Background: CIF converts NEDD8 glutamine 40 to glutamate, inactivating cullin-RING ubiquitin ligases (CRLs).
Results: CIF deamidates NEDD8 covalently attached to cullins, leading to NEDD8 system inhibition.
Conclusion: CIF directly inactivates neddylated CRL complexes and, subsequently, interferes with NEDD8 system function.
Significance: This newly described posttranslational mechanism highlights how a subtle amino acid change in a protein can have far-reaching and amplified biological effects.
Keywords: Signal Transduction, Ubiquitin, Ubiquitin Ligase, Ubiquitination, Yeast, CIF, NEDD8, Cullin-RING Ubiquitin Ligases, Ubiquitin-like Proteins
Abstract
The bacterial effector protein cycle inhibiting factor (CIF) converts glutamine 40 of NEDD8 to glutamate (Q40E), causing cytopathic effects and inhibiting cell proliferation. Although these have been attributed to blocking the functions of cullin-RING ubiquitin ligases, how CIF modulates NEDD8-dependent signaling is unclear. Here we use conditional NEDD8-dependent yeast to explore the effects of CIF on cullin neddylation. Although CIF causes cullin deneddylation and the generation of free NEDD8 Q40E, inhibiting the COP9 signalosome (CSN) allows Q40E to form only on NEDD8 attached to cullins. In the presence of the CSN, NEDD8 Q40E is removed from cullins more rapidly than NEDD8, leading to a decrease in steady-state cullin neddylation. As NEDD8 Q40E is competent for cullin conjugation in the absence of functional CSN and with overexpression of the NEDD8 ligase Dcn1, our data are consistent with NEDD8 deamidation causing enhanced deneddylation of cullins by the CSN. This leads to a dramatic change in the extent of activated cullin-RING ubiquitin ligases.
Introduction
Cullin-RING ubiquitin ligases (CRLs)2 are a large family of modular enzymes that promote the conditional ubiquitination of proteins, leading to their destruction by the proteasome (1). These enzymes are based on a conserved enzymatic core containing a cullin protein and a RING protein (RBX1 or RBX2) that serves as the binding site for ubiquitin-conjugating enzymes (E2s) (2–4). Variability within the amino-terminal region of cullins provides binding sites for subclasses of substrate receptors. Although only a limited number of CRL complexes have been definitively linked to substrates and have known cellular functions, several hundred CRL complexes may exist in humans on the basis of cullin-specific motifs in potential CRL receptors (5).
The ubiquitin-like protein NEDD8 has a critical role in regulating the activities of CRLs (6, 7). The covalent modification of a conserved cullin lysine by a single NEDD8 molecule is associated with structural rearrangements within the CRL catalytic core (8). This conformational change has been proposed to underlie the observed effects of NEDD8 on enhancing CRL substrate ubiquitination (8, 9). These studies suggest that NEDD8 modification (neddylation) promotes E2 recruitment and reduces the distance between E2 and lysine of the CRL-bound substrate to facilitate covalent ubiquitin transfer (10). This process is reversible through the deneddylase activity of the COP9 signalosome (CSN) (11, 12), providing a sensitive switch-like mechanism for regulating CRLs. It remains largely unresolved what controls the extent and timing of activation of a given CRL. Quantitative proteomic studies suggest, however, that NEDD8 may not actually regulate CRL assembly (13). Instead, more recent studies establish a reciprocal regulatory mechanism between CRLs and the CSN. Full CRL complex assembly may actually inhibit deneddylation, whereas CSN binding to unneddylated CRLs has been shown to prevent neddylation in a catalytically independent manner (14, 15).
Studies on the NEDD8-activating enzyme (NAE) inhibitor MLN4924 demonstrate that chemical inhibition of the NEDD8 system leads to the loss of neddylated cullins and the accumulation of CRL substrates. MLN4924 potently inhibits cancer cell proliferation prior to inducing cell death and slows tumor growth in animal models (16, 17). As a result of these promising studies, clinical trials testing MLN4924 on hematological malignancies such as lymphoma, multiple myeloma, leukemia, and advanced non-hematological malignancies are currently in progress (18). Exactly why cancers are susceptible to blocking neddylation and CRL inactivation is unclear, although this could reflect increased dependence on the different functions of NEDD8 to maintain proper CRL substrate levels in certain cancer cell types (19).
Recent work has identified an alternative mechanism to modulate the functions of NEDD8 in cells through a bacterial effector protein known as cycle-inhibiting factor (CIF) (20). CIF induces cytopathic effects such as cell cycle arrest, focal adhesion formation, and stress fiber accumulation and delays apoptosis (21–24). These correlate to inactivation of a subset of CRLs and have been attributed to CIF-mediated conversion of glutamine 40 of NEDD8 to glutamic acid (Q40E) (25, 26). Importantly, either CIF or NEDD8 Q40E is sufficient for inactivation of CRLs and the observed cytopathic effects (25).
The precise mechanism of action of CIF on NEDD8 and how this leads to CRL inactivation remain largely unresolved (Fig. 1). Here we use a molecular genetics approach to explore the effects of CIF on cullin neddylation dynamics in yeast and uncover a mechanism by which CIF modulates CRL homeostasis through enhanced deneddylation of NEDD8 Q40E by the CSN.
FIGURE 1.
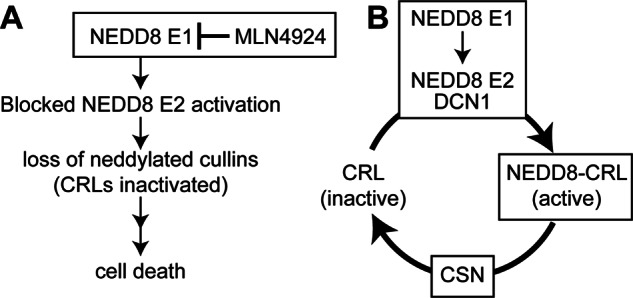
MLN4924 and CIF inhibit CRLs through different mechanisms. A, MLN4924 inhibits the NEDD8-activating enzyme (E1), resulting in general inhibition of the NEDD8 system. This inactivates CRLs (loss of neddylated cullins) and leads to cell death. B, NEDD8 is conjugated to CRLs by the NEDD8 system (E1, E2, and NEDD8 ligase DCN1) and removed by the CSN in a dynamic manner. Exactly how NEDD8 deamidation by CIF inactivates CRLs is unclear, although this is likely to be different from the specific effects of MLN4924 on the NEDD8 E1. Although the CIF product NEDD8 Q40E can be conjugated onto cullins in vitro, it does not appear to support NEDD8-dependent CRL activation (25, 26). Because CIF efficiently deamidates free NEDD8 and NEDD8 in different contexts in vitro (25), CIF could interfere with NEDD8 system function in cells. Finally, NEDD8 recognition and removal by the CSN could be altered by CIF and/or NEDD8 deamidation, changing the extent of activated CRLs.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Genetic Techniques
Yeast strains are described in supplemental Table 1. Strains used for viability assays were in the BY4743 background. Construction of the cdc34Δ/pCDC34 URA3 strain used as the starting strain for all viability experiments was described previously (27). The protease-deficient strain BJ5459 (ATCC) was the starting strain for experiments monitoring yCUL1 modification, NEDD8 deamidation, and Ubc12 charging. Genomic deletions were constructed by PCR-mediated gene replacement using cassettes containing either drug resistance genes to kanamycin (kan) or nourseothricin (nat) or a nutritional marker (LEU2) (28, 29). Strains were transformed using lithium acetate (30). Yeast strains were grown in synthetic dropout medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, and amino acids required to complement nutritional needs of a given strain).
For viability experiments, cells were grown to saturation in synthetic dropout medium, and 5-fold serial dilutions were plated. Cells were incubated at 30 °C, unless noted otherwise, until colonies formed. For the plasmid shuffle experiments shown in Figs. 2, A, C, and E, 3A, 4C, and 9C, cells were grown in synthetic dropout medium to saturation and then plated onto synthetic dropout medium with or without 0.75 g/liter 5-fluoroorotic acid (5-FOA). In the case of galactose induction, cells were grown in synthetic medium containing 2% raffinose instead of glucose and then induced by addition of 2% galactose to medium or plating onto agar plates containing synthetic medium with 2% galactose.
FIGURE 2.
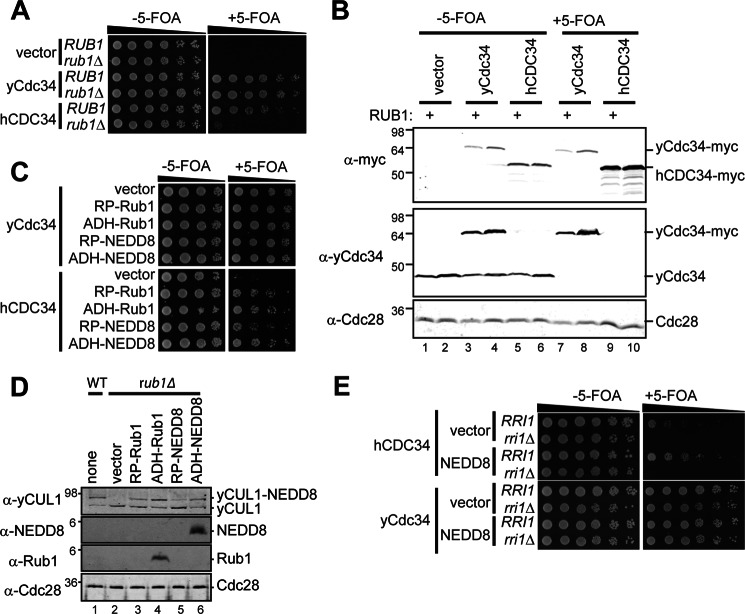
A yeast strain conditionally dependent on NEDD8 and cullin neddylation dynamics. A, haploid yeast strains that either have RUB1 (encodes yeast NEDD8) or the gene deleted (rub1Δ) were constructed so that the essential functions of the yeast ubiquitin-conjugating enzyme Cdc34 are provided by a counterselectable URA3-marked, plasmid-based copy (MPY101 or MPY143, respectively) and plasmids encoding yeast Cdc34-Myc13, human CDC34-Myc13, or vector control. Five-fold serial dilutions of cells were spotted onto medium in the presence or absence of 5-FOA. Cells on 5-FOA-containing medium harbor the indicated version of CDC34 as the sole source of the essential enzyme. B, lysates from the strains used in A with and without 5-FOA treatment were analyzed by immunoblotting to evaluate the expression of Myc-tagged human and yeast Cdc34, yeast Cdc34, and Cdc28 (loading control). C, the rub1Δ strains from A were provided Rub1 or NEDD8 from expression plasmids using either the Rub1 (RP) or ADH promoter and evaluated as in A. D, lysates from a rub1Δ strain (NEDD8-independent, MPY156) transformed with the same plasmids from C were analyzed by immunoblotting to evaluate the neddylation status of yeast CUL1 (Cdc53) and expression of monomeric NEDD8 and Rub1. A control strain containing endogenous RUB1 is shown for comparison (lane 1). E, the contribution of the CSN on the viability of yeast strains that either express ADH-NEDD8 or vector control was evaluated by comparing strains that are either RRI1+ or rri1Δ (MPY143 and MPY202, respectively) in the presence or absence of 5-FOA.
FIGURE 3.
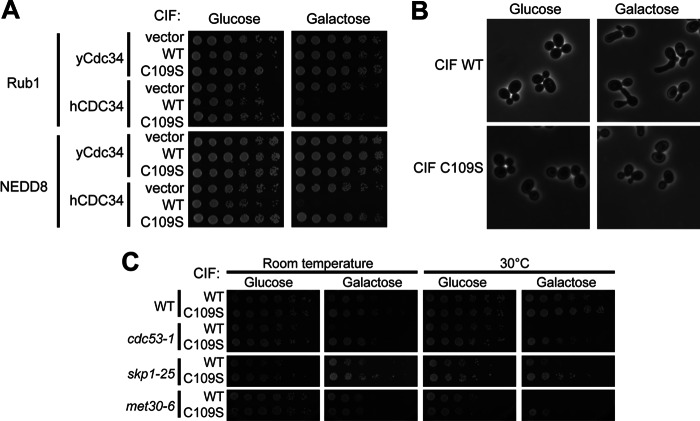
CIF inhibits NEDD8-dependent yeast proliferation. A, the yeast strains from Fig. 2D that express either Rub1 or NEDD8 from the ADH promoter and rely on either human or yeast CDC34 for viability were transformed with plasmids that express galactose-inducible FLAG-tagged CIF (WT), catalytically inactive CIF (C109S), or a vector control. Saturated cultures were serial-diluted and spotted onto medium containing either 2% glucose or galactose to repress or induce CIF expression, respectively. B, the yeast strains from A were diluted to log phase and then induced with either 2% glucose or galactose for 4 h at 30 °C prior to imaging using phase contrast microscopy. Representative fields of cells are shown. C, the plasmids that express galactose-inducible CIF and CIF C109S were transformed into temperature-sensitive strains with mutations in essential CRL pathway components, and cell growth was assessed under the indicated conditions.
FIGURE 4.
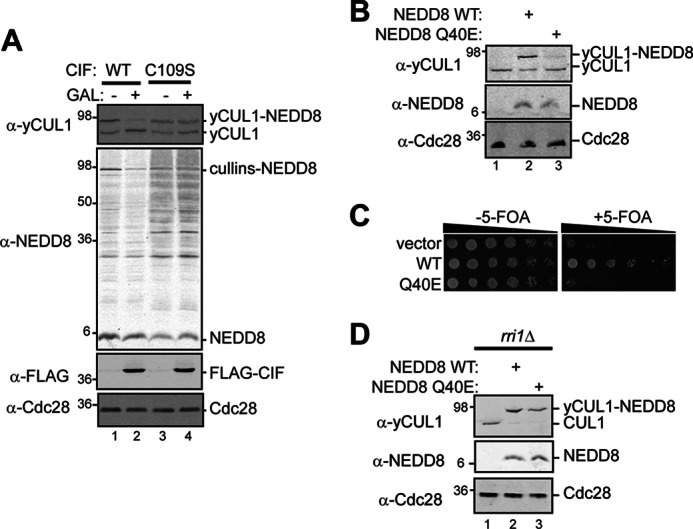
CIF and its product NEDD8 Q40E change the extent of cullin neddylation. A, the rub1Δ/pADH-NEDD8 strain (MPY174) coexpressing galactose-inducible CIF (WT or catalytically inactive C109S) was grown to saturation, diluted 1:3 in medium containing either 2% glucose or galactose (GAL) to repress or induce CIF expression, and incubated for 4 h at 30 °C. Lysates from these cells were analyzed by immunoblotting for yCUL1, NEDD8, and FLAG (detects CIF) with Cdc28 as a loading control. B, lysates from a rub1Δ strain (MPY156) expressing NEDD8, NEDD8 Q40E, or a vector control were analyzed by immunoblotting for yCUL1, NEDD8, and Cdc28. C, serial dilutions of the haploid cdc34Δ rub1Δ strain carrying counterselectable yeast Cdc34 and human CDC34 (MPY146) and expressing NEDD8 or NEDD8 Q40E were plated onto medium with 5-FOA to evaluate the effect of Q40E on NEDD8-conditional growth. D, haploid rub1Δ rri1Δ yeast strains (MPY201) coexpressing NEDD8 or NEDD8 Q40E were analyzed as described in B.
FIGURE 9.
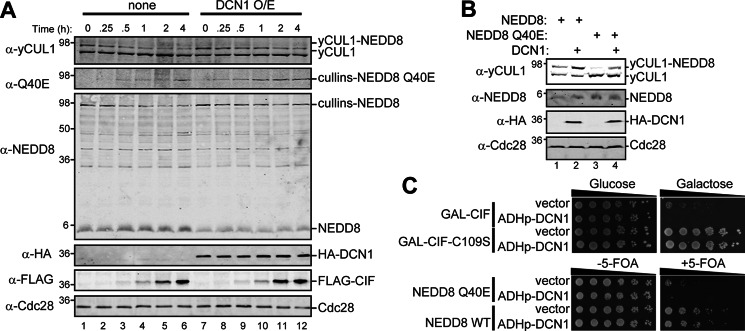
Overexpression of DCN1 alleviates CIF-mediated inhibition of neddylation but does not restore NEDD8-dependent cell proliferation. A, the rub1Δ yeast strains expressing NEDD8, galactose-inducible CIF (MPY231), and either ADH-HA-DCN1 (DCN1 overexpression (O/E)) or a vector control (none) were grown to late log phase. Galactose was added to 2%, and samples taken at the indicated times were analyzed by immunoblotting with the indicated antibodies. B, lysates from rub1Δ yeast strains (MPY156) coexpressing either NEDD8 (WT) or NEDD8 Q40E and ADH-HA-DCN1 or a vector control were analyzed by immunoblotting with the indicated antibodies. C, the effect of Dcn1 on NEDD8-dependent cell growth was evaluated in yeast strains expressing galactose-inducible CIF (WT or C109S, top panel) or NEDD8 (Q40E or WT; bottom panel). Serial dilutions were spotted onto glucose- or galactose-containing medium for inducible CIF expression and medium without or with 5-FOA for NEDD8 conditional growth.
Plasmids
A complete list of plasmids can be found in supplemental Table 2. Constructs were generated using conventional cloning procedures. Mutagenesis was done using QuikChange (Agilent).
Immunoblotting
Lysates were made by resuspending pelleted yeast cells in lysis buffer (50 mm NaHPO4 (pH 7.4), 1 mm EDTA, 4% SDS, 5% glycerol, 2 mm 1,10-orthophenathroline), adding silica beads and disrupting cells using a FastPrep instrument (MPBio). Lysates were clarified by centrifugation. Reducing sample buffer was added (except for experiments that specifically indicated non-reducing sample buffer), and lysates were subjected to SDS-PAGE. Immunoblot analyses were analyzed using a Licor Odyssey system.
The following commercially available antibodies were used: anti-Cdc53 (Santa Cruz Biotechnology, Inc., catalog no. sc-6716), anti-Cdc28 (Santa Cruz Biotechnology, Inc., catalog no. sc-6709), anti-NEDD8 (Invitrogen, catalog no. 34-1400), anti-Rub1 (Rockland, catalog no. 200-401-427), anti-FLAG (Sigma, catalog no. F 1804), and anti-HA (Santa Cruz Biotechnology, Inc., catalog no. sc-805). 9E10 sera (mouse monoclonal) were used to detect Ubc12-Myc. The NEDD8 Q40E antibody is a custom peptide antibody (rabbit) generated against the sequence C-EEKEGIPPQEQRLIY-amide and purified using positive and subtractive affinity chromatography.
Recombinant Protein Purification and Labeled NEDD8
Human NAE (ULA1 and UBA3-His6) was prepared as described previously (31). Human UBC12-His6, NEDD8, and NEDD8 Q40E were expressed in Rosetta 2 Escherichia coli (EMD Millipore) and purified by nickel-nitrilotriacetic acid (UBC12) or SP-Sepharose (NEDD8, NEDD8 Q40E) chromatography. NEDD8 and NEDD8 Q40E were amine-labeled using DyLight 680 N-hydroxysuccinimide ester (Thermo Scientific) under identical conditions at the same time according to the instructions of the manufacturer. After the labeling reaction, proteins were purified by size exclusion chromatography on a Superdex 75 column (GE Healthcare).
CSN Affinity Purification
The CSN2 ORF was PCR-amplified from the MegaMan library (Agilent) and cloned into pMP503 downstream of the CMV promoter and sequences that encode the FLAG epitope (DYKDDDDK) and a tobacco etch virus protease recognition site (NLYFQG) flanked on both sides by SGGGG linker. Stable cell lines derived from serum-free suspension 293-F cells (Invitrogen) were prepared according to the instructions of the manufacturer. The CSN was purified from clarified cell lysates prepared in 25 mm HEPES (pH 8.0), 250 mm NaCl, 0.5 mm EDTA, 1 mm DTT, and 0.1% Igepal CA-630 with FLAG-M2 beads (Sigma), and the CSN complex was eluted with 200 ng/μl FLAG peptide.
In Vitro Neddylation and Deneddylation Assays
Lysates from log phase yeast cells were generated in lysis buffer (20 mm HEPES (pH7.6), 50 mm NaCl, 0.2% Triton X-100, 1 mm DTT) using a FastPrep instrument (MPBio) with silica beads and clarified by centrifugation. Neddylation experiments used cells lacking both Rub1/NEDD8 and a functional CSN (MPY201), and deneddylation experiments used cells lacking a functional CSN and expressing either NEDD8 or NEDD8 Q40E (MPY257and MPY311, respectively).
Neddylation reactions (25 mm HEPES (pH 7.6), 1 mm DTT, 200 μm ATP, 100 nm NAE, 1.5 μm UBC12, 500 nm NEDD8 680, 2 μl MPY201 lysate (20 μg protein)) were incubated at 30 °C, and reducing sample buffer was added at the indicated times. Samples were boiled, and proteins were separated using 7.5% SDS-PAGE. NEDD8 680 was visualized by scanning gels on a LiCor Odyssey system. The identity of yCUL1 as the target for modification with labeled NEDD8 was determined from experiments comparing neddylation profiles between extracts with or without NEDD8/Rub1 expression (see Fig. 8C). A band consistent with the molecular weight of NEDD8-modified yCUL1 could only be detected in extracts lacking NEDD8/Rub1 expression. This modification depended upon the addition of NAE, UBC12, NEDD8, and ATP (data not shown).
FIGURE 8.
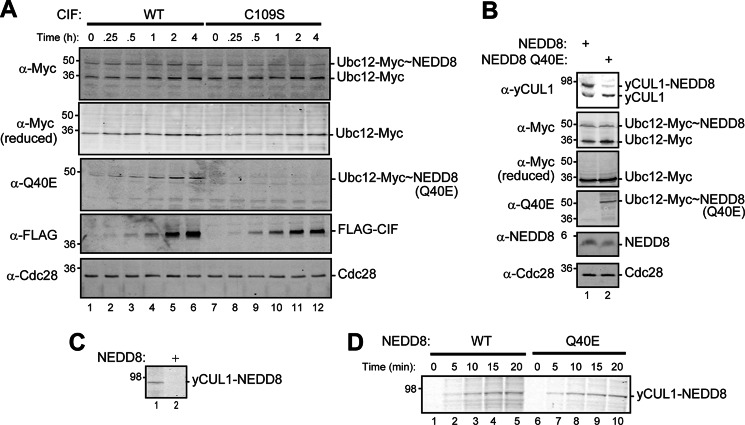
NEDD8 deamidation does not inhibit Ubc12 activation and cullin neddylation. A, lysates from the rub1Δ yeast strain expressing NEDD8 (MPY174) and Myc-tagged Ubc12 with galactose-inducible CIF (WT) or catalytically inactive CIF (C109S) were prepared at the indicated times after 2% galactose induction. Activated Ubc12 (Ubc12-Myc∼NEDD8) was detected by immunoblotting samples prepared under non-reducing conditions (omitting 2-mercaptoethanol from the sample buffer) with anti-Myc antisera and the anti-NEDD8 Q40E antibody. The same samples prepared with 2-mercaptoethanol were also analyzed for Myc-tagged Ubc12, FLAG-tagged CIF, and Cdc28. B, lysates from the rub1Δ yeast strain (MPY156) expressing either NEDD8 or NEDD8 Q40E and Myc-tagged Ubc12 were analyzed under non-reducing and reducing conditions by immunoblotting with the indicated antibodies. C, purified recombinant NEDD8-activating enzyme, UBC12, ATP, and fluorescently labeled NEDD8 were added to yeast lysates prepared from a rub1Δ rri1Δ strain (MPY201) with or without NEDD8 expression. After 30 min at 30 °C, samples were subjected to SDS-PAGE and analyzed for fluorescent NEDD8 by direct gel scanning on a Licor Odyssey. CUL1 neddylation was only detected in lysates lacking NEDD8. Because inactivation of the CSN through rri1Δ leads to complete cullin neddylation in cells containing NEDD8 (e.g. Fig. 4C), lysates from these cells do not have unmodified cullins that could serve as substrates for in vitro neddylation with fluorescent NEDD8. D, in vitro neddylation reactions using either fluorescent NEDD8 (WT) or NEDD8 Q40E were performed using yeast cell lysates from a rub1Δ rri1Δ strain (MPY201) as in C. The reactions were stopped at the indicated times with reducing sample buffer, and NEDD8-modified CUL1 was detected by SDS-PAGE and direct gel scanning using a Licor Odyssey.
For in vitro deneddylation experiments, an affinity-purified CSN (see above) was added to lysates (400 μg of protein) at a final concentration of 10 nm and incubated at 30 °C. Samples were taken at the indicated times, added to reducing sample buffer, and subjected to SDS-PAGE. Deneddylation of yeast CUL1 was visualized by Western blotting using anti-Cdc53 antibody.
RESULTS
Development of Conditional NEDD8-dependent Yeast
Unlike higher eukaryotes, budding yeast does not require NEDD8 (known as Rub1) for viability (32). The conservation of cullin neddylation and CSN-mediated deneddylation pathways in yeast, however, have allowed for the discovery of many fundamental regulatory mechanisms controlling CRLs in eukaryotes. With these considerations, we sought to devise a yeast strain that conditionally requires NEDD8 for vegetative growth. Because in vitro biochemical studies have demonstrated that cullin neddylation enhances the activity of the human ubiquitin-conjugating enzyme CDC34 (hCDC34) (33), we hypothesized that its replacement for the essential yeast Cdc34 homologue (yCdc34) may render cells dependent upon NEDD8.
Although yeast strains expressing yCdc34 grow equivalently in the presence or absence of Rub1, cells relying on hCDC34 for the essential functions of the enzyme do not (Fig. 2A). Yeast strains expressing hCDC34 with Rub1 are ∼5-fold growth-impaired compared with those utilizing yCdc34. Although the precise reason for this difference is currently unclear, it is not simply due to decreased expression of hCDC34 because its overall steady state abundance is higher than that of yCdc34 (Fig. 2B). Importantly, this strain phenocopies CRL1 temperature-sensitive mutant strains (34).
NEDD8 is the functional homolog of Rub1 because it can modify substrates in yeast and restore viability under certain conditions that require Rub1 (35). We tested whether NEDD8 and Rub1 are functionally interchangeable in this NEDD8-dependent strain and whether the extent of expression affects cell growth (Fig. 2C). Although Rub1 expressed from its own promoter and the constitutively strong alcohol dehydrogenase (ADH) promoter support growth equivalently, we observed a slight growth impairment associated with NEDD8 expressed from the Rub1 promoter relative to ADH-expressed NEDD8 under NEDD8-dependent conditions.
To determine whether these differences reflect differences in cullin neddylation, we assessed the neddylation status of yeast CUL1 (yCUL1, Cdc53) and the expression of NEDD8 and Rub1 by immunoblotting extracts from NEDD8-independent strains (Fig. 2D). Rub1 or NEDD8 expression from the strong ADH promoter allows for detection of monomeric forms of the proteins, whereas expression from the Rub1 promoter does not (Fig. 2D, compare lane 4 to lane 3 and lane 6 to lane 5). In examining yCUL1, we observed different extents of NEDD8 modifications. Overexpression of Rub1 from the ADH promoter results in a slight increase in yCUL1 neddylation relative to Rub1 expressed from its genomic locus (Fig. 2D, compare lane 4 to lane 1), whereas NEDD8 overexpression results in similar levels (Fig. 2D, compare lane 6 to lane 1). Furthermore, expression of NEDD8 from the Rub1 promoter leads to a decrease in yCUL1 neddylation (compare lane 5 to lane 1), correlating to the mild growth impairment observed under NEDD8-dependent growth conditions.
These observations are important in light of recent reports that NEDD8 overexpression in human cells leads to inappropriate activation of NEDD8 by the ubiquitin-activating enzyme UBA1 and the neddylation of proteins that may not be authentic NEDD8 system substrates (36–39). NEDD8 overexpression in yeast used here supports cullin neddylation and vegetative growth at levels similar to those relying on endogenous Rub1. It is also important to note that although yCUL1 is the only known neddylation substrate in budding yeast that provides an essential function (34, 40), other yeast cullins (Rtt101 and Cul3) are also reported to be modified (40).
Furthermore, NEDD8-dependent yeast strains require functional deneddylation machinery because NEDD8-dependent strains lacking RRI1 (the catalytic subunit of the CSN) are not viable (Fig. 2E). This observation reflects a dependence on cullin neddylation dynamics for cell proliferation.
CIF Inhibits NEDD8-dependent Cell Proliferation and Alters Cullin Neddylation
In human cells, the bacterial effector protein CIF triggers growth arrest and a variety of cytopathic effects that have been correlated to its unique NEDD8-directed deamidase activity (20). We reasoned that evaluating the effects of CIF on NEDD8- dependent and NEDD8-independent yeast provides an opportunity to gain mechanistic insight into CRL inactivation by CIF.
We found that galactose-induced expression of CIF (WT) impaired the growth of Rub1- or NEDD8-dependent yeast strains that rely on hCDC34, whereas a catalytically impaired form of CIF (C109S, Ref. 41) did not (Fig. 3A). This effect is directly attributable to the essential functions of NEDD8 because NEDD8 independent strains using yCdc34 are unaffected by CIF expression. In examining changes in cellular morphology associated with CIF (Fig. 3B), we found that its expression, but not expression of C109S, is associated with an elongated bud phenotype characteristic of mutants in the CRL1Cdc4 pathway (42, 43). Additionally, we tested whether CIF expression would synthetically impair the growth of previously characterized CRL1 temperature-sensitive strains because these strains require functional neddylation machinery (34). As expected, CIF expression (but not the C109S mutant form) resulted in synthetic growth defects with the cdc53-1, skp1-25, and met30-6 strains (Fig. 3C).
We next evaluated the steady-state levels of neddylated cullins with respect to CIF expression (Fig. 4A). CIF expression in yeast leads to a decrease in cullin neddylation with only a small fraction of neddylated yCUL1 persisting. This reduction depends on the catalytic activity of CIF because C109S has no effect on the NEDD8 modification of yCUL1 and on the steady-state levels of NEDD8-protein conjugates. The same decrease in yCUL1 neddylation levels is observed upon expression of NEDD8 Q40E (the CIF product) when compared with WT NEDD8 (Fig. 4B). Similar to CIF expression, this correlates to a growth defect in the NEDD8 dependent strain (Fig. 4C). To determine whether NEDD8 Q40E modifies yCUL1 in vivo, we expressed NEDD8 WT or Q40E in a NEDD8-independent strain in which RRI1 (encodes the CSN catalytic subunit, Refs. 11, 44, 45) is deleted. In the absence of deneddylation, essentially all available yCUL1 is modified with both WT and NEDD8 Q40E (Fig. 4D).
Collectively, these experiments establish a clear link between CIF and NEDD8 because only NEDD8-dependent yeast strains are growth-inhibited by the enzyme or its NEDD8 Q40E product. Our data are consistent with CIF causing CRL inactivation through the accumulation of cullins in their deneddylated state. The decrease in neddylated yCUL1 observed with both CIF and NEDD8 Q40E expression may be due to impaired neddylation and/or increased deneddylation upon NEDD8 deamidation.
CIF Deamidates NEDD8 in Yeast
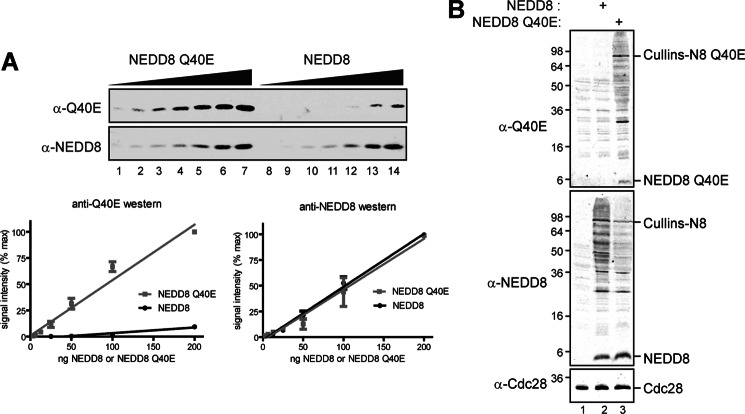
We next sought to address how the NEDD8-directed deamidase activity of CIF leads to cullin deneddylation. To facilitate these studies, we generated a rabbit polyclonal antibody that preferentially recognizes the CIF product NEDD8 Q40E (Fig. 5). In addition to validating the antibody by immunoblotting purified recombinant NEDD8 Q40E and NEDD8 (Fig. 5A), we found that the anti-Q40E antibody recognizes NEDD8 Q40E in both its unconjugated “free” state and when attached covalently to cullins, with no significant NEDD8 cross-reactivity in yeast cell extracts (Fig. 5B). Importantly, this antibody does not recognize ubiquitin (either purified recombinant protein or in extracts) or Rub1 in yeast cell extracts (data not shown).
FIGURE 5.
An antibody that preferentially recognizes NEDD8 Q40E. A, 2-fold serial dilutions of purified recombinant NEDD8 Q40E or NEDD8 (3.125–200 ng) were analyzed by immunoblot analysis with the indicated antibodies. Graphs of quantitative analyses comparing signal intensity detected using a Licor Odyssey are shown for each (n = 3). Error bars represent S.E. B, yeast lysates from a rub1Δ yeast strain (MPY156) expressing vector control, NEDD8, or NEDD8 Q40E were immunoblotted with the NEDD8 or NEDD8 Q40E antibodies.
We first used the anti-Q40E antibody to monitor the appearance of deamidated NEDD8 over time with respect to galactose-induced CIF expression (Fig. 6A). Interestingly, the majority of the observed decrease in NEDD8-modified yCUL1 occurs within the first 30 min of CIF-induction, whereas CIF levels are relatively low. This coincides with the appearance of deamidated NEDD8 in its free, unconjugated state. Over time, the steady-state abundance of free NEDD8 Q40E increases as yCUL1 neddylation decreases and CIF levels increase. Only a small amount of NEDD8 attached to cullins is detectable in its deamidated form.
FIGURE 6.
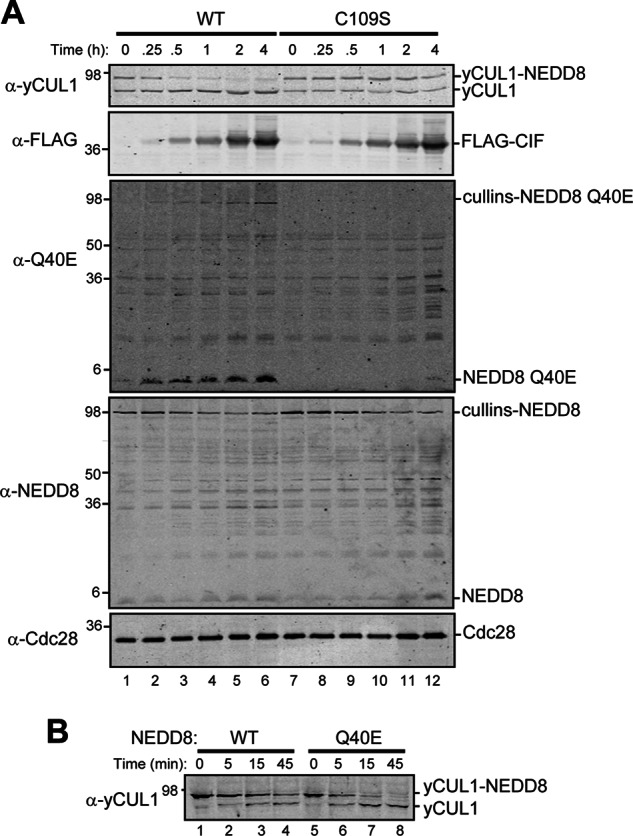
Deamidated NEDD8 is removed more efficiently by the CSN than NEDD8. A, haploid rub1Δ yeast strains expressing NEDD8 (MPY174) and galactose-inducible CIF (WT) or catalytically inactive CIF (C109S) were grown to late log phase and induced with galactose. Cell lysates prepared at the times after galactose addition were analyzed by immunoblotting with the indicated antibodies. B, an affinity-purified CSN was added to lysates from rub1Δ rri1Δ strains (MPY201) expressing NEDD8 or NEDD8 Q40E. Samples were analyzed at the indicated times by immunoblotting for yeast CUL1.
NEDD8 Q40E Is Removed from yCUL1 More Rapidly Than WT NEDD8
We next tested whether yCUL1 modified with NEDD8 Q40E is more susceptible to deneddylation by the CSN than the protein modified with WT NEDD8. Yeast extracts from strains containing either exogenous NEDD8 WT or Q40E and lacking functional deneddylation machinery (rri1-Δ, the majority of yCUL1 is neddylated) were incubated with the purified CSN. By monitoring CSN activity on yCUL1-NEDD8 over time, we found that NEDD8 Q40E was removed more rapidly than WT NEDD8 (Fig. 6B). This result suggests that the difference in steady-state neddylation levels with deamidated NEDD8 is at least partially due to an increase in deneddylation by the CSN.
Deamidation Occurs on NEDD8-modified Cullins
Although overexpression of NEDD8 does not alter the extent of yCUL1 neddylation (e.g. Fig. 2D), it may provide a large pool of free NEDD8 that serves as substrates for CIF in the context of the more limited amount of NEDD8 attached to cullins. An additional complication, however, relates to the dynamics of cullin neddylation and deneddylation. Although the rates of NEDD8 cycling on and off of cullins are unknown, they are likely extremely rapid on the basis of experiments treating human cells with the NAE inhibitor MLN4924. In these studies, all cullins are completely deneddylated within 10 min of NEDD8 system inhibition (46). Moreover, this effect is reversible because neddylation is restored within 15 min of removal of MLN4924 (47). Our data also suggest that this dynamic cycling is necessary for NEDD8-dependent yeast cell proliferation (Fig. 2).
To evaluate the importance of neddylation dynamics in the generation of NEDD8 Q40E, we inhibited cullin deneddylation by deleting RRI1 (Fig. 7). The fully neddylated yCUL1 persists independently of CIF induction, suggesting that CIF-associated deneddylation requires a CSN. In examining the generation of NEDD8 Q40E, we found that essentially all of the Q40E reactivity is associated with neddylated cullins, with very little (if any) deamidated free NEDD8. This is in sharp contrast with what we observed with unperturbed neddylation/deneddylation dynamics (Fig. 6). Importantly, the rri1Δ strain also overexpresses NEDD8 from the ADH promoter, and free NEDD8 is readily detectable, indicating that this observation is not attributable to the depletion of free NEDD8. Instead, our data are consistent with CIF preferentially deamidating NEDD8 attached to cullins, followed by CSN-mediated deneddylation. This leads to the accumulation of deamidated NEDD8 and unmodified cullins because of increased deneddylation.
FIGURE 7.
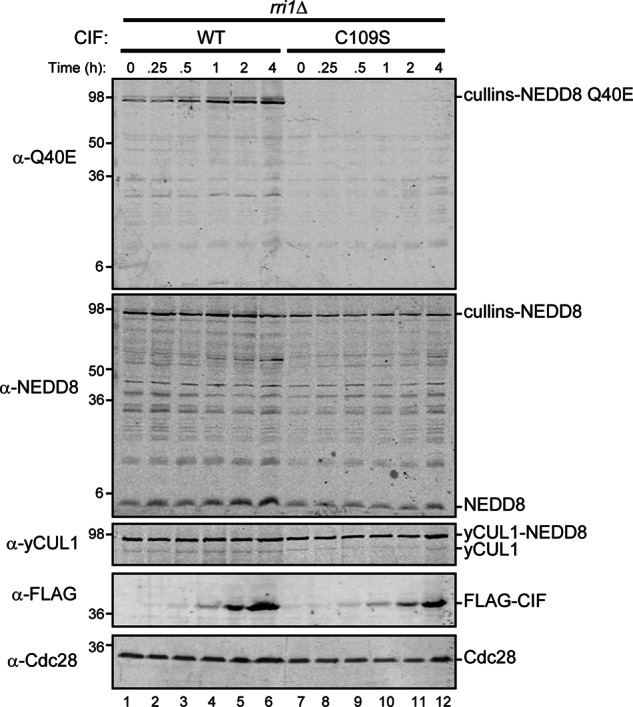
NEDD8 deamidation occurs primarily on cullins. Haploid rub1Δ rri1Δ yeast strains expressing NEDD8 (MPY257) and either WT or catalytically inactive (C109S) CIF were analyzed as described in Fig. 6A.
NEDD8 Q40E Is Not Impaired for Cullin Neddylation
Although NEDD8 Q40E is more rapidly removed from cullins by the CSN, it is also possible that NEDD8 Q40E may not be efficiently utilized by the NEDD8 system. To explore this possibility, we examined the extent of Ubc12 activation by NEDD8 with induced CIF expression (Fig. 8A). Ubc12, the only NEDD8-conjugating enzyme in yeast, is directly involved in covalent NEDD8 transfer onto cullins (32). We reasoned that CIF-mediated inhibition of its activation would implicate mechanisms at or upstream of NEDD8 activation. In contrast with this expectation, however, Ubc12 activation appears to be largely unaffected by induction of the enzyme. Moreover, as CIF levels increase, the amount of NEDD8 Q40E activated on Ubc12 increases correspondingly.
On the basis of our observation that genetic ablation of the CSN results in NEDD8 Q40E only covalently attached to cullins (Fig. 7), it is unlikely that the accumulation of deamidated NEDD8 on Ubc12 is due to the deamidation of NEDD8 in this context or during any of the other steps prior to cullin conjugation. However, it is possible that CIF could modulate the NEDD8 system independently of its NEDD8-directed deamidase activity. To address this, we tested whether NEDD8 Q40E expression is sufficient for the decrease in yCUL1 neddylation and associated accumulation of deamidated NEDD8 on Ubc12 (Fig. 8B). Like the effects of CIF, NEDD8 Q40E is transferred to Ubc12 to a similar extent as wild-type NEDD8 and is associated with a reduction in modified yCUL1.
To compare the rate of yCUL1 neddylation by NEDD8 and NEDD8 Q40E, purified components of the neddylation machinery (NAE, UBC12, ATP, and fluorescent NEDD8 or NEDD8 Q40E) were added to yeast lysates lacking both RUB1 and RRI1. This provides a completely unmodified pool of yCUL1, which, unlike wild-type lysates, can be modified in vitro with labeled NEDD8 transferred by recombinant NEDD8 system enzymes (Fig. 8C). In examining yCUL1 modification over time in this in vitro system with either NEDD8 or NEDD8 Q40E (Fig. 8D), we found very similar extents of NEDD8 incorporation with respect to time.
The NEDD8 Ligase Dcn1 Alleviates Inhibition of Cullin Neddylation Caused by CIF
Although Ubc12 directly modifies cullins with NEDD8 in vitro, recent work has identified an evolutionarily conserved protein in yeast known as Dcn1 (defective in cullin neddylation 1) that functions as a NEDD8 ligase to facilitate the covalent transfer of NEDD8 from Ubc12 onto yCUL1 (48–50) These studies further determined that overexpression of Dcn1 increases the steady-state levels of neddylated yCUL1, suggesting a rate-limiting role in the modification of cullins with NEDD8 (51).
We tested whether Dcn1 overexpression alleviates the loss of neddylated cullins found with CIF and NEDD8 Q40E expression (Fig. 9). Indeed, Dcn1 delays the loss of modified yCUL1 as the steady-state levels of CIF increase (Fig. 9A) and allows for NEDD8 Q40E to be conjugated on yCUL1 to a similar extent as NEDD8 without Dcn1 overexpression (Fig. 9B, compare lane 4 to lane 1). As NEDD8 Q40E is not markedly impaired for cullin neddylation in the absence of a functional CSN (Fig. 8D), this effect of DCN1 is likely indirect due to increased cullin neddylation relative to deneddylation. We next evaluated whether Dcn1 overexpression prevents growth arrest because of CIF or NEDD8 Q40E expression (Fig. 9C). Dcn1 was not sufficient in either case, suggesting that deamidated NEDD8 is unable to support CRL activation in yeast and that inhibited NEDD8-dependent growth is not simply due to impaired cullin neddylation.
DISCUSSION
Cycles of neddylation and deneddylation provide the requisite homeostasis of activated and inactivated CRLs necessary for cell survival (52). The precise dynamics of these processes, their regulation, and how they contribute to the activities, functions, and composition of CRLs are complex and remain highly significant to understand. The bacterial effector protein CIF, through its unique NEDD8-directed deamidase activity (25), represents an important new tool to investigate cullin neddylation in cells. By evaluating the effects of CIF and its product NEDD8 Q40E in yeast that are conditionally NEDD8-dependent, we have uncovered a mechanism by which NEDD8 deamidation by CIF disrupts normal CRL homeostasis to inactivate these enzymes, thereby effectively inhibiting cell proliferation (Fig. 10).
FIGURE 10.
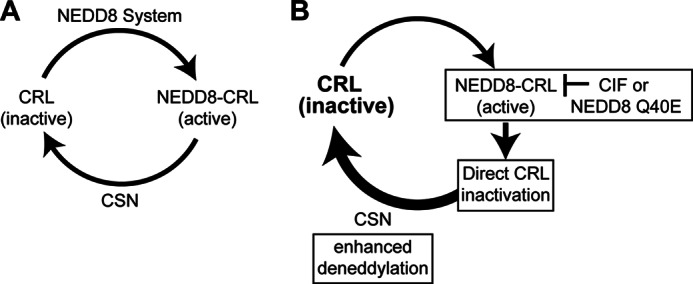
CIF alters CRL neddylation dynamics. A, CRLs typically cycle between active and inactive states, depending on cullin neddylation status. The NEDD8 system is responsible for the conjugation of NEDD8 to cullins, whereas the CSN juxtaposes this to provide the requisite homeostasis of activated CRLs. B, CIF deamidates NEDD8 attached to cullins (Fig. 7). This results in direct CRL inactivation because CRLs modified with NEDD8 Q40E are not active in vitro (25, 26) and do not support growth in yeast (Fig. 9). The CSN removes NEDD8 Q40E more efficiently than wild-type NEDD8 from CRLs, resulting in an increase in deneddylated CRLs (Fig. 6). Once removed, NEDD8 Q40E is competent for further rounds of CRL modification (Fig. 8). However, the enhanced deneddylation of deamidated NEDD8 results in reduced steady-state levels of neddylated CRLs (Fig. 4), rendering cells incompetent for vegetative growth (Fig. 3).
CIF Deamidates NEDD8 Attached to Cullins
In addition to providing rigorous genetic evidence that the effects of CIF on cell proliferation are through deamidation of glutamine 40 of NEDD8, our study demonstrates that NEDD8 attached to cullins, and not free NEDD8, is the preferred substrate of CIF. Although we observed the accumulation of free NEDD8 Q40E upon CIF induction in yeast (Fig. 6), inhibiting deneddylation allows Q40E to be detected only on NEDD8 associated covalently with cullins (Fig. 7). Importantly, genetic ablation of the CSN results in essentially all of yCUL1 converted to its neddylated form but does not deplete free NEDD8. Therefore, our results are not consistent with changes in the ratio of free and cullin-attached NEDD8 altering the preference of CIF for one form over the other. Instead, these data strongly suggest that CIF recognizes NEDD8 in the context of modified cullins.
Structures and sequence alignments of CIF from different bacterial pathogens have led to the identification of a cysteine-histidine-glutamine papain-like catalytic triad within the C-terminal domain that is distantly related to cysteine proteases and acetyltransferases (41, 53–55). Although the N-terminal domain has been proposed to function in substrate recognition, recent structural studies did not find specific residues important for the CIF and NEDD8 interaction (56). The authors instead proposed that shape complementarities rather than specific amino acid interactions are the driving force behind complex formation. Similar structural studies involving the CIF homolog in Burkholderia pseudomallei (CHBP) in a complex with NEDD8 identified several sites of contact over a large interaction interface, with surprising similarity to the interaction surface between NEDD8 and its E1-activating enzyme (57). In light of our data here and the copurification of neddylated cullins with CIF from human cells (26), it seems likely that NEDD8 deamidation in vivo requires aspects of CRL recognition by CIF, such as specific CIF:cullin contact residues or favorable positioning of NEDD8 for CIF binding. In contrast, CIF is capable of robustly deamidating free NEDD8 in vitro (25). It remains a high priority for future efforts to understand the molecular basis of NEDD8 recognition by CIF and how this involves its covalent attachment to cullins.
Deamidated NEDD8 Directly Inhibits the Functions of CRLs in Cells
In vitro experiments demonstrate that NEDD8 Q40E modifies cullins but is not competent for full CRL activation (25, 26) Overexpression of NEDD8 Q40E in human cells (46) and our data using yeast, however, found decreases in steady-state levels of neddylated cullins. These observations suggest that changes in the extent of cullin neddylation, rather than deamidation of the NEDD8 Q40E residue, may underlie the observed cellular phenotypes.
By overexpressing Dcn1, we uncoupled the decrease in steady-state neddylated cullins from NEDD8 Q40E expression (Fig. 9). Through this approach, we were able to determine that restoring cullin neddylation with NEDD8 Q40E is not sufficient for NEDD8-dependent growth. This suggests that the effects of NEDD8 deamidation are at least partially due to the inability of conjugated NEDD8 Q40E to activate CRLs and not exclusively through loss of neddylated cullins.
Exactly how conjugated NEDD8 Q40E inhibits CRL activation remains largely unknown. It is reasonable to propose that deamidated NEDD8 is not effective at promoting and/or stabilizing NEDD8-dependent CRL conformations necessary for activation. This could be through modulating non-covalent interactions between the Ile-44 hydrophobic patch on the surface of NEDD8 and cullins, by analogy to how the Q40E mutation in ubiquitin affects Ile-44 patch recognition by a ubiquitin associated (UBA) ubiquitin binding domain (46). Given the dramatic conformational changes associated with NEDD8 modification and the interactions of the Ile-44 patch with a short α helical region of cullins (8), it is remarkable that changes in this relatively small surface area could be transmitted across the large CRL protein complex.
CIF Alters Cullin Neddylation Dynamics
Sustained expression of CIF in yeast leads to cullin deneddylation. Interestingly, bacterial delivery of CIF into HeLa cells enhances cullin neddylation (26, 58). We reason that the difference is likely due to how CIF is expressed (galactose-induced constitutive expression versus direct protein delivery), the corresponding cellular concentrations of CIF, and extents of NEDD8 deamidation. In support of these possibilities and to reconcile our yeast data with human cell studies, overexpression of NEDD8 Q40E in human cells recapitulates the decrease in cullin neddylation that we observed in yeast cells with either galactose-induced CIF expression or constitutive NEDD8 Q40E expression (46). In addition, transfection of CIF-containing constructs into HEK293T cells did not result in increased cullin neddylation (46).
We found that NEDD8 Q40E accumulates on the active site of Ubc12 and modifies cullins with similar kinetics to WT NEDD8 (Fig. 8). Interestingly, the difference in steady-state neddylation levels is attributable to enhanced deneddylation of NEDD8 Q40E by the CSN (Fig. 6B). Dcn1 overexpression is able to counteract this effect, likely by increasing the rate of neddylation. In support of this, Dcn1 overexpression also increases the steady-state levels of neddylated cullins with wild-type NEDD8 (Ref. 51 and Fig. 9). Thus, the extent of Dcn1 expression can alter the homeostasis of neddylated cullins, suggesting a rate-limiting function in regulating CRL activation.
To our knowledge, the deamidation of NEDD8 by CIF is the only known posttranslational modification of a ubiquitin-like protein. The mechanism identified here highlights how a subtle amino acid change in a protein can have far-reaching and amplified biological effects. In addition to directly inactivating neddylated CRL complexes, NEDD8 Q40E is more efficiently removed from cullins by the CSN than NEDD8. This provides a second layer of control to ensure that CRLs are maintained in a quiescent state, which presumably confers a competitive advantage for the bacterial pathogen.
Supplementary Material
Acknowledgments
We thank C. Erec Stebbins for providing the CIF and CIF C109S open reading frames; Peter Kaiser for providing the cdc53-1, skp1-25, and met30-6 strains; and Dieter Wolf for helpful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by NCI National Institutes of Health Training Grant 5 T32 CA121949 (to T. B. T.). This work was also supported by institutional start-up funds (to M. D. P.), by a scholars award from the V Foundation for Cancer Research (to M. D. P.), and by American Cancer Society Research Scholars Grant RSG-11-224-01-DMC (to M. D. P.).

This article contains supplemental Tables 1 and 2.
- CRL
- cullin-RING ubiquitin ligase
- CSN
- COP9 signalosome
- NAE
- NEDD8-activating enzyme
- CIF
- cycle-inhibiting factor
- 5-FOA
- 5-fluoroorotic acid
- ADH
- alcohol dehydrogenase.
REFERENCES
- 1. Bosu D. R., Kipreos E. T. (2008) Cullin-RING ubiquitin ligases. Global regulation and activation cycles. Cell Div. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase. Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 3. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 4. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 5. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 6. Rabut G., Peter M. (2008) Function and regulation of protein neddylation. “Protein modifications. Beyond the usual suspects” review series. EMBO Rep. 9, 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K. (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997 [DOI] [PubMed] [Google Scholar]
- 8. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases. Conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 [DOI] [PubMed] [Google Scholar]
- 12. Wolf D. A., Zhou C., Wee S. (2003) The COP9 signalosome. An assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5, 1029–1033 [DOI] [PubMed] [Google Scholar]
- 13. Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emberley E. D., Mosadeghi R., Deshaies R. J. (2012) Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J. Biol. Chem. 287, 29679–29689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enchev R. I., Scott D. C., da Fonseca P. C., Schreiber A., Monda J. K., Schulman B. A., Peter M., Morris E. P. (2012) Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 17. Soucy T. A., Smith P. G., Rolfe M. (2009) Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin. Cancer Res. 15, 3912–3916 [DOI] [PubMed] [Google Scholar]
- 18. Wang M., Medeiros B. C., Erba H. P., DeAngelo D. J., Giles F. J., Swords R. T. (2011) Targeting protein neddylation. A novel therapeutic strategy for the treatment of cancer. Expert. Opin. Ther. Targets 15, 253–264 [DOI] [PubMed] [Google Scholar]
- 19. Soucy T. A., Dick L. R., Smith P. G., Milhollen M. A., Brownell J. E. (2010) The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer 1, 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taieb F., Nougayrède J. P., Oswald E. (2011) Cycle inhibiting factors (cifs). Cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins 3, 356–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchès O., Ledger T. N., Boury M., Ohara M., Tu X., Goffaux F., Mainil J., Rosenshine I., Sugai M., De Rycke J., Oswald E. (2003) Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 22. Nougayrède J. P., Boury M., Tasca C., Marchès O., Milon A., Oswald E., De Rycke J. (2001) Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect. Immun. 69, 6785–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Rycke J., Comtet E., Chalareng C., Boury M., Tasca C., Milon A. (1997) Enteropathogenic Escherichia coli O103 from rabbit elicits actin stress fibers and focal adhesions in HeLa epithelial cells, cytopathic effects that are linked to an analog of the locus of enterocyte effacement. Infect. Immun. 65, 2555–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samba-Louaka A., Nougayrède J. P., Watrin C., Oswald E., Taieb F. (2009) The enteropathogenic Escherichia coli effector Cif induces delayed apoptosis in epithelial cells. Infect. Immun. 77, 5471–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cui J., Yao Q., Li S., Ding X., Lu Q., Mao H., Liu L., Zheng N., Chen S., Shao F. (2010) Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jubelin G., Taieb F., Duda D. M., Hsu Y., Samba-Louaka A., Nobe R., Penary M., Watrin C., Nougayrède J. P., Schulman B. A., Stebbins C. E., Oswald E. (2010) Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 6, e1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petroski M. D., Kleiger G., Deshaies R. J. (2006) Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF-Cdc34 ubiquitin ligase complex. Mol. Cell 24, 523–534 [DOI] [PubMed] [Google Scholar]
- 28. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 29. Goldstein A. L., McCusker J. H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 30. Gietz R. D., Woods R. A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 31. Toth J. I., Yang L., Dahl R., Petroski M. D. (2012) A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 1, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liakopoulos D., Doenges G., Matuschewski K., Jentsch S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu K., Chen A., Pan Z. Q. (2000) Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324 [DOI] [PubMed] [Google Scholar]
- 34. Lammer D., Mathias N., Laplaza J. M., Jiang W., Liu Y., Callis J., Goebl M., Estelle M. (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liakopoulos D., Büsgen T., Brychzy A., Jentsch S., Pause A. (1999) Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl. Acad. Sci. U.S.A. 96, 5510–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L. R., Kurz T. (2012) Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hjerpe R., Thomas Y., Kurz T. (2012) NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 421, 27–29 [DOI] [PubMed] [Google Scholar]
- 38. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leidecker O., Matic I., Mahata B., Pion E., Xirodimas D. P. (2012) The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 11, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 40. Laplaza J. M., Bostick M., Scholes D. T., Curcio M. J., Callis J. (2004) Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem. J. 377, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao Q., Cui J., Zhu Y., Wang G., Hu L., Long C., Cao R., Liu X., Huang N., Chen S., Liu L., Shao F. (2009) A bacterial type III effector family uses the papain-like hydrolytic activity to arrest the host cell cycle. Proc. Natl. Acad. Sci. U.S.A. 106, 3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mathias N., Johnson S. L., Winey M., Adams A. E., Goetsch L., Pringle J. R., Byers B., Goebl M. G. (1996) Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16, 6634–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohta T., Michel J. J., Schottelius A. J., Xiong Y. (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541 [DOI] [PubMed] [Google Scholar]
- 44. Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D. A., Wei N., Shevchenko A., Deshaies R. J. (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 [DOI] [PubMed] [Google Scholar]
- 45. Wee S., Hetfeld B., Dubiel W., Wolf D. A. (2002) Conservation of the COP9/signalosome in budding yeast. BMC Genet. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boh B. K., Ng M. Y., Leck Y. C., Shaw B., Long J., Sun G. W., Gan Y. H., Searle M. S., Layfield R., Hagen T. (2011) Inhibition of cullin RING ligases by cycle inhibiting factor. Evidence for interference with Nedd8-induced conformational control. J. Mol. Biol. 413, 430–437 [DOI] [PubMed] [Google Scholar]
- 47. Milhollen M. A., Thomas M. P., Narayanan U., Traore T., Riceberg J., Amidon B. S., Bence N. F., Bolen J. B., Brownell J., Dick L. R., Loke H. K., McDonald A. A., Ma J., Manfredi M. G., Sells T. B., Sintchak M. D., Yang X., Xu Q., Koenig E. M., Gavin J. M., Smith P. G. (2012) Treatment-emergent mutations in NAEβ confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell 21, 388–401 [DOI] [PubMed] [Google Scholar]
- 48. Scott D. C., Monda J. K., Grace C. R., Duda D. M., Kriwacki R. W., Kurz T., Schulman B. A. (2010) A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell 39, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott D. C., Monda J. K., Bennett E. J., Harper J. W., Schulman B. A. (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., Peter M., Sicheri F. (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29, 23–35 [DOI] [PubMed] [Google Scholar]
- 51. Kurz T., Ozlü N., Rudolf F., O'Rourke S. M., Luke B., Hofmann K., Hyman A. A., Bowerman B., Peter M. (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435, 1257–1261 [DOI] [PubMed] [Google Scholar]
- 52. Deshaies R. J., Emberley E. D., Saha A. (2010) Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell. Biochem. 54, 41–56 [DOI] [PubMed] [Google Scholar]
- 53. Crow A., Race P. R., Jubelin G., Varela Chavez C., Escoubas J. M., Oswald E., Banfield M. J. (2009) Crystal structures of Cif from bacterial pathogens Photorhabdus luminescens and Burkholderia pseudomallei. PLoS ONE 4, e5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Samba-Louaka A., Taieb F., Nougayrède J. P., Oswald E. (2009) Cif type III effector protein: a smart hijacker of the host cell cycle. Future Microbiol. 4, 867–877 [DOI] [PubMed] [Google Scholar]
- 55. Hsu Y., Jubelin G., Taieb F., Nougayrède J. P., Oswald E., Stebbins C. E. (2008) Structure of the cyclomodulin Cif from pathogenic Escherichia coli. J. Mol. Biol. 384, 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crow A., Hughes R. K., Taieb F., Oswald E., Banfield M. J. (2012) The molecular basis of ubiquitin-like protein NEDD8 deamidation by the bacterial effector protein Cif. Proc. Natl. Acad. Sci. U.S.A. 109, E1830–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yao Q., Cui J., Wang J., Li T., Wan X., Luo T., Gong Y. N., Xu Y., Huang N., Shao F. (2012) Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis. Proc. Natl. Acad. Sci. U.S.A. 109, 20395–20400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morikawa H., Kim M., Mimuro H., Punginelli C., Koyama T., Nagai S., Miyawaki A., Iwai K., Sasakawa C. (2010) The bacterial effector Cif interferes with SCF ubiquitin ligase function by inhibiting deneddylation of Cullin1. Biochem. Biophys. Res. Commun. 401, 268–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.