Background: Compared with other auxiliary KChIPs that enhance Kv4 current, KChIP4a inhibits Kv4 function.
Results: We identified an ER retention motif and an adjacent VKL motif within the KChIP4a N terminus that reduces Kv4.3 surface expression and promotes closed-state inactivation (CSI), respectively.
Conclusion: ER retention and CSI enhancement are two distinct mechanisms by which the N terminus of KChIP4a suppresses Kv4 function.
Significance: This study provides mechanistic insight into auxiliary KChIP4a-induced inhibition of A-type Kv4 current.
Keywords: Confocal Microscopy, Electrophysiology, Endoplasmic Reticulum (ER), Gating, Neurobiology, Potassium Channels, Trafficking, KChIPs, Kv4, Closed-state Inactivation
Abstract
In the brain and heart, auxiliary Kv channel-interacting proteins (KChIPs) co-assemble with pore-forming Kv4 α-subunits to form a native K+ channel complex and regulate the expression and gating properties of Kv4 currents. Among the KChIP1–4 members, KChIP4a exhibits a unique N terminus that is known to suppress Kv4 function, but the underlying mechanism of Kv4 inhibition remains unknown. Using a combination of confocal imaging, surface biotinylation, and electrophysiological recordings, we identified a novel endoplasmic reticulum (ER) retention motif, consisting of six hydrophobic and aliphatic residues, 12–17 (LIVIVL), within the KChIP4a N-terminal KID, that functions to reduce surface expression of Kv4-KChIP complexes. This ER retention capacity is transferable and depends on its flanking location. In addition, adjacent to the ER retention motif, the residues 19–21 (VKL motif) directly promote closed-state inactivation of Kv4.3, thus leading to an inhibition of channel current. Taken together, our findings demonstrate that KChIP4a suppresses A-type Kv4 current via ER retention and enhancement of Kv4 closed-state inactivation.
Introduction
Surface expression and dynamic gating are fundamental properties of rapidly inactivating (A-type) Kv4 potassium channels that play a critical role in regulating firing frequency and shaping action potential waveforms (1, 2). Binding of auxiliary β-subunits can change the expression of Kv4 channels at the membrane surface as well as their intracellular trafficking and gating kinetics (3–7). Cytosolic Kv channel-interacting proteins (KChIPs),2 a class of β-subunits, co-assemble with pore-forming Kv4 α-subunits to encode the somatodendritic A-type K+ current (ISA) in neurons (3, 8–10) and the transient outward current (Ito) in cardiac myocytes (11, 12). Both ISA and Ito activate at subthreshold membrane potentials, inactivate rapidly, and recover quickly from inactivation (1). Neuronal ISA plays a critical role in regulating dendritic excitability, somatodendritic signal integration, and long term potentiation (13–17), whereas cardiac Ito is the major current for fast phase repolarization of the cardiac action potential (18, 19). Genetic disruption of human Kv4 function causes temporal lobe epilepsy and spinocerebellar ataxias as well as cardiac disorders, such as atrial fibrillation and Brugada syndrome (20–25), demonstrating the essential role of Kv4 channels in physiology and pathology.
Auxiliary KChIP1 to -4 subunits, belonging to the neuronal calcium sensor superfamily, consist of a conserved C-terminal core domain with four EF-hand-like calcium binding motifs (26) and a variable N-terminal domain that has been proposed to mediate diverse modulation on Kv4 function (27–30). Alternative splicing of the four KChIP genes generates a large number of variants with distinct N-terminal domains, making the KChIP class the most diverse among the neuronal calcium sensor protein family (31). Co-expression of different KChIPs with Kv4 in heterologous expression system leads to different effects on Kv4 channel function. The majority of the KChIP isoforms dramatically increase Kv4 surface expression and peak current, slow down fast inactivation, and accelerate the recovery from inactivation (3, 32–34). Consistently, many physiological studies have shown that the current amplitudes and gating kinetics of native Kv4 channels (both ISA and Ito) are primarily determined by the expression levels of KChIPs (8, 10, 11, 35, 36). It is of interest that the KChIP4 splice variant KChIP4a, which shares a conserved C-terminal core domain with other KChIPs, functions as an inhibitory subunit and exhibits a distinct modulation on Kv4 channel surface expression and gating kinetics via its unique N-terminal domain (4, 37–39). The first N-terminal 34 residues of KChIP4a, previously known as the KIS (Kv channel inactivation suppressor) domain, slow Kv4 channel activation, disrupt fast inactivation of opened channels, and abolish its core-mediated enhancement of Kv4 current and acceleration of recovery from inactivation (4). However, the mechanisms by which the KChIP4a N terminus functions as a specific Kv4 channel inhibitory domain (KID) to suppress A-type potassium current remains unknown.
In the present study, we show that auxiliary KChIP4a suppresses Kv4.3 function via an alternative N-terminal KID, which can overcome the positive modulatory effect on Kv4.3 channel function by KChIP1. Using confocal imaging, we find that the KID alone is sufficient to retain proteins in the endoplasmic reticulum (ER), and its retention ability is transferable and prominently dependent on its flanking location within the protein sequence. A novel ER retention motif (N-terminal residues 12–17, LIVIVL) within the KID overrides the KChIP4a core domain-mediated enhancement of Kv4.3 surface expression, leading to the reduction of peak current. Further experiments reveal a VKL motif (N-terminal residues 19–21), adjacent to the LIVIVL motif, that can also suppress Kv4.3 current by facilitating Kv4.3 closed-state inactivation (CSI). Taken together, our findings demonstrate that ER retention and CSI enhancement are two distinct mechanisms by which KChIP4a suppresses Kv4 channel function.
EXPERIMENTAL PROCEDURES
Plasmid Construction
For confocal imaging experiments, cDNAs of KChIP4a and KChIP4a mutants were subcloned into a pEGFP-N1 or pEGFP-C2 vector. DsRed2-ER is a red fluorescent protein tagged with the signal sequence from calreticulin at its N terminus and a KDEL sequence at its C terminus (Clontech). The C-terminal CAAX signal of H-Ras (GCMSCKCVLS), which targets Ras proteins to the plasma membrane (PM), was added to the C terminus of EGFP to construct a PM-targeting EGFP vector (pEGFP-CAAX-N1). For biochemistry experiments, cDNAs of Kv4.3, KChIP1, KChIP4a, and KChIP4a mutants were cloned into a pcDNA3.1 vector. For electrophysiological experiments, wild-type (WT) or mutant cDNA constructs of either Kv4.3 or KChIPs were transferred into a pBluescript KSM vector.
Confocal Microscopy and Imaging Analysis
For confocal imaging experiments, HEK 293 cells were reseeded on glass coverslips coated with poly-d-lysine for detection and transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After 24–48 h of transfection, cells were washed twice with phosphate-buffered saline (PBS) containing 1 mm MgCl2 and 0.1 mm CaCl2 and fixed in 4% paraformaldehyde at 4 °C for 15 min. Images were obtained using a confocal microscope (FV1000, Olympus). For staining of localization in the ER, HEK 293 cells were co-transfected with KChIP4a-EGFP or KChIP4a mutants-EGFP and DsRed2-ER. FV1000 Viewer software (Olympus) was used to calculate Pearson's correlation coefficient values for the co-localization of the fluorescence signals.
Electrophysiological Recordings
For whole-cell patch clamp recording in HEK293 cells, currents were recorded at room temperature using the EPC 10 UBS amplifier with PatchMaster software (HEKA Electronics). Patch pipettes were pulled from borosilicate glass and fire-polished to a resistance of 2–4 megaohms. The bath solution contained 135 mm NaCl, 2.5 mm KCl, 10 mm HEPES, 1 mm MgCl2, 1.5 mm CaCl2, and 10 mm glucose at pH 7.4, and the pipette solution contained 135 mm potassium gluconate, 10 mm KCl, 10 mm HEPES, 1 mm CaCl2, 1 mm MgCl2, and 10 mm EGTA at pH 7.3.
For two-electrode voltage clamp recordings in oocytes, all cRNAs were transcribed in vitro from linearized plasmids in pBluescript KSM vectors by using the T3 mMESSAGE Machine Kit (Ambion). Xenopus laevis oocytes (stage V-VI) were selected and injected with 46 nl of cRNA solution, containing 0.5–5.0 ng of the selected cRNA, using a microinjector (Drummond Scientific). Oocytes were then kept at 17 °C in ND96 solution (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH 7.4, adjusted with NaOH). 24 h after injection, oocytes were impaled with two microelectrodes (0.5–1.0 megaohms) filled with 3 m KCl in a 40-μl recording chamber. Currents were recorded in ND-96 solution at room temperature (22 ± 1 °C) using a GeneClamp 500B amplifier (Axon Instruments).
Data were acquired using PatchMaster software (HEKA Electronics) and digitized at 1 kHz with an LIH 8 + 8 computer interface (HEKA Electronics). OriginPro version 8.6 (OriginLab) was used to analyze the data. To measure the peak current amplitudes and the kinetics of open-state inactivation, the currents were evoked by a 2-s depolarizing pulse to +40 mV from a holding potential of −100 mV. The time constants of macroscopic inactivation were obtained by curve fitting with a single exponential function. The peak conductance-voltage (G-V) relationship was derived from peak current amplitudes evoked by depolarizing steps from −100 to +60 mV at 10-mV increments, and the calculation was based on the equation, G = I/(V − Vrev), where I is the peak current amplitude at the test potential V, and Vrev is the reversal potential. Steady-state inactivation (SSI) was assessed by determining the peak current amplitudes at +40 mV after 1-, 5-, or 10-s prepulses ranging from −120 to 0 mV. The voltage dependence of steady-state activation (G/Gmax) and inactivation (I/Imax) were fitted to the following single Boltzmann relationship, y = 1/(1 + exp((V − V½)/k)), where V is the test potential, V½ is the potential for half-maximal activation or inactivation, and k is the corresponding slope factor. CSI was monitored with a double-pulse protocol in which the current was evoked by a test pulse of +40 mV from −100 mV (Ipre) and from subsequent −50-mV prepulses of variable durations from 5 ms to 10.4 s (Ipost). Normalized currents for CSI were determined as the ratio of Ipost to Ipre. The kinetics of CSI were obtained by fitting the normalized current amplitudes as a function of prepulse duration. All holding potentials were −100 mV in this study unless specified.
Cell Surface Biotinylation and Western Blotting Assay
Transfected HEK 293 cells were washed three times with ice-cold PBS and incubated for 1 h at 4 °C with EZ-Link Sulfo-NHS-SS-biotin (0.5 mg/ml; Pierce) to biotinylate cell surface proteins. Excess biotin was quenched by incubating the cells for an additional 10 min with TBS containing 100 mm glycine. Cells were lysed with lysis buffer (150 mm NaCl, 20 mm Tris, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 10 mm EDTA, and proteinase inhibitor mixture (Roche Applied Science), pH 8.0) at 4 °C for 30 min. The cell lysates were then centrifuged at 13,000 × g for 15 min to yield the protein extract in the supernatant. One fraction containing 200 μg of protein was incubated with 20 μl of neutravidin beads (Pierce) for 4 h at 4 °C, and the other fraction was prepared as total protein. After incubation, the bead-binding surface proteins were washed four times with cell lysis buffer and eluted with loading buffer containing DTT. The total and biotinylated proteins were both subjected to Western blot analysis.
For Western blot assays, protein samples were loaded on 8% SDS-PAGE and transferred using gel electrophoresis to nitrocellulose membranes (Millipore). After blocking, nitrocellulose membranes were incubated with mouse monoclonal anti-Kv4.3 (1:2000; Abcam), rabbit polyclonal anti-actin (1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or mouse monoclonal anti-transferrin receptor (1:500; Invitrogen) antibodies at 4 °C overnight. For comparing expression levels of KChIP4a-3xFLAG and KChIP4a core-3xFLAG, mouse monoclonal anti-FLAG (1:4000; Sigma) antibody was used. The membranes were then incubated with their corresponding secondary HRP-conjugated antibodies and detected using an ECL Western blotting detection system (Millipore). Detection of signals was calculated using Quantity One software (Bio-Rad). For quantification, the signals from anti-Kv4.3 (or anti-FLAG) over the anti-actin (for total Kv4.3, KChIP4a-3xFLAG, or KChIP4aΔ2–34) or the anti-transferrin receptor (for surface Kv4.3) were normalized. The data are expressed as the mean ± S.E.; statistical differences between different groups were assessed using Student's t test.
RESULTS
KChIP4a Suppresses Kv4.3 Function via Its N-terminal KID
To investigate the underlying mechanism by which auxiliary KChIP4a inhibits Kv4 function, we started by examining the effect of KChIP4a coexpressed with Kv4.3 on the channel current in both mammalian cells and Xenopus laevis oocytes. By sequence alignment, KChIP4a reveals a unique N terminus, consisting of the first 34 amino acids, that has been shown to function as a KID to inhibit the channel activity (Fig. 1A). Whole-cell patch clamp recordings in HEK 293 cells and two-electrode voltage clamp recordings in X. laevis oocytes both showed that KChIP4a co-expression resulted in a reduction of Kv4.3 current (Fig. 1, B–D). In contrast, deleting the first 34 residues of the KChIP4a N terminus (KChIP4aΔ2–34, also named KChIP4a core) led to a dramatic increase in the current amplitude (Fig. 1, B–D). The inhibitory effect of KChIP4a KID on Kv4 current was further confirmed by fusing the KID to either the C terminus of WT Kv4.3 (Kv4.3-KID) or the N terminus of Kv4.3 N-terminal truncation mutant (KID-Kv4.3Δ2–24) (Fig. 1E). These results demonstrate that the inhibitory effect of KChIP4a on Kv4 is indeed mediated by its N-terminal KID.
FIGURE 1.

The N-terminal KID of KChIP4a suppresses Kv4.3 function. A, a schematic representation of the general domain structure of KChIPs. KChIPs consist of an NH2-terminal variable domain (multicolored box) and a conserved COOH-terminal core domain (light blue boxes) containing four EF-hands (light green boxes). The N-terminal domain of KChIP4a (designated KID in this study) is represented with the single-letter amino acid code in the protein sequence. The LIVIVL motif is marked in red, and the VKL motif is marked in black. B, representative current traces of Kv4.3 alone, Kv4.3-KChIP4a, and Kv4.3-KChIP4aΔ2–34 recorded from HEK 293 cells. The currents were evoked by a 1-s depolarizing pulse to +40 mV from a holding potential of −100 mV. C, quantitative analysis of peak current densities recorded from HEK 293 cells expressing the indicated channels or channel complexes. Values are mean ± S.E. (error bars); n = 6–10 cells; ***, p < 0.001. D, quantitative analysis of peak current amplitudes recorded from Xenopus oocytes expressing the indicated channels or channel complexes. Values are mean ± S.E.; n = 10–26 oocytes; ***, p < 0.001. E, the fusion of the KID to the C-terminal end of Kv4.3 (Kv4.3-KID) or the N terminus of Kv4.3 N-terminal truncation mutant (KID-Kv4.3Δ2–24) significantly suppressed the channel current. Values are mean ± S.E.; n = 5–10 oocytes. Statistical significance was assessed using Student's t test, and statistical significance was considered as p < 0.001 (***).
In neurons, KChIP4a co-exists with other KChIPs (10, 32), suggesting that KChIP4a may compete with other KChIPs to modulate Kv4 expression and gating (40). To test this hypothesis, we co-injected varying amounts of KChIP4a cRNAs with a fixed amount of Kv4.3 and KChIP1 cRNAs into Xenopus laevis oocytes and recorded the currents (Fig. 2A). As shown in Fig. 2B, with a fixed amount of KChIP1, increasing KChIP4a expression yielded a dose-dependent inhibition of Kv4.3 current amplitude. Consistent with previous reports (4), increasing amounts of KChIP4a gradually slowed the macroscopic open-state inactivation of Kv4.3 current (Fig. 2C). This dose-dependent, KChIP4a-mediated inhibition of Kv4.3-KChIP1 current was further confirmed by parallel biochemical experiments. We observed that total Kv4.3 protein expression was significantly reduced when co-expressed with KChIP4a (Fig. 2, D and E). However, KChIP4a co-expression resulted in a greater decrease in cell surface Kv4.3 protein, as compared with total expression (Fig. 2, F and G). Together, these results show that the N-terminal KID of KChIP4a acts as a dominant negative regulator to suppress the function of Kv4.3-KChIP1 channel complexes by inhibiting Kv4.3 expression as well as forwarding trafficking to the cell surface.
FIGURE 2.
KChIP4a dose-dependently inhibits the function Kv4.3-KChIP1 channel complexes. A, representative current traces of Xenopus oocytes co-injected with Kv4.3, KChIP1, and KChIP4a with varying mole ratios of 1:1:0 (left), 1:1:1 (middle), or 1:1:10 (right) for Kv4.3/KChIP1/KChIP4a (indicated as 1× or 10×), and outward potassium currents were evoked by 2-s step depolarization from −100 to +60 mV at 10-mV increments. B, quantitative analysis for suppression of Kv4-KChIP complex current by KChIP4a in a dose-dependent manner. Values are mean ± S.E. (error bars) of 12–24 oocytes. Statistical significance was determined by one-way analysis of variance followed by Tukey's post hoc test, and statistical significance was considered as p < 0.001 (***). C, quantitative analysis showed that KChIP4a dose-dependently slowed inactivation time constants of Kv4.3-KChIP complexes. Values are mean ± S.E.; n = 12–24 oocytes; ***, p < 0.001. D and F, Western blots of total (D) and surface (F) proteins from HEK 293 cells triple-transfected with Kv4.3 and KChIP1 at a 1:1 ratio (2 and 2 μg) and increasing amounts of KChIP4a cDNA as indicated (0–20 μg). For quantification, the intensities of the Kv4.3 bands were measured and normalized, first to the expression of endogenous actin or transferrin receptor (TransR) in the same lane and subsequently to Kv4.3 co-expressed with KChIP1 in the same blot. E and G, quantification analysis of total (E) and surface (G) expression levels of Kv4.3 co-expressed with various ratios of KChIP1 and KChIP4a. Values are mean ± S.E.; n = 3 blots; ***, p < 0.001.
The ER Retention Capacity of the KID Is Transferable and Dependent on Its Location
The above results show that the N-terminal KID of KChIP4a results in the decreased anterograde trafficking of Kv4 channels, and previous studies have suggested that the KChIP4a N terminus is required for ER localization of KChIP4a in heterologous cells (32, 37). To find potential ER retention motifs within the KID, we began testing whether the retention ability of the KID is transferable and dependent upon its location within the protein sequence. We created a series of constructs with insertions of the KID at the proximal N terminus, the intermediate region, and the C-terminal end of the entire fusion protein. As a positive control, N-terminal attachment of full-length KChIP4a to EGFP (KChIP4a-EGFP) completely altered the subcellular distribution of EGFP and led to co-localization with the ER marker DsRed2-ER (Fig. 3A). By contrast, the KID deletion mutant of KChIP4a (KChIP4aΔ2–34-EGFP) showed a diffuse distribution throughout the cytoplasm and nucleoplasm of HEK 293 cells (Fig. 3B). Transplanting the KID to the C terminus of EGFP (EGFP-KID) was sufficient to retain EGFP in the ER (Fig. 3C), whereas inserting the KID into the middle part of intact protein (EGFP-KChIP4a) disrupted its ER retention ability (Fig. 3D).
FIGURE 3.

The ER retention capacity of the KID is transferable and dependent on its location. A, robust co-localization of full-length KChIP4a with the ER marker DsRed2-ER. B, the N-terminal KID deletion mutant of KChIP4a (KChIP4aΔ2–34) showed a diffuse distribution. C, appending the KID to the C terminus of EGFP (EGFP-KID) is sufficient to retain EGFP in the ER. D, disruption of the KID-induced ER retention when the KID was inserted into the middle of the chimeric construct (EGFP-KChIP4a). E, ER retention of CD4 proteins by the KID fused to the C terminus of the chimeric proteins (CD4-EGFP-KID). F, adding CD4 to the N terminus of KChIP4a (CD4-KChIP4a-EGFP) disrupted the ER retention caused by the KID, resulting in the localization of the chimeric proteins to the PM. G, ER localization was quantified by co-localization with the ER marker DsRed2-ER, using Pearson's correlation. Values are mean ± S.E. (error bars); n = 12–31 cells; *, p < 0.001. Red squares, KID; yellow rectangles, KChIP4aΔ2–34 (the core domain of KChIP4a); green rectangles, EGFP; blue rectangles, CD4. Scale bar, 10 μm.
To establish whether the KID could redirect other membrane proteins to the ER, we fused the KID with EGFP to the C terminus of CD4 (CD4-EGFP-KID). Our results showed that CD4 fused with the KID caused localization of the chimeric proteins to the ER (Fig. 3E). However, fusing CD4 to the N terminus of KChIP4a (CD4-KChIP4a-EGFP, with equivalent insertion of the KID into the intermediate region of the intact protein) had no effect on trafficking and maintained the ability of CD4 proteins targetting to the PM instead of being retained in the ER (Fig. 3F). Quantitative analysis of fluorescence intensity for co-variance of WT KChIP4a or KChIP4a mutants against the ER marker DsRed2-ER further confirmed the effect of ER retention mediated by its KID (Fig. 3G). These results indicate that the KID functions as an ER retention signal when located at either the N or C terminus of a protein but not when buried in the intermediate region.
Identification of a Hydrophobic and Aliphatic ER Retention Motif, LIVIVL, within the N-terminal KID of KChIP4a
Because the N-terminal KID does not contain any canonical ER retention motif, the question then arises as to how the KID causes ER localization of KChIP4a. To identify specific residues critical for ER retention of KChIP4a, N-terminal truncations or mutations were introduced into the KID, and the fluorescence intensity of co-variance versus DsRed2-ER was quantified. With N-terminal deletions of residues 2–10 (KChIP4aΔ2–10-EGFP) or 23–34 (KChIP4aΔ23–34-EGFP) and alanine substitutions for residues 19–22 of the KID (KChIP4a 19–22A-EGFP), all of the proteins still co-localized with DsRed2-ER (Fig. 4, A, E, F, and G). In contrast, deleting N-terminal residues 2–15 (KChIP4aΔ2–15-EGFP) or substituting alanine for residues 11–14 (KChIP4a 11–14A-EGFP) or 15–18 (KChIP4a 15–18A-EGFP) eliminated ER retention (Fig. 4, B, C, D, and G). These data indicate that residues 11–18 of the KID are critical for ER retention.
FIGURE 4.

Identification of residues critical for the KID-mediated ER retention of KChIP4a. A, robust co-localization of KChIP4aΔ2–10-EGFP with the ER marker DsRed2-ER. B–D, diffuse distribution of KChIP4aΔ2–15-EGFP (B), KChIP4a 11–14A-EGFP (C), and KChIP4a 15–18A-EGFP (D). E and F, ER localization of KChIP4a 19–22A-EGFP (E), and KChIP4aΔ23–34-EGFP (F), with DsRed2-ER. G, ER localization was quantified by Pearson's co-localization with DsRed2-ER. Values are mean ± S.E. (error bars); n = 14–31 cells; *, p < 0.001. Scale bar, 10 μm.
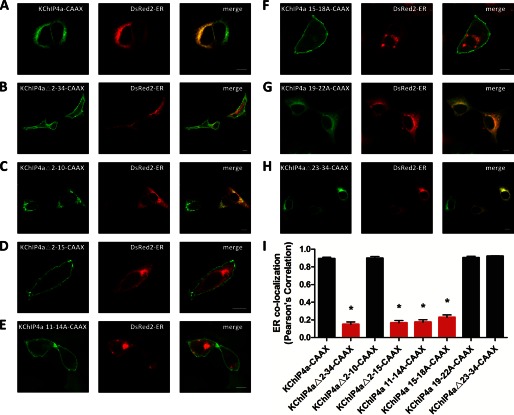
To understand the mechanism underlying the KID-mediated ER retention, we utilized a CAAX signal from the H-Ras proteins that functions as a membrane anchor to evaluate the retention ability of KChIP4a and its mutants. The CAAX signal was appended to the C terminus of KChIP4a-EGFP fusion proteins. We then tested the effect of the KID on CAAX signal-mediated forward trafficking to the cell surface. The result showed that the intact KID overcame the CAAX signal-mediated PM translocation (Fig. 5A). Consistent with our previous results shown in Figs. 3B and 4, the mutants KChIP4aΔ2–34-CAAX, KChIP4aΔ2–15-CAAX, KChIP4a 11–14A-CAAX, and KChIP4a 15–18A-CAAX lost their ability to stay in the ER as illustrated by their distribution at the PM (Fig. 5, B, D–F, and I), whereas the mutants KChIP4aΔ2–10-CAAX, KChIP4a 19-22A-CAAX, and KChIP4aΔ23–34-CAAX retained their ER retention ability (Fig. 5, C and G–I). These results demonstrate that residues 11–18 (VLIVIVLF) of the KID function as an ER retention signal that can override the H-Ras-CAAX-mediated classical vesicular trafficking.
FIGURE 5.
Identification of critical residues within the KID that mediate ER retention of KChIP4a with a C-terminal H-Ras-CAAX signal. A, C, G, and H, ER distribution of KChIP4a-CAAX (A), KChIP4aΔ2–10-CAAX (C), KChIP4a 19–22A-CAAX (G), and KChIP4aΔ23–34-CAAX (H). B and D–F, PM distribution of KChIP4aΔ2–34-CAAX (B), KChIP4aΔ2–15-CAAX (D), KChIP4a 11–14A-CAAX (E), and KChIP4a 15–18A-CAAX (F). I, ER localization was quantified by DsRed2-ER co-localization, using Pearson's correlation. Values are mean ± S.E. (error bars); n = 14–24 cells; *, p < 0.001. -CAAX indicates attachment of a PM-targeting CAAX signal from H-Ras to the C terminus of fusion proteins. Scale bar, 10 μm.
To determine whether the VLIVIVLF (residues 11–18) is sufficient for ER retention, we replaced the whole KID (34 amino acids) with VLIVIVLF and appended it to the KChIP4a core. Our results showed that VLIVIVLF alone is not only necessary but also sufficient to retain proteins in the ER, despite the presence of a CAAX signal (Fig. 6, A, F, and K). To further circumscribe the retention motif, we replaced the KID with VLIVI (residues 11–15), LIVIV (residues 12–16), IVIVL (residues 13–17), or VIVLF (residues 14–18). The VLIVI (residues 11–15) did not alter the cytoplasm and nucleoplasm-diffused distribution of the KChIP4a core domain, as compared with the core expressed alone (Fig. 6, B and K). The PM distribution of VLIVI-core-CAAX fusion proteins was also similar to the core-CAAX (KChIP4aΔ2–34-CAAX) (Fig. 6, G and K). Either LIVIV (residues 12–16) or IVIVL (residues 13–17) alone was sufficient to trap the chimeric proteins in the ER, with or without the CAAX signal (Fig. 6, C, D, H, I, and K). VIVLF (residues 14–18) allowed effective targeting and retention within the ER (Fig. 6, E and K) but was not sufficient to prevent the CAAX signal-mediated PM localization of the chimeric proteins (Fig. 6, J and K). To further narrow down residues critical for ER retention, we replaced the KID with different combinations of four amino acid sequences, such as VLIV (residues 11–14), LIVI (residues 12–15), IVIV (residues 13–16), VIVL (residues 14–17), and IVLF (residues 15–18), respectively. All chimeras carrying the four amino acid sequences, except VLIV (residues 11–14), stayed in the ER (Fig. 7, A–E and K), whereas those chimeras fused with CAAX signal traveled to the cell surface (Fig. 7, F–J and K), suggesting that the four-amino acid ER retention motifs are not strong enough to overcome the PM translocation mediated by the CAAX signal. These results reveal a hydrophobic and aliphatic motif, consisting of six residues 12–17 (LIVIVL), that functions as an ER retention signal responsible for KChIP4a localization in the ER.
FIGURE 6.
Identification of a hydrophobic and aliphatic ER retention motif, LIVIVL, within the KID. A and F, an insertion of VLIVIVLF (N-terminal residues 11–18 of the KID) caused a strong ER retention of the KChIP4a core (with or without CAAX signal). B and G, the VLIVI sequence alone was insufficient to cause ER retention of the core (with or without CAAX signal). C, D, H, and I, the LIVIV (C and H) and IVIVL (D and I) sequences caused the ER localization of the core (with or without CAAX signal). E and J, the VIVLF sequence retained the core in the ER (E) but not the core with CAAX signal (-core-CAAX) (J). K, ER localization was quantified by Pearson's co-localization with DsRed2-ER. Values are mean ± S.E. (error bars); n = 14–20 cells; *, p < 0.001. Scale bar, 10 μm.
FIGURE 7.
Four amino acids of residues 11–18 of KChIP4a are insufficient to cause ER retention. A and F, VLIV alone is insufficient to cause ER retention of core (with or without CAAX signal). B–E and G–J, LIVI (B and G), IVIV (C and H), VIVL (D and I) and IVLF (E and J) retained the core in the ER but not the core with CAAX signal (-core-CAAX). K, ER localization was quantified by DsRed2-ER co-localization, using Pearson's correlation. Values are mean ± S.E. (error bars); n = 8–30 cells; *, p < 0.001. Scale bar, 10 μm.
Enhanced CSI of Kv4.3 by the N-terminal KID of KChIP4a
To examine whether the ER retention signal within the N-terminal KID of KChIP4a affected total and surface expression of Kv4.3, surface biotinylation and Western blot were carried out on HEK 293 cells with Kv4.3 co-expressed with WT or mutated KChIP4a. The results showed that mutating residues 11–18 (11–14A and 15–18A) relieved ER retention and reversed the inhibitory effect of KChIP4a on total and surface expression of Kv4.3 (Fig. 8, A and C). The reduction of total Kv4.3 expression resulted from the decreased expression of KChIP4a, as compared with the KChIP4aΔ2–34 (Fig. 8, E and F), suggesting that the ER retention signal within the KID accelerates the rate of protein degradation. Quantitative analysis showed greater changes of Kv4.3 proteins in surface expression, as compared with total expression (Fig. 8, B and D), indicating that the KID inhibits the surface expression of Kv4.3 via reducing protein stability as well as anterograde trafficking. Electrophysiological recordings showed a dramatic increase in Kv4.3 current amplitudes when residues 11–18 were disrupted (Fig. 8G), consistent with their role in mediating Kv4.3 retention in the ER.
FIGURE 8.
The ER retention signal within the KID inhibits total and surface expression of Kv4.3. A and C, representative Western blots of total (A) and surface (C) proteins from HEK 293 cells transiently transfected with plasmids encoding Kv4.3 with WT KChIP4a or KChIP4a mutants. For quantification, the intensities of the Kv4.3 bands were measured and normalized, to endogenous actin (for total) or TransR (for surface) in the same lane and subsequently to Kv4.3 co-expressed with WT KChIP4a in the same blot. B and D, quantification analysis of total (B) and surface (D) expression levels of Kv4.3 co-expressed with WT KChIP4a or KChIP4a mutants. Values are mean ± S.E. (error bars); n = 4–6 blots. Statistical analysis was performed using one-way analysis of variance followed by Tukey's post hoc test, and statistical significance was assessed as follows. **, p < 0.01; ***, p < 0.001. E, representative Western blot of total proteins from HEK 293 cells transiently transfected with plasmids encoding KChIP4a-3xFLAG or KChIP4aΔ2–34-3xFLAG. F, quantification analysis of total expression levels of KChIP4a-3xFLAG and KChIP4aΔ2–34-3xFLAG, indicating reduced expression of KChIP4a as compared with KChIP4aΔ2–34. G, quantification analysis of relative current change in oocytes expressing Kv4.3 co-expressed with KChIP4a mutants normalized to Kv4.3 co-expressed with WT KChIP4a. Values are mean ± S.E.; n = 8–26 oocytes; ***, p < 0.001.
In addition to the identified residues 11–18 that are critical for ER retention, we were surprised to find that mutating residues 19–22 (19–22A) increased the current density of Kv4.3 without affecting its surface expression (Fig. 8, D and G). Recent pharmacological studies showed that some compounds can inhibit Kv4.3 function by accelerating channel CSI, suggesting that Kv4 inhibition might result from an increased number of unavailable channels that are in closed inactivated states (41–45). To test the gating effect of KChIP4a, we co-expressed Kv4.3 with the KChIP4a core (KChIP4aΔ2–34) and found that preferential CSI was reduced (Fig. 9, A and B). Depolarizing to +40 mV after holding for 10.4 s at −50 mV resulted in only about 38% of Kv4.3-core complex current being inactivated (Fig. 9B). In contrast, co-expression of Kv4.3 with KChIP4a increased CSI to about 90%, as compared with Kv4.3 alone (∼72%) and Kv4.3-core co-expression (∼38%) (Fig. 9B and Table 1). The SSI curve for Kv4.3 under a 5-s prepulse had a V½ of −52.0 ± 0.3 mV and a k of 5.5 ± 0.3 mV. The KChIP4a core significantly shifted the inactivation curve (V½) to the right in a depolarization direction (V½ of −46.4 ± 0.2 mV and k of 4.6 ± 0.2). Consistent with its effect on CSI, the KID overrode the right-shift effect of the core and caused the curve to shift in the hyperpolarization direction (V½ of −58.5 ± 0.5 mV and a k of 5.2 ± 0.4 mV) (Fig. 9C and Table 1). We also examined the conductance-voltage (G-V) curves and found no significant differences (Fig. 9D). These data suggest that the modulation of Kv4.3 inactivation by the KID is dominant over that by the KChIP4a core and reduces the number of Kv4.3 channels available for opening.
FIGURE 9.

KChIP4a directly inhibits Kv4.3 by enhancing CSI. A, CSI curve of Kv4.3 alone or co-expressed with either KChIP4a or KChIP4aΔ2–34 (core) measured by a double-pulse protocol as illustrated. B, mean current amplitudes evoked by the post pulse in protocol as shown in A, relative to the amplitudes obtained by the initial control pulse (Ipost/Ipre), were plotted against different durations of the conditioning pulses (n = 18–23 oocytes). C, voltage dependence of SSI for Kv4.3 alone or co-expressed with KChIP4a or KChIP4aΔ2–34. The SSI protocol consists of a 5-s prepulse that varies between −120 and 0 mV in 10-mV increments, followed by a test pulse of +40 mV. The fraction of available current (I/Imax) was plotted against the prepulse potential (n = 7–15 oocytes). D, steady-state activation curve of Kv4.3 alone or co-expressed with KChIP4a or KChIP4aΔ2–34 (n = 15–16 oocytes).
TABLE 1.
Inactivation kinetics of Kv4.3 alone and its co-expression with KChIP4a or its mutants
Values are means ± S.E.; n = number of oocytes. Mean data points obtained from the steady-state inactivation protocol with 5-s prepulse were best fitted by (OriginPro version 8.6 software) a standard single Boltzmann curve, in which V½ is the half-inactivation potential, and k is the slope factor. Inactivation time constants (τ) of closed-state inactivation were obtained by fitting single exponential function to the decaying phases of Kv4.3 current evoked by +40-mV depolarization after a subthreshold −50-mV prepulse with variable durations. Remaining current is the fraction of non-inactivated channel current.
| Steady-state inactivation |
Closed-state inactivation |
|||||
|---|---|---|---|---|---|---|
| n | V½ | k | n | τ | Remaining current | |
| mV | mV | s | ||||
| Kv4.3 | 15 | −52.0 ± 0.3 | 5.5 ± 0.3 | 19 | 3.722 ± 0.499 | 0.276 ± 0.041 |
| Kv4.3 + KChIP4a | 11 | −58.5 ± 0.5 | 5.2 ± 0.4 | 23 | 1.686 ± 0.144 | 0.105 ± 0.011 |
| Kv4.3 + Δ2–34 | 13 | −46.4 ± 0.2 | 4.6 ± 0.2 | 18 | 13.512 ± 1.588 | 0.616 ± 0.036 |
| Kv4.3 + Δ2–10 | 6 | −59.0 ± 0.2 | 4.5 ± 0.2 | 6 | 0.695 ± 0.078 | 0.118 ± 0.029 |
| Kv4.3 + Δ2–15 | 5 | −58.3 ± 0.1 | 3.9 ± 0.1 | 5 | 1.147 ± 0.038 | 0.102 ± 0.013 |
| Kv4.3 + 11–14A | 8 | −58.9 ± 0.4 | 5.3 ± 0.3 | 8 | 2.164 ± 0.203 | 0.136 ± 0.013 |
| Kv4.3 + 15–18A | 8 | −58.8 ± 0.3 | 5.0 ± 0.3 | 9 | 1.738 ± 0.069 | 0.087 ± 0.014 |
| Kv4.3 + 19–22A | 8 | −46.6 ± 0.5 | 5.9 ± 0.4 | 8 | 15.465 ± 2.648 | 0.646 ± 0.054 |
| Kv4.3 + 19A | 5 | −54.2 ± 0.4 | 5.4 ± 0.4 | 5 | 2.487 ± 0.176 | 0.220 ± 0.019 |
| Kv4.3 + 20A | 12 | −54.7 ± 0.5 | 6.1 ± 0.4 | 12 | 2.371 ± 0.347 | 0.262 ± 0.033 |
| Kv4.3 + 21A | 7 | −49.9 ± 0.3 | 4.7 ± 0.3 | 9 | 7.597 ± 0.298 | 0.363 ± 0.024 |
| Kv4.3 + 22A | 10 | −58.1 ± 0.2 | 4.1 ± 0.2 | 10 | 1.186 ± 0.091 | 0.115 ± 0.030 |
| Kv4.3 + 2xNt-core | 12 | −50.7 ± 0.5 | 5.1 ± 0.4 | 12 | 7.445 ± 0.389 | 0.332 ± 0.056 |
| Kv4.3 + 3xNt-core | 13 | −47.0 ± 0.4 | 4.8 ± 0.3 | 13 | 14.078 ± 0.632 | 0.594 ± 0.038 |
To determine which part of the KID is involved in modulating Kv4.3 CSI, we generated mutants and found that residues 19–22 of the KID played a critical role in facilitating the development of CSI and causing a hyperpolarizing shift of SSI (Fig. 10, A and B, and Table 1). The 19–22A mutant completely eliminated the enhancement effect of the KID on CSI, consistent with the effect of the KID deletion mutant (Δ2–34). Further dissection revealed that residues 19–21, VKL, are responsible for promoting CSI, and residue 21 (leucine) is the most critical (Fig. 10, C and D, and Table 1). A previous study suggested that either the N terminus of KChIP2x (2xNt) or KChIP3x (3xNt) functions like the KID (37). To test the effect of 2xNt or 3xNt on CSI, we replaced the KID with 2xNt or 3xNt, where their amino acid residues 19–21 are VKV or IAV, respectively. The VKV from 2xNt exerted an effect similar to that of the KChIP4a 21A mutant on CSI and SSI. The IAV from 3xNt, analogous to the KChIP4a 19–22A or Δ2–34 mutation, had no effect on promoting CSI (Fig. 10, E and F, and Table 1), confirming the specific effect of the VKL motif from the KID on CSI. Together, these results demonstrate that the VKL motif of the KID inhibits Kv4.3 function by facilitating the channel CSI.
FIGURE 10.

The VKL motif within the N-terminal KID of KChIP4a promotes CSI of Kv4.3. A, analysis for CSI of Kv4.3 co-expressed with KChIP4a or KChIP4a mutants (n = 8–13 oocytes). B, voltage dependence of SSI of Kv4.3 co-expressed with KChIP4a or KChIP4a mutants. C, CSI of Kv4.3 co-expressed with KChIP4a or KChIP4a 19–22 mutants (n = 5–23 oocytes). D, voltage dependence of SSI of Kv4.3 co-expressed with KChIP4a or KChIP4a 19–22 mutants (n = 5–12 oocytes). E, CSI of Kv4.3 co-expressed with KChIP4a, core (KChIP4aΔ2–34), 2xNt-core, or 3xNt-core (n = 12–23 oocytes). F, voltage dependence of SSI of Kv4.3 co-expressed with KChIP4a, core (KChIP4aΔ2–34), 2xNt-core, or 3xNt-core (n = 11–13 oocytes).
DISCUSSION
The goal of this study was to understand how auxiliary KChIP4a, which only differs from other KChIPs in the N terminus, inhibits Kv4 function. We found that KChIP4a acts as an inhibitory subunit to suppress Kv4 A-type current (ISA). The inhibitory property of KChIP4a on ISA is primarily mediated by its unique N-terminal KID. Our findings reveal two distinct mechanisms by which KChIP4a exerts its inhibition on Kv4 function. First, an LIVIVL motif (N-terminal residues 12–17) mediates ER retention of KChIP4a and Kv4 channels; second, the VKL motif (N-terminal residues 19–21) of the KID directly inhibits Kv4.3 function by increasing the number of channels in the closed inactivated state.
Previous studies on Kv4 modulation by KChIPs have mostly focused on the effects of a single KChIP on Kv4 function. Because all members of the KChIP family share a high homology in their C-terminal core regions and each can form two interfaces associated with two neighboring Kv4 N-terminal domains (46–48), this raises the possibility that different KChIPs may compete for common KChIP-binding sites, resulting in a diversity of modulation of Kv4 function by KChIPs. This hypothesis is supported by our observation that KChIP4a inhibits the surface expression and peak current of Kv4 coexpressed with KChIP1 in a dose-dependent and competitive manner by antagonizing the binding of KChIP1 to Kv4 (Fig. 2). Whether these observations imply that Kv4-KChIP complexes in native cells contain heteromeric KChIPs or only one KChIP isoform is presently unknown and deserves further investigation.
Cell surface expression of proteins depends on the delicate balance of forward trafficking and intracellular retention (49). ER retention/retrieval motifs are dominant trafficking signals that are often encoded in individual subunits of multiprotein complexes, thereby providing a negative regulatory mechanism for cell surface protein expression or a quality control mechanism to ensure proper oligomerization or correct folding (50). In most cases, ER retention/retrieval motifs are located in the α-subunits of ion channels and can be masked by subunit assembly (51–53) or antagonized by auxiliary subunits (54, 55) or regulatory proteins (56–58). Some accessory β-subunits or regulators contain ER retention motifs and down-regulate surface expression of α-subunits (59, 60). In the current study, we identified an LIVIVL sequence within the N-terminal KID of KChIP4a that functions as an ER retention motif to suppress Kv4 forward vesicular trafficking from the ER to the Golgi apparatus, which is different from the inhibition of Kv4 endocytosis induced by other KChIPs, such as KChIP2 (34). In addition, we utilized the CAAX signal from H-Ras that functions as a PM targeting signal to evaluate the retention ability of this new-found motif. The H-Ras-CAAX signal uses the exocytosis pathway in a similar fashion to the trafficking pathway of membrane proteins (61). Therefore, our results support that the ER retention motif identified in this study plays a dominant negative role in classical vesicular trafficking pathway, thus leading to the reduction of Kv4 surface expression.
Due to its hydrophobic nature, the LIVIVL motif identified and reported here differs from canonical motifs consisting of basic amino acids. A series of hydrophobic ER retention signals have been identified in different parts of proteins, including the N terminus, C terminus, transmembrane segments, and cytoplasmic linker of transmembrane segments (53, 62–68). Unlike well characterized basic motifs that consist of RXR, RR, and KK, the molecular identity of the hydrophobic motifs still remains to be further explored. The hydrophobic ER retention motifs vary in their location and composition, and the only common feature is membrane adjacency or association. Another interesting observation is that the retention ability of the LIVIVL motif is transferable and dependent on its flanking location within the protein sequence. This feature is similar to the H/KDEL motif that is often located in the C-terminal end of soluble ER-resident proteins, such as Bip (Grp78), calreticulin, endoplasmin (Grp94), protein-disulfide isomerase, etc. (50). The addition of the KDEL to the C terminus of various proteins can also lead to their ER retention (69–72). The H/KDEL motif is recognized by the KDEL receptor ERD2, which is mainly localized to the cis-Golgi and targets the KDEL-containing proteins into the retrograde COPI-mediated transport pathway (73, 74). However, how the LIVIVL motif causes ER retention of LIVIVL-containing proteins requires further investigation.
Previously, we proposed that the core of KChIPs increases total expression level of Kv4.3 by stabilizing Kv4.3 tetramerization via a clamping action (48). In the current study we show that the N-terminal KID of KChIP4a, a non-core region, exerts an inhibitory effect on total Kv4.3 and KChIP4a by reducing their protein stability (Fig. 8). Considering the ER retention effect of the KID, it is likely that the ER retention motif within the KID causes ER-associated protein degradation of KChIP4a itself and its associated proteins, such as Kv4.3. Kv4 expression and functional properties are regulated by KChIPs as well as dipeptidyl peptidase-like proteins containing DPP6 and DPP10 (5, 6). Considerable evidence suggests that native neuronal ISA channels function in macromolecular protein complexes composed of Kv4 pore-forming subunits together with accessory KChIPs and DPP6/10 subunits (75, 76). Different from cytosolic KChIPs, the accessory DPP6/10 subunits are transmembrane proteins and selectively increase surface expression of Kv4.2 but not total expression (77). In this study, we demonstrate that the ER retention of the KID is dominant over surface trafficking of multitransmembrane protein Kv4.3, single transmembrane protein CD4, and the secretory CAAX signal from H-Ras protein, so the DPP-mediated augmentation effect on Kv4 surface expression is likely to be inhibited when co-assembled with KChIP4a.
The molecular mechanism of Kv4 fast inactivation differs significantly from other rapidly inactivating Kv channels, such as Shaker and Kv1.4 (78–82). Kv4 channels exhibit a preferential CSI, implying that upon voltage-dependent activation, inactivation occurs preferentially from partially activated closed states that precede the open state (82, 83). In contrast to N-type and C-type inactivation, the molecular basis of CSI remains to be determined. The KID has been previously characterized as an open-state inactivation suppressor (so-called KIS domain) of Kv4 in heterologous cells and neurons (4, 84). Surprisingly, in this study, we found that the KID promotes CSI and shifts the voltage dependence of SSI in a hyperpolarization direction, whereas the core of KChIP4a-mediated Kv4 modulation on CSI and SSI kinetics is opposite to that of the KID (Fig. 9). Moreover, we demonstrated that the existence of the VKL motif (N-terminal residues 19–21) is responsible for the KID modulation on Kv4 CSI. Therefore, identification of a putative receptor site for the VKL motif binding to Kv4 channels may provide structural insights into the mechanism of Kv4 CSI.
Similar to Kv4 channels, Kv2 channels (Kv2.1 and Kv2.2) also inactivate preferentially from partially activated closed states, and its steady-state inactivation curve exhibits a U-type shape (85). The functional properties of Kv2 are regulated by forming heteromers with the otherwise “silent” α-subunits of the Kv5, Kv6, Kv8, or Kv9 subfamily. The modulatory effects of these “silent” α-subunits on Kv2 are analogous to those of KChIP4a on Kv4, such as a reduction in peak current amplitude, inhibition of open-state inactivation, acceleration of CSI, and promotion of hyperpolarized shifts in the voltage dependence of SSI (86–89). Altogether, these findings suggest that bidirectional modulation of Kv4 open-state and closed-state inactivation mediated by the KID may be a highly conserved function among various ion channels and ancillary subunits.
The channel subunit composition is a determinant in shaping the repertoire of A-type K+ channels present at the surface of neurons. Differential regulation of auxiliary KChIPs can modify the subunit composition of native Kv4 channel complexes, altering their biophysical and physiological properties as well as neuronal excitability. Recently, an interesting study has demonstrated that activation of noncoding RNA 38A induced by inflammatory stimuli causes a gene-specific alternative splicing shift from KChIP4bl to KChIP4a, leading to an inhibition of Kv4 function as well as altered amyloid production associated with neurodegenerative etiology (90). Therefore, it is possible that the expression level of KChIP4a may vary under pathological conditions that are affected by inflammation, pain, meningitis, multiple sclerosis, Alzheimer disease, Parkinson disease, or stroke-mediated neuronal dysfunction (91), resulting in perturbation of ISA and neuronal excitability. Furthermore, because KChIP4a can directly inhibit Kv4 function by promoting channel CSI, a specific disruption of the interaction between the KChIP4a KID and Kv4 channels aimed at enhancing ISA may provide a new strategy for treating hyperexcitable neurological disorders, such as pain and epilepsy.
Acknowledgments
We thank laboratory members Xu Cao, Jun Su, Yuanyuan Cui, and Hao Chen for discussion. We also thank Arun Sharma (Stanford University School of Medicine) for careful and critical reading of the manuscript. K. W. W. thanks J. M. Wang for consistent support during this research.
This work was supported by Ministry of Science and Technology of China Grant 2013CB531300 and National Science Foundation of China Grants 81221002 and 30970919 (to K. W. W.).
- KChIP
- Kv channel-interacting protein
- KID
- Kv4 channel inhibitory domain
- ER
- endoplasmic reticulum
- CSI
- closed-state inactivation
- PM
- plasma membrane
- EGFP
- enhanced green fluorescent protein
- SSI
- steady-state inactivation
- CAAX
- motif in which C represents cysteine, A represents an aliphatic residue, and X represents any amino acid residue.
REFERENCES
- 1. Birnbaum S. G., Varga A. W., Yuan L. L., Anderson A. E., Sweatt J. D., Schrader L. A. (2004) Structure and function of Kv4-family transient potassium channels. Physiol. Rev. 84, 803–833 [DOI] [PubMed] [Google Scholar]
- 2. Covarrubias M., Bhattacharji A., De Santiago-Castillo J. A., Dougherty K., Kaulin Y. A., Na-Phuket T. R., Wang G. (2008) The neuronal Kv4 channel complex. Neurochem. Res. 33, 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000) Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 [DOI] [PubMed] [Google Scholar]
- 4. Holmqvist M. H., Cao J., Hernandez-Pineda R., Jacobson M. D., Carroll K. I., Sung M. A., Betty M., Ge P., Gilbride K. J., Brown M. E., Jurman M. E., Lawson D., Silos-Santiago I., Xie Y., Covarrubias M., Rhodes K. J., Distefano P. S., An W. F. (2002) Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. U.S.A. 99, 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nadal M. S., Ozaita A., Amarillo Y., Vega-Saenz de Miera E., Ma Y., Mo W., Goldberg E. M., Misumi Y., Ikehara Y., Neubert T. A., Rudy B. (2003) The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron 37, 449–461 [DOI] [PubMed] [Google Scholar]
- 6. Jerng H. H., Qian Y., Pfaffinger P. J. (2004) Modulation of Kv4.2 channel expression and gating by dipeptidyl peptidase 10 (DPP10). Biophys. J. 87, 2380–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang K. (2008) Modulation by clamping. Kv4 and KChIP interactions. Neurochem. Res. 33, 1964–1969 [DOI] [PubMed] [Google Scholar]
- 8. Liss B., Franz O., Sewing S., Bruns R., Neuhoff H., Roeper J. (2001) Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 20, 5715–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rhodes K. J., Carroll K. I., Sung M. A., Doliveira L. C., Monaghan M. M., Burke S. L., Strassle B. W., Buchwalder L., Menegola M., Cao J., An W. F., Trimmer J. S. (2004) KChIPs and Kv4 α subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 24, 7903–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norris A. J., Foeger N. C., Nerbonne J. M. (2010) Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J. Neurosci. 30, 13644–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuo H. C., Cheng C. F., Clark R. B., Lin J. J., Lin J. L., Hoshijima M., Nguyêñ-Trân V. T., Gu Y., Ikeda Y., Chu P. H., Ross J., Giles W. R., Chien K. R. (2001) A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of Ito and confers susceptibility to ventricular tachycardia. Cell 107, 801–813 [DOI] [PubMed] [Google Scholar]
- 12. Guo W., Li H., Aimond F., Johns D. C., Rhodes K. J., Trimmer J. S., Nerbonne J. M. (2002) Role of heteromultimers in the generation of myocardial transient outward K+ currents. Circ. Res. 90, 586–593 [DOI] [PubMed] [Google Scholar]
- 13. Hoffman D. A., Magee J. C., Colbert C. M., Johnston D. (1997) K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature 387, 869–875 [DOI] [PubMed] [Google Scholar]
- 14. Bernard C., Anderson A., Becker A., Poolos N. P., Beck H., Johnston D. (2004) Acquired dendritic channelopathy in temporal lobe epilepsy. Science 305, 532–535 [DOI] [PubMed] [Google Scholar]
- 15. Cai X., Liang C. W., Muralidharan S., Muralidharan S., Kao J. P., Tang C. M., Thompson S. M. (2004) Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron 44, 351–364 [DOI] [PubMed] [Google Scholar]
- 16. Chen X., Yuan L. L., Zhao C., Birnbaum S. G., Frick A., Jung W. E., Schwarz T. L., Sweatt J. D., Johnston D. (2006) Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J. Neurosci. 26, 12143–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J., Jung S. C., Clemens A. M., Petralia R. S., Hoffman D. A. (2007) Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron 54, 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barry D. M., Xu H., Schuessler R. B., Nerbonne J. M. (1998) Functional knockout of the transient outward current, long-QT syndrome, and cardiac remodeling in mice expressing a dominant-negative Kv4 α subunit. Circ. Res. 83, 560–567 [DOI] [PubMed] [Google Scholar]
- 19. Hoppe U. C., Marbán E., Johns D. C. (2000) Molecular dissection of cardiac repolarization by in vivo Kv4.3 gene transfer. J. Clin. Invest. 105, 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh B., Ogiwara I., Kaneda M., Tokonami N., Mazaki E., Baba K., Matsuda K., Inoue Y., Yamakawa K. (2006) A Kv4.2 truncation mutation in a patient with temporal lobe epilepsy. Neurobiol. Dis. 24, 245–253 [DOI] [PubMed] [Google Scholar]
- 21. Giudicessi J. R., Ye D., Tester D. J., Crotti L., Mugione A., Nesterenko V. V., Albertson R. M., Antzelevitch C., Schwartz P. J., Ackerman M. J. (2011) Transient outward current (Ito) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm 8, 1024–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giudicessi J. R., Ye D., Kritzberger C. J., Nesterenko V. V., Tester D. J., Antzelevitch C., Ackerman M. J. (2012) Novel mutations in the KCND3-encoded Kv4.3 K+ channel associated with autopsy-negative sudden unexplained death. Hum. Mutat. 33, 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duarri A., Jezierska J., Fokkens M., Meijer M., Schelhaas H. J., den Dunnen W. F., van Dijk F., Verschuuren-Bemelmans C., Hageman G., van de Vlies P., Küsters B., van de Warrenburg B. P., Kremer B., Wijmenga C., Sinke R. J., Swertz M. A., Kampinga H. H., Boddeke E., Verbeek D. S. (2012) Mutations in potassium channel kcnd3 cause spinocerebellar ataxia type 19. Ann. Neurol. 72, 870–880 [DOI] [PubMed] [Google Scholar]
- 24. Olesen M. S., Refsgaard L., Holst A. G., Larsen A. P., Grubb S., Haunso S., Svendsen J. H., Olesen S. P., Schmitt N., Calloe K. (2013) Cardiovasc. Res. doi: 10.1093/cvr/cvt028 [DOI] [PubMed] [Google Scholar]
- 25. Lee Y. C., Durr A., Majczenko K., Huang Y. H., Liu Y. C., Lien C. C., Tsai P. C., Ichikawa Y., Goto J., Monin M. L., Li J. Z., Chung M. Y., Mundwiller E., Shakkottai V., Liu T. T., Tesson C., Lu Y. C., Brice A., Tsuji S., Burmeister M., Stevanin G., Soong B. W. (2012) Mutations in KCND3 cause spinocerebellar ataxia type 22. Ann. Neurol. 72, 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burgoyne R. D. (2007) Neuronal calcium sensor proteins. Generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takimoto K., Yang E. K., Conforti L. (2002) Palmitoylation of KChIP splicing variants is required for efficient cell surface expression of Kv4.3 channels. J. Biol. Chem. 277, 26904–26911 [DOI] [PubMed] [Google Scholar]
- 28. O'Callaghan D. W., Hasdemir B., Leighton M., Burgoyne R. D. (2003) Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J. Cell Sci. 116, 4833–4845 [DOI] [PubMed] [Google Scholar]
- 29. Decher N., Barth A. S., Gonzalez T., Steinmeyer K., Sanguinetti M. C. (2004) Novel KChIP2 isoforms increase functional diversity of transient outward potassium currents. J. Physiol. 557, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Hoorick D., Raes A., Snyders D. J. (2007) The aromatic cluster in KCHIP1b affects Kv4 inactivation gating. J. Physiol. 583, 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pruunsild P., Timmusk T. (2005) Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics 86, 581–593 [DOI] [PubMed] [Google Scholar]
- 32. Shibata R., Misonou H., Campomanes C. R., Anderson A. E., Schrader L. A., Doliveira L. C., Carroll K. I., Sweatt J. D., Rhodes K. J., Trimmer J. S. (2003) A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 278, 36445–36454 [DOI] [PubMed] [Google Scholar]
- 33. Cui Y. Y., Liang P., Wang K. W. (2008) Enhanced trafficking of tetrameric Kv4.3 channels by KChIP1 clamping. Neurochem. Res. 33, 2078–2084 [DOI] [PubMed] [Google Scholar]
- 34. Foeger N. C., Marionneau C., Nerbonne J. M. (2010) Co-assembly of Kv4 α subunits with K+ channel-interacting protein 2 stabilizes protein expression and promotes surface retention of channel complexes. J. Biol. Chem. 285, 33413–33422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panama B. K., Latour-Villamil D., Farman G. P., Zhao D., Bolz S. S., Kirshenbaum L. A., Backx P. H. (2011) Nuclear factor κB downregulates the transient outward potassium current Ito,f through control of KChIP2 expression. Circ. Res. 108, 537–543 [DOI] [PubMed] [Google Scholar]
- 36. Jeyaraj D., Haldar S. M., Wan X., McCauley M. D., Ripperger J. A., Hu K., Lu Y., Eapen B. L., Sharma N., Ficker E., Cutler M. J., Gulick J., Sanbe A., Robbins J., Demolombe S., Kondratov R. V., Shea S. A., Albrecht U., Wehrens X. H., Rosenbaum D. S., Jain M. K. (2012) Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jerng H. H., Pfaffinger P. J. (2008) Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating. J. Biol. Chem. 283, 36046–36059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwenk J., Zolles G., Kandias N. G., Neubauer I., Kalbacher H., Covarrubias M., Fakler B., Bentrop D. (2008) NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. J. Biol. Chem. 283, 18937–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang P., Wang H., Chen H., Cui Y., Gu L., Chai J., Wang K. (2009) Structural insights into KChIP4a modulation of Kv4.3 inactivation. J. Biol. Chem. 284, 4960–4967 [DOI] [PubMed] [Google Scholar]
- 40. Liang P., Chen H., Cui Y., Lei L., Wang K. (2010) Functional rescue of Kv4.3 channel tetramerization mutants by KChIP4a. Biophys. J. 98, 2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim H. J., Ahn H. S., Choi B. H., Hahn S. J. (2011) Inhibition of Kv4.3 by genistein via a tyrosine phosphorylation-independent mechanism. Am. J. Physiol. Cell Physiol. 300, C567–C575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim H. J., Ahn H. S., Choi J. S., Choi B. H., Hahn S. J. (2011) Effects of ranolazine on cloned cardiac kv4.3 potassium channels. J. Pharmacol. Exp. Ther. 339, 952–958 [DOI] [PubMed] [Google Scholar]
- 43. Jeong I., Choi B. H., Hahn S. J. (2011) Rosiglitazone inhibits Kv4.3 potassium channels by open-channel block and acceleration of closed-state inactivation. Br. J. Pharmacol. 163, 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeong I., Kim S. W., Yoon S. H., Hahn S. J. (2012) Block of cloned Kv4.3 potassium channels by dapoxetine. Neuropharmacology 62, 2261–2266 [DOI] [PubMed] [Google Scholar]
- 45. Choi J. S., Hahn S. J. (2012) Duloxetine blocks cloned Kv4.3 potassium channels. Brain Res. 1466, 15–23 [DOI] [PubMed] [Google Scholar]
- 46. Scannevin R. H., Wang K., Jow F., Megules J., Kopsco D. C., Edris W., Carroll K. C., Lü Q., Xu W., Xu Z., Katz A. H., Olland S., Lin L., Taylor M., Stahl M., Malakian K., Somers W., Mosyak L., Bowlby M. R., Chanda P., Rhodes K. J. (2004) Two N-terminal domains of Kv4 K+ channels regulate binding to and modulation by KChIP1. Neuron 41, 587–598 [DOI] [PubMed] [Google Scholar]
- 47. Pioletti M., Findeisen F., Hura G. L., Minor D. L., Jr. (2006) Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 13, 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. (2007) Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat. Neurosci. 10, 32–39 [DOI] [PubMed] [Google Scholar]
- 49. Ma D., Jan L. Y. (2002) ER transport signals and trafficking of potassium channels and receptors. Curr. Opin. Neurobiol. 12, 287–292 [DOI] [PubMed] [Google Scholar]
- 50. Ellgaard L., Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 51. Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 52. Margeta-Mitrovic M., Jan Y. N., Jan L. Y. (2000) A trafficking checkpoint controls GABAB receptor heterodimerization. Neuron 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 53. Wang J. M., Zhang L., Yao Y., Viroonchatapan N., Rothe E., Wang Z. Z. (2002) A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat. Neurosci. 5, 963–970 [DOI] [PubMed] [Google Scholar]
- 54. Bichet D., Cornet V., Geib S., Carlier E., Volsen S., Hoshi T., Mori Y., De Waard M. (2000) The I-II loop of the Ca2+ channel α1 subunit contains an endoplasmic reticulum retention signal antagonized by the β subunit. Neuron 25, 177–190 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Z. N., Li Q., Liu C., Wang H. B., Wang Q., Bao L. (2008) The voltage-gated Na+ channel Nav1.8 contains an ER-retention/retrieval signal antagonized by the β3 subunit. J. Cell Sci. 121, 3243–3252 [DOI] [PubMed] [Google Scholar]
- 56. Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 57. Girard C., Tinel N., Terrenoire C., Romey G., Lazdunski M., Borsotto M. (2002) p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 21, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O'Kelly I., Butler M. H., Zilberberg N., Goldstein S. A. (2002) Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111, 577–588 [DOI] [PubMed] [Google Scholar]
- 59. Renigunta V., Yuan H., Zuzarte M., Rinné S., Koch A., Wischmeyer E., Schlichthörl G., Gao Y., Karschin A., Jacob R., Schwappach B., Daut J., Preisig-Müller R. (2006) The retention factor p11 confers an endoplasmic reticulum-localization signal to the potassium channel TASK-1. Traffic 7, 168–181 [DOI] [PubMed] [Google Scholar]
- 60. Shruti S., Urban-Ciecko J., Fitzpatrick J. A., Brenner R., Bruchez M. P., Barth A. L. (2012) The brain-specific β4 subunit downregulates BK channel cell surface expression. PLoS One 7, e33429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hancock J. F. (2003) Ras proteins. Different signals from different locations. Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 62. Szczesna-Skorupa E., Kemper B. (2000) Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J. Biol. Chem. 275, 19409–19415 [DOI] [PubMed] [Google Scholar]
- 63. Kaether C., Capell A., Edbauer D., Winkler E., Novak B., Steiner H., Haass C. (2004) The presenilin C terminus is required for ER-retention, nicastrin-binding and γ-secretase activity. EMBO J. 23, 4738–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parker A. K., Gergely F. V., Taylor C. W. (2004) Targeting of inositol 1,4,5-trisphosphate receptors to the endoplasmic reticulum by multiple signals within their transmembrane domains. J. Biol. Chem. 279, 23797–23805 [DOI] [PubMed] [Google Scholar]
- 65. Zarei M. M., Eghbali M., Alioua A., Song M., Knaus H. G., Stefani E., Toro L. (2004) An endoplasmic reticulum trafficking signal prevents surface expression of a voltage- and Ca2+-activated K+ channel splice variant. Proc. Natl. Acad. Sci. U.S.A. 101, 10072–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meur G., Parker A. K., Gergely F. V., Taylor C. W. (2007) Targeting and retention of type 1 ryanodine receptors to the endoplasmic reticulum. J. Biol. Chem. 282, 23096–23103 [DOI] [PubMed] [Google Scholar]
- 67. Ciczora Y., Callens N., Séron K., Rouillé Y., Dubuisson J. (2010) Identification of a dominant endoplasmic reticulum-retention signal in yellow fever virus pre-membrane protein. J. Gen. Virol. 91, 404–414 [DOI] [PubMed] [Google Scholar]
- 68. Angelotti T., Daunt D., Shcherbakova O. G., Kobilka B., Hurt C. M. (2010) Regulation of G-protein coupled receptor traffic by an evolutionary conserved hydrophobic signal. Traffic 11, 560–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dayel M. J., Hom E. F., Verkman A. S. (1999) Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 76, 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Frigerio L., Pastres A., Prada A., Vitale A. (2001) Influence of KDEL on the fate of trimeric or assembly-defective phaseolin. Selective use of an alternative route to vacuoles. Plant Cell 13, 1109–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Raykhel I., Alanen H., Salo K., Jurvansuu J., Nguyen V. D., Latva-Ranta M., Ruddock L. (2007) A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Matsukawa S., Moriyama Y., Hayata T., Sasaki H., Ito Y., Asashima M., Kuroda H. (2012) KDEL tagging. A method for generating dominant-negative inhibitors of the secretion of TGF-β superfamily proteins. Int. J. Dev. Biol. 56, 351–356 [DOI] [PubMed] [Google Scholar]
- 73. Griffiths G., Ericsson M., Krijnse-Locker J., Nilsson T., Goud B., Söling H. D., Tang B. L., Wong S. H., Hong W. (1994) Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J. Cell Biol. 127, 1557–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. (1990) ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 75. Jerng H. H., Kunjilwar K., Pfaffinger P. J. (2005) Multiprotein assembly of Kv4.2, KChIP3, and DPP10 produces ternary channel complexes with ISA-like properties. J. Physiol. 568, 767–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Amarillo Y., De Santiago-Castillo J. A., Dougherty K., Maffie J., Kwon E., Covarrubias M., Rudy B. (2008) Ternary Kv4.2 channels recapitulate voltage-dependent inactivation kinetics of A-type K+ channels in cerebellar granule neurons. J. Physiol. 586, 2093–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Foeger N. C., Norris A. J., Wren L. M., Nerbonne J. M. (2012) Augmentation of Kv4.2-encoded currents by accessory dipeptidyl peptidase 6 and 10 subunits reflects selective cell surface Kv4.2 protein stabilization. J. Biol. Chem. 287, 9640–9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hoshi T., Zagotta W. N., Aldrich R. W. (1990) Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 250, 533–538 [DOI] [PubMed] [Google Scholar]
- 79. Choi K. L., Aldrich R. W., Yellen G. (1991) Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. U.S.A. 88, 5092–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Baukrowitz T., Yellen G. (1995) Modulation of K+ current by frequency and external [K+]. A tale of two inactivation mechanisms. Neuron 15, 951–960 [DOI] [PubMed] [Google Scholar]
- 81. Jerng H. H., Covarrubias M. (1997) K+ channel inactivation mediated by the concerted action of the cytoplasmic N- and C-terminal domains. Biophys. J. 72, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bähring R., Covarrubias M. (2011) Mechanisms of closed-state inactivation in voltage-gated ion channels. J. Physiol. 589, 461–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bähring R., Boland L. M., Varghese A., Gebauer M., Pongs O. (2001) Kinetic analysis of open- and closed-state inactivation transitions in human Kv4.2 A-type potassium channels. J. Physiol. 535, 65–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Baranauskas G. (2004) Cell-type-specific splicing of KChIP4 mRNA correlates with slower kinetics of A-type current. Eur. J. Neurosci. 20, 385–391 [DOI] [PubMed] [Google Scholar]
- 85. Klemic K. G., Shieh C. C., Kirsch G. E., Jones S. W. (1998) Inactivation of Kv2.1 potassium channels. Biophys. J. 74, 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Salinas M., de Weille J., Guillemare E., Lazdunski M., Hugnot J. P. (1997) Modes of regulation of Shab K+ channel activity by the Kv8.1 subunit. J. Biol. Chem. 272, 8774–8780 [DOI] [PubMed] [Google Scholar]
- 87. Salinas M., Duprat F., Heurteaux C., Hugnot J. P., Lazdunski M. (1997) New modulatory α subunits for mammalian Shab K+ channels. J. Biol. Chem. 272, 24371–24379 [DOI] [PubMed] [Google Scholar]
- 88. Kramer J. W., Post M. A., Brown A. M., Kirsch G. E. (1998) Modulation of potassium channel gating by coexpression of Kv2.1 with regulatory Kv5.1 or Kv6.1 α-subunits. Am. J. Physiol. 274, C1501–C1510 [DOI] [PubMed] [Google Scholar]
- 89. Kerschensteiner D., Stocker M. (1999) Heteromeric assembly of Kv2.1 with Kv9.3. Effect on the state dependence of inactivation. Biophys. J. 77, 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Massone S., Vassallo I., Castelnuovo M., Fiorino G., Gatta E., Robello M., Borghi R., Tabaton M., Russo C., Dieci G., Cancedda R., Pagano A. (2011) RNA polymerase III drives alternative splicing of the potassium channel-interacting protein contributing to brain complexity and neurodegeneration. J. Cell Biol. 193, 851–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Aktas O., Ullrich O., Infante-Duarte C., Nitsch R., Zipp F. (2007) Neuronal damage in brain inflammation. Arch. Neurol. 64, 185–189 [DOI] [PubMed] [Google Scholar]