Abstract
The mammalian target of rapamycin (mTOR) kinase is a master regulator of protein synthesis that couples nutrient sensing to cell growth and cancer. However, the downstream translationally regulated nodes of gene expression that may direct cancer development are poorly characterized. Using ribosome profiling, we uncover specialized translation of the prostate cancer genome by oncogenic mTOR signalling, revealing a remarkably specific repertoire of genes involved in cell proliferation, metabolism and invasion. We extend these findings by functionally characterizing a class of translationally controlled pro-invasion messenger RNAs that we show direct prostate cancer invasion and metastasis downstream of oncogenic mTOR signalling. Furthermore, we develop a clinically relevant ATP site inhibitor of mTOR, INK128, which reprograms this gene expression signature with therapeutic benefit for prostate cancer metastasis, for which there is presently no cure. Together, these findings extend our understanding of how the ‘cancerous’ translation machinery steers specific cancer cell behaviours, including metastasis, and may be therapeutically targeted.
It is unknown whether specialized networks of translationally controlled mRNAs can direct cancer initiation and progression, thereby mirroring cooperativity that has mainly been observed at the level of transcriptional control. This is an important question, as key oncogenic signalling molecules, such as the mTOR kinase, directly regulate the activity of general translation factors1,2. Downstream of the phosphatidylinositol-3-OH kinase (PI(3)K)–AKT signalling pathway, mTOR assembles with either raptor or rictor to form two distinct complexes: mTORC1 and mTORC2 (refs 3, 4). The major regulators of protein synthesis downstream of mTORC1 are 4EBP1 (also called EIF4EBP1) and p70S6K1/2 (refs 1, 2). 4EBP1 negatively regulates eIF4E, a key rate-limiting initiation factor for cap-dependent translation. Phosphorylation of 4EBP1 by mTORC1 leads to its dissociation from eIF4E, allowing translation initiation complex formation at the 5′ end of mRNAs5. The mTOR-dependent phosphorylation of p70S6K1/2 also promotes translation initiation as well as elongation6. At a genome-wide level, it remains poorly understood whether and how activation of these regulators of protein synthesis may produce specific changes in gene expression networks that direct cancer development. Here we use a powerful new technology known as ribosome profiling to delineate the translational landscape of the cancer genome at a codon-by-codon resolution upon pharmacological inhibition of mTOR7. Our findings provide genome-wide characterization of translationally controlled mRNAs downstream of oncogenic mTOR signalling and delineate their functional roles in cancer development. Moreover, we determine the efficacy of a novel clinically relevant mTOR inhibitor that we developed, INK128, which specifically targets this cancer program.
Ribosome profiling of the prostate cancer genome
mTOR is deregulated in nearly 100% of advanced human prostate cancers8, and genetic findings in mouse models implicate mTOR hyperactivation in prostate cancer initiation9–11. Given the critical role for mTOR in prostate cancer, we used PC3 human prostate cancer cells, where mTOR is constitutively hyperactivated, to delineate translationally controlled gene expression networks upon complete or partial mTOR inhibition. We optimized ribosome profiling to assess quantitatively ribosome occupancy genome-wide in cancer cells7. In brief, ribosome-protected mRNA fragments were deep-sequenced to determine the number of ribosomes engaged in translating specific mRNAs (Supplementary Fig. 1a and Methods). Treatment of PC3 cells with PP242 (refs 12, 13), an mTOR ATP site inhibitor, significantly inhibits the activity of the three primary downstream mTOR effectors 4EBP1, p70S6K1/2 and AKT. On the contrary, rapamycin, an allosteric mTOR inhibitor, only blocks p70S6K1/2 activity in these cells (Supplementary Fig. 1b). We used short 3-h drug treatments, which precede alterations in de novo protein synthesis, to capture direct changes in mTOR-dependent gene expression by ribosome profiling and to minimize compensatory feedback mechanisms (Supplementary Fig. 1c–f).
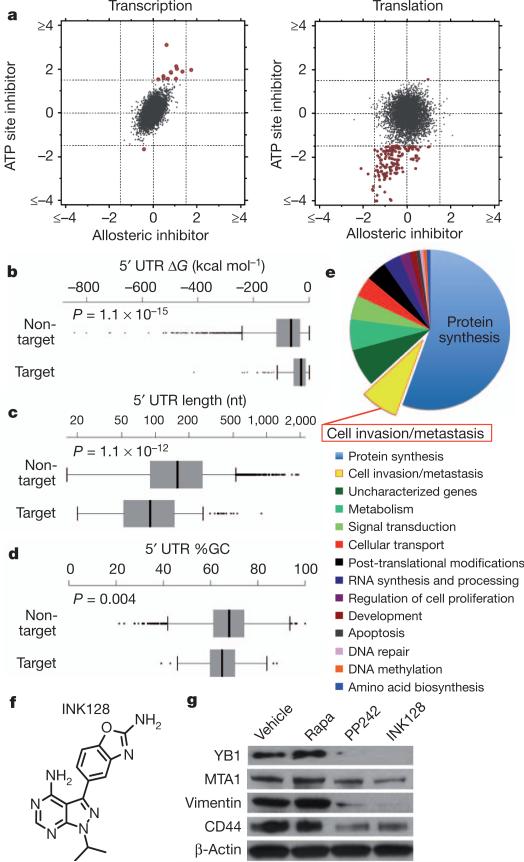
Ribosome profiling revealed 144 target mRNAs selectively decreased at the translational level upon PP242 treatment (log2 ≤–1.5 (false discovery rate <0.05)) compared to rapamycin treatment, with limited changes in transcription (Fig. 1a and Supplementary Figs 2a, b and 3–10). The fact that at this time point rapamycin treatment did not markedly affect gene expression is consistent with incomplete allosteric inhibition of mTOR activity (Supplementary Fig. 1b). By monitoring footprints of translating 80S ribosomes, our findings show that the effects of PP242 are largely at the level of translation initiation and not elongation (Supplementary Fig. 3). It has been proposed that mRNAs translationally regulated by mTOR may contain long 5′ untranslated regions (5′ UTRs) with complex RNA secondary structures. On the contrary, ribosome profiling revealed that mTOR-responsive 5′ UTRs possess less complex features (Fig. 1b–d), providing a unique data set to investigate the nature of regulatory elements that render these mRNAs mTOR-sensitive. It has been previously shown that some mTOR translationally regulated mRNAs, most notably those involved in protein synthesis, possess a 5′ terminal oligopyrimidine tract (5′ TOP)14,15 that is regulated by distinct transacting factors16,17. Of the 144 mTOR-sensitive target genes, 68% possess a 5′ TOP. However, as the 5′ TOP is not present in all mTOR-sensitive mRNAs, we next asked whether other 5′ UTR consensus sequences may exist. Strikingly, 63% of mTOR target mRNAs possess what we have termed a pyrimidine-rich translational element (PRTE) within their 5′ UTRs (P = 3.2 × 10–11). This element, unlike the 5′ TOP sequence, consists of an invariant uridine at position 6 flanked by pyrimidines and, importantly, does not reside at position +1 of the 5′ UTR (Supplementary Figs 2c and 7). We found that 89% of the mTOR-responsive genes possess a PRTE and/or 5′ TOP, making the presence of one or both sequences a strong predictor for mTOR sensitivity (Supplementary Figs 2d and 7). Notably, mRNA isoforms arising from distinct transcription start sites may possess both a 5′ TOP and a PRTE. Moreover, given the significant number of mRNAs that contain both the PRTE and 5′ TOP, a functional interplay may exist between these regulatory elements. Future studies are required to determine the regulatory logic for how these sequences either independently or coordinately confer mTOR responsiveness. Multiple cis-acting elements within specific 5′ UTRs could reflect regulation by distinct mTOR effectors. For example, our findings show that the PRTE imparts translational control specificity to 4EBP1 activity (see below).
Figure 1. Ribosome profiling reveals mTOR-dependent specialized translational control of the prostate cancer genome.
a, Representative comparison of mRNA abundance and translational efficiency after a 3-h treatment with an ATP site inhibitor (PP242) versus an allosteric inhibitor (rapamycin). b–d, Free energy, length and percentage G+C content of the 5′ UTRs of mTOR target versus non-target mRNAs (error bars indicate range, non-target n = 5,022, target n = 144, two-sided Wilcoxon). e, Functional classification of translationally regulated mTOR-responsive mRNAs. f, Chemical structure of INK128. g, Representative western blot from three independent experiments of mTOR-sensitive invasion genes in PC3 cells after a 48-h drug treatment. Rapa, rapamycin.
Surprisingly, mTOR-sensitive genes stratify into unique functional categories that may promote cancer development and progression, including cellular invasion (P = 0.009), cell proliferation (P = 0.04), metabolism (P = 0.0002) and regulators of protein modification (P = 0.01) (Fig. 1e). The largest fraction of mTOR-responsive mRNAs cluster into a node consisting of key components of the translational apparatus: 70 ribosomal proteins, 6 elongation factors, and 4 translation initiation factors (P = 7.5 × 10–82) (Fig. 1e and Supplementary Fig. 5). Therefore, this class of mTOR-responsive mRNAs may represent an important regulon that sustains the elevated protein synthetic capacity of cancer cells.
Notably, the second largest node of mTOR translationally regulated genes comprises bona fide cell invasion and metastasis mRNAs and putative regulators of this process (Fig. 1e). This group includes YB1 (Y-box binding protein 1; also called YBX1), vimentin, MTA1 (metastasis associated 1) and CD44 (Supplementary Fig. 11a). YB1 regulates the post-transcriptional expression of a network of invasion genes18. Vimentin, an intermediate filament protein, is highly upregulated during the epithelial-to-mesenchymal transition associated with cellular invasion19. MTA1, a putative chromatin-remodelling protein, is overexpressed in invasive human prostate cancer20 and has been shown to drive cancer metastasis by promoting neoangiogenesis21. CD44 is commonly overexpressed in tumour-initiating cells and is implicated in prostate cancer metastasis22. Consistent with their status as mTOR sensitive genes, YB1, vimentin, MTA1 and CD44 all possess a PRTE (Supplementary Fig. 5). Vimentin and CD44 also possess a 5′ TOP (Supplementary Fig. 7). To test the functional role of the PRTE in mediating translational control, we mutated the PRTE within the 5′ UTR of YB1, which rendered the YB1 5′ UTR insensitive to inhibition by 4EBP1 (Supplementary Fig. 11b). These findings highlight a novel cis-regulatory element that may modulate translational control of subsets of mRNAs upon mTOR activation. Moreover, ribosome profiling reveals unexpected transcript-specific translational control, mediated by oncogenic mTOR signalling, including a distinct set of pro-invasion and metastasis genes.
Translation of pro-invasion mRNAs by mTOR
We next extended the use of the mTOR pharmacological tools used in ribosome profiling towards functional characterization of the newly identified mTOR-sensitive cell invasion gene signature. To this end, we developed a new clinical-grade mTOR ATP site inhibitor, INK128, derived from the PP242 chemical scaffold (Fig. 1f). In brief, a structure-guided optimization of pyrazolopyrimidine derivatives was performed (see INK128 chemical synthesis in Supplementary Information) that improved oral bioavailability while retaining mTOR kinase potency and selectivity. INK128 was selected for clinical studies on the basis of its high potency (1.4 nM inhibition constant (Ki)), selectivity for mTOR, low molecular mass, and favourable pharmaceutical properties (Supplementary Figs 12 and 13).
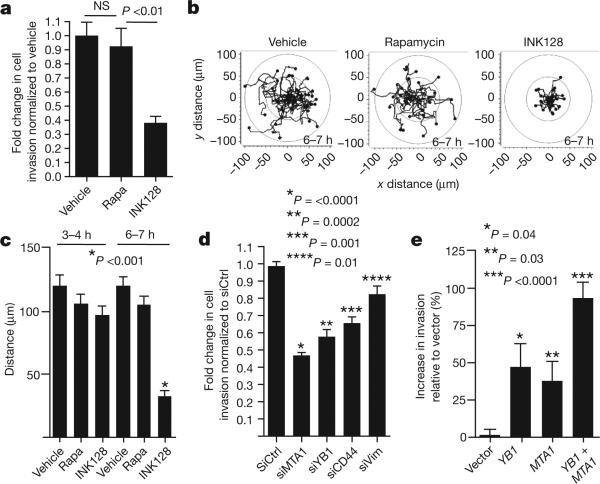
Using either PP242 or INK128, we observed a selective decrease in the expression of YB1, MTA1, vimentin and CD44 at the protein but not transcript level in PC3 cells starting at 6 h of treatment, which precedes any decrease in de novo protein synthesis (Fig. 1g and Supplementary Figs 1c, d, 14 and 15). In contrast, rapamycin treatment did not alter their expression (Fig. 1g and Supplementary Fig. 14a). Similar findings were observed using a broad panel of metastatic cell lines of distinct histological origins (Supplementary Fig. 16). The four-gene invasion signature is positively regulated by mTOR hyperactivation, as silencing PTEN expression increased their protein but not mRNA expression levels (Supplementary Fig. 17). We next investigated the effects of mTOR ATP site inhibitors on prostate cancer cell migration and invasion. We found that INK128, but not rapamycin, decreases the invasive potential of PC3 prostate cancer cells (Fig. 2a). Furthermore, INK128 inhibits cancer cell migration starting at 6 h of treatment, precisely correlating with when decreases in the expression of pro-invasion genes are evident, but preceding any changes in the cell cycle or overall global protein synthesis (Fig. 2b, c, and Supplementary Figs 1c, e, f, 14b and 18).
Figure 2. mTOR promotes prostate cancer cell migration and invasion through a translationally regulated gene signature.
a, Matrigel invasion assay in PC3 cells: 6-h pre-treatment followed by 6 h of cell invasion (n = 6, ANOVA). b, c, Migration patterns and average distance travelled by GFP-labelled PC3 cells during hours 3–4 and 6–7 of drug treatment (n = 34 cells per condition, ANOVA). d, Matrigel invasion assay in PC3 cells after 48 h of knockdown of YB1, MTA1, CD44, or vimentin followed by 24 h of cell invasion (n = 7, t-test). e, Matrigel invasion assay in BPH-1 cells after 48 h of overexpression of YB1 and/or MTA1, followed by cell invasion for 24 h (n = 7, t-test). Rapa, rapamycin. All data represent mean ± s.e.m. NS, not statistically significant.
Among the genes comprising the pro-invasion signature, YB1 has been shown to act directly as a translation factor that controls expression of a larger set of genes involved in breast cancer cell invasion18. Notably, YB1 translationally regulated target mRNAs, including SNAIL1 (also called SNAI), LEF1 and TWIST1, decreased at the protein but not transcript level upon YB1 knockdown in PC3 cells (Supplementary Figs 19 and 20). To determine the functional role of YB1 in prostate cancer cell invasion, we silenced YB1 gene expression in PC3 cells, and observed a 50% reduction in cell invasion (Fig. 2d). Similarly, knockdown of MTA1, CD44, or vimentin also inhibited prostate cancer cell invasion (Fig. 2d and Supplementary Fig. 19). These mTOR target mRNAs may be sufficient to endow primary prostate cells with invasive features, as overexpression of YB1 and/or MTA1 (Supplementary Fig. 21a) in BPH-1 cells, an untransformed prostate epithelial cell line, increased the invasive capacity of these cells in an additive manner (Fig. 2e). Notably, the effects of YB1 and MTA1 on cell invasion are independent from any effect on cell proliferation in both knockdown or overexpression studies (Supplementary Fig. 21b, c). Therefore, translational control of pro-invasion mRNAs by oncogenic mTOR signalling alters the ability of epithelial cells to migrate and invade, a key feature of cancer metastasis.
Dissecting mTOR translational effectors
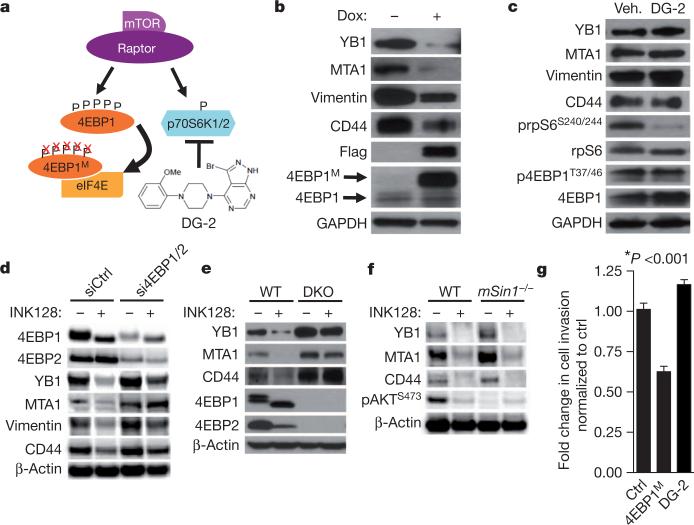
We sought to determine the molecular mechanism by which pro-invasion genes are regulated at the translational level and why these mRNAs are sensitive to INK128 but not rapamycin. To this end, we investigated whether the translational regulators downstream mTORC1, 4EBP1 and/or p70S6K1/2, control the expression of these mTOR-sensitive targets. We generated a human prostate cancer cell line that stably expresses a doxycycline-inducible dominant-negative mutant of 4EBP1 (4EBP1M) (Fig. 3a)13. This mutant binds to eIF4E, decreasing its hyperactivation without inhibiting general mTORC1 function (Supplementary Fig. 22a). Notably, expression of 4EBP1M does not alter global protein synthesis (Supplementary Fig. 22b), probably because endogenous 4EBP1 and 4EBP2 proteins retain their ability to bind to eIF4E (Supplementary Fig. 22c)13. Upon induction of 4EBP1M, YB1, vimentin, CD44 and MTA1 decrease at the protein but not mRNA level, whereas pharmacological inhibition of p70S6K1/2 with DG-2 (ref. 23) had no effect (Fig. 3b,c and Supplementary Fig. 22d). Next, we tested whether INK128 decreases expression of the four invasion genes through the 4EBP–eIF4E axis. Notably, knockdown of 4EBP1 and 4EBP2 in PC3 cells or using 4EBP1 and 4EBP2 double knockout mouse embryonic fibroblasts (MEFs)24 reduced the ability of INK128 to decrease expression of these pro-invasion mRNAs (Fig. 3d, e and Supplementary Fig. 23). Furthermore, ablation of mTORC2 activity25 had no effect on the expression of these mRNAs or responsiveness to INK128 (Fig. 3f and Supplementary Fig. 24a–c). Next, we determined the effect of 4EBP1M on human prostate cancer cell invasion. The expression of 4EBP1M resulted in a significant decrease in prostate cancer cell invasion without affecting the cell cycle, whereas DG-2 had no effect (Fig. 3g and Supplementary Fig. 24d). These findings demonstrate that eIF4E hyperactivation downstream of oncogenic mTOR regulates translational control of the pro-invasion mRNAs and provides an explanation for the selective targeting of this gene signature by mTOR ATP site inhibitors.
Figure 3. The 4EBP1–eIF4E axis controls the post-transcriptional expression of mTOR-sensitive invasion genes.
a, Schematic of the pharmacogenetic strategy to inhibit p70S6K1/2 or eIF4E hyperactivation. b, Representative western blot from three independent experiments of PC3 4EBP1M cells after 48-h doxycycline induction of 4EBP1M. c, Representative western blot from three independent experiments of PC3 cells after 48-h DG-2 treatment. d, Representative western blot from three independent experiments of PC3 cells after 48 h of 4EBP1/4EBP2 knockdown followed by 24-h INK128 treatment (see quantification of independent experiments in Supplementary Fig. 23a). e, Representative western blot from three independent experiments of wild type (WT) and 4EBP1/4EBP2 double knockout (DKO) MEFs treated with INK128 for 24 h. f, Representative western blot from two independent experiments of wild-type and mSin1–/– (also called Mapkap1tm1Bisu) MEFs after 24-h INK128 treatment. g, Matrigel invasion assay upon 48-h doxycycline induction of 4EBP1M, or treatment with DG-2 compared to control (n = 6 per condition, t-test). All data represent mean ± s.e.m.
Examining cell invasion networks in vivo
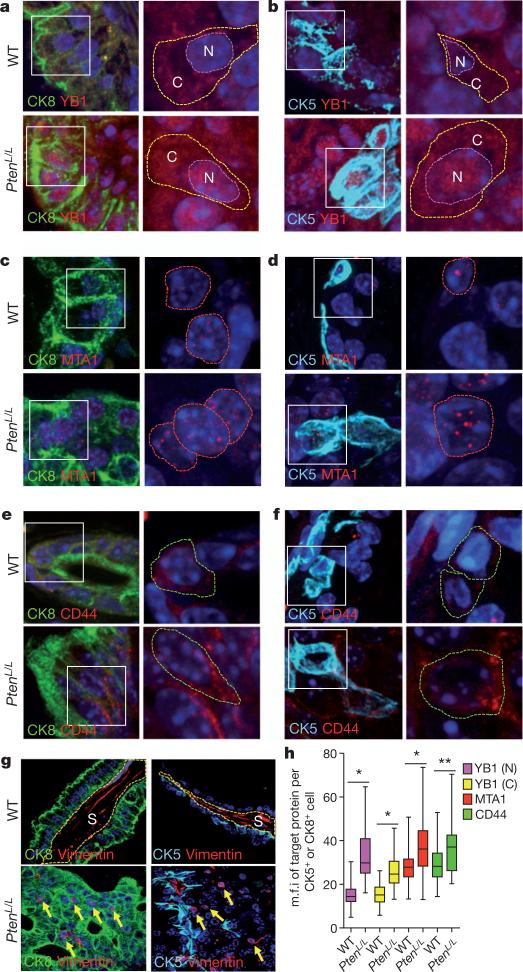
Both CK5+ and CK8+ prostate epithelial cells have been implicated in the initiation of prostate cancer upon loss of PTEN26,27. Ptenloxp/loxp;Pb-cre (PtenL/L) mice are an ideal model of prostate cancer because they display distinct stages of cancer development (prostatic intraepithelial neoplasia, invasive adenocarcinoma, and metastasis)28. However, the expression patterns of YB1, vimentin, CD44 and MTA1 in prostate basal (CK5+) and luminal (CK8+) epithelial cells have not been characterized. We therefore analysed their expression patterns in the PtenL/L prostate cancer mouse model, where mTOR is constitutively hyperactivated9,28. We found that YB1 localizes to the cytoplasm and nucleus of CK5+ and CK8+ prostate epithelial cells, consistent with its ability to shuttle between the two cellular compartments (Fig. 4a, b and Supplementary Fig. 25a, b)18,29. MTA1 expression is exclusively nuclear in both cell types (Fig. 4c, d). Of note, CD44, together with other cell-surface markers, has been used to isolate a rare prostate stem-cell population30. We observed expression of CD44 within a subset of CK5+ and CK8+ epithelial cells (Fig. 4e, f). In contrast, vimentin is not detected in either cell type (Fig. 4g). We next determined the impact of mTOR hyperactivation on the expression pattern of the pro-invasion gene signature. YB1, MTA1 and CD44 protein, but not transcript, levels were significantly increased in both PtenL/L luminal and basal epithelial cells compared to wild type (Fig. 4h and Supplementary Fig. 25c–e). Interestingly, a subset of PtenL/L luminal epithelial cells ectopically expresses vimentin at aberrantly high levels, with a perinuclear distribution (Fig. 4g and Supplementary Fig. 25f, g) suggesting that these cells may have acquired some mesenchymal-like features. Consistent with these findings, perinuclear vimentin localization is associated with invasive features of human prostate cancer cells31 and changes in cell polarity in actively moving fibroblasts32. These studies reveal a unique, translationally controlled signature of gene expression downstream of mTOR hyperactivation in a cancer-initiating subset of prostate epithelial cells.
Figure 4. mTOR hyperactivation augments translation of YB1, MTA1, CD44 and vimentin mRNAs in a subset of pre-invasive prostate cancer cells in vivo.
Left: immunofluorescent images of CK8/DAPI or CK5/DAPI with YB1 (a, b), MTA1 (c, d), or CD44 (e, f) co-staining in 14-month-old wild-type and PtenL/L mouse prostate epithelial cells. White boxes outline the area magnified in the right panel. Right: magnified immunofluorescent images of YB1 (a, b), MTA1 (c, d) and CD44 (e, f) co-stained with DAPI in wild-type and PtenL/L mouse prostate epithelial cells. Dotted lines encircle the cytoplasm (C) and/or the nucleus (N). g, Representative immunofluorescent images of CK5 or CK8 co-staining with vimentin in 14-month-old wild-type and PtenL/L mouse prostate epithelial cells. S, stroma; yellow arrows indicate perinuclear vimentin. h, Box plot of YB1 (N = nuclear, C = cytoplasmic), MTA1 and CD44 mean fluorescence intensity (m.f.i.) per CK5+ or CK8+ prostate epithelial cell in wild-type and PtenL/L mice (three mice per arm, n = 43–303 cells quantified per target gene, error bars indicate range (see Supplementary Fig. 25b); *P < 0.0001, **P = 0.0004, t-test).
Targeting prostate cancer metastasis
The most significant pre-clinical extension of this work would be to determine the therapeutic benefit of INK128 in reprogramming expression of the mTOR-dependent pro-invasion gene signature and prostate cancer metastasis directly in vivo. This is underscored by the clinical inefficacy of allosteric mTOR inhibition towards the lethal form of metastatic human prostate cancer33,34. Importantly, in our preclinical trial of RAD001 (rapalog) versus INK128 in PtenL/L mice, 4EBP1 and p70S6K1/2 phosphorylation was completely restored to wild-type levels after treatment with INK128, whereas RAD001 only decreased p70S6K1/2 phosphorylation levels (Supplementary Fig. 26a, b). We next determined the cellular consequences of complete versus partial mTOR inhibition during distinct stages of prostate cancer. INK128 treatment resulted in a 50% decrease in prostatic intraepithelial neoplasia (PIN) lesions in PtenL/L mice that was associated with decreased proliferation and a tenfold increase in apoptosis (Supplementary Fig. 26d–f). Notably, the unique cytotoxic properties of INK128 treatment in PtenL/L mice were evidenced by a marked reduction in prostate cancer volume. In addition, and consistent with these findings, INK128 induced programmed cell death in multiple cancer cell lines (Supplementary Fig. 27a, b). In contrast, RAD001 treatment mainly had cytostatic effects leading to only partial regression of PIN lesions associated with a limited decrease in cell proliferation and no significant effect on apoptosis (Supplementary Fig. 26c–f).
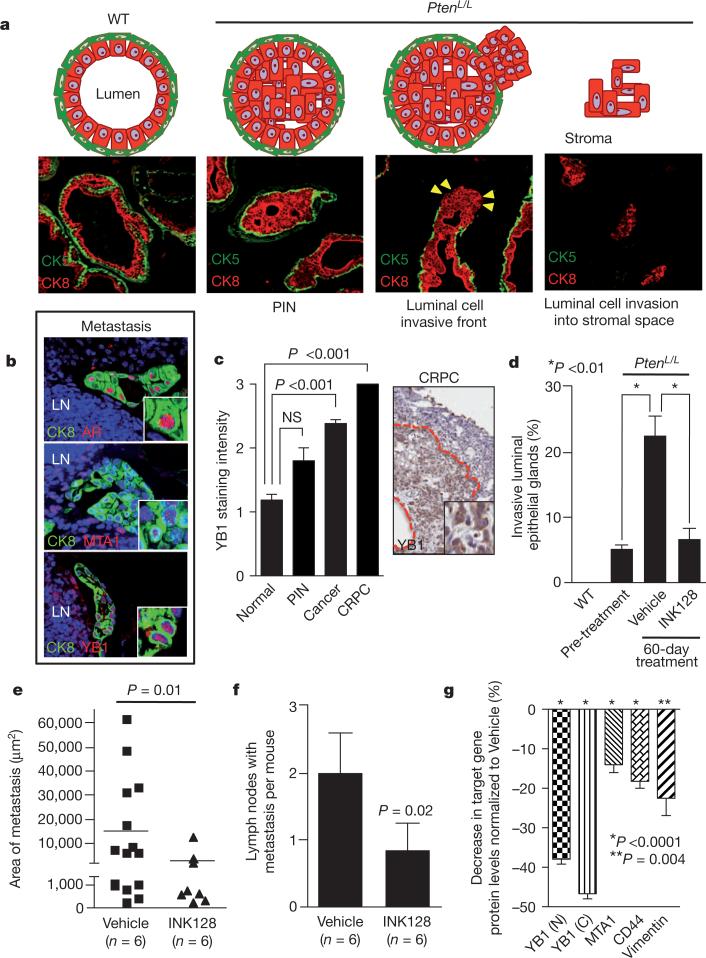
We extended the preclinical trial by examining the effects of INK128 treatment on the pro-invasion gene signature and prostate cancer metastasis, which is incurable and the primary cause of patient mortality. Cell invasion is the critical first step in metastasis, required for systemic dissemination. In PtenL/L mice after the onset of PIN, a subset of prostate glands show characteristics of luminal epithelial cell invasion by 12 months (Fig. 5a and Supplementary Fig. 27c)28. After 12 months of age, PtenL/L mice develop lymph-node metastases and these cells maintain strong YB1 and MTA1 expression (Fig. 5b). We further extended these findings directly to human prostate cancer patient specimens, observing that YB1 expression levels increase in a stepwise fashion from normal prostate to castration-resistant prostate cancer (CRPC), an advanced form of the disease associated with increased metastatic potential (Fig. 5c). MTA1 levels exhibit similar increases20. In human prostate cancer, high-grade primary tumours that display invasive features are more likely to develop systemic metastasis than low-grade non-invasive tumours35,36. Remarkably, treatment with INK128 completely blocked the progression of invasive prostate cancer locally in the prostate gland, and profoundly inhibited the total number and size of distant metastases (Fig. 5d–f). This was associated with a marked decrease in the expression of YB1, vimentin, CD44 and MTA1 at the protein, but not transcript, level in specific epithelial cell types within pre-invasive PIN lesions in PtenL/L mice (Fig. 5g and Supplementary Fig. 25c). Together, these findings reveal an unexpected role for oncogenic mTOR signalling in control of a pro-invasion translational program that, along with the lethal metastatic form of prostate cancer, can be efficiently targeted with clinically relevant mTOR ATP site inhibitors.
Figure 5. Complete mTOR inhibition by INK128 treatment prevents prostate cancer invasion and metastasis in vivo.
a, Diagram and images of normal prostate gland, pre-invasive PIN and invasive prostate cancer. CK8/CK5, luminal/basal epithelial cells, respectively. Yellow arrowheads indicate invasive front. b, Immunofluorescent images of 14-month-old PtenL/L lymph node (LN) metastasis co-stained with CK8/androgen receptor (AR), CK8/YB1 and CK8/MTA1. c, Left: human tissue microarray of YB1 protein levels in normal (n = 59), PIN (n = 5), cancer (n = 99) and CRPC (n = 3) (ANOVA). Right: immunohistochemistry of YB1 in human CRPC demarcated by the red line (inset shows nuclear and cytoplasmic YB1). d, Quantification of invasive prostate glands in wild-type and PtenL/L mice before (12-months old) and after (14-months old) 60 days of INK128 treatment (n = 6 mice per arm, ANOVA). e, f, Area and number of CK8/AR+ metastases in draining lymph nodes in 14-month-old PtenL/L mice after 60 days of INK128 treatment (n = 6 mice per arm, t-test). g, Percentage decrease of YB1 (N = nuclear, C = cytoplasmic), MTA1, CD44, or vimentin protein levels (determined by quantitative immunofluorescence, Supplementary Fig. 25b) in CK8+ or CK5+ prostate cells (CK8+ only for vimentin) in INK128-treated 14-month-old PtenL/L mice normalized to vehicle-treated mice (n = 3 mice per arm, t-test). All data represent mean ± s.e.m.
Discussion
Here we used ribosome profiling to generate a comprehensive map of translationally controlled mTOR targets in cancer that surprisingly stratify into specific cellular processes including proliferation, metabolism, protein synthesis and invasion (Fig. 1e). The effects of this translational control program are probably broad, converging on many subclasses of mRNAs that may cooperate towards distinct steps in cancer development and therapeutic response. This is supported by our in vivo findings where we developed a novel clinically relevant mTOR inhibitor, INK128, that significantly abrogates multiple aspects of prostate cancer development by inducing apoptosis as well as inhibiting cell proliferation, invasion and metastasis (Fig. 5d–g and Supplementary Fig. 26c–f). The superiority of INK128 as an mTOR inhibitor is also evident in its ability to reprogram the mTOR oncogenic translational program in prostate cancer, which is not achieved by rapalog treatment. Of note, however, the sensitivity of cells from distinct histological origins to ATP site versus allosteric inhibitors of mTOR may differ. For example, the Jurkat lymphoid cell line is particularly sensitive to rapamycin treatment37.
One of the most novel nodes of mTOR translationally controlled genes are those that cooperatively control, at least in part, the cellular invasive features of human prostate cancer cells (Figs 1g, 2 and 3b, g). Translational control of these mRNAs relies on the 4EBP1–eIF4E axis and is thereby specifically druggable with potent mTOR ATP site inhibitors, which, unlike rapamycin, target mTOR-dependent 4EBP1 phosphorylation (Figs 1g, 3d, e and 5g, and Supplementary Figs 1b, 23 and 26b). This has significant therapeutic implications not only for advanced prostate cancer but also for multiple metastatic cancers where we show that translational control of pro-invasion mRNAs is sensitive to ATP site inhibitors of mTOR (Supplementary Fig. 16). Thereby, these studies link translational regulation to the poorly understood mechanisms underlying cancer metastasis. Intriguingly, various components of the translation machinery, including oncogenic eIF4E38, localize to the leading edge of migrating fibroblasts39. This may allow spatially controlled translation of mRNAs important for cell migration, providing a rapid and specific response in transducing a migration program that could be co-opted at the invasive edge of metastatic cancer cells. Together, these studies reveal that the ability of mTOR to phosphorylate general translation factors results in exquisite transcript-specific translational control of key mRNAs that may cooperate in distinct steps of cancer initiation and progression, with significant implications for therapeutic intervention.
METHODS
Mice
Ptenloxp/loxp and Pb-cre mice where obtained from Jackson Laboratories and Mouse Models of Human Cancers Consortium (MMHCC), respectively, and maintained in the C57BL/6 background. Mice were maintained under specific pathogen-free conditions, and experiments were performed in compliance with institutional guidelines as approved by the Institutional Animal Care and Use Committee of UCSF.
Cell culture and reagents
Human cell lines were obtained from the ATCC and maintained in the appropriate medium with supplements as suggested by ATCC. Wild-type, mSin1–/– (provided by B. Su), and 4EBP1/4EBP2 double knockout MEFs (provided by N. Sonenberg) were cultured as previously described24,25. SMARTvector 2.0 (Thermo Scientific) lentiviral shRNA constructs were used to knock down PTEN (SH-003023-02-10). For generation of GFP-labelled PC3 cells, SMARTvector 2.0 lentiviral empty vector control particles that contain TurboGFP (S-004000-01) were used. Control (D-001810-01), YB1 (L-010213), MTA1 (L-004127), CD44 (L-009999), vimentin (L-003551), rictor (LL-016984), 4EBP1 (L-003005) and 4EBP2 (L-018671) pooled siRNAs were purchased from Thermo Scientific. Intellikine provided INK128 and PP242, which were used at 200 nM and 2.5 μM in cell-based assays unless otherwise specified. RAD001 was obtained from LC Laboratories. DG-2 was provided by K. Shokat and used at 20 μM in cell-based assays. Rapamycin was purchased from Calbiochem and used at 50 nM in cell-based assays. Doxycyline (Sigma) was used at 1 μg ml–1 in 4EBP1M induction assays. Lipofectamine 2000 (Invitrogen) was used to transfect cancer cell lines with siRNA. Amaxa Cell Line Nucleofector Kit R (Lonza) was used to electroporate BPH-1 cells with over expression vectors. The 4EBP1M has been previously described13.
Plasmids
pcDNA3-HA-YB1 was provided by V. Evdokimova. pCMV6-Myk-DDK-MTA1 was purchased from Origene. pGL3-Promoter was purchased from Promega. To clone the 5′ UTR of YB1 into pGL3-Promoter, the entire 5′ UTR sequence of YB1 was amplified from PC3 cDNA. PCR fragments were digested with HindIII and NcoI and ligated into the corresponding sites of pGL3-Promoter. The PRTE sequence at position +20–34 in the YB1 5′ UTR (UCSC kgID uc001chs.2) was mutated using the QuikChange Site-Directed Mutagenesis Kit following the manufacturer's protocol (Stratagene).
Ribosome profiling
PC3 cells were treated with rapamycin (50 nM; Calbiochem) or PP242 (2.5 μM; Intellikine) for 3 h. Cells were subsequently treated with cycloheximide (100 μg ml–1; Sigma) and detergent lysis was performed in the dish. The lysate was treated with DNase and clarified, and a sample was taken for RNA-seq analysis. Lysates were subjected to ribosome foot printing by nuclease treatment. Ribosome-protected fragments were purified, and deep sequencing libraries were generated from these fragments, as well as from poly(A) mRNA purified from non-nuclease-treated lysates. These libraries were analysed by sequencing on an Illumina GAII.
Each sequencing run resulted in approximately 20–25 million raw reads per sample, of which 5–12 million unique reads were used for subsequent analysis. Ribosome footprint and RNA-seq sequencing reads were aligned against a library of transcripts from the UCSC Known Genes database GRCh37/hg19. The first 25 nucleotides of each read were aligned using Bowtie and this initial alignment was then extended to encompass the full fragment-derived portion of the sequencing read while excluding the linker sequence. Read density profiles were then constructed for the canonical transcript of each gene, using only reads with 0 or 1 total mismatches between the read sequence and the reference sequence, comprised of the transcript fragment followed by the linker sequence. Footprint reads were assigned to an A site nucleotide at position +15 to +17 of the alignment, based on the total fragment length; mRNA reads were assigned to the first nucleotide of the alignment. The average read density per codon was then computed for the coding sequence of each transcript, excluding the first 15 and last 5 codons, which can display atypical ribosome accumulation.
Average read density was used as a measure of mRNA abundance (RNA-seq reads) and of protein synthesis (ribosome profiling reads). For most analyses, genes were filtered to require at least 256 reads in the relevant RNA-seq samples. Translational efficiency was computed as the ratio of ribosome footprint read density to RNA-seq read density, scaled to normalize the translational efficiency of the median gene to 1.0 after excluding regulated genes (log2 fold-change ±1.5 after normalizing for the all-gene median). Changes in protein synthesis, mRNA abundance and translational efficiency were similarly computed as the ratio of read densities between different samples, normalized to give the median gene a ratio of 1.0. This normalization corrects for differences in the absolute number of sequencing reads obtained for different libraries. 3,977 (replicate 1), and 5,333 (replicate 2) unique mRNAs passed a preset read threshold of 256 reads for single-gene quantification for all treatment conditions.
Western blot analysis
Western blot analysis was performed as previously described13 with antibodies specific to phospho-AKTS473 (Cell Signaling), AKT (Cell Signaling), phospho-p70S6KT389 (Cell Signaling), phospho-rpS6S240/244 (Cell Signaling), rpS6 (Cell Signaling), phospho-4EBP1T37/46 (Cell Signaling), 4EBP1 (Cell Signaling), 4EBP2 (Cell Signaling), YB1 (Cell Signaling), CD44 (Cell Signaling), LEF1 (Cell Signaling), PTEN (Cell Signaling), eEF2 (Cell Signaling), GAPDH (Cell Signaling), vimentin (BD Biosciences), eIF4E (BD Biosciences), Flag (Sigma), β-actin (Sigma), MTA1 (Santa Cruz Biotechnology), Twist (Santa Cruz Biotechnology), rpL28 (Santa Cruz Biotechnology), HA (Covance) and rictor (Bethyl Laboratory).
qPCR analysis
RNA was isolated using the manufacturer's protocol for RNA extraction with TRIzol Reagent (Invitrogen) using the Pure Link RNA mini kit (Invitrogen). RNA was Dnase-treated with Pure Link Dnase (Invitrogen). Dnase-treated RNA was transcribed to cDNA with SuperScript III First-Strand Synthesis System for RT–PCR (Invitrogen), and 1 μl of cDNA was used to run a SYBR green detection qPCR assay (SYBR Green Supermix and MyiQ2, Biorad). Primers were used at 200 nM.
5′ UTR analysis
5′ UTRs of the 144 downregulated mTOR target genes were obtained using the known gene ID from the UCSC Genome Browser (GRCh37/hg19). Target versus non-target mRNAs were compared for 5′ UTR length, %G+C content and Gibbs free energy by the Wilcoxon two-sided test. Multiple Em (expectation maximization) for Motif Elicitation (MEME) and Find Individual Motif Occurrences (FIMO) was used to derive the PRTE and determine its enrichment in the 144 mTOR-sensitive genes compared a background list of 3,000 genes. The Database of Transcriptional Start Sites (DBTSS Release 8.0) was used to identify putative 5′ TOP genes and putative transcription start sites in the 144 mTOR target genes.
Luciferase assay
PC3 4EBP1M cells were treated with 1 μg ml–1 doxycycline (Sigma) for 24 h. Cells were transfected with various pGL3-Promoter constructs using lipofectamine 2000 (Invitrogen). After 24 h, cells were collected. 20% of the cells were aliquoted for RNA isolation. The remaining cells were used for the luciferase assay per the manufacturer's protocol (Promega). Samples were measured for luciferase activity on a Glomax 96-well plate luminometer (Promega). Firefly luciferase activity was normalized to luciferase mRNA expression levels.
Kinase assays
mTOR activity was assayed using LanthaScreen Kinase kit reagents (Invitrogen) according to the manufacturer's protocol. PI(3)K α, β, γ and δ activity were assayed using the PI(3)K HTRF assay kit (Millipore) according to the manufacturer's protocol. The concentration of INK128 necessary to achieve inhibition of enzyme activity by 50% (IC50) was calculated using concentrations ranging from 20 μM to 0.1 nM (12-point curve). IC50 values were determined using a nonlinear regression model (GraphPad Prism 5).
Cell proliferation assay
PC3 cells were treated with the appropriate drug for 48 h, and proliferation was measured usingCellTiter-Glo Luminescent reagent (Promega) per the manufacturer's protocol. The concentration of INK128 necessary to achieve inhibition of cell growth by 50% (IC50) was calculated using concentrations ranging from 20.0 μM to 0.1 nM (12-point curve).
Mouse xenograft study
Nude mice were inoculated subcutaneously in the right subscapular region with 5 × 106 MDA-MB-361 cells. After tumours reached a size of 150–200 mm3, mice were randomly assigned into vehicle control or treatment groups. INK128 was formulated in 5% polyvinylpropyline, 15% NMP, 80% water and administered by oral gavage at 0.3 mg kg–1 and 1 mg kg–1 daily.
Pharmacokinetic analysis
The area under the plasma drug concentration versus time curves, AUC(0–tlast) and AUC(0–inf), were calculated from concentration data using the linear trapezoidal rule. The terminal t1/2 in plasma was calculated from the elimination rate constant (lz), estimated as the slope of the log-linear terminal portion of the plasma concentration versus time curve, by linear regression analysis. The bioavailability (F) was calculated using F = (AUC(0–last),poDi.v.)/(AUC(0–last),ivDp.o.)×100%, where Di.v. and Dp.o. are intravenous and oral doses, respectively. Cmax was a highest drug concentration in plasma after oral administration. Tmax was the time at which Cmax is observed after extravascular administration of drug. Tlast was the last time point a quantifiable drug concentration can be measured.
Metabolic stability assay
In vitro metabolic stability of INK128 was evaluated after incubation with liver microsomes or liver S9 fractions from various species in the presence of NADPH. The half-life of INK128 was estimated by log linear regression analysis.
CYP assay
INK128 inhibition of CYP450 isoforms in human liver microsomes was determined with isoform-specific substrates at concentrations approximately equal to the concentration at which the rate of the reaction is half-maximal (Km) for the individual isoforms: CYP1A2, CYP2C8, CYP2C9, CYP2C19, CYP2D6 and CYP3A4.
Pharmaceutical property assays
The percentage of protein binding of INK128 was determined in mouse, rat, dog, monkey and human plasma at CEREP. The IC50 for the inhibitory effect of INK128 on hERG potassium channel was determined at CEREP. A Bacterial Reverse Mutation Assay (Ames test) was conducted at BioReliance.
Polysome analysis
PC3 cells were treated for 3 h with either DMSO or INK128 (100 nM). Cells were re-suspended in PBS containing 100 μg ml–1 cycloheximide (Sigma) and incubated on ice for 10 min. Cells were centrifuged at 300g for 5 min at 4 °C and lysed in 10 mM Tris-HCl pH 8, 140 mM NaCl, 5 mM MgCl2, 640 U ml–1 Rnasin, 0.05% NP-40, 250 μg ml–1 cycloheximide, 20 mM DTT and protease inhibitors. Samples were incubated for 20 min on ice then centrifuged once for 5 min at 3,300g and once for 5 min at 9,300g, isolating the supernatant after each centrifugation. Lysates were loaded onto 10–50% sucrose gradients containing 0.1 mg ml–1 heparin and 2 mM DTT and centrifuged at 37,000 r.p.m. for 2.5 h at 4 °C. The sample was subsequently fractionated on a gradient fractionation system (ISCO). RNA was extracted from all fractions and run on a TBE-agarose gel to visualize 18S and 28S rRNA. Fractions 7–13 were found to correspond to the polysome fractions and were used for further qPCR analysis.
[35S] metabolic labelling
PC3 or PC3 4EBP1M cells with or without indicated treatment were incubated with 30 μCi of [35S]-methionine for 1 h after pre-incubation in methionine-free DMEM (Invitrogen). Cells were prepared using a standard protein lysate protocol, resolved on a 10% SDS polyacrylamide gel and transferred onto a PVDF membrane (Biorad). The membrane was exposed to autoradiography film (Denville) for 24 h and developed.
Cell cycle analysis
Appropriately treated PC3, BPH-1, or PC3-4EBP1M cells were fixed in 70% ethanol overnight at –20 °C. Cells were subsequently washed with PBS and treated with RNase (Roche) for 30 min. After this incubation, the cells were permeabilized and treated with 50 μg ml–1 propidium iodide (Sigma) in a solution of 0.1% Tween, 0.1% sodium citrate. Cell cycle data was acquired using a BD FACS Caliber (BD Biosciences) and analysed with FlowJo (v.9.1).
Apoptosis analysis
Appropriately treated LNCaP and A498 cells were labelled with Annexin V-FITC (BD Biosciences) and propidium iodide (Sigma) following the manufacturer's instructions. PI/Annexin data was acquired using a BD FACS Caliber (BD Biosciences) and analysed with FlowJo (v.9.1).
Matrigel invasion assay
BioCoat Matrigel Invasion Chambers (modified Boyden Chamber Assay; BD Biosciences) were used according to the manufacturer's instructions.
Real-time imaging of cell migration
Real-time imaging of GFP-labelled PC3 cells was performed in poly-d-lysine-coated chamber cover glass slides (Lab-Tek). PC3 GFP cells were plated and allowed to adhere for 24 h. Wells were wounded with a P200 pipette tip. The chamber slides were imaged with an IX81 Olympus wide-field fluorescence microscope equipped with a CO2 and temperature controlled chamber and time-lapse tracking system. Images from DIC and GFP channels were taken every 2 min and processed using ImageJ (http://rsb.info.nih.gov/ij/) and analysed for cell migration with Manual Tracking (http://rsbweb.nih.gov/ij/plugins/track/track.html), using local maximum centring correction to maintain a centroid xy coordinate for each cell per frame over time. Tracking data was subsequently processed with the Chemotaxis and Migration tool from ibidi (http://www.ibidi.de/applications/ap_chemo.html) to create xy coordinate plots, velocity and distance measurements.
Snail1 immunocytochemistry
Appropriately transfected or treated PC3 cells were plated on a poly-l-lysine-coated chamber slide (Lab-Tek) and cultured for 48 h. Cells were fixed with 4% paraformaldehye (EMS), rinsed with PBS and permeabilized with 0.1% Triton X-100. The samples were blocked in 5% goat serum and then incubated with anti-Snail1 antibody (Cell Signaling) in 5% goat serum for 2 h at room temperature. Cells were washed with PBS and incubated with Alexa 594 anti-mouse antibody (Invitrogen) and DAPI (Invitrogen) for 2 h at room temperature. Specimens were again washed with PBS and subsequently mounted with Aqua Poly/Mount (Polysciences). Image capture and quantification were completed as described below (see Immunofluorescence).
Cap-binding assay
PC3 4EBP1M cells were induced with doxycycline (1 μg ml–1, Sigma) for 48 h, then collected and lysed in buffer A (10 mM Tris-HCl pH 7.6, 150 mM KCl, 4 mM MgCl2, 1 mM DTT, 1 mM EDTA, and protease inhibitors, supplemented with 1% NP-40). Cell lysates were incubated overnight at 4 °C with 50 μl of the mRNA cap analogue m7GTP-sepharose (GE Healthcare) in buffer A. The beads were washed with buffer A supplemented with 0.5% NP-40. Protein complexes were dissociated using 1× sample buffer, and resolved by SDS–PAGE and western blotted with the appropriate antibodies.
Pharmacological treatment of PtenL/L mice and MRI imaging
Nine- and twelve-month-old PtenL/L mice were gavaged daily with either vehicle (see mouse xenograft study), RAD001 (10 mg kg–1; LC Laboratories), or INK128 (1 mg kg–1; Intellikine) for the indicated times. Weight measurements were taken every 3 days to monitor for toxicity. For the 28-day study, mice were imaged via MRI at day 0 and day 28 in a 14-T GE MR scanner (GE Healthcare).
Prostate tissue processing
Whole mouse prostates were removed from wild-type and PtenL/L mice, microdissected, and frozen in liquid nitrogen. Frozen tissues were subsequently manually disassociated using a biopulverizer (Biospec) and additionally processed for protein and mRNA analysis as described above.
Immunofluorescence
Prostates and lymph nodeswere dissected frommicewithin 2 h of the indicated treatment and fixed in 10% formalin overnight at 4 °C. Tissues were subsequently dehydrated in ethanol (Sigma) at room temperature, mounted into paraffin blocks, and sectioned at 5 μm. Specimens were de-paraffinized and rehydrated using CitriSolv (Fisher) followed by serial ethanol washes. Antigen unmasking was performed on each section using Citrate pH 6 (Vector Labs) in a pressure cooker at 125 °C for 10–30 min. Sections were washed in distilled water followed by TBS washes. The sections were then incubated in 5% goat serum, 1% BSA in TBS for 1 h at room temperature. Various primary antibodies were used including those specific for keratin 5 (Covance), cytokeratin 8 (Abcam and Covance), YB1 (Abcam), vimentin (Abcam), MTA1 (Cell signaling), CD44 (BD Pharmingen) and the androgen receptor (Epitomics), which were diluted 1:50–1:500 in blocking solution and incubated on sections overnight at 4 °C. Specimens were then washed in TBS and incubated with the appropriate Alexa 488 and 594 labelled secondary (Invitrogen) at 1:500 for 2 h at room temperature with the exception of YB1 which was incubated with biotinylated anti-rabbit secondary (Vector) followed by incubation with Alexa 594 labelled Streptavidin (Invitrogen). A final set of washes in TBS was completed at room temperature followed by mounting with DAPI Hardset Mounting Medium (Vector Lab). A Zeiss Spinning Disc confocal (Zeiss, CSU-X1) was used to image the sections at 40×–100×. Individual prostate cells were quantified for mean fluorescence intensity (m.f.i.) using the Axiovision (Zeiss, Release 4.8) densitometric tool.
Lymph node metastasis measurements
Mouse lymph nodes were processed as described above and stained for CK8 and androgen receptor. Lymph nodes were imaged using a Zeiss AX10 microscope. Metastases were identified and areas were measured using the Axiovision (Zeiss, Release 4.8) measurement tool.
Semi-quantitative RT–PCR
Whole prostates were removed from wild-type and PtenL/L mice, microdissected, dissociated into single-cell suspension, and stained for epithelial cell markers as previously described40 using fluorescence-conjugated antibodies for CD49f, Sca-1, CD31, CD45 and Ter119 (BD Biosciences). Luminal epithelial cells were sorted as previously described41 using a FACS Aria (BD Biosciences). Cell pellets were re-suspended in 500 μl TRIzol Reagent and RNA was isolated and transcribed into cDNA as described above. Semi-quantitative PCR analysis was performed using oligonucleotides for vimentin and β-actin at 200 nM in a 25 μl reaction with 12.5 μl GoTaq (Promega) for 32 and 33 cycles respectively, which were within the linear range (Supplementary Fig. 25f).
Immunohistochemistry
Immunohistochemistry was performed as described above (see immunofluorescence section) with the exception that immediately after antigen presentation and TBS washes, specimens were incubated in 3% hydrogen peroxide in TBS followed by TBS washes. The following primary antibodies were used: phospho-AKTS473 (Cell Signaling), phospho-rpS6S240/244 (Cell Signaling), phospho-4EBP1T37/46 (Cell Signaling), phospho-histone H3 (Upstate), and cleaved caspase 3 (Cell Signaling). This was followed by TBS washes and incubation with the appropriate biotinylated secondary antibody (Vector Lab) for 30 min at room temperature. An ABC-HRP Kit (Vector Lab) was used to amplify the signal, followed by a brief incubation in hydrogen peroxide. The protein of interest was detected using DAB (Sigma). Specimens were counterstained with haematoxylin (Thermo Scientific), dehydrated with Citrisolv (Fisher), and mounted with Cytoseal XYL (Vector Lab).
Haematoxylin and eosin staining
Paraffin-embedded prostate specimens were deparaffinized and rehydrated as described above (see immunofluorescence section), stained with haematoxylin (Thermo Scientific), and washed with water. This was followed by a brief incubation in differentiation RTU (VWR) and two washes with water followed by two 70% ethanol washes. The samples were then stained with eosin (Thermo Scientific) and dehydrated with ethanol followed by CitriSolv (Fisher). Slides were mounted with Cytoseal XYL (Richard Allan Scientific).
Oligonucleotides
YB1 5′ UTR cloning and site-directed mutagenesis oligonucleotides are as follows. YB1 5′ UTR cloning: forward 5′-GCTACAAGCTTGGGCTTATCCCGCCT-3′,reverse5′-TCGATCCATGGGGTTGCGGTGATGGT-3′; deletion (20–34): forward 5′-TGGGCTTATCCCGCCTGTCCTTCGATCGGTAGCGGGAGCG-3′, reverse 5′-CGCTCCCGCTACCGATCGAAGGACAGGCGGGATAAGCCCA-3′; transversion (20–34): forward 5′-TGGGCTTATCCCGCCTGTCCGCGGTAAGAGCGATCTTCGATCGGTAGCGGGAGCG-3′, reverse 5′-CGCTCCCGCTACCGATCGAAGATCGCTCTTACCGCGGACAGGCGGGATAAGCCCA-3′.
Human qPCR oligonucleotides are as follows. β-actin forward 5′-GCAAAGACCTGTACGCCAAC-3′, reverse 5′-AGTACTTGCGCTCAGGAGGA-3′; CD44 forward 5′-CAACAACACAAATGGCTGGT-3′, reverse 5′-CTGAGGTGTCTGTCTCTTTCATCT-3′; vimentin forward 5′-GGCCCAGCTGTAAGTTGGTA-3′, reverse 5′-GGAGCGAGAGTGGCAGAG-3′; Snail1 forward 5′-CACTATGCCGCGCTCTTTC-3′, reverse 5′-GCTGGAAGGTAAACTCTGGATTAGA-3′; YB1 forward 5′-TCGCCAAAGACAGCCTAGAGA-3′, reverse 5′-TCTGCGTCGGTAATTGAAGTTG-3′; MTA1 forward 5′-CAAAGTGGTGTGCTTCTACCG-3′, reverse 5′-CGGCCTTATAGCAGACTGACA-3′; PLAU forward 5′-TTGCTCACCACAACGACATT-3′, reverse 5′-GGCAGGCAGATGGTCTGTAT-3′; FGFBP1 forward 5′-ACTGGATCCGTGTGCTCAG-3′, reverse 5′-GAGCAGGGTGAGGCTACAGA-3′; ARID5B forward 5′-TGGACTCAACTTCAAAGACGTTC-3′, reverse 5′-ACGTTCGTTTCTTCCTCGTC-3′; CTGF forward 5′-CTCCTGCAGGCTAGAGAAGC-3′, reverse 5′-GATGCACTTTTTGCCCTTCTT-3′; RND3 forward 5′-AAAAACTGCGCTGCTCCAT-3′, reverse 5′-TCAAAACTGGCCGTGTAATTC-3′; KLF6 forward 5′-AAAGCTCCCACTTGAAAGCA-3′, reverse 5′-CCTTCCCATGAGCATCTGTAA-3′; BCL6 forward 5′-TTCCGCTACAAGGGCAAC-3′, reverse 5′-TGCAACGATAGGGTTTCTCA-3′; FOXA1 forward 5′-AGGGCTGGATGGTTGTATTG-3′, reverse 5′-ACCGGGACGGAGGAGTAG-3′; GDF15 forward 5′-CCGGATACTCACGCCAGA-3′, reverse 5′-AGAGATACGCAGGTGCAGGT-3′; HBP1 forward 5′-GCTGGTGGTGTTGTCGTG-3′, reverse 5′-CATGTTATGGTGCTCTGACTGC-3′; Twist1 forward 5′-CATCCTCACACCTCTGCATT-3′, reverse 5′-TTCCTTTCAGTGGCTGATTG-3′; LEF1 forward 5′-CCTTGGTGAACGAGTCTGAAATC-3′, reverse 5′-GAGGTTTGTGCTTGTCTGGC-3′; rpS19 forward 5′-GCTGGCCAAACATAAAGAGC-3′, reverse 5′-CTGGGTCTGACACCGTTTCT-3′; 5S rRNA forward 5′-GCCCGATCTCGTCTGATCT-3′, reverse 5′-AGCCTACAGCACCCGGTATT-3′; firefly luciferase forward 5′-AATCAAAGAGGCGAACTGTG-3′, reverse 5′-TTCGTCTTCGTCCCAGTAAG-3′.
Mouse qPCR oligonucleotides are as follows. β-actin forward 5′-CTAAGGCCAACCGTGAAAAG-3′, reverse 5′-ACCAGAGGCATACAGGGACA-3′; Yb1 forward 5′-GGGTTACAGACCACGATTCC-3′, reverse 5′-GGCGATACCGACGTTGAG-3′; vimentin forward 5′-TCCAGCAGCTTCCTGTAGGT-3′, reverse 5′-CCCTCACCTGTGAAGTGGAT-3′; Cd44 forward 5′-ACAGTACCTTACCCACCATG-3′, reverse 5′-GGATGAATCCTCGGAATTAC-3′; Mta1 forward 5′-AGTGCGCCTAATCCGTGGTG-3′, reverse 5′-CTGAGGATGAGAGCAGCTTTCG-3′.
siRNA/shRNA sequences are as follows. Control (D-001810-01) 5′-UGGUUUACAUGUCGACUAA-3′; vimentin (L-003551) 5′-UCACGAUGACCUUGAAUAA-3′, 5′-GGAAAUGGCUCGUCACCUU-3′, 5′-GAGGGAAACUAAUCUGGAU-3′, 5′-UUAAGACGGUUGAAACUAG-3′; YB1 (L-010213) 5′-CUGAGUAAAUGCCGGCUUA-3′, 5′-CGACGCAGACGCCCAGAAA-3′, 5′-GUAAGGAACGGAUAUGGUU-3′, 5′-GCGGAGGCAGCAAAUGUUA-3′; MTA1 (L-004127) 5′-UCACGGACAUUCAGCAAGA-3′, 5′-GGACCAAACCGCAGUAACA-3′, 5′-GCAUCUUGUUGGACAUAUU-3′, 5′-CCAGCAUCAUUGAGUACUA-3′; CD44 (L-009999) 5′-GAAUAUAACCUGCCGCUUU-3′, 5′-CAAGUGGACUCAACGGAGA-3′, 5′-CGAAGAAGGUGUGGGCAGA-3′, 5′-GAUCAACAGUGGCAAUGGA-3′; 4EBP1 (L-003005) 5′-CUGAUGGAGUGUCGGAACU-3′, 5′-CAUCUAUGACCGGAAAUUC-3′, 5′-GCAAUAGCCCAGAAGAUAA-3′, 5′-GAGAUGGACAUUUAAAGCA-3′; 4EBP2 (L-018671) 5′-GCAGCUACCUCAUGACUAU-3′, 5′-GGAGGAACUCGAAUCAUUU-3′, 5′-GCAAUUCUCCCAUGGCUCA-3′, 5′-UUGAACAACUUGAACAAUC-3′; rictor (LL-016984) 5′-GACACAAGCACUUCGAUUA-3′, 5′-GAAGAUUUAUUGAGUCCUA-3′, 5′-GCGAGCUGAUGUAGAAUUA-3′, 5′-GGGAAUACAACUCCAAAUA-3′; PTEN SH-003023-01-10 5′-GCTAAGAGAGGTTTCCGAA-3′, SH-003023-02-10 5′-AGACTGATGTGTATACGTA-3′.
Supplementary Material
Acknowledgements
We thank M. Barna for critical discussion and reading of this manuscript; T. Wilson for support and advice; T. Sanders and E. Llagostera-Martin for technical support with confocal microscopy; L. Li, E. Ulm, L. Kessler, J. Kucharski and L. Darjania for technical support for the discovery and development of INK128. J. Kurhanewicz and R. Bok of the Surbeck Institute for Advanced Imaging for technical support and MRI images; N. Sonenberg for providing the 4EBP1/2 double knockout mouse embryonic fibroblasts; J. M. Shen for support; and K. Tong for editing the manuscript. A.C.H. is supported in part by the American Cancer Society (119084-PF-10-233-01-TBE), and is a Prostate Cancer Foundation Young Investigator, and a recipient of the DOD Prostate Cancer Training Award. This work is supported by NIH R01 CA154916 (D.R.), NIH R01 CA140456 (D.R.) and the Phi Beta Psi Sorority (D.R.). D.R. is a Leukemia & Lymphoma Society Scholar.
Footnotes
Author Contributions A.C.H. and D.R. conceived the experiments. A.C.H., M.P.E., M.R.J., A.S., E.Y.S., C.R.S., C.C. and S.W. performed the experiments, PtenL/L preclinical trials, and collected the data. N.T.I. and J.S.W. contributed to ribosomal profiling data analysis. M.J.B. provided pathology support. Y.L., P.R., M.M., S.W., K.J., M.E.F., K.M.S. and C.R. developed and/or supported development of INK128, conducted pharmacokinetic, pharmacodynamic and preclinical studies. A.C.H. and D.R. analysed the data and wrote the manuscript. All authors discussed results and edited the manuscript.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Small-RNA sequencing data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE35469. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Brown EJ, et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 2.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E–BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 6.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nardella C, et al. Differential requirement of mTOR in postmitotic tissues and tumorigenesis. Sci. Signal. 2009;2:ra2. doi: 10.1126/scisignal.2000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guertin DA, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furic L, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl Acad. Sci. USA. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, et al. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 16.Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Evdokimova V, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Lahat G, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS ONE. 2010;5:e10105. doi: 10.1371/journal.pone.0010105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Hofer MD, et al. The role of metastasis-associated protein 1 in prostate cancer progression. Cancer Res. 2004;64:825–829. doi: 10.1158/0008-5472.can-03-2755. [DOI] [PubMed] [Google Scholar]
- 21.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1α protein by recruiting histone deacetylase 1. EMBO J. 2006;25:1231–1241. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuzumi T, et al. Inhibitor hijacking of Akt activation. Nature Chem. Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulholland DJ, et al. Lin–Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555–8562. doi: 10.1158/0008-5472.CAN-08-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland BW, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 30.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–818. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 31.Lang SH, et al. Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate. 2002;52:253–263. doi: 10.1002/pros.10088. [DOI] [PubMed] [Google Scholar]
- 32.Helfand BT, et al. Vimentin organization modulates the formation of lamellipodia. Mol. Biol. Cell. 2011;22:1274–1289. doi: 10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amato RJ, Jac J, Mohammad T, Saxena S. Pilot study ofrapamycin inpatients with hormone-refractory prostate cancer. Clin. Genitourin. Cancer. 2008;6:97–102. doi: 10.3816/CGC.2008.n.015. [DOI] [PubMed] [Google Scholar]
- 34.George DJ, et al. A phase II study of RAD001 in men with hormone refractory metastatic prostate cancer (HRPC). Am. Soc. Clin. Oncol. Genitourin. Cancers Symp. 2008 Abstract 181. [Google Scholar]
- 35.Pontes JE, Wajsman Z, Huben RP, Wolf RM, Englander LS. Prognostic factors in localized prostatic carcinoma. J. Urol. 1985;134:1137–1139. doi: 10.1016/s0022-5347(17)47659-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhou P, et al. Predictors of prostate cancer-specific mortality after radical prostatectomy or radiation therapy. J. Clin. Oncol. 2005;23:6992–6998. doi: 10.1200/JCO.2005.01.2906. [DOI] [PubMed] [Google Scholar]
- 37.Grolleau A, et al. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- 38.Ruggero DR, et al. The translation factor eIF-4F promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 39.Willett M, Brocard M, Davide A, Morley SJ. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem. J. 2011;438:217–227. doi: 10.1042/BJ20110435. [DOI] [PubMed] [Google Scholar]
- 40.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nature Protocols. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl Acad. Sci. USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.