Background: Antibiotics produce errors in bacterial protein synthesis, but their effects on translation in mammalian cells are poorly understood.
Results: Antibiotics lead to high levels of mistranslation of selenocysteine-encoding UGA codons in mammalian cells.
Conclusion: Antibiotics specifically disrupt selenoprotein expression and function in mammalian cells in culture.
Significance: Most selenoproteins are oxidoreductases, implicating antibiotics in dysregulation of cellular redox control.
Keywords: Protein Synthesis, Selenocysteine, Selenoprotein, Transfer RNA (tRNA), Translation, Misreading, UGA Codon
Abstract
Antibiotics target bacteria by interfering with essential processes such as translation, but their effects on translation in mammalian cells are less well characterized. We found that doxycycline, chloramphenicol, and Geneticin (G418) interfered with insertion of selenocysteine (Sec), which is encoded by the stop codon, UGA, into selenoproteins in murine EMT6 cells. Treatment of EMT6 cells with these antibiotics reduced enzymatic activities and Sec insertion into thioredoxin reductase 1 (TR1) and glutathione peroxidase 1 (GPx1). However, these proteins were differentially affected due to varying errors in Sec insertion at UGA. In the presence of doxycycline, chloramphenicol, or G418, the Sec-containing form of TR1 decreased, whereas the arginine-containing and truncated forms of this protein increased. We also detected antibiotic-specific misinsertion of cysteine and tryptophan. Furthermore, misinsertion of arginine in place of Sec was commonly observed in GPx1 and glutathione peroxidase 4. TR1 was the most affected and GPx1 was the least affected by these translation errors. These observations were consistent with the differential use of two Sec tRNA isoforms and their distinct roles in supporting accuracy of Sec insertion into selenoproteins. The data reveal widespread errors in inserting Sec into proteins and in dysregulation of selenoprotein expression and function upon antibiotic treatment.
Introduction
Selenocysteine (Sec)2 is the 21st protein amino acid in the genetic code and is decoded by UGA, which is normally used as a termination codon (1–3). Unlike any other known amino acid in eukaryotes, Sec is biosynthesized on its tRNA (4). Cys was recently shown to replace Sec in the Sec biosynthetic pathway, inserting Cys in the selenium-containing proteins (selenoproteins), thioredoxin reductase 1 (TR1; Txnrd1) and 3 (TR3; Txnrd2) (5). The level of Cys/Sec replacement in vivo was found to be dependent on the selenium status in the diet of mice, wherein selenium-deficient diets resulted in a 1:1 ratio of Sec to Cys inserted into proteins. Diets containing normal amounts of selenium had approximately a 9:1 ratio. A significant presence of Cys, even under conditions of sufficient dietary selenium, suggested that this amino acid naturally replaces Sec in a subset of selenoprotein molecules. Furthermore, this observation suggested that Cys may play a substantial role in selenoprotein function under conditions of selenium deficiency.
The selenoprotein population in mammals is comprised of two subclasses, housekeeping (e.g. TR1 and TR3) and stress-related selenoproteins (e.g. glutathione peroxidase 1 (GPx1) and selenoprotein W (SelW)) (6). These subclasses are synthesized by two different Sec tRNA[Ser]Sec isoforms, differing from each other by a single methyl group, Um34 (7). Some selenoproteins, such as glutathione peroxidase 4 (GPx4) and selenoprotein P, appear to be synthesized by both isoforms (8). Interestingly, the non-Um34-containing isoform, methylcarboxymethyl-5′-uridine, apparently must have Sec attached to it for the Um34 methylase to be synthesized (9). This phenomenon would rule out Cys/Sec replacement in those selenoproteins that are synthesized exclusively by the Um34 isoform, methylcarboxymethyl-5′-uridine-2′-O-methylribose.
Antibiotics, such as Geneticin (G418), doxycycline (Dox), and chloramphenicol (Cp), are widely used in mammalian cell culture and also have cytotoxic effects on the host cell. The mechanisms of action of these antibiotics have not been fully characterized. Although all three have roles in inhibiting protein synthesis, their effects have been studied principally in bacteria (see Refs. 10 and 11 for reviews). G418 is an aminoglycoside, and aminoglycosides bind primarily to the small ribosomal subunit promoting readthrough of termination codons (Refs. 12 and 13 and references therein). These compounds, including G418, have been used to rescue the p53 tumor suppressor protein by suppressing a premature stop codon in a mutated form of p53 (13). G418 has also been shown to cause mistranslation of GPx1 by inserting Arg at the UGA-Sec codon in mammalian cells (14). Dox is a tetracycline that also binds to the small ribosomal subunit but, unlike aminoglycosides, blocks the attachment of aminoacyl-tRNA to the A-site (10, 11). In addition, Dox has nonantibiotic roles such as antitumor activity (15–17). Cp binds to the large ribosomal subunit and blocks the peptidyl transfer step during elongation of protein synthesis (10, 11). This antibiotic has been reported to induce translational misreading in bacteria (18). Both Dox and Cp are used in treating infections in humans. However, Cp is also known to be toxic to bone marrow tissue, which can result in aplastic anemia (see Ref. 19 and references therein).
We undertook the present study to examine the effect of G418, Dox, and Cp on the expression and function of selenoproteins, specifically TR1, GPx1, and GPx4. These selenoproteins play much different roles in their regulation and requirement for cell growth and development. For example, the targeted removal of TR1 and GPx4 from the mouse genome is embryonic lethal (20, 21), whereas knock-out of GPx1 in mice has no phenotype unless the animals are stressed (22).
Mouse breast cancer cells, EMT6, were used in the current study to elucidate the effects of G418, Dox, and Cp as this cell line expresses TR1, GPx1, and GPx4 in high amounts. Another feature of these selenoproteins is that they are synthesized by different mechanisms, i.e. as noted above, TR1 utilizes the methylcarboxymethyl-5′-uridine isoform, GPx1 utilizes the methylcarboxymethyl-5′-uridine-2′-O-methylribose isoform, and GPx4 appears to use both isoforms (8). We observed that the activities of TR1 and GPx1 were reduced substantially by G418, Dox, and Cp. This reduction in activity was due to substitution of Sec with Arg, premature termination of TR1 at UGA, and insertion of Cys and Trp in the latter protein. Both GPx1 and GPx4 had significant levels of Cys in place of Sec following G418 treatment, but no detectable amounts of Trp following treatment with any of these antibiotics. In each case, the substitution of Arg, Trp, or Cys was apparently due to misreading and not due to replacement of Sec with another amino acid that arose through the Sec biosynthetic pathway. This study exposed an unexpected level of vulnerability in Sec insertion under conditions of antibiotic treatment.
EXPERIMENTAL PROCEDURES
Materials
Coomassie Blue staining solution, NADPH, 5,5′-dithiobis(2-nitrobenzoic acid), Dox, Cp, and iodoacetamide were purchased from Sigma. Biotin-conjugated iodoacetamide (N-(biotinoyl)-N′-(iodoacetyl)-ethylenediamine (BIAM)), PVDF membranes, NuPAGE 4–12% Bis-Tris gel, Dulbecco's modified Eagle's medium (DMEM), Waymouth's medium, methionine/cysteine-free DMEM, HEPES, antibiotic-antimycotic solution (10,000 units/ml penicillin, 10,000 μg/ml streptomycin, and 25 μg/ml amphotericin B), fetal bovine serum, and puromycin were from Life Technologies. LipoD293 was from SignaGen Laboratories, GP2-293 cells were from Clontech, primary antibodies for GPx1 and GPx4 were from Abcam, TR1 was from Epitomics, GAPDH was from Sigma, β-actin was from Cell Signaling Technology, and G418 was from Cellgro. BCA protein assay reagent, SuperSignal West Dura extended duration substrate, and streptavidin-HRP primary antibody were from Thermo Fisher Scientific. EasyTagTM Express 35S protein labeling mix was from PerkinElmer Life Sciences, nickel-nitrilotriacetic acid-agarose was from Qiagen, GPx activity assay kit was from Enzo Life Sciences, and anti-rabbit HRP conjugated secondary antibody was from Cell Signaling Technology. 75Se was obtained from the Research Reactor Facility, University of Missouri, Columbia, MO.
Cell Culture
EMT6 cells were grown at 37 °C, 5% CO2 in Waymouth's medium supplemented with 15% (v/v) fetal bovine serum, 20 mm HEPES, and with or without antibiotic/antimycotic solution (100 units/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B). The antibiotic/antimycotic solution was used in all experiments in this study. GP2-293 cells were grown in DMEM supplemented with 10% (v/v).
75Se Labeling and Western Blotting
Murine breast cancer, EMT6, cells (American Type Culture Collection) were seeded onto a 6-well plate (1 × 105 cells/well), incubated for 48 h without or with Dox, Cp, and G418, and then 75Se (15 μCi; 20 nm/well) was added. After a 24-h incubation, EMT6 cells were washed twice with PBS and then harvested with lysis buffer (20 mm Tris-HCl, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mm sodium fluoride, 5 mm EDTA, and proteinase inhibitor mixture). Protein levels were quantified using BCA protein assay reagent, and 30 μg of each protein sample was separated by electrophoresis on NuPAGE 4–12% Bis-Tris gels, transferred to a PVDF membrane, and then incubated initially with primary antibody (anti-TR1, anti-GPx1, or anti-GPx4) and finally with HRP-conjugated secondary antibody. Membranes were treated with SuperSignal West Dura extended duration substrate and exposed to x-ray film. Proteins in gels were stained with Coomassie Blue staining solution, and the gel was dried and exposed to a PhosphorImager (GE Healthcare). 75Se-labeled selenoproteins on exposed membranes were identified by autoradiography. The band intensities on Western blots and 75Se-labeled selenoproteins were quantified using ImageJ software (obtained from the National Institutes of Health).
Constructs and Retroviral Transductions
Mouse TR1 cDNA was cloned as described previously (23). GPx1 and GPx4 cDNAs were amplified by PCR using the primers 1+2 (GPx1) or 3+4 (GPx4) (supplemental Table 1) and cloned into BamHI-XhoI (GPx1) or XhoI-XhoI sites (GPx4) of the retroviral vector pRV IRES Puro (23). A His tag was inserted at the N terminus of TR1 and at the C terminus of GPx1 and GPx4. GP2-293 cells were plated in 60-mm dishes and transfected using LipoD293 following the manufacturer's instructions. 1.5 μg of each retroviral vector and 1 μg of an amphotropic envelope expression vector (pVSV-G) were used. Medium was replaced with fresh medium 24 h following transfection. Cell culture supernatants were harvested after an additional 24 h, filtered with 0.22-μm filters, and diluted (1:2) with fresh medium. Diluted viral supernatants were then added to the cells (plated at a low confluency 24 h prior to the infection) and incubated overnight at 37 °C.
Purification of TR1, GPx1, and GPx4
To isolate the recombinant TR1, GPx1, and GPx4, cells were cultured with or without 10 μg/ml Dox, 25 μg/ml Cp, or 100 μg/ml G418 and 2 μg/ml puromycin for 72 h, harvested, and lysed in 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 20 mm imidazole on ice. Each His tag protein was mixed with nickel-nitrilotriacetic acid-agarose as described (4, 24).
Enzymatic Activities of TR1 and GPx1
To measure TR1 activities, samples were mixed with 100 mm potassium phosphate (pH 7.0), 10 mm EDTA, 0.24 mm NADPH, and 3 mm 5,5′-dithiobis(2-nitrobenzoic acid), and the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) by thioredoxin reductase was measured with absorbance at 412 nm spectrophotometrically (GE Healthcare) (25, 26). GPx1 activities were measured using a glutathione activity assay kit according to the manufacturer's instructions. The purified proteins were quantified using BCA protein assay reagent.
BIAM Labeling Assay
The biotin labeling of reactive residues with BIAM was used to assess the amount of Sec moiety in selenoproteins as described (27). Briefly, reduced proteins were mixed with 100 μm BIAM in 100 mm Tris-HCl and 1 mm EDTA (pH 6.5) and incubated at 37 °C for 15 min. Alternatively, alkylation of Sec was achieved by the addition of freshly prepared 250 μm iodoacetamide. The samples were denatured in SDS sample buffer and applied onto NuPAGE 4–12% Bis-Tris gels, and Western blot analysis was carried out as described above. Biotin was detected by streptavidin-HRP.
Analyses of TR1, GPx1, and GPx4 by Mass Spectrometry
Recombinant TR1, GPx1, and GPx4 were isolated from cells grown in the presence or absence of Dox (10 and 20 μg/ml), Cp (25 and 50 μg/ml), or G418 (100 and 200 μg/ml), purified, and reduced with DTT followed by alkylation of the Sec residues with iodoacetamide as described (28). Alkylated proteins were applied onto NuPAGE 4–12% Bis-Tris gels and stained with Coomassie Blue staining solution. Protein bands were detected, cut out, and subjected to in-gel tryptic digestion, and the resulting peptides were measured as described (5).
35S Labeling
Cells were seeded onto a 6-well plate (1 × 105 cells/well), incubated without or with Dox (10 and 20 μg/ml), Cp (25 and 50 μg/ml), or G418 (100 and 200 μg/ml), for 72 h, and then washed with methionine/cysteine-free DMEM. The cells were further incubated in methionine/cysteine-free DMEM supplemented with 10% dialyzed FBS and 25 μCi/ml EasyTag Express 35S protein labeling mix. After a 1-h incubation, cells were washed twice with PBS and harvested with lysis buffer (as above), and protein was quantified using BCA protein assay reagent. Thirty μg of each protein sample was separated by electrophoresis on NuPAGE 4–12% Bis-Tris gels. Proteins in gels were stained with Coomassie Blue staining solution, and the gel was dried and exposed to an autoradiography film.
RESULTS
Dox, Cp, and G418 Affect Sec Insertion
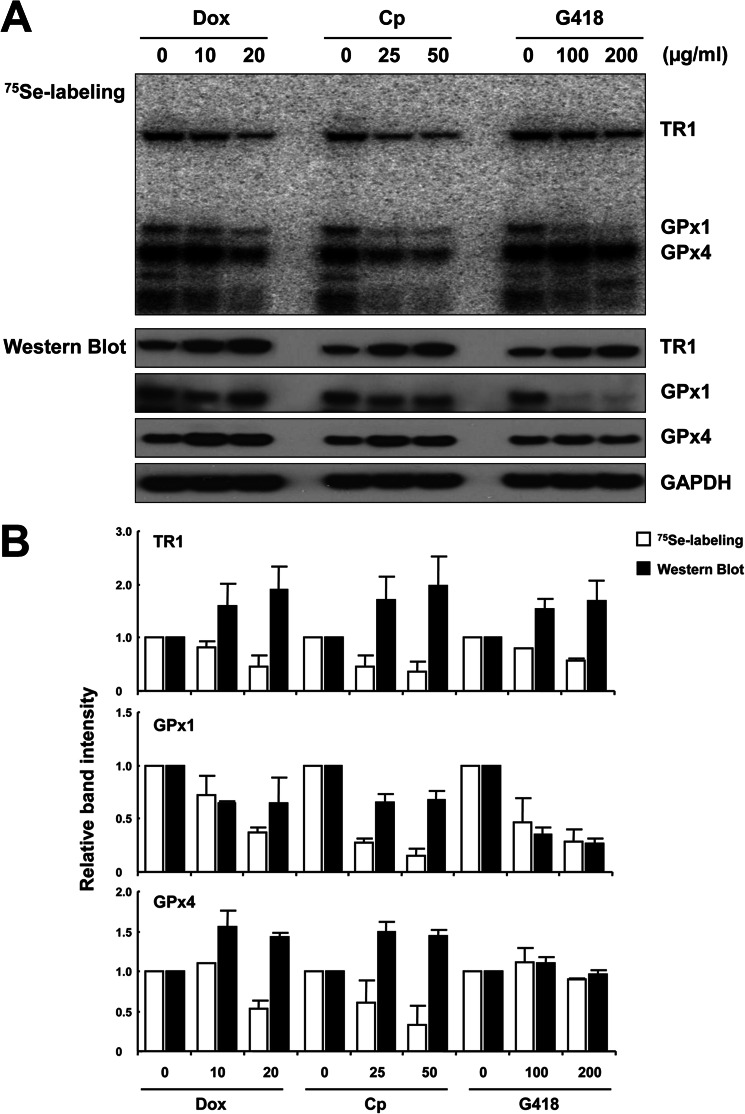
To assess the effect of Dox, Cp, and G418 on selenoprotein synthesis, EMT6 cells were labeled with 75Se, grown in the presence of these antibiotics, and compared with cells grown in their absence (Fig. 1A, upper panel). Insertion of Sec into selenoproteins was noticeably reduced with the increase in antibiotic concentration, with GPx1 being the most significantly affected. GPx1 levels examined by Western blotting were also reduced (Fig. 1A, middle panels). In contrast, the levels of TR1 and GPx4 examined by Western blotting were increased in the presence of antibiotics with the possible exception of GPx4 in the presence of G418. Thus, although the 75Se and Western blot signals correlated in the case of GPx1, they followed opposite trends in the case of TR1 and GPx4. Quantification of these data is shown in Fig. 1B, which revealed up to a 4-fold difference in Sec insertion and protein expression. The observed differences among selenoproteins, and especially the contrasting patterns of protein levels and 75Se labeling in these selenoproteins, suggested that amino acids other than Sec were inserted in the presence of antibiotics.
FIGURE 1.
The antibiotics Dox, Cp, or G418 differentially affect Sec insertion and selenoprotein expression in EMT6 cells. EMT6 cells were grown in the presence and absence of Dox, Cp, and G418, and the levels of selenoproteins were assessed. A, determination of selenoprotein levels by 75Se labeling and Western blotting. Cells were cultured in the presence or absence of Dox (10 or 20 μg/ml), Cp (25 or 50 μg/ml), or G418 (100 or 200 μg/ml) and labeled with 75Se (15 μCi/well), protein extracts were prepared and electrophoresed, labeled selenoproteins were identified by autoradiography (upper panel), and TR1, GPx1, and GPx4 levels were further assessed by Western blotting (four lower panels). GAPDH is shown as a loading control. B, quantification of 75Se labeling and expression of TR1, GPx1, and GPx4. Values shown are the relative intensities of 75Se labeling or Western blot signals of proteins in treated cells when compared with those in cells grown in the absence of antibiotics. Values are the means ± S.D. of three independent experiments. Experimental details are given under ”Experimental Procedures.“
TR1 and GPx1 Activities Are Inhibited by Antibiotics
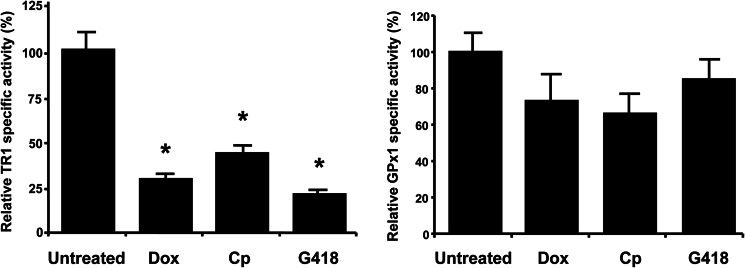
To characterize the observed translational defects involving selenoproteins, we quantified their catalytic activities, examined Sec insertion, and analyzed amino acids inserted in positions corresponding to UGA codons in TR1, GPx1, and GPx4. To prepare sufficient amounts of these proteins, we overexpressed them in cultured cells (supplemental Fig. 1). We noticed that the elevated expression of TR1 and GPx4 decreased GPx1 levels, probably because of competition for Sec insertion machinery. TR1 and GPx1 were purified from the cells grown in the absence or presence of Dox, Cp, or G418, and their catalytic activities were examined (Fig. 2). TR1 activity was significantly reduced in the presence of antibiotics, whereas GPx1 activity was only slightly reduced. These data were consistent with the observed trends in protein and 75Se signals from cells treated or not treated with antibiotics. It appeared that only 20–40% of TR1 molecules had a 75Se signal and possessed TR1 activity following treatment with antibiotics. On the other hand, the majority of GPx1 molecules were active under these conditions (although the levels of GPx1 were substantially reduced).
FIGURE 2.
Antibiotics regulate selenoprotein activities. EMT6 cells carrying recombinant TR1 or GPx1 were grown in the absence or presence of Dox (10 μg/ml), CP (25 μg/ml), or G418 (100 μg/ml), TR1 and GPx1 were purified, and their activities were measured spectrophotometrically. Specific activities are expressed as the percentage of that found in cells grown in the absence of antibiotics (measured specific activities in control cells were: TR1, 409.6 ± 57.2 units; and GPx1, 460.2 ± 18.1 units). Values are the means ± S.D. of three independent experiments. *, p < 0.01. Experimental details are given under ”Experimental Procedures.“
Sec Insertion into TR1 and GPx1 Is Reduced by Antibiotics
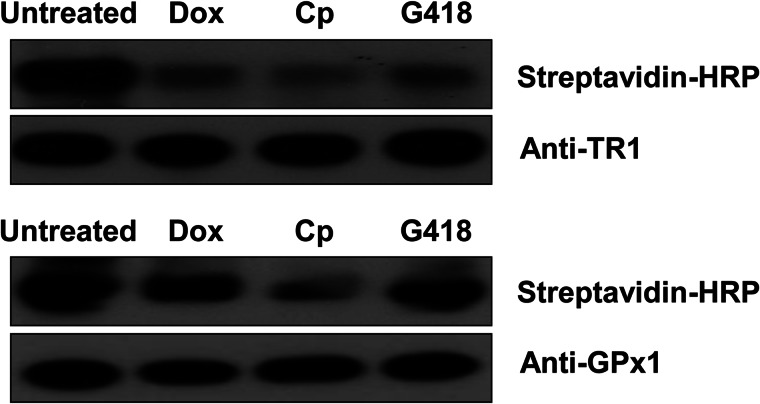
To further examine Sec insertion into selenoproteins, we subjected purified TR1 and GPx1 to alkylation under conditions that target only the most reactive nucleophilic residues, thereby primarily modifying the Sec residue (Fig. 3). The alkylated species were then examined by Western blotting, which revealed significantly lower alkylation levels of TR1 preparations from cells grown in the presence of antibiotics. GPx1 alkylation was also affected by antibiotics, although to a lesser extent. These data suggest that the residues that could not be alkylated were inserted in place of Sec in TR1, and to a lower degree in GPx1, in the presence of antibiotics.
FIGURE 3.
Growth of EMT6 cells in the presence of antibiotics reduces the levels of reactive Sec. Cells carrying the recombinant vector for (upper panel) TR1 or (lower panel) GPx1 were grown in the absence or presence of Dox (10 μg/ml), Cp (25 μg/ml), or G418 (100 μg/ml). Proteins were purified from treated cells, reduced, and alkylated with BIAM. The labeled TR1 and GPx1 were then subjected to Western blotting with HRP-conjugated streptavidin to measure reactive nucleophilic residues and with TR1 or GPx1 antibodies to detect the total protein. Experimental details are given under ”Experimental Procedures.“
Antibiotics Induce Misreading of the Sec UGA Codon
To elucidate which amino acids were inserted into TR1 in place of Sec and/or whether the protein was truncated at the penultimate (UGA) codon, we digested the purified samples and subjected them to MS/MS analyses (Table 1). As expected, the presence of Sec was maximal in untreated cells and decreased upon treatment with antibiotics. In the presence of antibiotics, the Sec-containing form in TR1 decreased 2.5–3-fold when compared with untreated cells, whereas the form with Arg in place of Sec increased more than 5-fold, and the truncated form increased more than 3-fold. In the case of G418, we also observed a 7-fold increase of Cys insertion, and Trp insertion increased 6- and 9-fold in TR1 when cells were cultured in the presence of G418 or Cp, respectively. The decrease in Sec insertion corresponds closely with the data on 75Se labeling, protein expression, alkylation, and catalytic activity, suggesting that only about one-third of TR1 molecules had Sec inserted in cells treated with antibiotics.
TABLE 1.
Characterization of TR1 isolated from EMT6 cells by mass spectrometry
Cells were cultured without or with 10 μg/ml Dox, 25 μg/ml Cp, or 100 μg/ml G418 for 72 h. Peptides were normalized to an internal control peptide from TR1 (R.FLIATGERPR.Y, where the period indicates where the peptide is cleaved by trypsin) to account for differences in protein expression between the different treatments. AA, amino acid.
| Peptidea | AA | Untreatedb | Dox | Cp | G418 |
|---|---|---|---|---|---|
| R.SGGDILQSGCUG. | Sec | 1.0 | 0.4 | 0.4 | 0.3 |
| R.SGGDILQSGCCG. | Cys | 1.0 | 0.5 | 0.9 | 7.4 |
| R.SGGDILQSGCR.G | Arg | 1.0 | 10.9 | 5.8 | 8.0 |
| R.SGGDILQSGCWG. | Trp | 1.0 | 0.2 | 9.6 | 6.4 |
| R.SGGDILQSGC. | Stop | 1.0 | 7.1 | 7.5 | 3.9 |
a The period in each peptide designates where the peptide is cleaved by trypsin. The underline indicates the position of Sec or misincorporated amino acid.
b Signals for each TR1 form found in the untreated sample were normalized to 1.0 and were used as a reference for quantifying these forms in samples treated with antibiotics. The different amino acid replacements and Sec-containing peptides should not be directly compared with each other because the Arg-containing peptide was of shorter length and gave a different response. However, the relative frequency (%) of each detected peptide in the untreated samples was Sec, 92.0; Cys, 1.4; Arg, 3.2, Trp, 0.02; Stop (i.e. truncated peptide), 3.3.
The Sec forms of GPx1 and GPx4 were also at maximal levels in untreated cells (Tables 2 and 3, respectively). Antibiotics reduced Sec in proteins, but to a lesser degree when compared with TR1. GPx1 was least affected with regard to replacement of Sec with other amino acids as 70–80% of the Sec-containing form was still present following antibiotic treatment. However, an increase in insertion of both Arg and Cys was observed in response to antibiotic treatment. GPx4 had greater Arg insertion than GPx1 in the presence of the three antibiotics but similar decrease with respect to Sec-containing peptides (Table 3). It is not surprising that truncated peptides were not observed with GPx1 or GPx4 as these truncated peptides are unstable (29).
TABLE 2.
Characterization of the purified GPx1 by mass spectrometry
Cells were cultured without or with 10 μg/ml Dox, 25 μg/ml Cp, or 100 μg/ml G418 for 72 h. Peptides were normalized to an internal control peptide from GPx1 (K.AHPLFTFLR.N, where the period indicates where the peptide is cleaved by trypsin) to account for differences in protein expression between the different treatments. AA, amino acid.
| Peptidea | AA | Untreatedb | Dox | Cp | G418 |
|---|---|---|---|---|---|
| K.VLLIENVASLUGTTIR.D | Sec | 1.0 | 0.8 | 0.7 | 0.8 |
| K.VLLIENVASLCGTTIR.D | Cys | 1.0 | NQc | NQc | 2.9 |
| K.VLLIENVASLR.G | Arg | 1.0 | 1.4 | 2.3 | 1.1 |
a The period in each peptide designates where the peptide is cleaved by trypsin. The underline indicates the position of Sec or misincorporated amino acid.
b Signals for each GPx1 form found in the untreated sample were normalized to 1.0 and were used as a reference for quantifying these forms in samples treated with antibiotics. The different amino acid replacements and Sec-containing peptides should not be directly compared with each other because the Arg-containing peptide was of shorter length and gave a different response. However, the relative frequency (%) of each detected peptide in the untreated samples was Sec, 84.7; Cys, 7.1; Arg, 5.5.
c NQ, not quantifiable (signal:noise < 10).
TABLE 3.
Characterization of the purified GPx4 by mass spectrometry
Cells were cultured without or with 10 μg/ml Dox, 25 μg/ml Cp, or 100 μg/ml G418 for 72 h. Peptides were normalized to an internal control peptide from GPx4 (R.YAECGLR.I, where the period indicates where the peptide is cleaved by trypsin) to account for differences in protein expression between the different treatments. AA, amino acid.
| Peptidea | AA | Untreatedb | Dox | Cp | G418 |
|---|---|---|---|---|---|
| R.GFVCIVTNVASQUGK.T | Sec | 1.0 | 0.8 | 0.7 | 0.6 |
| R.GFVCIVTNVASQCGK.T | Cys | NQc | NQc | NQc | —d |
| R.GFVCIVTNVASQR.G | Arg | 1.0 | 1.7 | 7.9 | 2.2 |
a The period in each peptide designates where the peptide is cleaved by trypsin. The underline indicates the position of Sec or misincorporated amino acid.
b Signals for each GPx4 form found in the untreated sample were normalized to 1.0 and were used as a reference for quantifying these forms in samples treated with antibiotics. The different amino acid replacements and Sec-containing peptides should not be directly compared with each other because the Arg-containing peptide was of shorter length and gave a different response. However, the relative frequency (%) of each detected peptide in the untreated samples was Sec, 88.5; Arg, 11.5.
c NQ, not quantifiable (signal:noise < 10).
d Peptides with Cys decoded by UGA were detected, but could not be quantified as they were missing in the untreated control.
Antibiotics and Protein Expression
To assess whether other proteins were affected by the antibiotics used herein, EMT6 cells were labeled with a [35S]Met-Cys mix in the presence and absence of G418, Dox, or Cp, protein extracts were electrophoresed, and an autoradiogram was generated (supplemental Fig. 2). Cells grown in the presence of each antibiotic showed similar patterns, but we also observed some differences in [35S]Met-Cys-labeled proteins when compared with the labeled proteins isolated from cells grown in the absence of antibiotics. Total proteins and the two housekeeping proteins, GAPDH and β-actin, which were used as loading controls (see supplemental Fig. 2), did not appear to manifest significant differences in patterns in cells grown in the presence and absence of antibiotics. Whether antibiotics cause mistranslation of certain proteins is unclear, and they may affect protein synthesis through various mechanisms.
DISCUSSION
A mouse breast cancer cell line, EMT6, was used in the present study to examine the effect of three widely used antibiotics, Dox, Cp, and G418, on selenoprotein synthesis. EMT6 cells were selected as they highly express TR1, GPx1, and GPx4, which are three of the most studied selenoproteins (30). The amounts of TR1 measured by 75Se labeling steadily declined with increasing concentrations of the three antibiotics, and the enzymatic activity pattern followed this trend, whereas the amount of TR1 protein measured by Western blotting steadily increased. GPx4 manifested similar patterns of decline with 75Se labeling and similar patterns of protein enrichment with Western blotting. GPx1, on the other hand, showed a loss of 75Se labeling in cells grown in the presence of Dox and Cp and a similar drop in protein levels.
The most likely explanation for the opposing patterns in TR1 and GPx1 expression between 75Se labeling and Western blotting was that Sec was replaced by another amino acid (or amino acids). This replacement must have occurred by misreading of the Sec UGA codon by another aminoacyl-tRNA because the TR1 level, as measured by Western blotting, was increased. Presumably, the aminoacyl-tRNA could not involve tRNA[Ser]Sec because this step in decoding the UGA Sec codon is rate-limiting in generating the selenoprotein product (31, 32). Furthermore, the only likely amino acid that can replace Sec in selenoproteins by decoding the Sec UGA codon with tRNA[Ser]Sec is Cys (5).
The amount of Sec identified by mass spectrometry in purified selenoproteins from cells grown in the presence of antibiotics varied considerably. Following antibiotic treatment, Sec levels in TR1 were as low as 30% of the level found in TR1 from untreated cells, whereas Sec levels in GPx1 and GPx4 were >60% of the levels found in untreated cells. Upon treatment of cells with antibiotics, the truncated form was only detected in TR1 because truncated GPx1 and GPx4 peptides would be too short and were likely unstable, and therefore, readily degraded (29). In general, TR1 and GPx1 showed the opposite patterns, with GPx4 being in the middle, with regard to misincorporation of Arg and Cys UGA codons. This trend corresponded well to the biochemical analyses, including enzymatic activity, 75Se insertion, alkylation of reactive residues, and protein expression. This finding agrees with the observation that insertion of Sec into TR1 and GPx1 is supported by two different tRNA[Ser]Sec isoforms, whereas both isoforms support Sec insertion into GPx4. As the two isoforms differ by a single methyl group, our data suggest that methylation increases accuracy of Sec insertion, and thus, reduces misreading.
Our data showing that G418 induces the replacement of Sec with Arg in GPx1 confirm and extend an earlier study by Handy et al. (14), who reported Sec/Arg replacement in GPx1 from COS7 cells treated with this antibiotic. We also found that G418 induced Cys insertion in place of Sec in GPx1 and that all three selenoproteins were affected, and not only by G418, but by other antibiotics as well. Interestingly, preferential misreading induced by antibiotics occurred at the 5′-position of the codon, inserting Arg in place of Sec. There are two Arg-tRNA isoforms that could conceivably be involved: the isoform that normally decodes CGA and the one that decodes AGA. Because the G in the 3′-position of the anticodon of the isoform that decodes CGA would presumably base pair more favorably with the 5′-U in UGA than the U in the 3′-position of anticodon of the isoform that decodes AGA, Arg is most likely inserted in place of Sec by the former tRNA. In addition, Arg-tRNA that decodes AGA occurs in lower levels when compared with the isoform decoding CGA in mammalian cells (33). This observation further suggested that UGA is misread by the Arg-tRNA that decodes CGA.
The misinsertion of Cys and Trp is consistent with mispairing in the 3′-position of UGA as these amino acids are encoded by the UGX group of codons (UGU and UGC code for Cys, and UGG codes for Trp). Both Cys and Trp were found in place of Sec in TR1, whereas Cys was the only amino acid found at the UGA Sec codon in GPx1 and GPx4, and only from cells grown in the presence of G418. The variation in levels of misreading at the 3′-position is likely a positional effect in these three selenoproteins, wherein misincorporation is more favorable at the penultimate Sec codon in TR1 than at the more proximal, N-terminal Sec codon in GPx1 and GPx4.
The overexpression of recombinant TR1 in EMT6 cells resulted in lower expression of GPx1, GPx4, and other selenoproteins, whereas overexpression of recombinant GPx1 and GPx4 caused a slight down-regulation in the synthesis of TR1. Contrasting patterns of TR1 and GPx1 expression in malignant cells, wherein enhanced synthesis of TR1 resulted in reduced expression of GPx1 and vice versa, were reported in different cancer cell lines several years ago (34). Clearly, there is competition between different selenoproteins for limited amounts of cofactors involved in translation of these selenium-containing proteins (31, 32). However, contrasting patterns of selenoprotein expression may also result from oxidative stress, which represses GPx1 and induces the expression of TR1 (34).
Overall, our study exposed a very significant vulnerability of Sec insertion in mammalian cells treated with antibiotics. Although these compounds affect the translation accuracy of other termination codons, UGA codons designating Sec appear to be far more significantly affected. Several selenoproteins in mammals are essential in development, including TR1 (21) and GPx4 (22), and such high levels of misinsertion at their catalytic residue in the presence of antibiotics undoubtedly affects their function, resulting in selenoprotein and selenium deficiencies. In light of the findings reported herein, studies worthy of pursuit would appear to be whether prolonged usage of the antibiotics Dox and Cp in treating human infections, as well as the usage of Dox to induce transgenic mouse models dependent on this antibiotic for expression, may promote selenoprotein and selenium deficiencies.
Supplementary Material
This work was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research (to D. L. H.) and by National Institutes of Health Grants GM065204 and CA080946 (to V. N. G.).

This article contains supplemental Figs. 1 and 2 and Table 1.
- Sec
- selenocysteine
- Cp
- chloramphenicol
- Dox
- doxycycline
- TR
- thioredoxin reductase
- GPx
- glutathione peroxidase
- BIAM
- N-(biotinoyl)-N′-(iodoacetyl)-ethylenediamine
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Birringer M., Pilawa S., Flohé L. (2002) Trends in selenium biochemistry. Nat. Prod. Rep. 19, 693–718 [DOI] [PubMed] [Google Scholar]
- 2. Hatfield D. L., Gladyshev V. N. (2002) How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stadtman T. C. (2002) Discoveries of vitamin B12 and selenium enzymes. Annu. Rev. Biochem. 71, 1–16 [DOI] [PubMed] [Google Scholar]
- 4. Xu X. M., Carlson B. A., Mix H., Zhang Y., Saira K., Glass R. S., Berry M. J., Gladyshev V. N., Hatfield D. L. (2007) Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 5, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu X. M., Turanov A. A., Carlson B. A., Yoo M. H., Everley R. A., Nandakumar R., Sorokina I., Gygi S. P., Gladyshev V. N., Hatfield D. L. (2010) Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc. Natl. Acad. Sci. U.S.A. 107, 21430–21434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlson B. A., Xu X. M., Gladyshev V. N., Hatfield D. L. (2005) Um34 in selenocysteine tRNA is required for the expression of stress-related selenoproteins in mammals. Top. Curr. Genet. 12, 431–438 [Google Scholar]
- 7. Hatfield D. L., Carlson B. A., Xu X. M., Mix H., Gladyshev V. N. (2006) Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 81, 97–142 [DOI] [PubMed] [Google Scholar]
- 8. Carlson B. A., Moustafa M. E., Sengupta A., Schweizer U., Shrimali R., Rao M., Zhong N., Wang S., Feigenbaum L., Lee B. J., Gladyshev V. N., Hatfield D. L. (2007) Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 282, 32591–32602 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. Y., Carlson B. A., Xu X. M., Zeng Y., Chen S., Gladyshev V. N., Lee B. J., Hatfield D. L. (2011) Inhibition of selenocysteine tRNA[Ser]Sec aminoacylation provides evidence that aminoacylation is required for regulatory methylation of this tRNA. Biochem. Biophys. Res. Commun. 409, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohanski M. A., Dwyer D. J., Collins J. J. (2010) How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCoy L. S., Xie Y., Tor Y. (2011) Antibiotics that target protein synthesis. RNA 2, 209–232 [DOI] [PubMed] [Google Scholar]
- 12. Hermann T. (2007) Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell. Mol. Life Sci. 64, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Floquet C., Deforges J., Rousset J. P., Bidou L. (2011) Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 39, 3350–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Handy D. E., Hang G., Scolaro J., Metes N., Razaq N., Yang Y., Loscalzo J. (2006) Aminoglycosides decrease glutathione peroxidase-1 activity by interfering with selenocysteine incorporation. J. Biol. Chem. 281, 3382–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fife R. S., Rougraff B. T., Proctor C., Sledge G. W., Jr. (1997) Inhibition of proliferation and induction of apoptosis by doxycycline in cultured human osteosarcoma cells. J. Lab. Clin. Med. 130, 530–534 [DOI] [PubMed] [Google Scholar]
- 16. Gilbertson-Beadling S., Powers E. A., Stamp-Cole M., Scott P. S., Wallace T. L., Copeland J., Petzold G., Mitchell M., Ledbetter S., Poorman R. (1995) The tetracycline analogs minocycline and doxycycline inhibit angiogenesis in vitro by a non-metalloproteinase-dependent mechanism. Cancer Chemother. Pharmacol. 36, 418–424 [DOI] [PubMed] [Google Scholar]
- 17. Son K., Fujioka S., Iida T., Furukawa K., Fujita T., Yamada H., Chiao P. J., Yanaga K. (2009) Doxycycline induces apoptosis in PANC-1 pancreatic cancer cells. Anticancer Res. 29, 3995–4003 [PubMed] [Google Scholar]
- 18. Thompson J., O'Connor M., Mills J. A., Dahlberg A. E. (2002) The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. J. Mol. Biol. 322, 273–279 [DOI] [PubMed] [Google Scholar]
- 19. Tan L. K. (1999) Chloramphenicol-induced aplastic anaemia–should its topical use be abandoned? Singapore Med. J. 40, 445–446 [PubMed] [Google Scholar]
- 20. Jakupoglu C., Przemeck G. K., Schneider M., Moreno S. G., Mayr N., Hatzopoulos A. K., de Angelis M. H., Wurst W., Bornkamm G. W., Brielmeier M., Conrad M. (2005) Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol. 25, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seiler A., Schneider M., Förster H., Roth S., Wirth E. K., Culmsee C., Plesnila N., Kremmer E., Rådmark O., Wurst W., Bornkamm G. W., Schweizer U., Conrad M. (2008) Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 8, 237–248 [DOI] [PubMed] [Google Scholar]
- 22. Ho Y. S., Magnenat J. L., Bronson R. T., Cao J., Gargano M., Sugawara M., Funk C. D. (1997) Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J. Biol. Chem. 272, 16644–16651 [DOI] [PubMed] [Google Scholar]
- 23. Naranjo-Suarez S., Carlson B. A., Tsuji P. A., Yoo M. H., Gladyshev V. N., Hatfield D. L. (2012) HIF-independent regulation of thioredoxin reductase 1 contributes to the high levels of reactive oxygen species induced by hypoxia. PLoS One 7, e30470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson B. A., Xu X. M., Kryukov G. V., Rao M., Berry M. J., Gladyshev V. N., Hatfield D. L. (2004) Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 12848–12853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arnér E. S., Zhong L., Holmgren A. (1999) Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 300, 226–239 [DOI] [PubMed] [Google Scholar]
- 26. Smith A. D., Levander O. A. (2002) High-throughput 96-well microplate assays for determining specific activities of glutathione peroxidase and thioredoxin reductase. Methods Enzymol. 347, 113–121 [DOI] [PubMed] [Google Scholar]
- 27. Carvalho C. M., Lu J., Zhang X., Arnér E. S., Holmgren A. (2011) Effects of selenite and chelating agents on mammalian thioredoxin reductase inhibited by mercury: implications for treatment of mercury poisoning. FASEB J. 25, 370–381 [DOI] [PubMed] [Google Scholar]
- 28. Turanov A. A., Su D., Gladyshev V. N. (2006) Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J. Biol. Chem. 281, 22953–22963 [DOI] [PubMed] [Google Scholar]
- 29. Jung J. E., Karoor V., Sandbaken M. G., Lee B. J., Ohama T., Gesteland R. F., Atkins J. F., Mullenbach G. T., Hill K. E., Wahba A. J., et al. (1994) Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J. Biol. Chem. 269, 29739–29745 [PubMed] [Google Scholar]
- 30. Hatfield D. L., Berry M. J., Gladyshev V. N. (2012) Selenium: Its Molecular Biology and Role in Human Health, Springer Science, New York [Google Scholar]
- 31. Copeland P. R., Fletcher J. E., Carlson B. A., Hatfield D. L., Driscoll D. M. (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Squires J. E., Stoytchev I., Forry E. P., Berry M. J. (2007) SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol. Cell. Biol. 27, 7848–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hatfield D., Matthews C. R., Rice M. (1979) Aminoacyl-transfer RNA populations in mammalian cells chromatographic profiles and patterns of codon recognition. Biochim. Biophys. Acta 564, 414–423 [DOI] [PubMed] [Google Scholar]
- 34. Gladyshev V. N., Factor V. M., Housseau F., Hatfield D. L. (1998) Contrasting patterns of regulation of the antioxidant selenoproteins, thioredoxin reductase, and glutathione peroxidase, in cancer cells. Biochem. Biophys. Res. Commun. 251, 488–493 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.