represents 10 μM 4PA peptide and 10 μM FeIII protoporphyrin IX. Dashed line represents 10 μM FeIII protoporphyrin IX.
UV-visible spectroscopy provides an easy method to test the specificity of the 4PA. There is an excellent fit between the designed porphyrin-binding sites in 4PA and the structure of the targeted DPP-FeII/III cofactor, including specific interactions with the functional groups extending from the porphyrin ring. The complex displays uniform and favorable van der Waals packing, with nearly complete burial of the hydrophobic groups of the DPP-FeII/III cofactor. Only the terminal polar carboxylates of the cofactor are exposed to solvent. To test the specificity of 4PA, we examined its binding to a natural porphyrin, FeIII protoporphyrin IX. The spectra of FeIII protoporphyrin IX remain largely unaffected by addition of a stoichiometric amount of the 4PA peptide (Figure 3b). Thus, 4PA is able to discriminate between different porphyrins, showing a strong interaction with DPP-FeII/III but not a natural heme. These experiments illustrate how incorporating repeating structural units can easily modulate the number of bound porphyrins without detracting from the cofactor selectivity specified by the original design.
Molecular weight determination
Size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) of the 4PA indicate that it exists as a homogeneous tetrameric species in both the apo and cofactor-bound states (Supporting Info S2 & S3). SEC showed a slight degree of aggregation occurs upon addition of cofactor, but this could be minimized by heating the sample above the Tm followed by cooling the sample to room temperature. Both the apo and cofactor-bound 4PA elute as single species at apparent molecular weights of 29,700 D and 29,100 D, respectively, consistent with a tetrameric assembly of α-helices.
Sedimentation equilibrium AUC of the 4PA in the absence of cofactor indicate that it behaves as a single molecular species with an apparent MW = 23,500±160 D, in excellent agreement with the value expected for a tetramer (25,328 D); the protein also sedimented as a tetramer when reconstituted with cofactor (MWobs = 27,900 ±130 D; MWcalc = 27,988 D). Sedimentation velocity experiments were conducted to determine the overall shape of the structure. Coefficients of 1.9 s and 2.5 s were obtained for the apo and cofactor-bound state and were fit to respective prolate ellipsoid axis ratios of 6.0 and 4.8, using the Teller method,12 suggesting both the apo and cofactor-bound protein conformations are elongated, and the coiled-coil widens to accommodate the DPP-FeIII cofactor.
Removal of the capping sequences affects the stability of the apo state, and the 4PAwithout cap elutes as a monomeric species when analyzed by SEC. Addition of stoichiometric amounts of DPP-FeIII induces oligomerization, and the cofactor-bound 4PAwithout cap elutes at an apparent molecular weight consistent with a tetrameric assembly of α-helices (Supporting Info S2). The capping sequences, therefore, contribute to stability but are not required for assembly into the four-helix bundle in the presence of cofactor.
CD spectroscopy
In the apo form, the 2PA exists predominantly as a random coil (Figure 4a). Extension of this sequence through the insertion of the three-heptad repeat enforces a helical conformation, observed by the minima at 208 and 222 nm in the circular dichroism (CD) spectrum of the apo 4PA (Figure 4b). Removal of the capping sequences on the 4PA destabilizes the apo state, and random coil monomers were observed at ambient temperature in the CD spectrum of the apo 4PAwithout cap (Figure 4c).
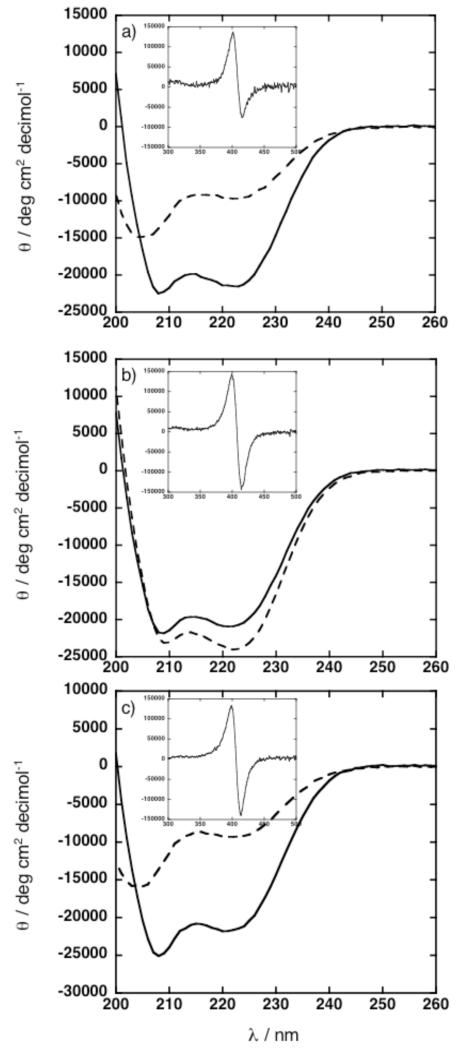
Figure 4.
CD spectra of peptides in 20 mM sodium phosphate, pH 7.5 acquired at 25 °C. (a) Solid line represents 50 μM 2PA peptide and 25 μM DPP-FeIII. Dashed line represents 50 μM 2PA peptide. (b) Solid line represents 50 μM 4PA peptide and 50 μM DPP-FeIII. Dashed line represents 50 μM 4PA peptide. (c) Solid line represents 50 μM 4PAwithout cap peptide and 50 μM DPP-FeIII. Dashed line represents 50 μM 4PAwithout cap. Insets (a – c) highlights the Soret region. θ is normalized for cofactor concentration.
In the cofactor-bound state, the 2PA, the 4PA, and 4PAwithout cap are all helical (Figure 4a, b, and c). The small decrease in ellipticity at 222 nm, observed upon addition of cofactor to 4PA, cannot be interpreted as a change in helical content because the porphyrins also have transitions in this region. In the presence of stoichiometric amounts of DPP-FeIII, the 4PA and 4PAwithout cap possess identical CD and UV-vis spectra in the Soret region, each exhibiting maxima and minima at 399 and 414 nm in the CD spectra of the Soret region (Figure 4b and c, Supporting Info S5). The negative and positive Cotton effects observed in the Soret region are consistent with the confinement of the metalloporphyrins within an intramolecular framework.15
EPR spectroscopy
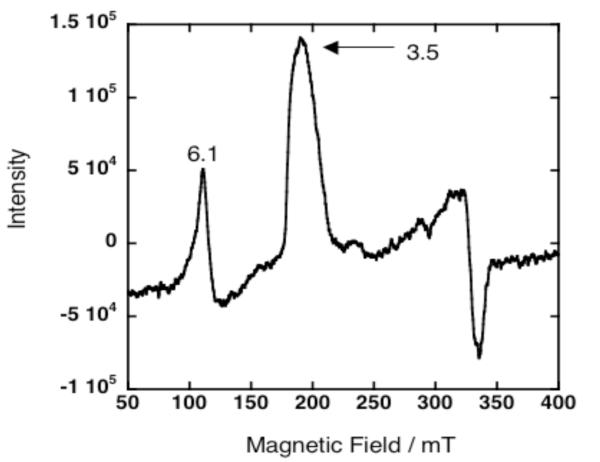
Probing the geometry of the 4PA binding site with EPR spectroscopy reveals the presence of a highly anisotropic low spin system (HALS) demonstrated by the signal at g = 3.5 (Figure 5).16 The signals at g values 6.1 and 3.5 are assigned to high spin, unbound DPP-FeIII and a low spin DPP-FeIII coordinated to the imidazole rings within the protein interior. During sample preparation, if the protein is not frozen rapidly enough, a small amount of unbound cofactor is generated causing the appearance of high spin unbound DPP-FeIII cofactor at g = 6.1. The intrinsic intensity of the high spin component is approximately five times greater than the intensity of an equivalent amount of the low spin component, so even a small amount of this species can easily be detected. The EPR spectra of the 4PA and the 2PA3 show nearly identical ligation environments. Comparing the HALS spectra of model compounds,16 the slight broadening of the signal at g = 3.5 probably reflects the quasi-equivalence of the cofactor sites within the 4PA. The EPR data suggest that extending the coiled-coil framework by insertion of a three-heptad repeat preserves the geometry of the binding site specified by the original design.
Figure 5.
EPR spectrum of 250 μM 4PA peptide and 225 μM DPP-FeIII in 20 mM sodium phosphate, 100 mM NaCl, pH 7.5 acquired at 7 K.
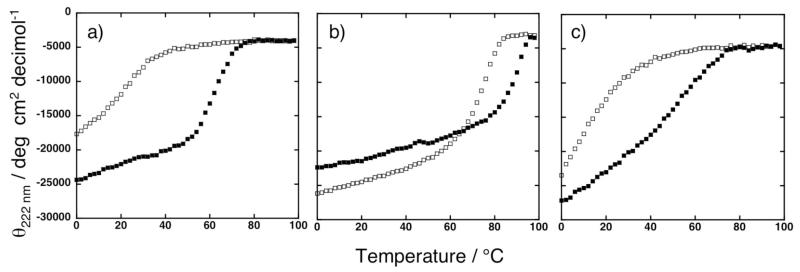
Stability
Thermal denaturation curves exhibit highly cooperative unfolding transitions for both the cofactor-bound 2PA and 4PA. Reconstituted 4PA exhibits a higher Tm compared to the 2PA version (87 °C vs. 63 °C, respectively) (Figure 6a and b). The cofactor further stabilizes the apo 4PA, as the Tm of the protein increases from 77 °C to 87 °C upon stoichiometric addition of DPP-FeIII (Figure 6b). The capping sequences provide additional stability, and removal of the capping sequences reduces the Tm of the cofactor-bound 4PAwithout cap to 50 °C. The apo 4PAwithout cap is even less stable, showing a midpoint near room temperature (Figure 6c). It is interesting to note that the cofactor-bound forms of both 4PA and 2PA show sigmoidal unfolding curves typical of cooperatively folded proteins. By contrast, the transition of 4PAwithout cap is less steep, and the midpoint occurs at lower temperature than either 4PA or 2PA. These data suggest that the addition of the capping sequences contribute more to stability than the addition of the three-heptad repeat in 4PA.
Figure 6.
Thermal denaturation curves of proteins in 20 mM sodium phosphate, pH 7.5. (a) Solid squares represent 50 μM 2PA peptide and 25 μM DPP-FeIII. Open squares represent 50 μM 2PA peptide. (b) Solid squares represent 50 μM 4PA peptide and 50 μM DPP-FeIII. Open squares represent 50 μM 4PA peptide. (c) Solid squares represent 50 μM 4PAwithout cap peptide and 50 μM DPP-FeIII. Open squares represent 50 μM 4PAwithout cap peptide. Temperature ramp from 0 to 98 °C shown.
Although the folding is reversible, given sufficient incubation time, the equilibration at each time point is so slow that sample degradation at elevated temperatures and instrument drift make it impractical to perform thermal unfolding curves under fully reversible conditions. No attempt, therefore, was made to extract thermodynamic parameters from these curves (Figure 6).
Although these capping sequences affect the stability of both the apo and cofactor-bound protein, they do not impact cofactor binding or orientation, as probed by their UV-vis and CD spectra. Future designs, therefore, may remove or alter the capping sequences to modify protein stability. The combined CD, SEC, and AUC data demonstrate how the addition of a three-heptad repeat stabilizes the cofactor-bound conformations while promoting the formation of elongated structures and avoiding non-controlled polymerization. The stability data suggest that the modular section of the coiled-coil can be applied to construct metalloporphyrin arrays of extended lengths.
CONCLUSIONS
Using a modular α-helical coiled-coil repeat, computationally designed peptides can assemble into structures of defined lengths and selectively accommodate arrays of non-biological metalloporphyrins. The simple incorporation of a three-heptad repeat enables the assembly of a four-porphyrin array without detracting from the cofactor specificity or disrupting the coiled-coil structure imparted by the original de novo design. The elongated structures attained by this approach demonstrate the feasibility of forming protein nanowires with precisely spaced metalloporphyrins, providing a powerful approach to the design of one-dimensional arrays of non-biological cofactors. By varying the length of the coiled-coil or the spacing between adjacent metalloporphyrins, it should be possible to strategically engineer the properties of electrically and optically responsive multi-porphyrin arrays.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Robyn Broach for assistance with EPR measurements. We thank Dr. Ben North for assistance with the Crick parameterization. We acknowledge support from NIH (GM54616 & GM-071628). JKB, WFD, JGS & MJT received partial support from the DOE grant DE-FG02-04ER46156. This research was partially supported by the Nano/Bio Interface Center through the National Science Foundation NSEC DMR-0425780.
Footnotes
SUPPORTING INFORMATION AVAILABLE
UV-vis, SEC, AUC, HPLC, MALDI-TOF MS data. Complete reference 6.
Contributor Information
Karen A. McAllister, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104
Hongling Zou, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104.
Frank V. Cochran, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104
Gretchen M. Bender, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104
Alessandro Senes, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104.
H. Christopher Fry, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104.
Vikas Nanda, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104.
Patricia A. Keenan, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104
James D. Lear, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104
Jeffery G. Saven, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104
Michael J. Therien, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104
J. Kent Blasie, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104.
William F. DeGrado, Department of Biochemistry and Molecular Biophysics, Johnson Foundation, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104; Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104.
REFERENCES
- (1).DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu. Rev. Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- (2).Lu Y, Berry SM, Pfister TD. Chem. Rev. 2001;101:3047–3080. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]
- (3).Cochran FV, Wu SP, Wang W, Nanda V, Saven JG, Therien MJ, DeGrado WF. J. Am. Chem. Soc. 2005;127:1346–1347. doi: 10.1021/ja044129a. [DOI] [PubMed] [Google Scholar]
- (4).North B, Summa CM, Ghirlanda G, DeGrado WF. J. Mol. Biol. 2001;311:1081–1090. doi: 10.1006/jmbi.2001.4900. [DOI] [PubMed] [Google Scholar]
- (5).Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- (6).MacKerrell AD, et al. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- (7).Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- (8).Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- (9).Vetterling WT, Teukolsky SA, Press WH, Flannery BP. Numerical Recipes. 2nd Edition Cambridge University Press; Cambridge: 2002. [Google Scholar]
- (10).Berry EA, Trumpower BL. Analytical Biochemistry. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- (11).Brooks IS, Soneson KK, Hensley P. Biophys. J. 1993;64:A244. [Google Scholar]
- (12).Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- (13).Schuck P, Rossmanith P. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- (14).Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- (15).Pescitelli G, Gabriel S, Wang Y, Fleischhauer J, Woody RW, Berova N. J. Am. Chem. Soc. 2003;125:7613–7628. doi: 10.1021/ja030047v. [DOI] [PubMed] [Google Scholar]
- (16).Walker FA. Chem Rev. 2004;104:589–615. doi: 10.1021/cr020634j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.