Abstract
Genome-wide association studies (GWAS) have achieved great success in identifying common variants associated with increased risk of developing breast cancer. However, GWAS do not typically provide information about the broader context in which genetic variants operate in different subtypes of breast cancer. The objective of this study was to determine whether genes containing single nucleotide polymorphisms (SNPs, herein called genetic variants) are associated with different subtypes of breast cancer. Additionally, we sought to identify gene regulator networks and biological pathways enriched for these genetic variants. Using supervised analysis, we identified 201 genes that were significantly associated with the six intrinsic subtypes of breast cancer. The results demonstrate that integrative genomics analysis is a powerful approach for linking GWAS information to distinct disease states and provide insights about the broader context in which genetic variants operate in different subtypes of breast cancer.
Keywords: GWAS subtypes breast cancer
Introduction
Genome-wide association studies (GWAS) have made it possible to identify single nucleotide polymorphisms (SNPs), herein called genetic variants, that are associated with an increased risk of developing breast cancer.1,2 Results from GWAS are providing valuable clues about allelic architectures and are improving our understanding of the emerging genetic susceptibility landscape of breast cancer. However, despite these remarkable achievements, significant challenges remain. Although many compelling genetic variants have been found and replicated in multiple independent GWAS,1,2 they explain only a small fraction of the variation. Importantly, GWAS do not typically inform the broader context in which the genetic variants operate, leading to the development of different breast cancer subtypes. As a consequence, they provide limited insights about the molecular mechanisms determining the different subtypes of breast cancer.
The advent of microarray technology has made possible the identification of molecular signatures and molecular classifications of subtypes of breast cancer based on mRNA expression profiles.3,4 However, although these primary analyses have identified clinically actionable biomarkers, they have been unsuccessful in determining which genes have causal roles as opposed to merely being consequences of disease states.1 Few genetic association studies have systematically evaluated the association between putative common susceptibility alleles and specific subtypes of breast cancer. The largest studies providing a comprehensive catalogue of such genetic variants have been published recently.1,2 The association of common low-penetrance genetic variants with subtypes of breast cancer has also been reported.5–7 However, to date, there is little information associating GWAS information with intermediate phenotypes of different subtypes of breast cancer. Subtypes of breast cancer originate from a complex interplay between a constellation of changes in DNA (both common and rare variants) and a broad range of environmental factors. These complex multidimensional interactions are believed to affect entire network states and biological pathways that in turn increase or decrease the risk of breast cancer.8 In fact, the emerging picture from large-scale genomic studies is that subtypes of breast cancer are emergent properties of networks whose states are affected by functionally related genes interacting in complex gene regulatory networks and biological pathways.1,8
Integrating GWAS information with gene expression data holds promise not only for identifying the molecular networks and biological pathways that are enriched for SNPs associated with increased risk of developing breast cancer, but also for causally associating the genetic variants with different subtypes. There are potentially two routes through which clinically actionable biomarkers can be identified using integrative genomics approaches. One route is based on using patterns of gene expression profiles to identify SNP-containing genes that are functionally related and are associated with specific breast cancer subtypes. The second involves identification of gene regulatory networks and biological pathways enriched for genetic variants. The functionally-related genes interacting in complex gene regulatory networks and multi-gene biological pathways produced from this type of integrative genomics analysis can help link GWAS information to intermediate phenotypes of breast cancer. This approach would provide an alternative path for understanding the broader context in which genetic variants operate, leading to different disease states. In addition, this approach could lead to the identification of molecular markers for potential risk prediction of different subtypes of breast cancer and to the development of new effective therapies.
The objectives of this study were 2-fold. Firstly, we wished to determine whether genes containing SNPs associated with increased risk of developing breast cancer are associated with different subtypes of breast cancer. Secondly, we sought to identify functionally related genes, gene regulatory networks and biological pathways enriched for SNPs associated with increased risk of developing subtypes of breast cancer. We hypothesized that molecular perturbations in genes containing SNPs associated with increased risk of developing breast cancer differ between subtypes and benign controls, as well as between individual tumor subtypes. We further hypothesized that genes containing SNPs associated with an increased risk of developing breast cancer are functionally related and interact with each other in complex gene regulatory networks and biological pathways. We have tested these hypotheses using an integrative genomics approach, which combines GWAS information with publicly available gene expression data on six intrinsic subtypes of breast cancer. Throughout this report, we have used the terms SNP and genetic variant interchangeably and we have also assumed the gene as the unit of association. This holistic approach was undertaken to understand the broader context in which genetic variants operate, leading to different subtypes of breast cancer.
Methods
Source of SNP data
The methods for GWAS data collection were based on the guidelines proposed by the Human Genome Epidemiology Network for systematic review of genetic association studies and follow the preferred reporting items for systematic reviews and meta-analysis (PRISMA).9–13 We mined SNP data and gene information from the published reports on GWAS for breast cancer. GWAS were eligible to be included if they met the following criteria: First, publications must have been from peer-reviewed journals, in print or online and published in English before October 2012. Second, the study design must have been a case-control, cohort or cross-sectional association study conducted using human populations. Third, cancers must have been diagnosed by histological examination. In addition, studies were eligible if they were based on unrelated individuals, examined the association between breast cancer and the polymorphic phenotype and had a sample size of greater than 500 in the cases and greater than 500 in the controls. Only studies published as full-length articles or letters in peer-reviewed journals in English were included in the analysis. In addition, the study must have provided sufficient information such that genotype frequencies for both breast cancer cases and controls could be determined without ambiguity.
To identify all relevant publications, we used two search strategies. First, we queried PubMed with the terms GWAS, GWA, WGAS, WGA, genome-wide, genomewide, whole genome, and all terms plus association or scan in combination with breast cancer, to find all the GWAS published before October 2012. This search yielded publications that were screened by title, abstract and full text review to identify studies that met our eligibility criteria. The data was manually extracted from reported GWAS that met our eligibility criteria. To obtain additional detailed information about these studies, we searched the websites containing supplementary data on the studies that met our eligibility criteria. The search yielded 500 SNPs mapped to 203 genes from a population of over 450,000 cases and over 450,000 controls. A list of publications which met our eligibility criteria along with genetic variants and associated genes is presented in Table A, provided as supplementary data to this report.
To address publication bias, we catalogued all of the available SNPs that showed significant (P < 0.05) associations with an increased risk of developing breast cancer. The rationale for including all significant SNPs is that relatively few SNPs have “strong” evidence of association (ie, P-values being sufficiently small enough, P < 10−8 to give conclusive evidence of association). Generally, there are several hundred SNPs with moderate (P ~ 10−5 to 10−7) or weak evidence of association (P ~ 10−3 to 10−4). While some of the genetic variants would likely be false-positives, it is conceivable that others may contain genuine effects of small magnitude. We reasoned that the presence of a number of associated SNPs mapped to genes with similar biological functions interacting in gene regulatory networks and multi-gene biological pathways gives a degree of confidence that the associations may be genuine, even if none of the SNPs are individually highly significant. The SNP, IDs (rs-ID), locations and gene names were verified using the database of genetic variation (dbSNP) http://www.ncbi.nlm.nih.gov/projects/SNP/database with chromosome report build 37.7 and the Human Genome Nomenclature (HGNC) database. SNPs were matched with gene names using SNP IDs (rs-IDs) information in the database (dbSNP). For SNPs replicated in multiple independent studies, we combined the P-values to estimate the overall effect size using Fisher’s methods as described in our previous study.1
Characteristics of gene expression data
The goal of this study was to use gene expression data as a first step in linking GWAS information with the breast cancer intermediate phenotypes. In clinical practice, tumors are routinely classified according to their expression of estrogen receptor alpha (ERα), progesterone receptor A (PR), and HER2/nue to guide treatment. Gene expression has been used as the standard for classifying breast tumors into intrinsic subtypes.3,4 Using this standard, we used 6 intrinsic subtypes, treating each subtype as a distinct disease entity. The 6 subtypes included luminal A, luminal B, ERBB-2, normal-like, basal and basal-like. We further subdivided the 6 subtypes into 2 subgroups based on response to treatment. Subgroup 1 included tumors responsive to targeted therapy. This group included the subtypes luminal A, luminal B and ERBB-2. Subgroup 2 included the more aggressive subtypes of breast cancer which are treated primarily with cytotoxic chemotherapy, commonly known as triple-negative breast cancers (TNBC).14 TNBC are often classified as basal-like or basaloid breast cancers if they demonstrate expression of basal-like cytokeratins.14 Basal-like breast cancer represents 10%–25% of all tumors, depending on the demographics of the population, and make up about 50%–75% of the TNBC subset.14 However, they can less commonly fall into other intrinsic subtypes including the normal-like and the basal types.14 For this reason, the TNBC subgroup in this study included normal-like, basal and basal-like. Recently, 6 subtypes of TNBC were reported, but these subtypes have not been replicated.15 We did not include the more recently identified TNBC subtype, the Claudin-low subtype,14 because we did not find a suitable data set to match the other subtypes. We acknowledge this weakness in our investigation.
Gene expression data consisted of 429 samples distributed as follows: Luminal A (N = 89), Luminal B (N = 49), ERBB-2 (N = 24), Normal-like (N = 29), Basal (N = 75) and basal-like (N = 20) and cancer-free controls (N = 143). These sample sizes were sufficiently large to identify the significant differential expression at P < 0.05 with a statistical power of 99%. All gene expression data was derived from populations of European ancestry to reflect the populations used in GWAS studies. All samples were assessed for global gene expression profiles using the Affymetrix platform on U133PLUS 2.0 Human GeneChip. The microarray data from these samples, including the raw probe-level hybridization intensities, were downloaded from the NCBI’s Gene Expression Omnibus (GEO) database16 under accession numbers GSE2990 and GSE17705 for cancer and control groups, respectively.17,18 Methods of sample collection, preparation and processing have been fully described by the data originators.17,18 For each data set, the entries in the data matrix were expression values generated by Affymetrix’s Microarray platform normalized using the RMA suite on a log scale (log2). We preprocessed the data to remove spiked control genes.
Data analysis
To obtain a more robust analysis on the gene expression data, we performed both supervised and unsupervised analysis followed by network and pathway analysis and visualization. First we compared gene expression profiles between each subtype of breast cancer and cancer-free controls. In this analysis, each subtype of breast cancer was treated as a distinct disease entity. This approach was based on the commonly accepted theory that different subtypes of breast cancer originate from different cellular populations (eg, bipotent mammary stem cells, luminal precursors, myoepitherial precursors), and thus present a distinct pathological process. The significant differences in gene expression profiles of SNP-containing genes between each subtype of breast cancer and cancer-free controls were tested using a t-test. This approach eliminated SNP-containing genes that were not associated with any subtype of breast cancer and narrowed the focus, highlighting the set of genes that were highly significantly associated with each subtype of breast cancer. To assess variability and differences in gene expression profiles among all the six subtypes of breast cancer, we performed an analysis of variance (ANOVA).
Second, we performed an analysis comparing gene expression profiles between the 2 clinically defined subgroups to determine whether SNP-containing genes significantly differ in their expression profiles between the subtypes responsive to targeted therapy and those responsive to chemotherapy (TNBC). We performed additional analyses comparing gene expression profiles of SNP-containing genes between and among the subtypes of breast cancer within each clinically defined subgroup of breast cancer using a t-test and an ANOVA, respectively. We used permutation tests to calculate empirical P-values. The empirical P-values and those from the t-test (ANOVA) did not differ appreciably. We used a false discovery rate (FDR) to correct for multiple hypotheses testing.19 Due to small sample sizes for some subtypes of breast cancer, we did not divide the data into test and validation sets; instead, we used an out-of-sample validation approach to identify genes with predictive power.20 For each analysis conducted, genes were ranked based on estimated P-values and FDR. Those showing highly significant differences in expression profiles were selected. Supervised analysis was performed using Pomello II and GenePattern software packages.21,22
To determine whether SNP-containing genes have similar patterns of expression profiles and are functionally related, we performed unsupervised analysis based on hierarchal clustering using the complete linkage method and the Pearson correlation coefficient as the measure of distance between pairs of genes. First, we performed an unbiased screen by subjecting all the SNP-containing genes to hierarchical clustering. This analysis produced spurious and overlapping patterns of gene expressions. To address this problem, we next performed subclass mapping focusing on genes that were highly significantly associated with each subtype of breast cancer. This analysis was then recapitulated using all 6 breast cancer subtypes. Gene expression data was normalized using median normalization. The data were standardized and centered prior to clustering.23 Hierarchical clustering was performed using GenePattern.22
To further assess functional relationships among SNP-containing genes, we performed additional analyses using the gene ontology (GO) information.24 The GO Consortium has developed 3 separate categories including molecular function, biological process and cellular component, to describe the attributes of gene products. Molecular function defines what a gene product does at the biochemical level without specifying where or when the event actually occurs or its broader context. Biological process describes the contribution of the gene product to the biological objective. Cellular component refers to where in the cell a gene product functions. Because our goal in this study was to understand the broader context in which genetic variants associated with increased risk of developing the subtypes of breast cancer operate, we considered all 3 GO categories.
Finally, we performed pathway prediction, network modeling and visualization using the Ingenuity pathway analysis (IPA) program (http://www.ingenuity.com).25 The goal was to identify gene regulatory networks and biological pathways that are enriched for genetic variants associated with an increased risk of developing breast cancer. We hypothesized that genes containing SNPs associated with an increased risk for developing different subtypes of breast cancer interact with each other and other genes within biological pathways enriched for genetic variants. Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) identifiers were mapped onto networks and pathways available in the Ingenuity System database, which were ranked by score. The score indicates the likelihood of the genes in a network being found together by random chance. Using a 99% confidence interval, scores of ≥3 are considered significant. Additional information, validation of predicted pathways and identification of other downstream target genes was achieved through the literature and database-mining module built in the Ingenuity System, which allowed identification of other functionally related genes that were not identified by GWAS. The distribution of the overall effect of SNPs in the pathway was calculated using the procedure we have previously reported.1 Genes showing spurious interactions were pruned from the networks to ensure reliability of the identified networks.
Results
Assessment of evidence and credibility of associations
We mined the literature and associated websites on GWAS to identify genetic variants and genes associated with an increased risk of developing breast cancer. Evidence and credibility of associated loci were assessed at three levels which included the amount and level of evidence as determined by the SNP association P-value, the extent of replication, and protection from bias.9 The level of evidence was further assessed as strong (P < 10−8), moderate (P ~ 10−5–10−7) and weak (P ~ 10−3–10−4) associations along with replication. We identified 500 SNPs mapped to 203 genes associated with an increased risk of developing breast cancer. The results showing all 500 SNPs and associated P-values, the genes and chromosome positions they map to, along with references indicating sources of GWAS information are presented in Table A, provided as supplementary data. Out of the total number of SNPs identified, 45 SNPs had strong associations (P < 10−8). In addition, 46 SNPs were replicated in multiple independent studies (Table A). The remaining SNPs had small to moderate effects (P ~ 10−3–10−7; Table A).
Genetic susceptibility to breast cancer varied markedly, reflecting the heterogeneity inherent in breast cancer and suggesting potential functional diversity of identified loci. Among the identified genetic variants and associted genes were rs2046210(ESR1), rs12662670(ESR1), rs3803662(TOX3) and rs999737(RAD51L1), which have been associated with TNBC.26,27 In addition, we identified genetic variants and genes rs1045485(CASP8), rs17468277(CASP8) and rs1982073(TGFB1), which have been associated with progesterone receptor negative tumors28 and SNPs mapped to genes FGFR2 and TNRC9 which have been shown to have stronger associations for estrogen receptor positive than estrogen receptor negative tumors.28 Further evaluations of the genetic variants and genes using accumulated literature information revealed genetic variants mapped to genes FGFR2, TOX3, LSP1, MAP3K1, TGFB1 and ESR1 which have been associated with both ER-positive and ER-negative breast cancers.6 This suggests that functional diversity may exist across associated loci. For example, SNPs in ESR1 may result in loss of expression (thereby producing an ER− tumor) or related altered function with retained expression (hence producing an ER+ tumor). These results may also reflect the biological origins of the subtypes of breast cancer, and suggest that tumor stratification might help in the identification and characterization of novel risk factors for breast cancer subtypes. The overwhelming large number of genetic variants with small to moderate associations (or small to moderate effect sizes) suggests that the functional effects of identified genetic variants are likely to be subtle. However, as demonstrated later in this report and supported by the literature,29 the presence of associated SNPs mapped to functionally related genes interacting in networks and pathways gives a degree of confidence that the associations may be genuine even if none of the SNPs individually are highly significant.
Association of SNP-containing genes with subtypes of breast cancer
To determine whether SNP-containing genes are associated with individual subtypes of breast cancer, we compared gene expression levels between groups with each subtype of breast cancer and the control group as explained in the Methods section. The results showing estimates of P-values along with FDR for all the 203 genes containing SNPs associated with increased risk of developing breast cancer for each subtype of breast cancer are presented in Table B, provided as supplementary data. We identified 201 SNP-containing genes that are significantly (P < 0.05) associated with the subtypes of breast cancer. A comparison of gene expression values in breast cancer patients diagnosed as luminal A, luminal B, ERBB2, normal-like, basal and basal-like to cancer-free control subjects identified 181, 171, 162, 149, 164 and 178 significantly (P < 0.05) differentially-expressed genes, respectively (Table B). These results confirm that SNP-containing genes are associated with intrinsic subtypes of breast cancer, and that their expression profiles vary by tumor characteristics. However, there was considerable overlap in associations between the subtypes of breast cancer. The overlap in gene expression levels is consistent with the composition of the breast cancer subtypes.14 Further analysis of gene expression among the six intrinsic subtypes of breast cancer using an ANOVA produced 197 significantly differentially expressed genes (Table B).
In GWAS, evidence and the credibility of association are usually assessed by the strength of the statistical association as determined by the P-value (P < 10−8) and replication in multiple independent studies.30,31 Using these criteria, we evaluated the genes containing SNPs with strong association and SNPs replicated in multiple independent studies. The results of genes containing SNPs with strong association are presented in Table 1. The results of genes containing SNPs replicated in multiple independent studies are shown in Table 2. We identified 23 genes containing SNPs with strong evidence of associations (P < 10−8), which were significantly associated with individual subtypes of breast cancer (Table 1). In addition, we identified 42 genes containing SNPs replicated in multiple independent studies that were significantly associated with individual subtypes of breast cancer (Table 2). These results further confirm our hypothesis that genes containing SNPs associated with increased risk of developing breast cancer are strongly associated with intrinsic subtypes of breast cancer, and that this variation significantly varies across tumor subtypes (see Tables 1 and 2). A complete list of estimates of P-values for genes containing SNPs with strong associations and genes containing SNPs replicated in multiple independent studies for each subtype of breast is presented in Table B provided as supplementary data.
Table 1.
Estimates of P-values in different subtypes of breast cancer for genes containing SNPs with strong associations (P < 10−7) estimated from GWAS.
| Gene symbol | SNP ID (rs-ID) | SNP (PV) | Expression P-value | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| LMA | LMB | ERBB | NLK | BASAL | BLK | |||
| ANKLE1 | rs8170 | 2 × 10−9 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 0.267116 | 5.00 × 10−6 |
| ANKLE1 | rs2363956 | 6 × 10−9 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 0.267116 | 5.00 × 10−6 |
| ANKRD16 | rs2380205 | 5 × 10−7 | 5.00 × 10−6 | 0.046552 | 0.885408 | 0.456038 | 0.00151 | 5.00 × 10−6 |
| BRCA1 | rs9397435 | 1.3 × 10−8 | 5.00 × 10−6 | 0.318183 | 0.034513 | 5.00 × 10−6 | 5.00 × 10−6 | 0.022438 |
| BRCA1 | rs2046210 | 4.5 × 10−9 | 5.00 × 10−6 | 0.318183 | 0.034513 | 5.00 × 10−6 | 5.00 × 10−6 | 0.022438 |
| CASP8 | rs1045485 | 1.1 × 10−7 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| CCND1 | rs614367 | 3 × 10−15 | 5.45 × 10−4 | 5.00 × 10−6 | 2.50 × 10−5 | 0.096103 | 0.626906 | 5.00 × 10−6 |
| CDKN2B | rs1011970 | 3 × 10−8 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| CHEK2 | rs17879961 | 4.76 × 10−8 | 2.00E−05 | 0.024433 | 0.018789 | 5.00 × 10−6 | 0.197886 | 5.00 × 10−6 |
| COL1A1 | rs2075555 | 8.3 × 10−8 | 5.00 × 10−6 | 0.209955 | 0.068825 | 0.07671 | 0.275921 | 0.397922 |
| ECHDC1 | rs6569480 | 6.1 × 10−8 | 5.00 × 10−6 | 5.00 × 10−6 | 8.5 × 10−3 | 9.5 × 10−5 | 3.5 × 10−5 | 0.002005 |
| ECHDC1 | rs7776136 | 6.6 × 10−8 | 5.00 × 10−6 | 5.00 × 10−6 | 0.008539 | 9.5 × 10−5 | 3.5 × 10−5 | 0.002005 |
| FGFR2 | rs2981575 | 1.2 × 10−8 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.15 × 10−4 | 0.982236 | 5.00 × 10−6 |
| FGFR2 | rs2981582 | 2.0 × 10−78 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.15 × 10−4 | 0.982236 | 5.00 × 10−6 |
| FGFR2 | rs2981579 | 1.8 × 10−31 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.15 × 10−4 | 0.982236 | 5.00 × 10−6 |
| LOC643714 | rs3803662 | 1.0 × 10−36 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| LSP1 | rs3817198 | 3.0 × 10−9 | 0.100053 | 6.8 × 10−3 | 0.676883 | 0.06848 | 0.597843 | 0.134961 |
| LSP1 | rs909116 | 7.3 × 10−7 | 0.100053 | 6.8 × 10−3 | 0.676883 | 0.06848 | 0.597843 | 0.134961 |
| NEK10 | rs1357245 | 1.9 × 10−7 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| RAD51L1 | rs999737 | 1.7 × 10−7 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 0.025663 | 5.00 × 10−6 |
| RNF146 | rs6569479 | 1.2 × 10−7 | 0.212725 | 0.029158 | 0.529813 | 0.766361 | 6.59 × 10−3 | 0.976052 |
| RNF146 | rs2180341 | 2.9 × 10−8 | 0.212725 | 0.029158 | 0.529813 | 0.766361 | 6.59 × 10−3 | 0.976052 |
| SLC4A7 | rs4973768 | 4.0 × 10−23 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| STXBP4 | rs6504950 | 1.4 × 10−8 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 1.65 × 10−4 | 5.00 × 10−6 |
| TERT | rs10069690 | 1.0 × 10−10 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| TOX3 | rs12443621 | 2.0 × 10−19 | 5.00 × 10−6 | 2.6 × 10−4 | 8.65 × 10−4 | 0.419581 | 0.090054 | 5.00 × 10−6 |
| TOX3 | rs3803662 | 5.9 × 10−19 | 5.00 × 10−6 | 2.6 × 10−4 | 8.65 × 10−4 | 0.419581 | 0.090054 | 5.00 × 10−6 |
| TOX3 | rs8051542 | 1.0 × 10−36 | 5.00 × 10−6 | 2.6 × 10−4 | 8.65 × 10−4 | 0.419581 | 0.090054 | 5.00 × 10−6 |
| ZMIZ1 | rs704010 | 4 × 10−9 | 5.00 × 10−6 | 1.00 × 10−5 | 0.022029 | 0.048337 | 5.00 × 10−6 | 0.227309 |
| ZNF365 | rs10822013 | 5.87 × 10−9 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| ZNF365 | rs10995190 | 5 × 10−15 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 | 5.00 × 10−6 |
| H19 | rs2107425 | 2.0 × 10−7 | 0.042847 | 1.14 × 10−3 | 2.58 × 10−3 | 0.027343 | 8 × 10−5 | 9.3 × 10−3 |
| MAP3K1 | rs889312 | 4.6 × 10−20 | 2.7 × 10−3 | 5.00 × 10−6 | 3.5 × 10−4 | 4.9 × 10−4 | 9.3 × 10−3 | 5.00 × 10−6 |
Note: rs-ID is the SNP id, SNP(Pv) is the SNP P-value derived from GWAS.
Abbreviations: LMA, luminal A; LMB, luminal B; ERBB, ; NLK, normal-like, BLK, basal-like.
Table 2.
Estimates of P-values in different subtypes of breast cancer for genes containing SNPs replicated in multiple independent studies obtained from GWAS.
| Gene symbol | SNP ID (rs-ID) | Number of studies | LMA | LMB | ERBB | NLK | BASAL | BLK |
|---|---|---|---|---|---|---|---|---|
| CASP8 | rs1045485 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020314 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020390 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020394 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs1884051 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs2228480 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020396 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020400 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020401 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3798577 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.18 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| FGFR2 | rs2981582 | 9 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5.15 × 10−4 | 0.982236 | 5 × 10−6 |
| FGFR2 | rs2981579 | 7 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5.15 × 10−4 | 0.982236 | 5 × 10−6 |
| FGFR2 | rs2420946 | 5 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5.15 × 10−4 | 0.982236 | 5 × 10−6 |
| FGFR2 | rs1219648 | 5 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5.15 × 10−4 | 0.982236 | 5 × 10−6 |
| LSP1 | rs3817198 | 6 | 0.100053 | 6.8 × 10−3 | 0.676883 | 0.06848 | 0.597843 | 0.134961 |
| STXBP4 | rs6504950 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 1.65 × 10−4 | 5 × 10−6 |
| TGFB1 | rs1800470 | 3 | 5 × 10−6 | 3.5 × 10−5 | 2.5 × 10−5 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| TOX3 | rs12443621 | 3 | 5 × 10−6 | 2.6 × 10−4 | 8.6 × 10−4 | 0.419581 | 0.090054 | 5 × 10−6 |
| ADH1B | rs1042026 | 3 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| SORBS1 | rs10450393 | 2 | 5 × 10−6 | 5.7 × 10−3 | 2.5 × 10−4 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| ICAM5 | rs1056538 | 2 | 5 × 10−6 | 0.387298 | 0.189892 | 0.045497 | 5 × 10−6 | 1.72 × 10−3 |
| RB1 | rs198580 | 2 | 0.084304 | 2.7 × 10−4 | 0.168073 | 3.5 × 10−3 | 0.681552 | 1 × 10−5 |
| RNF146 | rs2180341 | 2 | 0.212725 | 0.029158 | 0.529813 | 0.766361 | 0.00659 | 0.976052 |
| RB1 | rs2854344 | 2 | 0.084304 | 2.75 × 10−4 | 0.168073 | 3.5 × 10−3 | 0.681552 | 1 × 10−5 |
| IGFBP3 | rs2854744 | 4 | 0.103283 | 0.058821 | 0.919706 | 0.067505 | 0.864765 | 2.24 × 10−3 |
| CDKN1A | rs3176336 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| CDKN1B | rs34330 | 2 | 0.107648 | 0.795089 | 3.25 × 10−4 | 2.2 × 10−3 | 5.15 × 10−4 | 3 × 10−5 |
| CDKN2A | rs3731239 | 2 | 0.28172 | 0.154304 | 0.196626 | 0.602623 | 0.039622 | 5 × 10−6 |
| LOC643714 | rs3803662 | 12 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| EHMT1 | rs4634736 | 2 | 5 × 10−6 | 0.007025 | 0.13664 | 1 × 10−5 | 0.035818 | 5 × 10−6 |
| SOD2 | rs4880 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 0.032173 | 5 × 10−6 |
| CCND1 | rs678653 | 2 | 5.45 × 10−4 | 5 × 10−6 | 2.5 × 10−5 | 0.096103 | 0.626906 | 5 × 10−6 |
| HCN1 | rs981782 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| CCNE1 | rs997669 | 2 | 0.463623 | 5 × 10−6 | 2.5 × 10−5 | 0.440789 | 0.00761 | 5 × 10−6 |
| RAD51L1 | rs999737 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 0.025663 | 5 × 10−6 |
| CDKN2B | rs1011970 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| CASP8 | rs10931936 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| ZNF365 | rs10995190 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| FGFR2 | rs11200014 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5.15 × 10−4 | 0.982236 | 5 × 10−6 |
| COX11 | rs1156287 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 3.7 × 10−4 | 5 × 10−6 |
| WRN | rs1346044 | 2 | 0.0167838 | 0.64972 | 0.894917 | 0.433395 | 0.00761 | 5 × 10−6 |
| GSTP1 | rs1695 | 3 | 5 × 10−6 | 5 × 10−6 | 2.75 × 10−3 | 0.082619 | 5 × 10−6 | 5 × 10−6 |
| RELN | rs17157903 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| CHEK2 | rs17879961 | 2 | 2 × 10−5 | 0.024433 | 0.018789 | 5 × 10−6 | 0.197886 | 5 × 10−6 |
| CDKN1A | rs1801270 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| H19 | rs2107425 | 2 | 0.042847 | 0.00114 | 2.58 × 10−3 | 0.027343 | 8 × 10−5 | 5 × 10−6 |
| IFNG | rs2430561 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| ESR1 | rs3020377 | 2 | 5 × 10−6 | 3 × 10−5 | 5 × 10−6 | 3.1 × 10−3 | 5 × 10−6 | 5 × 10−6 |
| COMT | rs4818 | 2 | 8.4 × 10−3 | 0.860185 | 5.27 × 10−3 | 7.13 × 10−3 | 0.304889 | 0.240923 |
| SLC4A7 | rs4973768 | 4 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 |
| RNF146 | rs6569479 | 2 | 0.2127251 | 0.029158 | 0.529813 | 0.766361 | 6.59 × 10−3 | 0.976052 |
| ZMIZ1 | rs704010 | 2 | 5 × 10−6 | 1 × 10−5 | 0.022029 | 0.048337 | 5 × 10−6 | 0.227309 |
| VDR | rs731236 | 2 | 2.2 × 10−3 | 0.022014 | 0.159809 | 0.476042 | 0.624336 | 9.16 × 10−3 |
| RAD51L1 | rs8009944 | 2 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 5 × 10−6 | 0.025663 | 5 × 10−6 |
| TOX3 | rs8051542 | 4 | 5 × 10−6 | 2.6 × 10−4 | 8.6 × 10−4 | 0.419581 | 0.090054 | 5 × 10−6 |
| MAP3K1 | rs889312 | 4 | 2.7 × 10−3 | 5 × 10−6 | 3.5 × 10−5 | 4.9 × 10−4 | 9.3 × 10−3 | 5 × 10−6 |
Notes: rs-ID is the SNP id, SNP(Pv) is the SNP P-value derived from GWAS. Due to the large number of studies in column 3, the SNP P-values are provided in Table A as supplementary data.
Abbreviations: LMA, luminal A; LMB, luminal B; ERBB, ; NLK, normal-like; BLK, basal-like.
Interestingly, among the genes exhibiting strong associations with subtypes of breast cancer included the genes containing SNPs TOX3(rs3803662), RAD51L1 (rs999737), ESR1(rs2046210), CASP8 (rs17468277) and ANKLE1 (rs8170, rs8100241) associated with increased risk of developing the TNBC subtypes.26,27 Another set of SNP-containing genes found to be associated with subtypes of breast cancer in this study included the genes P53, PTEN, RB1, BRCA1, BRCA2, ATR, ATM, MAP3K1, CDKN2A, ATR, CHEK1, CCND1 and NOTCH2. These genes were found to be frequently mutated in breast cancer.32
One of the major concerns with GWAS is that the credible genetic variants (P < 10−8) explain only a proportion of the phenotypic variation. This has raised questions of whether there are many more DNA variants with smaller effects that are not being reliably identified in GWAS because of limited statistical power. To address this problem, we evaluated the association of genes containing SNPs with small to moderate effects (P ~ 10−3–10−7) with the subtypes of breast cancer. We reasoned that such associations offer a thread of evidence from which to build functional validation, although they do not necessarily mean that the genes cause the clinical phenotype. We identified many genes containing SNPs with small to moderate effect sizes that were significantly associated with different subtypes of breast cancer (Table B, Supplementary data). This is a significant finding given that only a small number of statistically unimpeachable, common low-penetrance breast cancer susceptibility loci have been reported and confirmed in different breast cancer subtypes.5–7
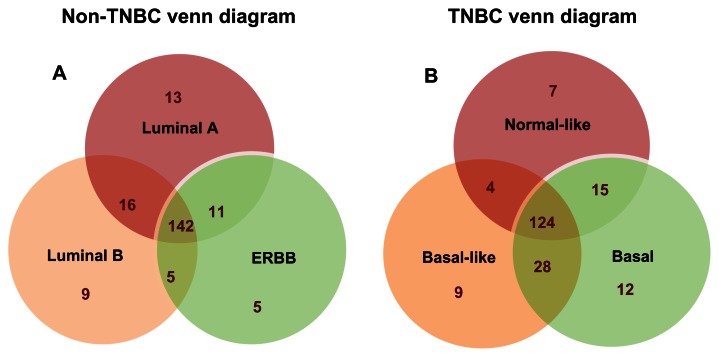
Although each of the SNP-containing genes analyzed in this report showed independent associations with intrinsic subtypes of breast cancer, there was considerable overlap in associations. Therefore, to discern the degree of overlap in association, we used a Venn diagram delineating three subtypes of breast within each clinically defined subgroup. The results showing SNP-containing genes exhibiting subtype-specific and overlapping associations within each subgroup of breast cancer are presented in Figure 1. Within the subgroup responsive to targeted therapy, 142 genes exhibited overlapping associations across all the three subtypes (Fig. 1A), whereas 124 genes exhibited overlapping associations in the TNBC subtypes (Fig. 1B), indicating that TNBC is more heterogeneous than the other subgroup, which is consistent with the literature reports.14,15 These results suggest that molecular subtyping may be necessary to identify subtype-specific genetic risk factors.
Figure 1.
Overlap in association of SNP-containing genes with the intrinsic subtypes of breast cancer within each clinically defined subgroup. Group (A) depicts the subgroup comprising the three subtypes responsive to targeted therapy. Group (B) represents the subgroup responsive to chemotherapy (ie, TNBC subtypes).
To discern the degree of variability in gene expression levels within each subgroup of breast cancer, we performed an ANOVA. We performed an additional analysis to determine whether gene expression profiles significantly differ between subtypes of breast cancer within each molecularly defined subgroup. The results showing estimates of P-values and FDR based on the ANOVA and t-tests are presented in Table C, provided as supplementary data. Comparison of gene expression profiles between the clinical breast cancer subgroups produced 143 significantly (P < 0.05) differentially expressed genes (Table C). Within the subgroup of breast cancer responding to targeted therapy, the ANOVA produced 111 genes that exhibited significant variation (Table C). Additional analysis comparing gene expression between luminal A and luminal B, between luminal A and ERBB-2 and between luminal B and ERBB-2 produced 84, 72, and 72 highly significantly differentially expressed genes (Table C). Within the TNBC subgroup ANOVA produced 180 genes that exhibited significant variations among the three subtypes studied (Table C). A comparison of gene expression between normal-like and basal-like, normal-like and basal, and basal-like and basal produced, 156, 117, and 133 significantly differentially expressed genes, respectively (Table C). These results confirmed our hypothesis that gene expression levels of SNP-containing genes significantly vary in subtypes of breast cancer within each clinically defined group, and that gene expression levels in TNBC subtypes vary more than in the subtypes responsive to targeted therapy.
Patterns of gene expression profiles for SNP-containing genes
Our second goal in this study was to understand the broader context in which genes containing SNPs associated with increased risk of developing breast cancer operate in different breast cancer subtypes. We hypothesized that SNP-containing genes have similar patterns of expression profiles and are functionally related. The rationale is that genes with similar patterns of expression that are functionally related are not only likely to be regulated via the same molecular mechanisms, but are also more likely to have their promoter regions bound by common transcription factors.33 As a first step, we subjected all the 203 genes to unsupervised hierarchical clustering. This analysis produced spurious and overlapping patterns of gene expression profiles (results not presented). For these reasons, we performed a further pattern recognition analysis using subclass mapping, focusing on SNP-containing genes that were highly significantly (P < 10−6) associated with each subtype of breast cancer.
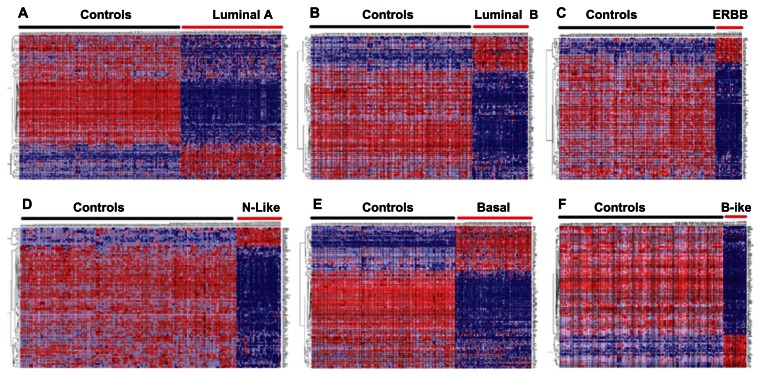
The results based on subclass mapping are presented in Figure 2. We identified functionally-related genes with similar patterns of expression profiles. Interestingly, genes containing SNPs with strong evidence of association (Table 1) and genes containing SNPs replicated in multiple independent studies (Table 2) were found to be co-expressed and their expression profiles contained similar patterns (Fig. 2). This is a significant finding given that genetic variants reported thus far explain only a small proportion of the phenotypic variation and that some genetic variants have not been replicated, but map to genes co-regulated with those containing genetic variants replicated in multiple independent studies. Most notably, the results suggest that the role of SNP-containing genes as potential biomarkers may largely depend on their collective actions, discernible through functional co-regulation. The results also suggest that when it comes to insights into disease pathogenesis, locus effect size may be almost immaterial, because even loci with modest effects, once confirmed as genuine, can reveal novel causal mechanisms.34 Overall, these results demonstrate that the strategy of applying the “genetics of gene expression” approach offers an appealing and straightforward way of initiating the complicated task of connecting risk variants to their target genes and phenotypes of intrinsic subtypes of breast cancer.
Figure 2.
Subclass mapping of gene expression signatures for the 6 intrinsic subtypes of breast cancer relative to the control subjects.
Notes: Genes are shown in rows and samples in columns. Red indicates up regulated and blue indicates down regulated. Association with each subtype of breast cancer was determined at P < 10−6. The controls are indicated by a black bar on top of each figure. Similarly, the subtypes are indicated by a red bar on top of each figure and include luminal A, luminal B, ERBB, normal-like (N-Like), Basal and basal-like (B-Like).
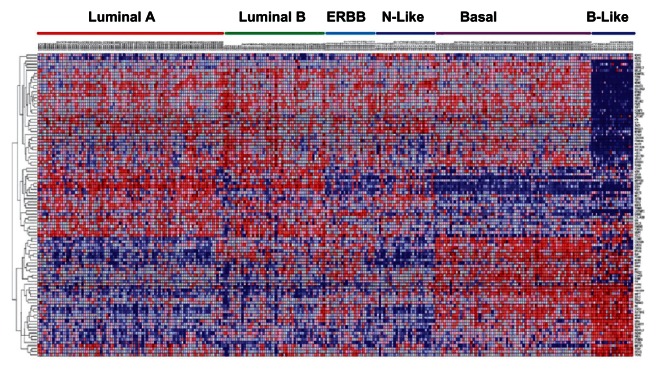
In order to assess similarity in patterns of gene expression profiles in both the subtypes responsive to targeted therapy and the TNBC subtypes, we performed an unsupervised analysis focusing on the 143 genes that exhibited significant differences in expression levels between the two subgroups of breast cancer. The results of this analysis are presented in Figure 3. Luminal A, luminal B, ERBB and normal-like had similar patterns of expression profiles and clustered together (Fig. 3). Basal-like exhibited distinct patterns of expression from the other subtypes (Fig. 3). Non-luminal basal tended to cluster with both subgroups indicating that this group is the most heterogeneous (Fig. 3). These results are consistent with literature reports.14
Figure 3.
Patterns of gene expression profiles in 6 intrinsic subtypes of breast cancer.
Notes: Analysis if based on 147 SNP-containing genes with the strongest association (P < 10−6) with individual subtypes (those responsive to targeted therapy which include: luminal A, luminal B, ERBB, normal-like) and TNBC subtypes (basal and basal-like). The color bars on top of the figure indicate individual subtypes. The rows and columns indicate genes and patients; respectively. Red and blue in the hit map indicate up and down regulation; respectively.
To gain further insights about the biology and the functional relationships of genes containing SNPs associated with the subtypes of breast cancer, we performed a GO analysis as explained in the methods section in this report. The GO analysis revealed that genes containing SNPs associated with increased risk of developing subtypes of breast cancer are functionally related (Table D supplementary data). The results of the GO analysis further revealed that SNP-containing genes are involved in multiple overlapping molecular functions, biological processes and cellular components, suggesting that genetic susceptibility to breast cancer subtypes may involve many genes acting together to produce the phenotypes (Table D).
Network and pathway analysis
Identifying genetic variants that are associated with breast cancer, as well as intermediate molecular phenotypes that respond more proximally to these genetic variants and in turn cause disease, are excellent first steps to uncovering the drivers of breast cancer subtypes. However, the emerging view from large-scale genomic studies is that breast cancer subtypes are emergent properties of networks and biological pathways whose states are affected by complex interactions of genetic and environmental factors, each contributing a small effect.1,8,35 Therefore, to understand the broader context in which genetic variants operate, genetic variants and associated genes must be understood in the context of molecular networks and biological pathways that define the disease states. Based on this reasoning, we performed network and pathway analysis to identify gene regulatory networks and biological pathways enriched for genetic variants associated with breast cancer subtypes. We hypothesized that genes containing SNPs associated with an increased risk of developing subtypes of breast cancer and their downstream targets interact with each other in gene regulatory networks and biological pathways. The rationale is that through these complex arrays of interacting genes the genetic variants affect entire network states and biological pathways that in turn increase the risk of developing a subtype of breast cancer or affect the severity of the disease.
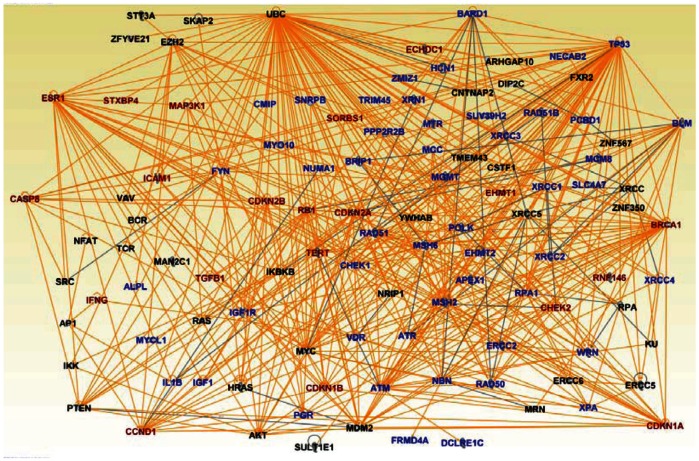
Network analysis produced five multi-gene networks with scores ranging from 20 to 47. These networks were enriched for SNPs, confirming our hypothesis. We consolidated the networks into one large network using the design and overlay features implemented in Ingenuity IPA. The results of consolidated network analysis are presented in Figure 4. The network was pruned to remove genes showing spurious interactions to ensure the reliability of the networks. In the network the nodes represent SNP-containing genes and vertices represent interactions. Network analysis revealed that genes containing SNPs associated with an increased risk of developing subtypes of breast cancer interact with each and their downstream targets in gene regulatory networks confirming our hypothesis (Fig. 4). Interestingly, network analysis also revealed novel genes not reported in GWAS (Fig. 4).
Figure 4.
Graphical representation of gene regulatory networks enriched for SNP-containing genes.
Notes: The nodes indicate the genes represented by gene symbols and the vertices represent the interactions or functional relationships. The gene symbols in dark red fonts represent genes containing SNPs with strong statistical associations with increased risk of developing breast cancer. The gene symbols in blue font represent the genes containing SNPs replicated in multiple independent studies, some of which have low to moderate association. The gene symbols in black font represent novel genes that have not been reported in GWAS.
Of particular interest, was the revelation through network analysis that genes containing SNPs with strong associations and SNPs replicated in multiple independent studies were found to interact with each other and with genes containing SNPs with weak to moderate associations (Fig. 4). This is a significant finding given that relatively fewer SNPs have P-values that are sufficiently small or that are replicated in multiple independent studies to give conclusive evidence of association. The results demonstrate that SNP-containing genes, regardless of effect size, tend to act in concert to produce the breast cancer phenotype. The identification of many genes interacting in complex gene regulatory networks suggests that the great majority of breast cancer cases may not be associated with only the mutated genes with high penetrance such as BRCA1, BRCA2, PTEN and P53. SNP-containing genes of moderate penetrance (ATM, BRIP1, CHEK2, PALB2, RAD50) and low penetrance (FGFR2, LSP1, MAP3K1, TGFB1, TOX3) frequently mutated in the general population may play an important role in the pathogenesis of breast cancer.
To gain biological insights about the functional relationship of the genes in the networks, we used the Ingenuity system to classify genes according to molecular and cellular functions. Network analysis revealed functional relationships among the SNP-containing genes and novel genes. Many of the identified genes have multiple overlapping functions and are involved in a multitude of biological processes and cellular components. About 81 genes were highly significantly (8.84E-40-1.05E-09) associated with DNA replication, recombination, and repair. Another set of 111 genes were highly significantly (4.88E-30-1.13E-09) associated with cell death and survival, whereas 107 genes were highly significantly (1.83E-26-1.10E-09) associated with cell growth and proliferation. Further examination or the results revealed 80 genes highly significantly (2.01E-26-1.14E-09) associated with cell cycle and 104 genes highly significantly (3.01E-26-1.10E-09) associated with cellular development. The identified genes included TP53, E2F1, JUN and ESR1, which are upstream transcriptional regulators. In addition, network analyses revealed SNP-containing genes that have been implicated in TNBC including P53, ATM, BLM, BRCA1, CHEK1, TERT, CCCND1, CHEK2 and RB1.32 Complete information about the molecular function, biological process and cellular components in which all 201 are involved is provided in Table D of the Supplementary Data section of this report.
To further refine the genetic susceptibility landscape and understand the broader context in which genetic variants operate, we mapped the genes onto the canonical pathways. We hypothesized that genes containing SNPs associated with increased risk of developing subtypes of breast cancer interact with each other in biological pathways. The goal was to identify pathways enriched for SNPs that are associated with increased risk of developing subtypes of breast cancer. We identified many biological pathways enriched for SNPs. The Figures 5 and 6 show the identified pathways. Among the identified pathways included: the role of BRCA in DNA damage, p53, NF-kB, Kinase, ATM, ATR, apoptosis, DNA repair, DNA mismatch repair, hereditary breast cancer signaling and the DNA double-strand break repair by non-homologous end joining pathways.
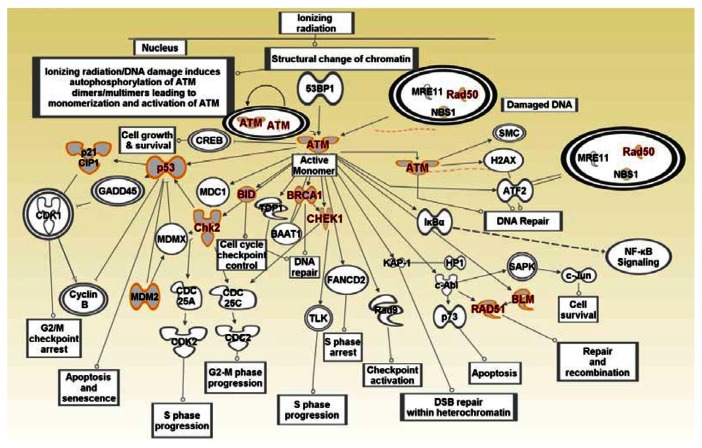
Figure 5.
Graphical representation of the ATM biological pathway and its crosstalk with other biological pathways (P53, BRCA1, NF-kB) involved in (DNA damage, repair, apoptosis, cell-cycle) which are enriched for SNPs associated with increased risk of developing breast cancer.
Notes: Gene symbols in red font represent genes containing SNPs associated with an increased risk of developing breast cancer. Gene symbols in black font represent novel genes not reported in GWAS. Dual ring circles indicate complex regulation involving many genes. Lines and arrows indicate direction of regulation. Biological activities are indicated in the text mapped to rectangular shapes. Information on association of individual SNP-containing genes with individual subtypes of breast cancer is provided in Table B as supplementary material.
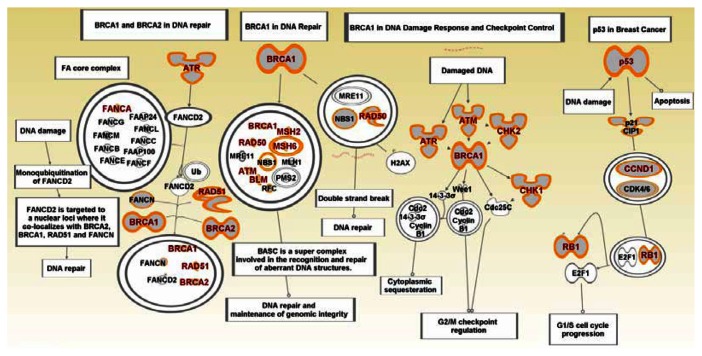
Figure 6.
Graphical representation of the hereditary breast cancer signaling pathway and other pathways (P53, BRCA1, ATR) involved in DNA damage and repair that are enriched for SNPs associated with increased risk of developing breast cancer.
Notes: Gene symbols in red font represent genes containing SNPs associated with an increased risk of developing breast cancer. Gene symbols in black font represent novel genes not reported in GWAS. Dual ring circles indicate complex regulation involving many genes. Lines and arrows indicate direction of regulation. Biological activities are indicated in the text mapped to rectangular shapes. Information on association of individual SNP-containing genes with individual subtypes of breast cancer is provided in Table B as supplementary material.
Pathway analysis revealed many SNP-containing genes implicated in both the subtypes responsive to targeted therapy and TNBC including P53, ATM, BRCA1, CHEK1, CHEK2, RAD51, RA50, BLM, BID, ATR, MSH2, MUSH6, FANCA, RAB1 and CCND1. (Figs. 5 and 6). In addition, pathway analysis revealed novel genes that regulate SNP-containing genes including 53BP1, NBS1, MRE11, MDM2, CDK1, CCNB1, GADD45, FANCD2, FNCN, P21 and E2F1 (Figs. 5 and 6). This is a significant finding given that may of the identified variants and associated genes may not have a direct causal association, but instead their actions may be mediated by other genes as demonstrated in Figures 5 and 6.
Many of the genes and pathways identified have been implicated in breast cancer. The SNP-containing gene ATM (Fig. 5) is a DNA damage-signaling kinase that is aberrantly reduced or lost in BRCA1 and BRCA2-difficent and triple-negative breast cancer.35 Loss of heterozygosity at the ATM locus has been reported in 30%–40% of breast tumors and 50%–70% show altered ATM protein levels.36,37 P53 is well known for its oncosuppressive role and its involvement in DNA repair mechanisms. The study of P53 status in tumors has revealed that ATM/P53 signaling is frequently altered either by a very low ATM expression or by the presence of a mutated P53.36 The SNP-containing gene ATR (Fig. 6) has been shown to phosphorylate several tumor suppressors including BCRCA1, BRCA2, CHEK1 and P53.38 All of these genes have been implicated in TNBC.32,39
Many of the identified genes in (Figs. 5 and 6) are involved in DNA damage response and repair. The DNA damage response pathway plays a key role in determining how individual cancers respond to radiation and chemotherapy.40 Defects in specific DNA repair pathways play key roles in the pathogenesis of TNBC; therefore, these pathways are potential therapeutic targets. For example, BRCA1-deficient breast cancers, most of which are high grade TNBC, display a defect in homology-mediated DNA repair that renders them exquisitely sensitive to cross-linking agents such as cis-platinum.40 The P53 binding protein 1 (53BP1) (Fig. 5) is a protein involved in DNA-damage checkpoint activation and DNA repair that is involved in both nonhomologous end-joining and homology-mediated repair of double-strand DNA breaks.40 As demonstrated in Figure 5, it transmits DNA damage signals from sensor proteins NBS1, MRE11 and RAD50 to transducer proteins ATM, CHEK2, CHEK1 and then to effectors P53, BRCA1, BRCA2, BRIP1, PALB2 and CASP8, all of which contain SNPs associated with an increased risk of developing breast cancer (Table A, Supplementary Data). Depletion of 53BP1 abrogates the ATM-dependent checkpoint response and G2 cell-cycle arrest triggered by the accumulation of DNA breaks in BRCA1-deleted cells.41 Studies have shown that loss of 53BP1 leads to resistance to cis-platinum and PARP inhibitors in BRCA1-difficient cells.40 Importantly, when both BRCA1 and 53BP1 are lost, sensitivity to DNA damage is reduced and homology-mediated repair is restored.40 The MRE11, RAD50 and NBS1 genes encode proteins of the MRE11-RAD50-NBS1 (MRN) complex (Figs. 5 and 6) which is critical for proper maintenance of genomic integrity and tumor suppression.42 Interestingly, mutations in two SNP-containing genes ATM and CHEK2 whose products are functionally intimately linked with MRN complex (Figs. 5 and 6) are associated with subtypes of breast cancer. For example, the moderately breast cancer-predisposing c.1100delC variant is mapped to ATM-activated CHEK2 kinase.42
BRCA1 has been shown to play a direct role in the repair of DNA by homologous recombination, by interacting with RAD51 protein and facilitating the formation of RAD51 aggregates at the site of DNA damage.43 The absence of BRCA2 results in chromosome instability, which is likely secondary to the defect in DNA repair.43BRCA1 one plays a role in sensing DNA damage and replication stress and mediating the signaling response.43 Therefore, in addition to its role in mediating DNA repair by homologous recombination via BRCA2, it also signals cell cycle checkpoints and mediates other transcription responses to DNA damage.43 The SNP-containing cyclin D1 (CCND1) gene (Fig. 6) belongs to the family of three closely related D-type cyclins, termed cyclin D1, D2 and D3. D-cyclins collectively control cell cycle progression by activating their cyclin-dependent kinase partners, CDK4 and CDK6 (Fig. 6), which leads to phosphorylation of the retinoblastoma (RB1) protein (Fig. 6), and in turn to the advance through the G1 phase of the cell cycle.44,45 The identification of the BRCA1 pathway is of particular interest, because disease-causing genetic variants in BRCA1 and BRCA2 confer a high risk of breast cancer, approximately 10- to 20-fold relative risk.46 Importantly, almost all BRCA1 breast cancers are diagnosed as TNBC. These breast cancers are early-onset and have higher relative risk.46
Overall, network and pathway analyses revealed the broader context in which genetic variants operate and provide functional bridges between GWAS findings and the disease state in different subtypes of breast cancer. However, we did not identify subtype-specific networks and pathways. The lack of identifying subtype-specific pathways suggests that subtype-specific genotyping and sequencing for mutational analysis may be warranted to uncover subtype-specific genetic risk factors. Such work was beyond the scope of this study. The identification of multi-gene pathways enriched for genetic variants suggests that pathway-crosstalk is probably involved in the development and progression of subtypes of breast cancer.
Discussion
This investigation shows that integrating GWAS, gene expression and biological information holds the promise of not only associating GWAS information with subtypes of breast cancer but also identifying gene regulatory networks and biological pathways that are enriched for genetic variants. GWAS have uncovered many loci associated with breast cancer, but two fundamental limitations have hampered our ability to translate GWAS results into clinically useful predictors of breast cancer subtypes and identification of potential targets for the development of novel, more effective therapies. First, the genetic loci identified thus far explain only a small proportion of the variation.8 Second, the SNP-trait associations alone do not necessarily lead directly to the identification of the causal genes, much less elucidate the broader context in which the cause genes operate in different subtypes breast cancer.8 The integrative genomics approach presented in this study addresses those longstanding questions and provides the basis for understanding the biological context in which genetic variants operate, which is a necessary step in identifying potential drug targets. This is the first study to infer the causal association between gene expression and different subtypes of breast cancer.
In the published literature on GWAS, a few individual genetic variants have been associated with subtypes of breast cancer.5–7 The main difference between reported GWAS-associating genetic variants with subtypes of breast cancer and the results reported here is that, this study takes a holistic approach by focusing on multi-gene networks and biological pathways rather than looking into a single genome location for a single SNP driving the breast cancer subtype, a classic reductionist approach to elucidating a complex disease. Most notably, this study also identified novel genes that have not been reported by GWAS. Indeed, pathway-based approaches have been previously reported by our group1 and others.47,48 These approaches have shown that integrating GWAS information with gene expression data is useful in linking GWAS information to disease state.1 However, this is the first study to associate GWAS information with the six intrinsic subtypes of breast cancer and to identify gene regulatory networks and key biological pathways enriched for SNPs.
In the published literature on GWAS, most replication efforts have focused on genetic variants with the strongest statistical evidence of association.30 However, efficient identification of additional susceptibility variants (both common and rare) might benefit from the integration of statistical evidence with some estimates of functional candidacy as demonstrated in this study. Because breast cancer is a complex disease, susceptibility effects are likely mediated through remote regulatory elements, and the causal variant could lie beyond the interval of maximal association in another gene or biological pathway.
The integrative genomics approach presented in this study provides a unified approach for linking susceptibility loci with distinct subtypes of breast cancer, and allows for identification of associated networks and biological pathways enriched for genetic variants. However, limitations must be acknowledged. GWAS and gene expression data used in this study was based on populations of European ancestry. There is a need to extend the analysis to other populations with differing mutational rates. It is conceivable that some loci may confer-population specific risk. For example, TNBC disproportionately affects African American women.14 We did not address this problem in this study; therefore, these results cannot be generalized to other populations.
Both the GWAS information and gene expression data used in this study were obtained from the public domain. It is conceivable that some of the GWAS findings may be statistical artifacts. Moreover, the populations used for GWAS maybe admixed. Verification of such artifacts and controlling for admixture were beyond the scope of this study. However, although caution is warranted in placing weight on use of publicly available data, it can provide a more cost-effective and rapid route towards identification of candidate genes and pathways for targeted sequencing and functional analysis. Another limitation worth mentioning is that we did not perform allele-specific expression analysis, a weakness that we readily acknowledge. However, allelic variation and allele-specific differences in human gene expression has been reported.49–51 In fact, allele-specific up-regulation of FGFR2, the most replicated gene and a critical biomarker in ER-positive breast cancer, has been shown to increase susceptibility.52
In conclusion, the results in this study demonstrate the power of using an integrative genomics approach to dissect the emerging genetic susceptibility landscape of breast cancer subtypes. The results based on this approach provide insights about the broader context in which genetic variants operate leading to different subtypes of breast cancer, a critical step towards identification of potential clinically actionable biomarkers. However, more research is needed to understand how genetic variants directly regulate molecular perturbation in different subtypes of breast cancer and different ethnic populations.
Supplementary Material
Table A. List of single nucleotide polymorphisms (SNPs) and associated genes, and corresponding estimates of P-values derived from GWAS, along with references of genome-wide association studies (GWAS) from which the information used in this study was extracted.
Table B. Estimates of P-values and FDR based on t-test (comparing gene expression profiles between each subtype of breast cancer and controls) and ANOVA (comparing gene expression profiles across all the six intrinsic subtypes of breast cancer) for all the 203 SNP-containing genes indicating their level of association with the six intrinsic subtypes of breast cancer.
Table C. Estimates of P-values and FDR based on t-test (comparing gene expression profiles between subtypes of breast cancer) and ANOVA (comparing gene expression profiles among the intrinsic subtypes of breast cancer within subgroup) for all the 203 SNP-containing genes indicating their level of differential expression within each clinically defined subgroup of breast cancer. NONTBC = subtypes responsive to targeted therapy, TNBC = triple-negative cancer (subgroup responsive to chemotherapy).
Table D. List of all the 203 SNP-containing genes including information on the biological processes, molecular functions and cellular components in which they are involved as determined by the Gene Ontology (GO) nomenclature.
Footnotes
Author Contributions
Conceived and designed the experiments: CH, LM, ASB, JM, KB. Analyzed the data: CH, TK. Wrote the first draft of the manuscript: CH, TK, ASB, JM, KB, LM. All authors contributed to the writing of the manuscript: CH, TK, ASB, JM, KB, LM. Agree with manuscript results and conclusions: CH, TK, ASB, JM, KB, LM. Jointly developed the structure and arguments for the paper: CH, TK, LM. Made critical revisions and approved final version CH, TK, ASB, JM, KB, LM. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
The authors wish to thank the University of Mississippi Cancer Institute for providing funding support for the project.
References
- 1.Hicks C, Asfour R, Pannuti A, Miele L. An integrative genomics approach to biomarker discovery in breast cancer. Cancer Inform. 2011;10:185–204. doi: 10.4137/CIN.S6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B, Beeghly-Fadiel A, Long J, Wei Z. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet. 2011;12:477–88. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T. Molecular portraits of Breast cancer: tumour subtypes as distinct disease entities. Eur J. Cancer. 2004;40:2667–75. doi: 10.1016/j.ejca.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Mavaddat N, Dunning AM, Ponder BA, Easton DF, Pharoah PD. Common genetic variation in candidate genes and susceptibility to subtypes of breast cancer. Cancer Epidem Biomarker Prev. 2009;18(1):255–9. doi: 10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broeks A, Schmidt MK, Sherman ME, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: Findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:1–15. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XR, Chang-Claude J, Goode EL, et al. Associations of breast cancer risk factors with tumor subtypes: A pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–23. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: Interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 10.Khoury MJ, Bertram I, Boffetta P, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation in human diseases. Am J Epidemiol. 2009;170:269–79. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28. doi: 10.1371/journal.pmed.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 13.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perou CM. Molecular stratification of triple-negative breast cancer. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann BD, Bauer JA, Chen X, et al. Identification of triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2012;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabatier R, Finetti P, Cervera N, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126:407–20. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119(2):335–46. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 1995;57(1):289–300. [Google Scholar]
- 20.RadMacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9(3):505–11. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 21.Morrissey ER, Diaz-Uriarte R. Pomello II: Finding differentially expressed genes. Nucleic Acids Res. 2009;37:W581–6. doi: 10.1093/nar/gkp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich M, Liefield T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Nat Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontolgy Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingenuity Pathways Analysis (IPA) system. Ingenuity Incorporated; California: [Google Scholar]
- 26.Stevens KN, Vachon CM, Lee AM, et al. Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res. 2011;71(19):6240–9. doi: 10.1158/0008-5472.CAN-11-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens KN, Fredericksen Z, Vachon CM, et al. 19p13.1 Is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72(7):1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Closas M, Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor (ER) status. Cancer Res. 2008;14(24):8000–9. doi: 10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmans P, Green EK, Pahwa JS, et al. Gene ontology analysis of GWA study data sets provides insights into biology of bipolar disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci of human diseases and traits. Proc Nat Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 32.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 35.Tommiska J, Bartkova J, Heinonen M, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27(17):2501–6. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- 36.Angèle S, Hall J. The ATM gene and breast cancer: is it really a risk factor? Mutat Res. 2000;462(2–3):167–78. doi: 10.1016/s1383-5742(00)00034-x. [DOI] [PubMed] [Google Scholar]
- 37.Hall J. The Ataxia-telangiectasia mutated gene and breast cancer: gene expression profiles and sequence variants. Cancer Lett. 2005;227(2):105–14. doi: 10.1016/j.canlet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Durocher F, Labrie Y, Soucy P, et al. Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families. BMC Cancer. 2006;29(6):230. doi: 10.1186/1471-2407-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hicks C, Kumar R, Pannuti A, et al. An integrative genomics approach for associating GWAS information with triple-negative breast cancer. Cancer Inform. 2013;12:1–20. doi: 10.4137/CIN.S10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neboori HJ, Haffty BG, Wu H, et al. Low p53 binding protein 1 (53BP1) expression is associated with increased local recurrence in breast cancer patients treated with breast-conserving surgery and radiotherapy. Int J Radiat Oncol Biol Phys. 2012;83(5):e677–83. doi: 10.1016/j.ijrobp.2012.01.089. [DOI] [PubMed] [Google Scholar]
- 41.Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartkova J, Tommiska J, Oplustilova L, et al. Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol Oncol. 2008;2(4):296–316. doi: 10.1016/j.molonc.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell SN, Kachnic LA. Therapeutic exploitation of tumor cell defects in homologous recombination. Anticancer Agents Med Chem. 2008;8(4):448–60. doi: 10.2174/187152008784220267. [DOI] [PubMed] [Google Scholar]
- 44.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–21. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 45.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 46.Stratton MS, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 47.Menashe I, Maeder D, Garcia-Closas M, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;70(11):4453–9. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haiman CA, Hsu C, de Bakker PI, et al. comprehensive association testing of common genetic variation in DNA repair pathways genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17(6):825–34. doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 49.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 50.Buckland PR. Allele-specific gene expression differences in humans. Hum Mol Genet. 2004;13(2):R255–60. doi: 10.1093/hmg/ddh227. [DOI] [PubMed] [Google Scholar]
- 51.Paracios R, Gazave E, Goni J, et al. Allelle-specific gene expression is wide-spread across the genome and biological processes. PLoS One. 2009;4(1):e4150. doi: 10.1371/journal.pone.0004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer KB, Maja A-T, O’Reilly M, et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6(5):e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. List of single nucleotide polymorphisms (SNPs) and associated genes, and corresponding estimates of P-values derived from GWAS, along with references of genome-wide association studies (GWAS) from which the information used in this study was extracted.
Table B. Estimates of P-values and FDR based on t-test (comparing gene expression profiles between each subtype of breast cancer and controls) and ANOVA (comparing gene expression profiles across all the six intrinsic subtypes of breast cancer) for all the 203 SNP-containing genes indicating their level of association with the six intrinsic subtypes of breast cancer.
Table C. Estimates of P-values and FDR based on t-test (comparing gene expression profiles between subtypes of breast cancer) and ANOVA (comparing gene expression profiles among the intrinsic subtypes of breast cancer within subgroup) for all the 203 SNP-containing genes indicating their level of differential expression within each clinically defined subgroup of breast cancer. NONTBC = subtypes responsive to targeted therapy, TNBC = triple-negative cancer (subgroup responsive to chemotherapy).
Table D. List of all the 203 SNP-containing genes including information on the biological processes, molecular functions and cellular components in which they are involved as determined by the Gene Ontology (GO) nomenclature.