Introduction
Aberrant lymphoma epigenome is the result of a combinatorial contribution of changes in DNA methylation, histone modifications, and noncoding RNA (ncRNA) expression in lymphoma cells. All of the components of epigenome have been the focus of intense studies in various subtypes of lymphoma, which resulted in the deeper understanding of novel mechanisms of lymphomagenesis and revealed novel therapeutic targets. Some of the changes are common to all lymphomas and even other types of cancers, whereas others are subtype specific. In this review, we attempt to summarize the current state of knowledge in the field of lymphoma epigenomics with particular emphasis on subtype-specific changes, differences between adult and pediatric lymphomas, and novel epigenetic therapies.
DNA methylation
DNA methylation patterning and the role of DNA methyltransferases in normal B-cell development and lymphomagenesis
DNA methylation is part of epigenetic programming that is required for normal B-cell development and is disrupted during lymphomagenesis.1 To understand how aberrant methylation contributes to lymphomagenesis, it is important to understand the patterns of methylation in normal B cells. Deaton et al2 showed that not only differentially methylated regions (DMRs) around transcriptional start sites are of significance, but also methylation patterns in the intergenic areas have cell and tissue specificity. The dynamic nature of methylome during hematopoietic development was studied by Ji et al in mouse multipotent progenitors using comprehensive high-throughput array-based relative methylation analysis, which examined 4.6 million CpGs in the genome.3 This study demonstrated DMRs and revealed that lymphoid lineage commitment requires more DNA methylation than myeloid lineage. Challen et al demonstrated that DNMT3a expression is higher in primitive long-term hematopoietic stem cells than in progenitors and differentiated cells.4 Importantly, the same group demonstrated that loss of DNMT3a in mice resulted in significant expansion of HSCs because of their reduced differentiation ability and was associated with broad changes in the distribution of methylated CpGs. Shaknovich et al addressed changes in epigenome during germinal center (GC) transit and revealed that transition from naïve B cells (NB) to centroblasts (CB) is associated with predominant loss of methylation in 235 differentially methylated genes that affect NF-κB and mitogen-activated protein kinase pathways.5 These studies set the stage for interpretation of epigenetic changes in pre-GC and GC-derived lymphomas.
The key factors responsible for DNA methylation are members of the DNA methyltransferase (DNMT) family: DNMT1, DNMT3a, and DNMT3b, which have complex patterns of expression in peripheral B cells and during GC transit.5,6 DNMT expression is highly compartmentalized within the GC, with DNMT1 and DNMT3b being the most highly expressed within GC B cells, but not in NB cells. GC formation is dependent on the amount of DNMT1 with significant diminution of GCs in Dnmt1 hypomorphic mice.5 The important role of DNMTs in lymphomagenesis is underscored by the evidence from Amara et al7 that DNMT1, DNMT3a, and DNMT3b are overexpressed in 48%, 13%, and 45% of 81 de novo diffuse large B-cell lymphomas (DLBCLs) and correlate with advanced clinical stages. Importantly, concomitant expression of DNMT1 and DNMT3b correlated with resistance to treatment, whereas DNMT3b overexpression correlated with shorter overall and progression-free survival. Somatic mutations in DNMTs may contribute to chromosomal instability, as supported by the observation of increased mutation rates in patients with germline mutations in DNMT3b.8 Mutational analysis of DNMT3a by Kim et al in 401 hematologic malignancies identified that mutations do occur infrequently in pre–B-acute lymphocytic leukemia (ALL), but allelic loss is more frequent (48.1%) in lymphomas.9 Van Vlierberghe et al identified DNMT3a mutations in 2 of 57 patients with adult immature T-ALL.10 It appears that DNMT mutations in precursor B and T neoplasms, albeit infrequent, result in aberrant pathogenic function, whereas the overall DNMT expression levels play a role in more mature lymphoid malignancies. These findings suggest that the development of specific DNMT inhibitors is warranted for the treatment of lymphomas.
Aberrant DNA methylation patterning in B-cell lymphomas
DNA methylation in DLBCLs.
DLBCL is the most common aggressive lymphoma in adults, and many studies so far focused on the changes in DLBCL epigenome elucidating the patterns of aberrant methylation.
Two key changes in DNA methylation seem to be at play in DLBCLs and possibly in follicular lymphoma (FL): (a) site-directed changes affecting specific oncogenes or tumor suppressor genes and (b) increasing epigenomic instability and heterogeneity. Hypermethylation of gene regulatory regions can lead to transcriptional silencing, in part because of recruitment of methylation-dependent repressor proteins, whereas hypomethylation can lead to increased gene expression, and genomic instability when affecting large regions of the genome.1 P16 (INK4A) is an example of a known tumor suppressor gene hypermethylated in lymphomas. Even though P16 methylation in lymphomas has been thought to impart a worse prognosis, Zainuddin et al studied 113 cases of primary DLBCL using pyrosequencing and showed no association with worse outcome.11 KLF4 is another example of a tumor suppressor gene reported to be aberrantly hypermethylated in many subtypes of lymphoma, including FL, DLBCL, Burkitt’s lymphoma (BL) and classical Hodgkin lymphoma (HL) by Guan et al, suggesting common subtype-independent mechanisms of lymphomagenesis.12
Recent reports suggest that regulation of gene expression by DNA methylation patterning is complex and is not a simple “on” or “off” switch for gene expression. For example, hypermethylation of a CpG-rich region within the first intron of BCL6 was reported to induce higher levels of BCL6 expression, at least in part by blocking binding of CCCTC-binding factor.13 Accordingly, BCL6 levels were decreased in lymphoma cell lines exposed to the DNMT inhibitor decitabine.13 These findings illustrate the need for more comprehensive studies characterizing the mechanisms of epigenetic regulation.
Additional insight into the pathobiology of lymphomas came from the study of 83 mature aggressive B-cell non-HL by Martin-Subero et al that revealed 56 genes de novo methylated in all lymphoma subtypes and 22 genes with a subtype-specific pattern of methylation.14 Remarkably, de novo methylated genes were enriched for Polycomb (PcG) targets in embryonic stem cells, highlighting the complex epigenetic mechanisms of lymphomagenesis. Velichutina et al demonstrated that in normal GC B cells, DNA methylation and H3K27me3 mark created by the PcG family protein EZH2 were mutually exclusive, whereas this epigenetic segregation was disrupted in DLBCLs.15 These aberrant epigenetic events may be caused by mutations in the SET domain of EZH2 that have been detected in as much as 12% of FLs and 9.7% of DLBCLs.16,17 The effect of SET domain mutations on the physical interaction between EZH2 and DNMTs is not known but might contribute to aberrant overlap of gene silencing marks, as stated before.18 EZH2 inhibitors promise to be effective therapeutic options for patients with mutated EZH2. Knutson et al reported the discovery of EPZ005687, a potent inhibitor of EZH2 with Tyr641 or Ala677 mutations.19,20 Treatment of DLBCL cell lines with mutant EZH2 with this novel inhibitor induced apoptosis and eventually cell death.
In addition to individual genes that are aberrantly methylated in lymphomas, a new concept of epigenetic instability has emerged as contributing to the pathogenesis of some lymphoma subtypes. De et al clearly demonstrated that increased variance in DNA methylation, as compared with normal B cells, correlated with worse overall and progression-free survival in DLBCLs and in FLs.21,22 The intrasample and intersample heterogeneity reflect possible clonal heterogeneity within lymphoma and heterogeneity between individuals respectively. Normal B cells appear to have bimodal distribution of methylation, whereas lymphomas have much greater variance of methylation at each individual CpG, and this variance is increasing from low grade to high grade FLs and further to GCB-like DLBCLs and ABC-like DLBCLs, resulting in unimodal distribution. The source of such growing epigenetic instability is not entirely clear but has been linked to the events in the GC. Shaknovich et al demonstrated that GC B cells have higher epigenetic heterogeneity than NB cells and that this phenomenon may be dependent on activation-induced cytidine deaminase (AID).5 It appears that AID may contribute to epigenetic instability in lymphomas through its role as a demethylase, similar to its contribution to genomic instability through its role in somatic hypermutation (SHM).23-27 Another potential contributor to epigenetic instability is CCCTC-binding factor—an insulator protein that may prevent the spread of aberrant methylation in lymphomas.28
DNA methylation in FL.
The first truly genome-wide methylome analysis in FL was performed by Choi et al, interrogating 726 003 CpGs in FL cell line RL and 1.3 million CpGs in CD19+ B cells using 454 sequencing technology.29 Comparison of the methylation maps between CD19+ B cells and FL cell line revealed the overall differences in the distribution of CpG methylation: in normal B cells, methylation was predominantly associated with inter- and intragenic regions enriched for repetitive sequences, whereas in FL there was increased methylation in promoter regions and decreased methylation of intra- and intergenic regions. This finding supports the idea of increased chromosomal fragility in lymphomas as a result of hypomethylation and aberrant methylation of tumor suppressor genes. Significant differences were observed in methylation in HOX genes as well as SOX and FRIZZLED gene families. Earlier studies by O’Riain et al and Benett et al30 discovered that, similarly to DLBCLs, there is hypermethylation of PRC2 targets in FL.31 Hypermethylation of EZH2 targets along with EZH2 mutations in 7.2% of FLs16 opens the possibility for effective epigenetic treatments with EZH2 inhibitors in combination with demethylating agents.
DNA methylation of MCL.
Methylation profiling of mantle cell lymphoma (MCL) by Enjuanes et al revealed 252 hypermethylated genes. Five genes (SOX9, HOXA9, AHR, NR2F2, and ROBO1) correlated with higher proliferation, increased number of chromosomal abnormalities, and shorter survival of the patients.32 Methylation of HOX genes was also reported in MCL.33 Leschenko et al performed DNA methylation profiling using the HELP assay and NimblegenRoche methylation arrays and revealed differences in methylation between 22 primary MCLs and 10 purified fractions of IgD+ B-cell controls, which are the putative normal precursors to MCL.34 The study revealed several aberrantly methylated genes, of which CD37 was confirmed to be aberrantly overexpressed in MCL and served as a novel therapeutic target for small modular immunotherapeutic (CD37-SMIP), resulting in significant loss of viability in cell lines and in synergism with epigenetic therapy with decitabine.
DNA methylation in CLL.
Chronic lymphocytic leukemia (CLL) displays concomitant genome-wide loss of methylation and site-specific gain of methylation. Hypomethylation of the genome has been demonstrated using a variety of techniques from high-performance liquid chromatography35 to whole-genome bisulphate sequencing.36 Kulis et al identified DMRs between unmutated CLL and mutated CLL and their putative normal B-cell precursors NB cells and memory B cells and confirmed that although majority of CpGs were hypomethylated and centered in gene bodies, hypermethylated CpGs were enriched in 5′ regulatory regions, CpG islands, and 5′ regions of introns.36 Aberrantly hypermethylated genes in CLL that serve as biomarkers and shed light into disease biology include CDKN2A and CDKN2B, ZAP70, DAPK1, and ID4 among others.37-40 Another significant epigenetic event in CLL is silencing of Wnt pathway inhibitors such as CDH1, DKK1, DKK2, DKK3, SFRP1, SFRP2, SFRP3, SFRP4, SFRP5, and WIF1 genes, which leads to activation of the pathway and overexpression of WNT family members and its receptor Frizzled (Fzd).41,42
Recent study by Kulis et al36 revealed that 3 epigenetic subtypes of CLL have different clinical course and time to first treatment, underscoring the importance of epigenome in the pathogenesis of the disease and promising novel disease biomarkers and therapeutic targets.
DNA methylation in pediatric lymphoid neoplasms.
The most common pediatric malignancy is pre–B-cell ALL, and, consequently, most of the relevant studies in pediatric lymphomas/leukemias come from this diagnostic category. The biology of childhood ALL is different from adult ALL with different characteristic leukemogenic events.43 There are limited data suggesting that epigenetic lesions may be similar in the 2 categories.43 Garcia-Manero et al studied promoter-associated CpG islands in MDR1 (multidrug resistance gene 1), P15, C-ABL, CD10, P16, and P73 in 16 pediatric patients and concluded that there was no difference in the methylation levels of those genes between pediatric and adult patients.44 Wong et al identified a common methylation signature for all genetic subtypes of pediatric ALL that could differentiate disease-free bone marrow specimens from malignant specimens.43 Davidsson et al profiled the 2 most common genetic subgroups of B-ALL: hyperdiploid and t(12;21)(p13;q22) or EVT6/RUNX1 fusion–associated cases and detected subtype-specific methylation differences.45 Methylation hotspots were associated with chromosomal bands containing imprinted genes. Hyperdiploid ALL had more hypermethylated genes (7650 vs 3983), and the top candidates were enriched for genes from cell signaling, transcription regulation, and apoptosis gene ontology (GO) categories. There is already emerging molecular evidence that epigenetic treatments can be effective in pediatric leukemia: histone deacetylase (HDAC) inhibitors may reverse aberrant hypomethylation of MYC, SET, RUNX1, and RAN, as well as the MLL-AF4 fusion product in t(4;11)-positive primary infant ALL cells, leading to cell death.45
Histone modifications
Histone code in normal lymphoid development
A major part of the epigenetic regulation is conveyed through “the histone code,” which serves as a platform for the assembly of the appropriate transcription regulatory machinery.46,47 Cells use a large army of histone-modifying enzymes—“writers,” “erasers,” and chromatin “remodelers”—to edit the histone code,48,49 and then they use another set of enzymes (“readers”), which contain specific domains that recognize different modifications, to interpret this code and initiate additional chromatin modification and/or relevant biological processes.49,50 This tightly-woven network plays an essential role in lymphoid development, and genetic abnormalities involving readers, writers, erasers, and remodelers have been implicated in many lymphoid malignancies. For example, PAX5, a transcription factor (TF) that is required for B-cell identity commitment and maintenance,51 was shown to activate its target genes by binding to promoters and putative enhancers that were enriched in active histone marks, such as H3K9ac, H3K4me2, and H3K4me352,53 in mice pro–B cells. However, Pax5−/− pro–B cells lost those epigenetic marks at these promoters and enhancers. Recently, it was demonstrated that PAX5 can recruit histone modifiers, such as the histone acetyltransferase (HAT) CREBBP, the subunit RbBP5 of the MLL-containing methyltransferase complex and the chromatin-remodeling BAF complex to its activated genes, and the HDAC3-containing NCOR corepressor complex to its repressed genes in pro–B cells.53 The recruitment of these enzymes by PAX5 may establish the epigenetic signature required for B-cell commitment. Interestingly, somatic mutations in these enzymes, including CREBBP and ARID1A (encodes BAF250a), have been discovered in different subtypes of lymphomas, suggesting that deregulation of this epigenetic circuit is crucial for lymphomagenesis.54-59
Mutations in histone-modifying enzymes
Owing to the advances in next-generation sequencing technology, including RNA-seq, exome-seq, and whole-genome-seq, recent years brought about the discovery of many novel mutations in cancers. Next we summarize recent findings of novel mutations within histone modifying enzymes in lymphoid malignancies and discuss the potential implications of these mutations for lymphomagenesis (Table 1).13
Table 1.
Chromatin modifying enzymes deregulated in lymphoid neoplasms
| Genes | Activity | Genetic Deregulation | Disease | Implications |
|---|---|---|---|---|
| EZH2 | H3K27 methyltransferase (PRC2 complex) | Mutation | FL | Increasing H3K27me3 level (FL, DLBCL, PMBL); Loss-of-function (T-ALL) |
| GCB-DLBCL | ||||
| PMBL | ||||
| T-ALL | ||||
| MLL2 | H3K4 methyltransferase | Mutation | FL | May result in reduced H3K4me3 level |
| Frame-shift | DLBCL | |||
| KDM2B | H3K36 methyltransferase | Mutation | DLBCL | Not known |
| UTX | H3K27 demethylase | Mutation | MM | Slowing of proliferation |
| MMSET | Histone methyltransferase | Activating translocation | MM | Affecting H3K27me3 and H3K36me2/3 levels |
| JMJD2C | H3K9 demethylase | Amplification | PMBL | Not known |
| HL | ||||
| MLL1 | H3K4 methyltransferase | Translocation | ALL | Not known |
| EED | PRC2 complex | Mutation | T-ALL | Loss-of-function, may result in reduced H3K27me3 |
| SUZ12 | PRC2 complex | Mutation | T-ALL | Loss-of-function, may result in reduced H3K27me3 |
| CREBBP | Histone acetyltransferase | Mutation | FL | Disrupting acetyltransferase and transcription co-activation activities |
| Deletion | DLBCL | |||
| ALL | ||||
| EP300 | Histone acetyltransferase | Mutation | FL | Disrupting acetyltransferase and transcription coactivation activities |
| Deletion | DLBCL | |||
| ARID1A | Chromatin remodeling SWI/SNF complex | Mutation | Pediatric-BL | May give malignant B-cell growth advantages |
| CLL | ||||
| CHD2 | Chromatin remodeling CHD family member | Mutation | CLL | Deregulation of histone variant H3.3 deposition |
Mutations affecting histone methylation marks.
Much focus has been given to H3K4 methylation and H3K27 methylation as they pose opposite effects (H3K4 and H3K27 methylation are associated with transcriptional activation and repression, respectively), yet they coexist at the promoters of genes controlling lineage differentiation in embryonic stem cells (bivalent genes).60,61 H3K36 and H3K79 methylation at gene bodies also reflects active transcription.61 In lymphomas, genetic abnormalities observed in enzymes that “write” or “erase” methylation of these lysines could potentially destroy the balance between the active and repressive histone marks.
For example, several studies discovered mutations that could affect the H3K4 and H3K27 methylation balance in FL and DLBCL.16,17,54,55,62 Morin et al first identified a heterozygous point mutation within EZH2, a PRC2 complex component that methylates H3K27.54 This mutation occurs within the SET domain, which has methyltransferase activity.17,55,62 Interestingly, in the very same cohorts, these investigators also discovered missense, nonsense, and frame-shift mutations within MLL2 gene that encodes a histone H3K4 methyltransferase.16,55,62 The frequencies of MLL2 mutations were 23% to 32%55,63 in the 3 cohorts of DLBCLs16 and was 89% in FLs.16 EZH2 plays an important role in B-cell development. It is required for VDJ recombination in pro–B cells.63 In GC B cells, which may give rise to GCB-DLBCLs and FLs, EZH2 epigenetically silences expression of genes involved in cell growth and proliferation, such as CDKN1A, CDKN1B, and CDKN2A.15 Although wide-type EZH2 preferentially methylates unmethylated H3K27 to produce H3K27me1, biochemical experiments revealed that PRC2 complexes containing mutant forms of EZH2 preferentially methylates H3K27me1 and H3K27me2, resulting in elevated H3K27me2 and H3K27me3 levels.64-66 Given this shift of enzymatic activity, one can imagine that there might be an increased level of the repressive H3K27me3 mark in lymphoma cells that harbor this gain-of-function mutation. On the other hand, the MLL2 mutations discovered in FL and DLBCL are all predicted to disrupt the methyltransferase SET domain of the MLL2 protein and cause reduced levels of H3K4me3. Therefore, it appears that both EZH2 and MLL2 mutations would establish an aberrant histone methylation code to further repress gene expression in FL and DLBCL. However, the existence of this aberrant histone methylation code and its consequence to FL and DLBCL development are still not clear. Modeling these mutations in B cells is crucial to address these questions. In addition to EZH2 and MLL2, Pasqualucci et al also discovered KDM2B (H3K36 demethylase) mutations in 7.4% of the DLBCLs, suggesting there might be an altered H3K36me3 pattern in these lymphomas.62
The key epigenetic changes in multiple myeloma (MM) involve H3K36me3 and H3K27me3. Van Haaften et al identified inactivating mutations in UTX, a histone H3K27 demethylase, in 10% of MM cases.67 Interestingly, UTX mutation is mutually exclusive with MMSET-activating translocation t(4;14),67 another hallmark of MM. MMSET is a SET domain–containing histone methyltransferase. About 15% to 20% of MM patients carry t(4;14) translocation that places MMSET and FGFR3 genes under the control of IGH enhancers.68 However, only MMSET overexpression is thought to be important for MM pathogenesis.69,70 Martinez-Garcia et al showed that overexpression of MMSET correlated with an increase in H3K36me3 and a decrease in H3K27me3.71 On the contrary, Kuo et al only observed reduction of H3K36me2 after MMSET knock down in a MM cell line (KMS11), suggesting MMSET is primarily an H3K36me2 methyltransferase.72 Furthermore, they demonstrated that overexpression of MMSET could disrupt the physiological genomic distribution of H3K36me2.72 Although the enzymatic function of MMSET needs to be further clarified, it is obvious that deregulation of MMSET affects H3K36 methylation. Therefore, the disrupted balance between H3K27me3 and H3K36me2/3 may play an important role in the molecular pathogenesis of MM. To further complicate the situation, UTX is also shown to be associated with MLL2/3 complexes,73 raising the possibility that UTX mutations may also tip the balance between H3K27me3 and H3K4me3. In addition, mutations in MLL family members have also been identified in several MM patients.74 However, no correlation between the change in global H3K27me3 and H3K4me3 levels was observed in Utx null cells compared with wide-type cells.67
In addition to FL, DLBCL, and MM, genetic alterations of factors affecting histone methylation have been found in other lymphoid malignancies. For example, H3K9 demethylase JMJD2C was amplified along with JAK2 in primary mediastinal B-cell lymphoma (PMBL) and HL.75 It is proposed that JAK2 and JMJD2 cooperatively remodel the PMBL and HL epigenome to deregulate oncogenes such as MYC.75 MLL1 translocations are seen in 8% to 10% of ALL, and more rarely in T-ALL, all of which result in a loss of its histone methyltransferase SET domain.76 More interestingly, 2 groups have recently discovered loss-of-function mutations of PRC2 complex genes EZH2, EED, and SUZ12 in T-ALL. Zhang et al reported that 42% of the ETP ALL (early T-cell precursor ALL) cases and 12% of non-ETP T-ALL cases harbored a deletion and/or mutation in these genes.77 Similarily, Ntziachristos et al reported 25% of T-ALL had loss-of-function mutations and deletions of the EZH2 and SUZ12 genes.78 By using a NOTCH1-dependent mouse model of T-ALL, they further suggested a dynamic interplay between oncogenic NOTCH1 and tumor-suppressing PRC2 function for gene expression regulation and malignant transformation.78 It is intriguing that genetic profiling revealed 2 opposite roles of EZH2 in lymphoid malignancies. It is hoped that further mechanistic studies of the PRC2 complex in normal and malignant B cells will shed some light on this paradox.
Mutations affecting histone acetylation marks.
Histone acetylation is often associated with a more “open” chromatin conformation that facilitates transcription, translation, and DNA repair. It is distributed at actively transcribed promoters and their nearby enhancers.79,80 In particular, acetylation of H3K27 (opposing mark to H3K27me3 on the same lysine that is trimethylated by PRC2 complex) is dynamically regulated at enhancers of genes involved in development and differentiation and can predict developmental state.81 Histone acetylation is written by HATs and erased by HDACs. In addition, it can be read by proteins containing bromodomains and/or PHD fingers.
Recurrent mutations in HATs, such as CREBBP and/or EP300, have been identified in 16% to 39% of DLBCLs and 41% to 62% of FLs.54-56,82 In addition, focal deletion and mutations involving CREBBP were found in 18% of relapsed ALL.57 Mutations in CREBBP and EP300 are often heterozygous and may disrupt their acetyltransferase activity, affecting the cellular acetylome and promoting the oncogenic phenotype. Indeed, mutations affecting the HAT activity of CREBBP and EP300 promote the activation of BCL6 and HSP90 while destabilizing P53.56,82 Therefore, loss of BCL6 acetylation in CREBBPHAT or EP300HAT mutant cells may exacerbate the oncogenic activity of BCL6 and HSP90 in these lymphomas. Although the balance between HAT and HDACs can be pharmacologically manipulated by using HDAC inhibitors (HDACi), DLBCL with CREBBPHAT and/or EP300HAT mutations tend to be resistant to this strategy.82 The combination of HDACi with molecules that inactivate EP300 substrate proteins such as HSP90 and BCL6 may offer a way to circumvent this problem.82 Interestingly, BCL6 itself can repress EP300 in DLBCLs, and treatment with RI-BPI, a BCL6 peptide inhibitor, can induce EP300 expression, which is required for killing of DLBCL cells by RI-BPI,82 providing an alternative way to restore the HAT/HDAC balance in lymphomas with wild-type enzymes. Currently, it is important to establish a mechanistic link between CREBBP/EP300 mutations, aberrant histone acetylation, impaired target regulation, and lymphomagenesis to design a targeted therapy specific for lymphomas with CREBBP/EP300 mutations.
Mutations affecting chromatin remodeling.
In addition to histone modifications, remodeling of chromatin can also epigenetically regulate transcription, replication, and DNA repair. Chromatin remodelers use the energy from ATP hydrolysis to change the location of nucleosomes, or to deposit histone variants, resulting in change of chromatin conformation and composition.83 Based on the difference of the core adenosine triphosphatase subunits, chromatin remodelers can be divided into the SWI/SNF (SWItch/Sucrose NonFermentable), ISWI (Imitation of SWItch), CHD (Chromodomain Helicase DNA-binding), and INO (INOsitol) families.
Mutations in ARID1A, which encodes BAF250a, a subunit of the SWI/SNF chromatin remodeling complex, has been found in 17% of pediatric BL58 and 2 of 105 CLL patients.59 ARID1A was shown to have tumor suppressor functions in gastric adenocarcinoma by slowing down cell proliferation through repressing expression of EF1 and CCND1.84 Other ARID1A targets include cell cycle regulator CDKN1A85 and ES cell self-renewal factors SOX2 and OCT4.86 Although the role of ARID1A mutations in lymphomagenesis has not been explored, it is possible that mutant ARID1A may give malignant B cells growth advantages.
CHD2 mutations were also identified in 4.8% of CLL cases.59 CHD2 belongs to the CHD family of chromatin remodelers. Chd2-deficient mice showed defects in erythrocyte differentiation and developed lymphomas, probably because of the deregulation of activated T cells.87 Recently, Harada et al demonstrated that CHD2 can deposit histone variant H3.3 at genes important for differentiation.88 Histone H3.3 has been shown to localize at promoters of actively transcribed genes as well as some silent genes presumably poised to be expressed. In addition, H3.3 has been proposed to take part in the epigenetic transmission of active chromatin states.89 Therefore, it will be interesting to see whether CHD2 mutants could induce an aberrant gene transcription programming by deregulating H3.3 deposition in malignant B cells.
Although the discovery of mutations within histone-modifying enzymes led us one step closer to the understanding of molecular pathogenesis of lymphoid malignancies, the lack of the knowledge about how these mutations lead to disease development still hinders us from developing effective targeted therapies. To date, it is still unclear whether these mutations are “driver” mutations that initiate malignant transformation, “facilitator” mutations that promote disease progression, or just merely “passenger” mutations that have no effect on disease pathogenesis. Recently, Green et al established a hierarchy model of FL somatic evolution by comparing and contrasting the exome mutational profiles of FL in subpopulations at diagnosis and relapse, based on CD20 expression levels.90 Their model suggested that CREBBP mutations were early “drivers” of FL disease evolution, and because these mutations were found in the CD20 subpopulations of FLs at both diagnosis and relapse, MLL2 mutations were likely later “accelerators” because MLL2 mutations were only found in one of the subpopulations. Although the observation was interesting, the case number was limited. Moreover, the mechanistic explanations of the CREBBP driving effects and MLL2 accelerating effects on lymphomagenesis are still missing. We will not be able to offer rationalized and targeted therapies until we fill in these blanks.
MicroRNAs and ncRNAs in lymphoid malignancies
Shortly after the discovery of human microRNAs (miRNAs) in 2000,91 the first studies emerged linking miRNA genes and disease. The finding that miR-15 and miR-16-1 were frequently deleted or repressed in CLL92 was the first of several studies to uncover the critical role of deregulation of miRNA and ncRNA expression in the pathogenesis of several tumors, including leukemia and lymphoma. It gradually became clear that aberrant gene expression of both precursor and mature miRNAs occurred as a result of genomic alterations, deregulated processing machinery, or epigenetic mechanisms, with deregulated miRNAs functioning as oncogenes (oncomiRs) or tumor suppressors (Table 2). The miR-17-92 cluster and miR-155 were the first oncomiRs discovered, and their role in the pathogenesis of hematologic malignancies was validated using gain- and loss-of-function mouse models.93,94 He et al showed that overexpression of miR-17-92 accelerated C-MYC–induced tumorigenesis in a B-cell lymphoma model.93 Overexpression of the miR17-92 cluster is observed in several lymphoid malignancies95 and its oncogenic role is a result of the miRNAs within the cluster targeting critical genes (including PTEN, BIM, CDKN1A, and E2F) inducing lack of differentiation and increased proliferation and angiogenesis.95-98 Furthermore, deregulation of miR-17-92 either because of amplification or aberrant targeting by MLL proteins has been associated with MLL-rearranged ALL.29 Another notable oncomiR, miR-155, is amplified in several tumors,99,100 and specific overexpression in B cells leads to an expansion of the pre–B-cell compartment followed by the development of leukemia or high-grade lymphoma in transgenic mice.95-101 Other oncomiRs include miR-125a and miR-125b, which aberrantly target TNFAIP3, leading to constitutive NF-κB activation.102
Table 2.
Deregulated miRNAs in lymphoid neoplasms
| miRNA | Disease | Target Genes | Genetic/Epigenetic deregulation |
|---|---|---|---|
| miR-17-92 cluster (includes miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a) | DLBCL, MCL, Burkitt, FL, ALL | PTEN, BIM, CDKN1A, E2F TFs | Amplification upregulated by MLL |
| miR-155 | CLL, HD, pediatric Burkitt, DLBCL | Amplification | |
| miR-125a/miR-125b | DLBCL | TNFAIP3 | Amplification Upregulation |
| p15AS | AML, ALL | p15 | Upregulation |
| Tumor supressor miRNAs | |||
| miR-15a/miR-16-1 | CLL, NHL | BCL2, MCL-1 | Deleted Repressed by HDACs |
| miR-26a | Burkitt | EZH2 | Repressed by MYC |
| miR-29/miR-29b | CLL, MCL,DLBCL, Burkitt | CDK6,IGF-1R | Repressed by MYC |
| miR-31 | ATL | NIK | Repressed by PRC2 |
| miR-203 | MALT, ALL, CLL, MM | ABL1 | Promoter hypermethylation |
| miR-124 | ALL, NHL | CDK6 | Promoter hypermethylation |
ATL, adult T-cell leukemia.
On the contrary, several key miRNAs that target antiapoptotic genes are downregulated or deleted in lymphoid tumors. For example, miR-15a/16-1 acts as a tumor suppressor in CLL by targeting BCL2.103 More recently, epigenetic silencing of several miRNAs resulting from aberrant DNA methylation and altered histone modification patterns has been associated with the pathogenesis of lymphomas and leukemias. HDAC complexes mediate repression of miR-15a, miR-16, and miR-29b in CLL and in NHL.104,105 Treatment with HDACi induced the expression of these miRNAs, antagonizing survival protein MCL-1 and triggering cell death.104 Deregulated PRC2 complexes repress miR-31 in adult T-cell leukemia and lead to activation of NF-κB, triggering oncogenic signaling.106 DNA hypermethylation led to miR-124 and miR-203 silencing in several lymphoid malignancies, providing a basis for therapeutic targeting with demethylating agents.107-112 Moreover, genome-wide miRNA profiling studies in ALL further revealed an EZH2 targeted and aberrantly methylated miRNA signature, suggesting that these cases might benefit from EZH2 inhibition.107-111,113 Along the same lines, miR-26a and miR-29 are repressed by MYC during lymphomagenesis.114,115 EZH2 targeting by miR-26a might lead to global deregulation of gene expression, whereas MYC recruits HDAC3 and EZH2 complexes to aberrantly repress miR-29. Combined pharmacologic inhibition of HDAC and PRC2 complexes restored miR-29 levels, suppressing lymphoma growth. Alternatively, the ncRNA p15AS was shown to induce heterochromatin formation to epigenetically silence its target P15, a cyclin-dependent kinase inhibitor, in acute leukemia.116 Finally, leukemogenesis has also been associated with deregulation of some highly-conserved ncRNAs, namely transcribed ultraconserved regions, with unclear functions in CLL.117 As genome-wide ncRNA and epigenetic profiles of both normal and malignant cells become available, the integration of these datasets will further uncover the underlying mechanisms of epigenetic deregulation of miRNAs and facilitate the design of targeted therapeutic approaches in leukemia and lymphoma.
Future challenges
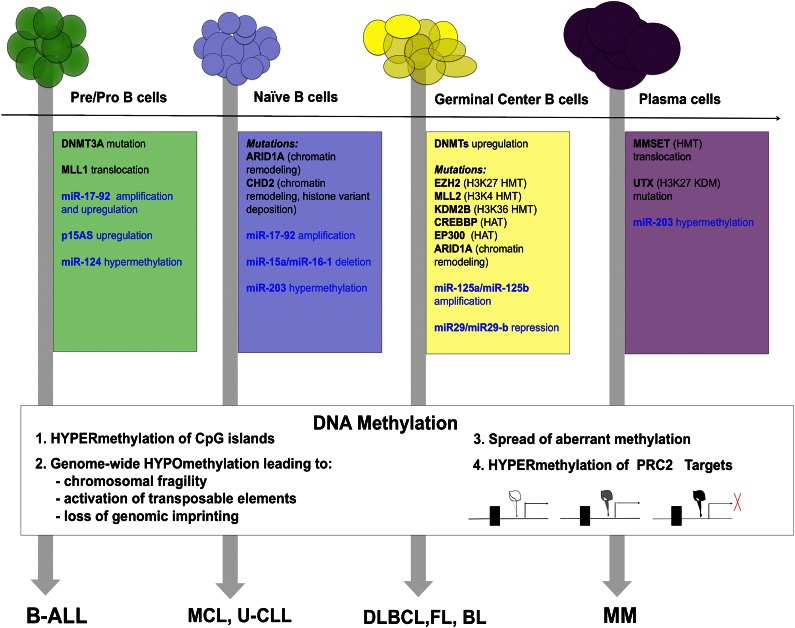
Complete understanding of the role epigenome plays in lymphomagenesis will require systematic study of a large number of cases using high-resolution techniques. Because the genome of each subtype of lymphoma proved to be unique, the epigenome similarly is emerging as a dynamic set of marks that is deregulated in a unique fashion in each lymphoid malignancy (Figure 1). In addition, a causal link is still missing between the deregulated epigenome and the development of the diseases. Moreover, given the increasing scale of the genome-wide genetic and epigenetic studies, a new generation of computer-savvy biologists is needed to provide solutions to the challenging data analysis. Until then, we only have small glimpses into the nature of epigenomic regulation.
Figure 1.
Summary of epigenetic abnormalities that contribute to neoplastic transformation of normal B-cell precursors (on the top) to specific lymphoma subtypes (on the bottom). Abnormalities in histone- and DNA-modifying enzymes are within colored boxes in black, abnormalities in miRNAs are within colored boxes in blue. DNA methylation changes are summarized in the white box.
The study of DNA methylation in lymphoid malignancies led to the accumulation of a large body of correlative studies linking changes in patterns and expression levels of methylation machinery to the disease. However, thus far, the amount of direct evidence linking changes in DNA methylation or its machinery to lymphomagenesis is insufficient. Further functional evidence including animal models is necessary to establish the cause and effect among deregulated DNA methylation machinery, aberrant DNA methylation pattern, and malignant transformation.
The challenge in solving the “malignant histone code” lies in the diverse nature of histone marks and their complex combinatorial effect that is difficult to interrogate. Moreover, careful functional characterization of newly discovered mutations within histone-modifying enzymes to identify true “driver” and/or “facilitator” mutations is in urgent need. The interplay between the aberrant DNA methylation pattern and the “malignant histone code” is yet another complicated question waiting to be addressed.
Finally, the ultimate goal is to integrate the genomic and epigenomic data to identify new therapeutic targets. High-throughput screening of therapeutic molecules promises to deliver many new candidate therapies. Their preclinical and clinical testing will require an increasing agility from regulatory agencies.
Acknowledgments
The authors thank Dr L. Cerchietti for editing the manuscript and Dr N. Weinhold for help with the figure design and graphics.
This work was supported by grants from the National Institutes of Health (K08 CA127353) and The Leukemia & Lymphoma Society (LLS 6304-11) (R.S.) and by an ASH Scholar Award (Y.J.).
Authorship
Contribution: Y.J., K.H., and R.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rita Shaknovich, Department of Medicine, Weill Cornell Medical College, Building C Room 620C, 1300 York Ave, New York, NY 10065; e-mail: ris9004@med.cornell.edu.
References
- 1.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Deaton AM, Webb S, Kerr AR, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21(7):1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaknovich R, Cerchietti L, Tsikitas L, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118(13):3559–3569. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuno S, Chijiwa T, Okamura T, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97(5):1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 7.Amara K, Ziadi S, Hachana M, Soltani N, Korbi S, Trimeche M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101(7):1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J, Kim TY, Jung Y, et al. DNA methyltransferase 3B mutant in ICF syndrome interacts non-covalently with SUMO-1. J Mol Med (Berl) 2008;86(11):1269–1277. doi: 10.1007/s00109-008-0392-5. [DOI] [PubMed] [Google Scholar]
- 9.Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS. 2013;121(2):85–94. doi: 10.1111/j.1600-0463.2012.02940.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208(13):2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zainuddin N, Kanduri M, Berglund M, et al. Quantitative evaluation of p16(INK4a) promoter methylation using pyrosequencing in de novo diffuse large B-cell lymphoma. Leuk Res. 2011;35(4):438–443. doi: 10.1016/j.leukres.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Guan H, Xie L, Leithäuser F, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116(9):1469–1478. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 13.Lai AY, Fatemi M, Dhasarathy A, et al. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med. 2010;207(9):1939–1950. doi: 10.1084/jem.20100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martín-Subero JI, Kreuz M, Bibikova M, et al. Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. New insights into the biology and origin of mature aggressive B-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113(11):2488–2497. doi: 10.1182/blood-2008-04-152900. [DOI] [PubMed] [Google Scholar]
- 15.Velichutina I, Shaknovich R, Geng H, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bödör C, O’Riain C, Wrench D, et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia. 2011;25(4):726–729. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 18.Viré E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 19.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 20.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 21.De S, Shaknovich R, Riester M, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PloS Genet. 2013;9(1):e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010;24(19):2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng W. AID in reprogramming: quick and efficient: identification of a key enzyme called AID, and its activity in DNA demethylation, may help to overcome a pivotal epigenetic barrier in reprogramming somatic cells toward pluripotency. Bioessays. 2010;32(5):385–387. doi: 10.1002/bies.201000014. [DOI] [PubMed] [Google Scholar]
- 25.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188(8):3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 28.Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139(6):1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 29.Choi JH, Li Y, Guo J, et al. Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PloS One. 2010;5(9) doi: 10.1371/journal.pone.0013020. Pii:e13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett LB, Schnabel JL, Kelchen JM, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48(9):828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Riain C, O’Shea DM, Yang Y, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;23(10):1858–1866. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enjuanes A, Fernàndez V, Hernández L, et al. Identification of methylated genes associated with aggressive clinicopathological features in mantle cell lymphoma. PloS ONE. 2011;6(5):e19736. doi: 10.1371/journal.pone.0019736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halldórsdóttir AM, Kanduri M, Marincevic M, et al. Mantle cell lymphoma displays a homogenous methylation profile: a comparative analysis with chronic lymphocytic leukemia. Am J Hematol. 2012;87(4):361–367. doi: 10.1002/ajh.23115. [DOI] [PubMed] [Google Scholar]
- 34.Leshchenko VV, Kuo PY, Shaknovich R, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlfors J, Hiltunen H, Heinonen K, Hämäläinen E, Alhonen L, Jänne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992;80(8):2074–2080. [PubMed] [Google Scholar]
- 36.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 37.Claus R, Lucas DM, Stilgenbauer S, et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J Clin Oncol. 2012;30(20):2483–2491. doi: 10.1200/JCO.2011.39.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen SS, Claus R, Lucas DM, et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117(3):862–871. doi: 10.1182/blood-2010-05-284638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raval A, Tanner SM, Byrd JC, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129(5):879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosialls AM, Santidrián AF, Coll-Mulet L, et al. Epigenetic profile in chronic lymphocytic leukemia using methylation-specific multiplex ligation-dependent probe amplification. Epigenomics. 2012;4(5):491–501. doi: 10.2217/epi.12.40. [DOI] [PubMed] [Google Scholar]
- 41.Gandhirajan RK, Staib PA, Minke K, et al. Small molecule inhibitors of Wnt/beta-catenin/lef-1 signaling induces apoptosis in chronic lymphocytic leukemia cells in vitro and in vivo. Neoplasia. 2010;12(4):326–335. doi: 10.1593/neo.91972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66(2):653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 43.Wong NC, Ashley D, Chatterton Z, et al. A distinct DNA methylation signature defines pediatric pre-B cell acute lymphoblastic leukemia. Epigenetics. 2012;7(6):535–541. doi: 10.4161/epi.20193. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Manero G, Jeha S, Daniel J, et al. Aberrant DNA methylation in pediatric patients with acute lymphocytic leukemia. Cancer. 2003;97(3):695–702. doi: 10.1002/cncr.11090. [DOI] [PubMed] [Google Scholar]
- 45.Davidsson J, Lilljebjörn H, Andersson A, et al. The DNA methylome of pediatric acute lymphoblastic leukemia. Hum Mol Genet. 2009;18(21):4054–4065. doi: 10.1093/hmg/ddp354. [DOI] [PubMed] [Google Scholar]
- 46.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 47.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 48.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 52.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27(1):49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 53.McManus S, Ebert A, Salvagiotto G, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30(12):2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giulino-Roth L, Wang K, MacDonald TY, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120(26):5181–5184. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 61.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su IH, Basavaraj A, Krutchinsky AN, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 64.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majer CR, Jin L, Scott MP, et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012;586(19):3448–3451. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 67.Van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–3034. [PubMed] [Google Scholar]
- 69.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101(6):2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 70.Keats JJ, Reiman T, Maxwell CA, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101(4):1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 71.Martinez-Garcia E, Popovic R, Min DJ, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117(1):211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44(4):609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 74.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Cancer Biol. 2005;15(3):175–188. doi: 10.1016/j.semcancer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 77.Zhang M, Xu C, Yan H, Zhao N, von Wettstein D, Liu B. Limited tissue culture-induced mutations and linked epigenetic modifications in F hybrids of sorghum pure lines are accompanied by increased transcription of DNA methyltransferases and 5-methylcytosine glycosylases. Plant J. 2009;57(4):666–679. doi: 10.1111/j.1365-313X.2008.03719.x. [DOI] [PubMed] [Google Scholar]
- 78.Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18(2):298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 80.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cerchietti LC, Hatzi K, Caldas-Lopes E, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010. Pii:42869. [DOI] [PMC free article] [PubMed]
- 83.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 84.Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 85.Guan B, Wang TL, Shih IM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105(18):6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 2009;28(8):1053–1062. doi: 10.1038/onc.2008.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harada A, Okada S, Konno D, et al. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012;31(13):2994–3007. doi: 10.1038/emboj.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21(3):421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 92.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mi S, Li Z, Chen P, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA. 2010;107(8):3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24(1):1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kluiver J, Haralambieva E, de Jong D, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45(2):147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 100.Kluiver J, Poppema S, de Jong D, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 101.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci USA. 2012;109(20):7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sampath D, Liu C, Vasan K, et al. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood. 2012;119(5):1162–1172. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X, Chen X, Lin J, et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene. 2012;31(24):3002–3008. doi: 10.1038/onc.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamagishi M, Nakano K, Miyake A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-κB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21(1):121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 107.Agirre X, Martínez-Climent JA, Odero MD, Prósper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. 2012;26(3):395–403. doi: 10.1038/leu.2011.344. [DOI] [PubMed] [Google Scholar]
- 108.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 109.Chim CS, Wong KY, Leung CY, et al. Epigenetic inactivation of the has-miR-203 in haematological malignancies. J Cell Mol Med. 2011;15(12):2760–2767. doi: 10.1111/j.1582-4934.2011.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Craig VJ, Cogliatti SB, Rehrauer H, Wündisch T, Müller A. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 2011;71(10):3616–3624. doi: 10.1158/0008-5472.CAN-10-3907. [DOI] [PubMed] [Google Scholar]
- 111.Roman-Gomez J, Agirre X, Jiménez-Velasco A, et al. Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J Clin Oncol. 2009;27(8):1316–1322. doi: 10.1200/JCO.2008.19.3441. [DOI] [PubMed] [Google Scholar]
- 112.Wong KY, So CC, Loong F, et al. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE. 2011;6(4):e19027. doi: 10.1371/journal.pone.0019027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cao Q, Mani RS, Ateeq B, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20(2):187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang X, Zhao X, Fiskus W, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22(4):506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Sander S, Bullinger L, Klapproth K, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 116.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]