Background: The IL-6R stalk region is uncharacterized.
Results: Deletions within the IL-6R stalk selectively block ectodomain shedding by ADAM10 and ADAM17. The stalk length regulates IL-6 classic signaling.
Conclusion: ADAM10/ADAM17-mediated shedding depends on regions within the stalk, and a minimal 22-amino acid long stalk is needed for IL-6 signaling.
Significance: These results show the engineering of a protease-resistant, but still functional IL-6R.
Keywords: ADAM ADAMTS, Interleukin, Proteolytic Enzymes, Receptor Structure-Function, Signal Transduction, ADAM10, ADAM17, IL-6 Signal Transduction, Interleukin-6 Receptor, Interleukin-6
Abstract
Signaling of the pleiotropic cytokine Interleukin-6 (IL-6) is coordinated by membrane-bound and soluble forms of the IL-6 receptor (IL-6R) in processes called classic and trans-signaling, respectively. The soluble IL-6R is mainly generated by ADAM10- and ADAM17-mediated ectodomain shedding. Little is known about the role of the 52-amino acid-residue-long IL-6R stalk region in shedding and signal transduction. Therefore, we generated and analyzed IL-6R stalk region deletion variants for cleavability and biological activity. Deletion of 10 amino acids of the stalk region surrounding the ADAM17 cleavage site substantially blocked IL-6R proteolysis by ADAM17 but only slightly affected proteolysis by ADAM10. Interestingly, additional deletion of the remaining five juxtamembrane-located amino acids also abrogated ADAM10-mediated IL-6R shedding. Larger deletions within the stalk region, that do not necessarily include the ADAM17 cleavage site, also reduced ADAM10 and ADAM17-mediated IL-6R shedding, questioning the importance of cleavage site recognition. Furthermore, we show that a 22-amino acid-long stalk region is minimally required for IL-6 classic signaling. The gp130 cytokine binding sites are separated from the plasma membrane by ∼96 Å. 22 amino acid residues, however, span maximally 83.6 Å (3.8 Å/amino acid), indicating that the three juxtamembrane fibronectin domains of gp130 are not necessarily elongated but somehow flexed to allow IL-6 classic signaling. Our findings underline a dual role of the IL-6R stalk region in IL-6 signaling. In IL-6 trans-signaling, it regulates proper proteolysis by ADAM10 and ADAM17. In IL-6 classic-signaling, it acts as a spacer to ensure IL-6·IL-6R·gp130 signal complex formation.
Introduction
The pleiotropic cytokine Interleukin 6 (IL-6) is critically involved in health and disease (1) and activates downstream signaling pathways such as Janus kinase/signal transducer and activator of transcription (Jak/STAT), phosphatidylinositol 3-kinase cascade, and the mitogen-activated protein kinase cascade through a homodimer of glycoprotein 130 (gp130)2 (2). To achieve specificity and avoid unwanted cellular activation, IL-6 has to bind initially to the non-signaling interleukin-6 receptor α (IL-6R), which shows a distinct expression pattern on only a limited number of cell types including hepatocytes and some leukocytes (2).
The IL-6R is expressed as a type-I transmembrane protein. The IL-6R consists of an Ig-like domain (D1), the cytokine binding module (CBM) domains (D2 and D3), and a 52-amino acid-residue-long flexible stalk region (3) followed by the transmembrane and intracellular domains. The crystal structure suggests a hexameric signal transducing complex (4) containing two molecules of IL-6, IL-6R and gp130 each. IL-6 binding is achieved through three conserved epitopes (named site I, site II, and site III), whereas site I binds initially to the CBM of the IL-6R. Contact to gp130 is then facilitated by site II and site III of IL-6. The IL-6·IL-6R complex formation subsequently leads to the recruitment of two gp130 receptor proteins, as neither IL-6 nor the IL-6R on their own have any measureable affinity toward gp130 (5). The stalk region of the IL-6R does not directly take part in signal transduction but might act as a spacer, positioning the three extracellular domains in a defined distance from the plasma membrane, thereby allowing complex formation of IL-6·IL-6R/gp130. The cytokine binding area of gp130 is separated from the plasma membrane by three FNIII domains (2), which in a stretched conformation span about 96 Å, or about 83 Å if arranged in a bend conformation. The 52 amino acids of the IL-6R stalk region span maximally about 198 Å. Therefore, we have characterized the minimal length of the IL-6R to span this distance and to maintain biological activity of the IL-6R. IL-6 signaling via the membrane-bound IL-6R was named classic signaling and has to be distinguished from IL-6 trans-signaling, where IL-6 signals in complex with the soluble IL-6R (sIL-6R) (6). Importantly, IL-6 binds with the same affinity to the membrane-bound and the sIL-6R. Because gp130 but not the IL-6R is ubiquitously expressed, the IL-6/sIL-6R complex can virtually activate all cells of the human body. Classic signaling is involved in regenerative, anti-inflammatory activities, whereas IL-6 trans-signaling is considered to be proinflammatory and causally involved in various autoimmune and chronic diseases (1, 6). Soluble gp130 is the natural antagonist of IL-6 trans-signaling (5), although under conditions when the molar ratio of sIL-6R exceeds those of IL-6, soluble gp130 can also interfere with IL-6 classic signaling (7). Furthermore, soluble gp130 is able to block IL-30/sIL-6R signaling (8).
The soluble IL-6R can be generated by two distinct mechanisms (9). Besides alternative splicing of the mRNA, a process that leads to excision of the exon coding for the transmembrane region (10), the sIL-6R is predominantly (∼90%) generated via limited proteolysis of the membrane-bound precursor, also called ectodomain shedding. The responsible proteases that cleave the human IL-6R are two members of a disintegrin and metalloprotease family ADAM10 and ADAM17 (11–13). ADAM17 can be activated by different stimuli, for example phorbol esters like phorbol-12-myristate-13-acetate (PMA), bacterial metalloproteinases, and apoptosis, whereas calcium influx, mediated for example by activation of P2-family receptors or the ionophore ionomycin, leads to specific activation of ADAM10 (13). ADAM17 has been shown to cleave the human IL-6R between amino acid residues Gln-357 and Asp-358 (11), which is part of the stalk region and in close proximity of the plasma membrane. Whether ADAM10 cleavage occurs at the same site in the IL-6R has not been investigated so far. In this study the human IL-6R was engineered to be protease-resistant but still IL-6 signal transducing-competent.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Ba/F3-gp130 cells were obtained from Immunex (Seattle, WA) (14)), and HEK293 cells were from DSMZ GmbH (Braunschweig, Germany). All cells were grown under standard conditions (DMEM high glucose culture medium (PAA Laboratories, Cölbe, Germany) supplemented with 10% fetal bovine serum, penicillin (60 mg/liter) and streptomycin (100 mg/liter), 37 °C, 5% CO2, water-saturated atmosphere). Ba/F3-gp130 cells were cultured using 10 ng/ml recombinant Hyper-IL-6. Its expression and purification was done according to Refs. 15 and 16. Ba/F3-gp130, which were stable-transduced with different hIL-6R cDNAs, were cultured with 10 ng/ml recombinant human IL-6 instead of Hyper-IL-6. Human IL-6 was expressed and purified as described previously (17). The anti-hIL-6R mAb 4–11 was described previously (18), and the anti-β-actin mAb (sc-47778) was from Santa Cruz Biotechnology (Santa Cruz, CA). Phosphatidylethanolamine-coupled anti-mouse/rat CD126 (IL-6R) mAb and its isotype control were from BioLegend (San Diego, CA). The peroxidase-conjugated secondary antibodies were purchased from Pierce. The FITC-conjugated goat anti-mouse IgG was from Jackson ImmunoResearch Laboratories (Dianova GmbH, Hamburg, Germany). PMA and ionomycin were purchased from Sigma. The two metalloprotease inhibitors GI254023X (GI, selective for ADAM10) and GW280264X (GW, selective for both ADAM10 and ADAM17) were generous gifts from GlaxoSmithKline (Stevenage, UK) (19, 20).
Construction of Plasmid Coding for hIL-6R and Deletion Variants
Expression plasmids for hIL-6R have been described (13, 18). Deletion variants of the IL-6R stalk regions were constructed using standard cloning procedures via splicing by overlapping extension PCRs. Constructs were cloned with a 5′ KpnI site and a 3′ NotI site into pcDNA3.1. Subcloning into pMOWS was achieved by digestion of the pcDNA3.1 plasmids with PmeI (21).
Transient Transfection of HEK293 Cells and Ectodomain Shedding Assays
2 × 106 HEK293 cells were seeded on 10-cm culture dishes 24 h before transfection. Cells were transiently transfected with pcDNA3.1 expression plasmids using Turbofect (Fermentas) according to the manufacturer's instructions. Transfection efficiency of this method was ∼80% as seen by transfection of a pEGFP-N1 plasmid. Ectodomain shedding assays have been performed as described previously (13). To analyze ectodomain shedding in stable transduced Ba/F3-gp130 cells, cells were washed one time with PBS and diluted to a concentration of 2 × 106/ml. Shedding assays were performed analogous to HEK293 cells.
Precipitation of Soluble Cytokine Receptors
Transiently transfected HEK293 cells were incubated with PMA or ionomycin for the indicated time points. Cell supernatants were centrifuged to remove cellular debris, and cleared supernatants were transferred to fresh tubes. 1 ml of each sample was then incubated with 50 μl of a concanavalin A-Sepharose conjugate (Sigma) overnight at 4 °C under constant agitation to bind the shed IL-6R variants onto the Sepharose beads. Afterward, the Sepharose was collected by centrifugation, and the supernatant was removed. The Sepharose beads were washed two times with ice-cold PBS and directly boiled in Laemmli buffer, which was subsequently used for Western blotting analysis.
Enzyme-linked Immunosorbent Assay
The ELISA for human IL-6R was described previously (18). Accordingly, microtiter plates were coated with the mouse anti-hIL-6R mAb 4–11 (diluted to 1 μg/ml in PBS) overnight at room temperature. After blocking, cell culture supernatants were added at an appropriate dilution. The biotinylated goat anti-hIL-6R antibody BAF227 (R&D Systems, Minneapolis, MN) was used as secondary antibody. The ELISA was performed with streptavidin-horseradish peroxidase (R&D Systems, Minneapolis, MN). The enzymatic reaction was performed with the peroxidase substrate BM blue POD (Roche Applied Science), and the absorbance was read at 450 nm on a Tecan infinite M200 PRO reader (Tecan, Maennedorf, Switzerland).
Shedding Assay Data Analysis and Calculation
We used two different mathematical methods to analyze ectodomain shedding of the different IL-6R variants. First, to analyze whether activation of ADAM10 or ADAM17 results principally in an increased sIL-6R generation, we set the amount that was shed without stimulus (DMSO) to 1 and calculated the x-fold increase after PMA or ionomycin treatment. To show differences in the total amount of sIL-6R that was generated through ectodomain shedding between the IL-6R variants, we set the amount of sIL-6R generated after PMA or ionomycin treatment of wild-type IL-6R to 100% and calculated the percentage of unstimulated shedding (DMSO) or ionomycin-induced shedding of the different IL-6R constructs according to this value.
Retroviral Transduction of Murine Cells
Retroviral transduction of the murine pre-B cell line Ba/F3-gp130 was described previously (21). In brief, Phoenix-Eco cells were transiently transfected with the pMOWS plasmids encoding the different IL-6R cDNAs. Cell supernatants containing the retroviruses were collected 24 h after transfection. Ba/F3-gp130 cells were washed once with PBS, and 1 × 105 cells were mixed with 250 μl of retroviral supernatant and centrifuged for 2 h at room temperature at a speed of 1800 rpm. After this, cells were resuspended in DMEM containing 10 ng/ml Hyper-IL-6. Selection of transduced cells was achieved by the addition of puromycin (1.5 μg/ml) 48 h after transduction. Cells were cultured with Hyper-IL-6 (10 ng/ml) during the time of selection and switched afterward to IL-6 (both 10 ng/ml) where appropriate.
Flow Cytometry
For detection of cell surface expression of the different IL-6R variants, 1 × 106 HEK293 or Ba/F3-gp130 cells were washed one time with FACS buffer (PBS, 0.5% BSA) and incubated in 100 μl of FACS buffer containing 1:100 diluted anti-hIL-6R 4–11 mAb for 60 min on ice. After two subsequent washing steps in FACS buffer, cells were incubated in 100 μl of FACS buffer containing a 1:100 dilution of FITC-conjugated anti-mouse mAb (Dianova). Cells were washed twice in FACS buffer, suspended in 500 μl of FACS buffer, and analyzed by flow cytometry on a BD Biosciences FACS Canto II and FCS Express (De Novo Software, Los Angeles, CA).
Proliferation Assays
Proliferation of the different Ba/F3-gp130 cell lines was determined as described previously (7) using the Cell Titer Blue Cell viability assay reagent (Promega, Karlsruhe, Germany) following the manufacturer's protocol. The extinction was measured using a Tecan infinite M200 PRO reader (excitation 530 nm, emission 590 nm, gain 90, i-control 1.7 software, Tecan AG). Normalization of relative light units was achieved by subtraction of negative control values. All values were measured in triplicates per experiment.
Western Blot
48 h after transfection adherent cells were lysed in mild lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, complete protease inhibitor mixture tablets). Protein lysates were separated by SDS-PAGE under reducing conditions. After transfer to a PVDF membrane (GE Healthcare), membranes were blocked with 5% skim milk powder in TBS-T (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 0.05% Tween 20). The membrane was probed with anti-hIL-6Rα (4–11) mAb at 4 °C overnight. After several washing steps, membranes were probed with an anti-mouse-antibody conjugated to horseradish peroxidase, and proteins were detected with ECL Prime Western blotting detection reagent (GE Healthcare) according to the manufacturer's instructions. Before probing for β-actin as loading control, membranes were stripped with stripping buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 0.1% β-mercaptoethanol, for 30 min at 60 °C), washed 3 times, and blocked again.
Statistical Analysis
Data are expressed as mean values ± S.D. calculated from three independent experiments unless otherwise stated.
RESULTS
Deletion of Ser-353 to Val-362 within the Human IL-6R Stalk Abolishes Human ADAM17- but Not Human ADAM10-mediated Shedding
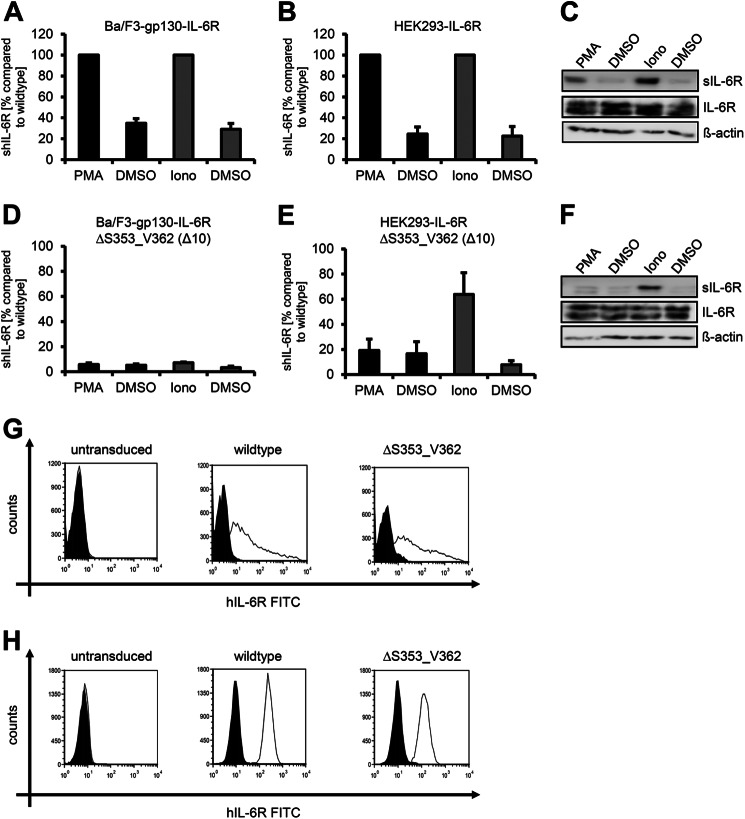
Treatment of cells with either the phorbol-ester PMA or ionomycin selectively induces ADAM17 or ADAM10-mediated IL-6R shedding, respectively (11–13). To determine whether activation of ADAM10 or ADAM17 results in an increased sIL-6R generation, we set the amount that was shed without stimulus (DMSO) to 1 and calculated the x-fold increase after PMA or ionomycin treatment according to this. We performed all experiments with murine Ba/F3-gp130 cells that were stably transduced with the individual IL-6R variants as well as transiently transfected human HEK293 cells. After PMA or ionomycin treatment, Ba/F3-gp130-hIL-6R cells showed a 2.9 ± 0.4-fold or a 3.6 ± 0.7-fold increase of the sIL-6R, respectively, as determined by ELISA (Fig. 1A and Table 1). We observed the same pattern in HEK293 cells transiently transfected with human IL-6R, which showed an 4.3 ± 1.2-fold and after ionomycin treatment an 5.7 ± 2.7-fold increase of the sIL-6R as determined by ELISA and Western blotting (Fig. 1, B and C, Table 2, and Ref. 13). Cleavage of the IL-6R by ADAM17 occurs within the extracellular 52-amino acid-residue-long juxtamembrane stalk region between Gln-357 and Asp-358, whereas the cleavage site of ADAM10 is not known (Fig. 2A). The hIL-6RΔS353_V362 variant, which has a deletion of 10 amino acids surrounding the cleavage site of ADAM17, is resistant to ADAM17-mediated proteolysis (22). Indeed, Ba/F3-gp130-hIL-6RΔS353_V362 cells showed no increase in sIL-6R after PMA stimulation (1.2 ± 0.4-fold, Fig. 1D), and the same was true for HEK293 cells transiently transfected with a cDNA coding for hIL-6RΔS353_V362 (1.3 ± 0.4-fold; Fig. 1, E and F). Whereas also ionomycin-induced ADAM10-mediated shedding was largely absent in Ba/F3-gp130-hIL-6RΔS353_V362 cells (2.3 ± 0.7-fold, Fig. 1D, Table 1), HEK293 cells transfected with hIL-6RΔS353_V362 showed an 8.9 ± 2.3-fold increase of sIL-6R after ionomycin-induced activation of ADAM10. Both wild-type and hIL-6RΔS353_V362 were equally expressed at the cell surface of HEK293 (Fig. 1G) and Ba/F3-gp130 cells (Fig. 1H) as analyzed by flow cytometry. To show differences in the total amount of sIL-6R that was generated by ectodomain shedding, we next set the amount of sIL-6R generated after PMA or ionomycin treatment of wild-type IL-6R to 100% and calculated the percentage of unstimulated shedding (DMSO) and PMA or ionomycin-induced shedding of the IL-6R deletion variant according to this value. Compared with wild-type IL-6R in HEK293 cells, the total amount of sIL-6RΔS353_V362 generated by ADAM10 was reduced to 63.7 ± 17.4% and by ADAM17 to 19.2 ± 9.0% (Fig. 1, E and F, Table 2), indicating that hIL-6RΔS353_V362 is cleaved with less efficiency than wild-type IL-6R. This reduction was even stronger in Ba/F3-gp130 cells, with a reduction to 5.7 ± 1.3% (ADAM17) and 7.2 ± 0.7% (ADAM10). Previously, we showed that murine and human ADAM proteins differentially shed murine IL-6R (13); therefore, it is not surprising that these proteases also differ in their behavior to shed human IL-6R. Moreover, these results suggest that the ADAM10 cleavage site used by human ADAM10 is not located between Ser-353 and Val-362 of the stalk region of the human IL-6R and might differ from the cleavage site used by murine ADAM10.
FIGURE 1.
ADAM17 and ADAM10 use different cleavage sites of the human IL-6R. A, Ba/F3-gp130-hIL-6R cells were treated for 2 h with PMA (100 nm) or for 1 h with ionomycin (Iono, 1 μm). Soluble IL-6R was measured by ELISA. The amount of soluble cytokine receptor without stimulation was considered as constitutive shedding and set to 1. Based on this, the increase of soluble receptors was calculated. B, HEK293 cells were transfected with an expression plasmid encoding wild-type human IL-6R. Cells were treated as described in panel A, sIL-6R was measured by ELISA, and values were calculated accordingly. C, transiently transfected HEK293 cells were treated as described under panel A. To determine the soluble cytokine receptors via Western blotting, they were precipitated from conditioned media with concanavalin A-covered Sepharose beads and visualized with 4-11 antibody (IL-6R). Cells were lysed after stimulation, and lysates were subsequently analyzed via Western blotting, whereas β-actin served as loading control. D, Ba/F3-gp130-IL-6RΔS353_V362 cells were treated as described in panel A. E and F, HEK293 cells were transfected with an expression plasmid encoding hIL-6RΔS353_V362. The experiment was performed as described in panels A and B. ELISA data are the mean (±S.D.) from three independent experiments, and Western blotting shows one representative experiment. G, cell surface expression of wild-type IL-6R and IL-6RΔS353_V362 on transiently transfected HEK293 cells was determined via flow cytometry as described under “Experimental Procedures.” H, cell surface expression on stably transduced Ba/F3-gp130-hIL-6R and Ba/F3-gp130-IL-6RΔS353_V362 cells was determined via flow cytometry as described under “Experimental Procedures.” One representative experiment of three performed is shown. The expressed IL-6R variant is given above the respective FACS plot.
TABLE 1.
Proteolysis of IL-6R and its deletion variants in Ba/F3-gp130 cells
Calculation of the values was done as described under “Material and Methods.” The values shown are the mean ± S.D. of three independent experiments. ND indicates that values needed for calculation were beyond the detection limit of the hIL-6R ELISA.
| IL-6R Construct | x-Fold increase after PMA (ADAM17) | x-Fold increase after ionomycin (ADAM10) | % of ADAM17-mediated proteolysis (compared to wt) | % of ADAM10 mediated proteolysis (compared to wt) |
|---|---|---|---|---|
| Wildtype | 2.9 ± 0.4 | 3.6 ± 0.7 | 100 | 100 |
| ΔS353_V362 | 1.2 ± 0.4 | 2.3 ± 0.7 | 5.7 ± 1.3 | 7.2 ± 0.7 |
| ΔI343_T352 | 6.7 ± 2.9 | 9.9 ± 3.9 | 79.3 ± 61.4 | 53.9 ± 31.3 |
| ΔA333_N342 | 5.6 ± 0.9 | 4.3 ± 0.4 | 76.2 ± 28.1 | 44.8 ± 6.8 |
| ΔS353_F367 | 1.2 ± 0.4 | 1.4 ± 0.6 | 5.1 ± 0.2 | 4.1 ± 1.2 |
| ΔA333_V362 | ND | ND | 5.0 ± 0.8 | 4.1 ± 0.8 |
| ΔE317_T352 | ND | ND | 5.9 ± 2.5 | 3.1 ± 0.7 |
| ΔA323_V362 | ND | ND | ND | 0.9 ± 0.5 |
| ΔA323_F367 | ND | ND | 3.6 ± 2.6 | 4.7 ± 2.3 |
| ΔE317_V362 | ND | ND | 1.1 ± 0.3 | 1.3 ± 0.4 |
TABLE 2.
Proteolysis of IL-6R and its deletion variants in HEK293 cells
Calculation of the values was done as described under “Material and Methods.” The values shown are the mean ± S.D. of three independent experiments.
| IL-6R Construct | x-Fold increase after PMA (ADAM17) | x-Fold increase after ionomycin (ADAM10) | % of ADAM17-mediated proteolysis (compared to wt) | % of ADAM10 mediated proteolysis (compared to wt) | % of constitutive proteolysis (compared to wt) |
|---|---|---|---|---|---|
| Wildtype | 4.3 ± 1.2 | 5.7 ± 2.7 | 100 | 100 | 100 |
| ΔS353_V362 | 1.3 ± 0.4 | 8.9 ± 2.3 | 19.2 ± 9.0 | 63.7 ± 17.4 | 32.7 ± 9.1 |
| ΔI343_T352 | 5.5 ± 2.6 | 3.0 ± 0.7 | 52.4 ± 19.3 | 42.6 ± 14.8 | 61.0 ± 3.0 |
| ΔA333_N342 | 3.5 ± 0.9 | 4.3 ± 1.8 | 63.8 ± 29.2 | 67.7 ± 21.8 | 68.1 ± 13.8 |
| ΔS353_F367 | 2.4 ± 0.9 | 2.0 ± 0.2 | 17.0 ± 3.6 | 14.5 ± 5.1 | 50.2 ± 10.4 |
| ΔA333_V362 | 1.7 ± 0.5 | 2.9 ± 1.2 | 5.4 ± 2.0 | 13.3 ± 4.1 | 17.6 ± 2.6 |
| ΔE317_T352 | 2.0 ± 0.5 | 5.3 ± 1.6 | 4.1 ± 1.6 | 6.7 ± 4.5 | 10.8 ± 1.8 |
| ΔA323_V362 | 2.1 ± 1.1 | 5.1 ± 3.7 | 0.2 ± 0.05 | 0.3 ± 0.08 | 3.1 ± 0.4 |
| ΔA323_F367 | 1.1 ± 0.1 | 2.7 ± 0.7 | 1.1 ± 0.3 | 4.0 ± 0.8 | 11.9 ± 2.3 |
| ΔE317_V362 | 2.1 ± 0.8 | 3.7 ± 2.3 | 0.4 ± 0.3 | 0.9 ± 0.7 | 5.0 ± 0.5 |
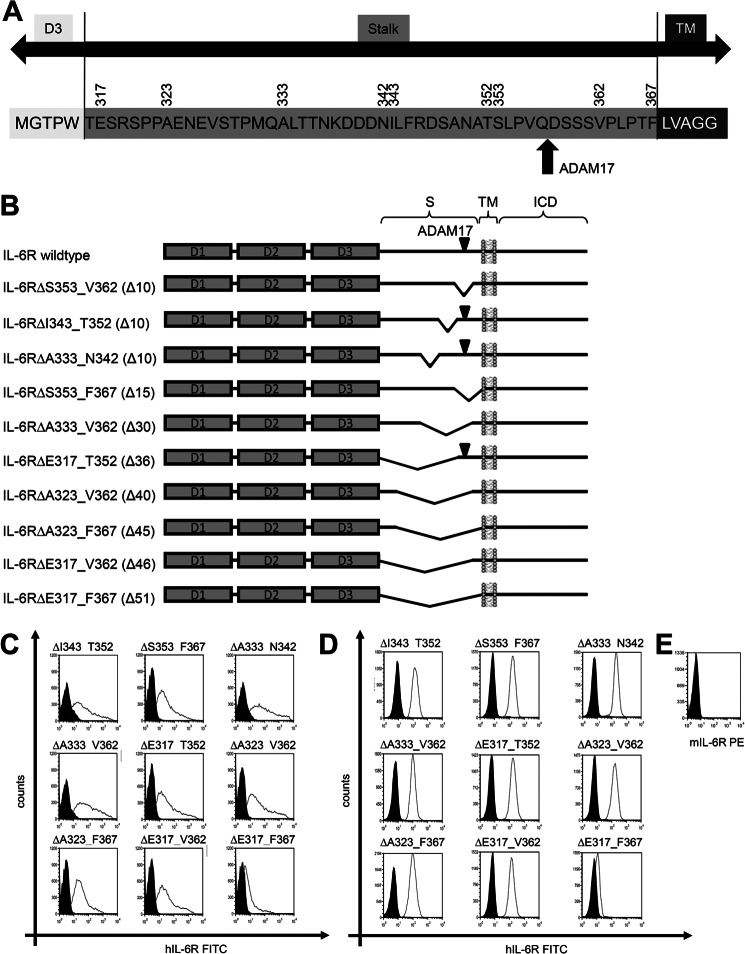
FIGURE 2.
Schematic overview of the human IL-6R constructs. A, shown is a schematic overview of the human IL-6R stalk region. The amino acid sequence of the human IL-6R from Met-311 to Gly-372 is shown. Amino acids belonging to the D3 region are shown in white, and amino acids belonging to the intracellular domain (ICD) are in dark gray. The stalk region is shown in light gray. Numbers above the sequence denote amino acids important for construction of deletion variants. The ADAM17 cleavage site is marked with a red arrow. B, the three extracellular domains of the human IL-6R are shown as dark gray-filled boxes (D1, D2, and D3). The other abbreviations used are stalk region (S), transmembrane region (TM), and intracellular domain (ICD). The known ADAM17 cleavage site Gln-357/ Asp-358 is marked with a black triangle in all constructs where this cleavage site is present. Deletions within the stalk regions are shown as kinks. The range of deleted amino acids is written in front of the schematic drawing, and the number of deleted amino acids given enclosed in parentheses. C, cell surface expression on transiently transfected HEK293 cells was determined via flow cytometry as described under “Experimental Procedures.” D, cell surface expression on stably transduced Ba/F3 cells was determined via flow cytometry as described under “Experimental Procedures.” E, the absence of endogenous IL-6R expression on Ba/F3-gp130 was stained with a murine IL-6R-specific antibody. One representative experiment of three performed is shown. The expressed IL-6R variant is given above the respective FACS plot. PE, phosphatidylethanolamine.
The Human ADAM10 Cleavage Site Differs from ADAM17 and Is Located in Close Proximity to the Plasma Membrane
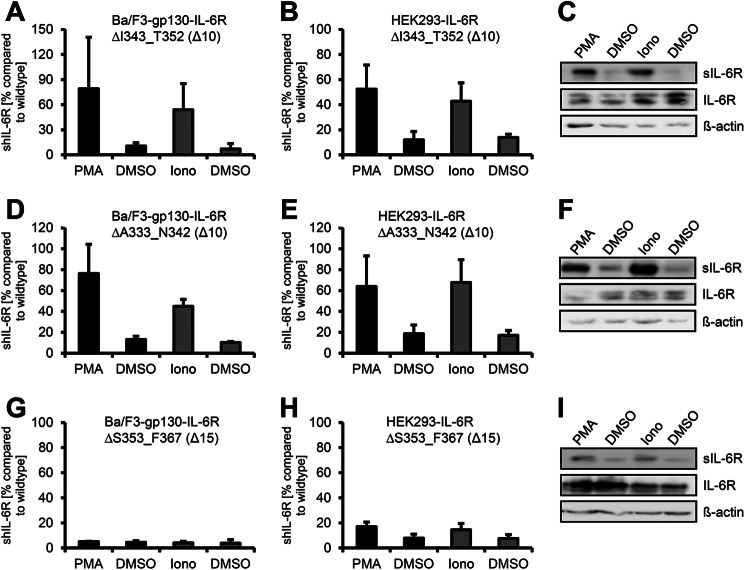
To identify IL-6R variants that are not shed by human ADAM10 and ADAM17, we generated in total 10 different IL-6R stalk region deletion variants. A schematic overview of all IL-6R deletion variants used in this study is shown in Fig. 2B, and all values concerning limited proteolysis by ADAM10 and ADAM17 are given in Tables 1 and 2. The two IL-6R variants hIL-6RΔI343_T352 and IL-6RΔA333_N342 are also based on deletions of 10 amino acid residues in, however, greater distance from the transmembrane region compared with hIL-6RΔS353_V362. Experiments with Ba/F3-gp130 cells revealed that hIL-6RΔI343_T352 was efficiently shed by ADAM17 (6.7 ± 2.9-fold increase of sIL-6R; Table 1, Fig. 3A) and ADAM10 (9.9 ± 3.9-fold increase of sIL-6R) (Fig. 3A, Table 1). These results were confirmed in HEK293 cells (ADAM17, 5.5 ± 2.6-fold increase; ADAM10, 3.0 ± 0.7-fold increase; Fig. 3, B and C, and Table 2). Next, we tested Ba/F3-gp130-IL-6RΔA333_N342 and found shedding by ADAM17 (5.6 ± 0.9-fold increase of sIL-6R) as well as ADAM10 (4.3 ± 0.4-fold increase of sIL-6R) (Fig. 3D and Table 1). Again, we observed similar results in HEK293 cells (ADAM17, 3.5 ± 0.9-fold increase; ADAM10, 4.3 ± 1.8-fold increase; Fig. 3, E and F, and Table 2). We concluded from these findings that the ADAM10 cleavage site is not located upstream of the ADAM17 cleavage site (amino acid position Ala-333 to Thr-352). Shortening of the IL-6R stalk by 10 amino acids does not abrogate ADAM17 substrate recognition as long as the cleavage site is retained, but again, an overall reduction of the total amount of shed IL-6R compared with wild type IL-6R to 52.4 ± 19.3% (IL-6RΔI343_T352) and 63.8 ± 29.2% (IL-6RΔA333_N342) was detected in HEK293 cells. Both variants were detected at the cell surface of transiently transfected HEK293 cells (Fig. 2C), although the expression seemed slightly reduced in comparison to wild-type IL-6R, which besides shedding also might contribute to a smaller extent to the reduction in sIL-6R generation. In line with this, also the respective Ba/F3-gp130 cell lines showed a significantly reduced generation of sIL-6R compared with wild-type (Table 1). Again, no differences in protein expression or cell surface localization of IL-6RΔI343_T352 and IL-6RΔA333_N342 compared with wild-type IL-6R were observed (Fig. 2, C and D, and Fig. 3, C and F).
FIGURE 3.
The ADAM10 cleavage site of the IL-6R is located close to the plasma membrane. A, Ba/F3-gp130-IL6R-ΔI343_T352 cells were treated for 2 h with PMA (100 nm) or for 1 h with ionomycin (Iono, 1 μm). Soluble IL-6R was measured by ELISA. The amount of soluble cytokine receptor without stimulation was considered as constitutive shedding and set to 1. Based on this, the increase of soluble receptors was calculated. B, HEK293 cells were transfected with an expression plasmid encoding IL-6RΔI343_T352. Cells were treated as described in panel A, sIL-6R was measured by ELISA, and values were calculated accordingly. C, transiently transfected HEK293 cells were treated as described in panel A. To determine the soluble cytokine receptors via Western blotting, they were precipitated from conditioned media with concanavalin A-covered Sepharose beads and visualized with 4–11 antibody (IL-6R). Cells were lysed after stimulation, and lysates were subsequently analyzed via Western blotting, whereas β-actin served as the loading control. D, Ba/F3-gp130-IL-6RΔA333_N342 cells were treated as described in panel A. E and F, HEK293 cells were transiently transfected with an expression plasmid encoding IL-6RΔA333_N342, and experiments were performed as described under panels (B and C). G, Ba/F3-gp130-IL-6RΔS353_F367 cells were treated as described in panel A. H and I, HEK293 cells were transiently transfected with an expression plasmid encoding IL-6RΔS353_F367, and experiments were performed as described in panels B and C.
Our first deletion variant hIL-6RΔS353_V362 still contained five juxtamembrane located amino acids of the stalk region before beginning of the transmembrane region (Fig. 2A). To analyze if this area is needed for ADAM10-mediated IL-6R shedding in HEK293 cells, we generated the hIL-6RΔS353_F367 variant in which these 5 amino acids plus the 10 amino acids from hIL-6RΔS353_V362 were deleted. We observed no induced shedding by either ADAM10 (1.4 ± 0.6-fold) or ADAM17 (1.2 ± 0.4-fold increase in sIL-6R) in Ba/F3-gp130 cells (Fig. 3G and Table 1). Importantly, also in HEK293 cells, both ADAM10 and ADAM17-mediated shedding was significantly reduced (2.0 ± 0.2-fold increase of sIL-6R after ionomycin and 2.4 ± 0.9-fold increase after PMA, respectively) (Fig. 3, H and I, and Table 2). Moreover, we found a total reduction of the sIL-6R amount to 17.0 ± 3.6% (ADAM17, PMA) and 14.5 ± 5.1% (ADAM10, ionomycin) compared with wild-type IL-6R. Even though in HEK293 cells PMA and ionomycin-shedding of IL-6RΔS353_V362 appeared to be slightly induced, the total amount of induced sIL-6R shedding was even lower than total wild-type sIL-6R released by constitutive shedding without any stimulus. In conclusion, our data showed that deletion of the 15 amino acids from Ser-353 to Phe-367 efficiently abrogates shedding of the IL-6R by ADAM10 and ADAM17. These findings underline the importance of specific substrate cleavage sites for ADAM17 and ADAM10.
Larger Deletions within the Stalk Region Also Reduced Shedding by ADAM10 and ADAM17
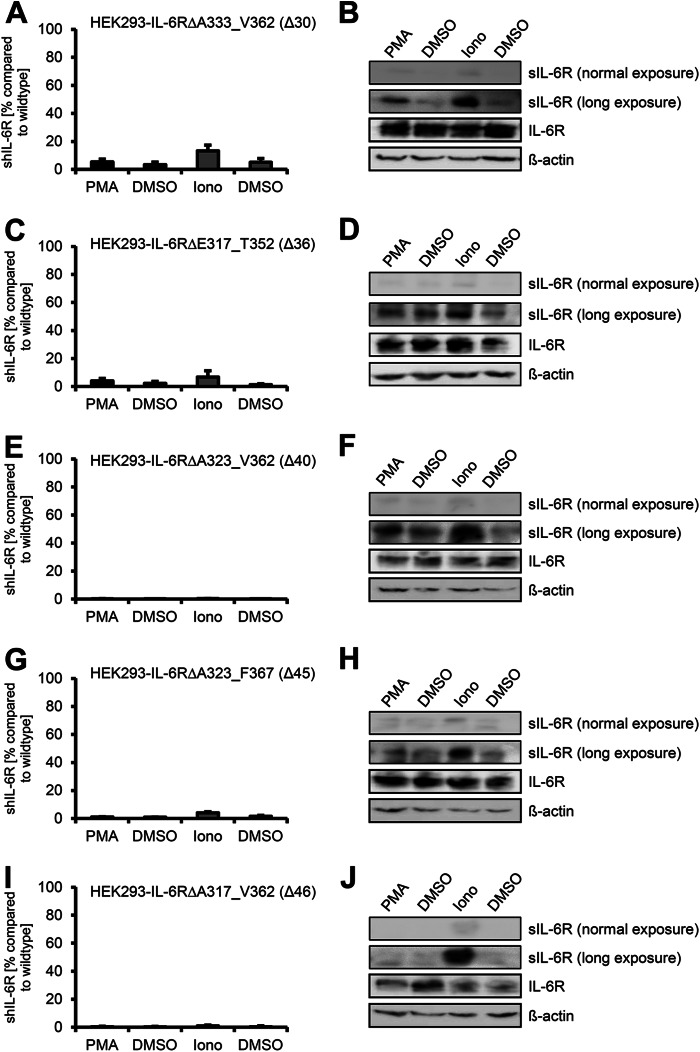
To test whether shedding of the IL-6R can be prevented by deletion of alternative amino acids within the stalk region, which are not necessarily part of ADAM10 and/or ADAM17 cleavage sites, we generated IL-6R variants with larger deletions within the stalk region. First, we combined the three 10-amino acid deletion variant encoding cDNAs, which resulted in the hIL-6RΔA333_C362 cDNA lacking the ADAM17 cleavage site, but still retained the hypothetical ADAM10 cleavage site. In transiently transfected HEK293 cells, hIL-6RΔA333_V362 showed nearly no PMA-induced ADAM17-mediated shedding (1.7 ± 0.5-fold increase) and a 2.9 ± 1.2-fold increase of proteolysis after activation of ADAM10 (Fig. 4, A and B, and Table 2). Compared with wild-type hIL-6R, the amount of totally generated sIL-6R was, however, drastically reduced to 13.3 ± 4.1 and 5.4 ± 2.0% after ionomycin or PMA treatment, respectively (Fig. 4A). Precipitated sIL-6R could only be made visible by Western blotting after overexposure of the membrane, whereas after normal exposure hardly any protein was detectable (Fig. 4B). Again, we observed no reduction in expression of hIL-6RΔA333_V362 as shown by Western blotting (Fig. 4B) and no reduction of cell-surface expression by flow cytometry in transiently transfected HEK293 cells (Fig. 2C). The amount of sIL-6R released from Ba/F3-gp130-hIL-6RΔA333_V362 without stimulus was below the detection limit of our IL-6R ELISA (∼30 pg/ml), and we, therefore, were not able to calculate the increase after activation of ADAM10 or ADAM17, respectively (Table 1), which was also true for all other variants described below. Again, there were no alterations in cell-surface expression of these IL-6R variants (Fig. 2D).
FIGURE 4.
Larger deletions within the stalk region abrogate ADAM10 and ADAM17 mediated shedding. HEK293 cells were transfected with expression plasmids encoding B IL-6RΔA333_V362 (A), IL-6RΔE317_T352 (C and D), IL-6RΔA323_V362 (E and F), IL-6RΔA323_F367 (G and H), or IL-6RΔE317_V362 (I and J). Soluble IL-6R was measured by ELISA. Precipitation and Western blotting was performed as described in the legend of Fig. 1. ELISA data are the mean ± S.D. from three independent experiments. Western blotting shows one representative experiment. Iono, ionomycin.
Next, in hIL-6RΔE317_T352 we removed 36 amino acid residues but retained the ADAM17 and the hypothetical ADAM10 cleavage sites. Again, we observed no differences in protein expression as judged by Western blotting (Fig. 4D) and cell-surface expression by flow cytometry in HEK293 cells (Fig. 2C) compared with wild-type IL-6R. Even though the ADAM17 cleavage site was conserved in hIL-6RΔE317_T352, PMA treatment led only to a 2.0 ± 0.5-fold increase of IL-6R compared with untreated control. In contrast, ionomycin treatment induced a significant 5.3 ± 1.6-fold increase in sIL-6R, indicating a functional ADAM10 cleavage site (Fig. 4, C and D, and Table 2). However, compared with wild-type IL-6R, proteolysis by ADAM17 was reduced to only 4.1 ± 1.6% and by ADAM10 to 6.7 ± 4.5% (Table 2), and we were, as in the previous described IL-6R variant hIL-6RΔA333_V362, only able to detect any sIL-6R by Western blotting after massive overexposure. We conclude from this that, although shedding of hIL-6RΔE317_T352 appears to be induced by PMA and ionomycin, close inspection of the total sIL-6R amounts as compared with wild-type IL-6R revealed that hIL-6RΔE317_T352 did not really serve as a substrate, and therefore, the observed induced shedding effect of IL-6R is misleading. This phenomenon is also true for the remaining IL-6R variants. We conclude from these data that although both protease cleavage sites are present, deletion of 36 amino acid residues prevented ADAM17 cleavage and drastically reduced ADAM10 cleavage.
We speculated that further shortening of the IL-6R stalk would completely diminish ADAM-mediated shedding. Therefore, we generated two additional variants containing deletions of 40 (IL-6RΔA323_V362) and 45 (IL-6RΔA323_F367) amino acids. Both variants lacked the ADAM17 cleavage site, and in accordance with this, we observed no significant increase after PMA stimulation (ΔA323_V362: 2.1 ± 1.1-fold increase, ΔA323_F367: 1.1 ± 0.1-fold increase), and the overall shed IL-6R was reduced to 0.2 ± 0.05% (ΔA323_V362) and 1.1 ± 0.3% (ΔA323_F367) compared with wild-type IL-6R (Fig. 4, E and F, and Fig. 4, G and H). Stimulation with ionomycin revealed inducible shedding by ADAM10 of ΔA323_V362 (5.1 ± 3.7-fold increase), whereas ΔA323_F367 was only weakly susceptible toward ADAM10-mediated shedding (2.7 ± 0.7-fold increase). It is important to note that although we were able to detect an sIL-6R increase after ionomycin stimulation, the amount of sIL-6R was only 0.3 ± 0.08% (ΔA323_V362) and 4.0 ± 0.8% (ΔA323_F367) compared with wild-type IL-6R (Table 2).
Finally, we created IL-6RΔE317_V362, lacking 46 amino acids and IL-6RΔE317_F367 where the complete stalk region was removed. IL-6RΔE317_V362 showed nearly no proteolytic processing by both ADAM10 and ADAM17 (Fig. 4, I and J), and the overall amount of sIL-6R was below 1 % compared with wild-type IL-6R (Table 2). Nevertheless, also this variant was equally well expressed at the cell surface of HEK293 cells (Fig. 2C), highlighting once more that the observed differences indeed results from altered proteolytic processing and not from impaired expression or transport to the plasma membrane. The last variant with a complete deletion of the stalk (IL-6RΔE317_F367) showed only weak cell surface expression on HEK293 (Fig. 2C) and stably transduced Ba/F3-gp130 cells (Fig. 2D) and was, therefore, excluded from further analysis.
In conclusion, we show that besides the distinct cleavage sites, the three-dimensional extracellular structure of the IL-6R also contributes to substrate recognition by ADAM10 and ADAM17 in constitutive and induced shedding. As a result, deletions within the stalk region, thereby moving the three extracellular domains of the IL-6R closer toward the plasma membrane, reduced proteolysis by both proteases.
Deletions within the Stalk Reduce Constitutive IL-6R Shedding
Besides induced shedding, the IL-6R is also constitutively released from cells, and this non-induced shedding has been attributed mainly to ADAM10 (12, 13). To study constitutive shedding of the IL-6R deletion variants, we monitored sIL-6R generation over 24 h in transiently transfected HEK293 cells and set the amount of constitutively shed wild-type IL-6R to 100% (Fig. 5A). As mentioned before, all IL-6R variants showed reduced shedding after ADAM10 activation by ionomycin, and the same was true for constitutive, non-induced shedding (Fig. 5, A and B, and Table 2). The three IL-6R variants with subsequent deletions of 10 amino acid residues each showed a reduction to 32.7 ± 9.1% (hIL-6RΔS353_V362), 61.0 ± 3.0% (hIL-6RΔI343_T352), and 68.1 ± 13.8% (hIL-6RΔA333_N342). As expected, we also observed diminished constitutive sIL-6R generation of hIL-6RΔS353_F367 (50.2 ± 10.4%) (Fig. 5A and Table 2).
FIGURE 5.
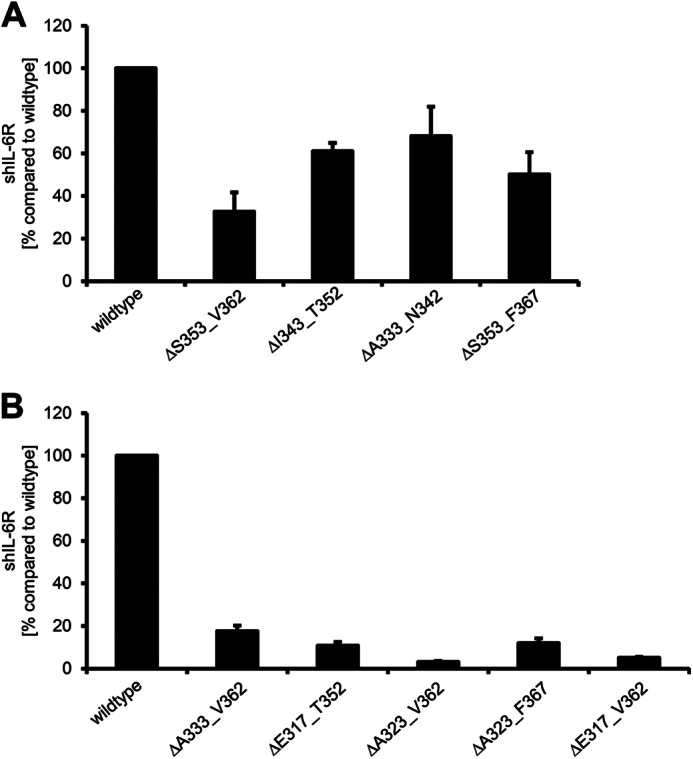
Deletions within the stalk region differentially influence constitutive IL-6R shedding. A and B, HEK293 cells were transfected with the indicated IL-6R constructs. 48 h after transfection medium was replaced with serum-free medium. The amount of constitutive shed IL-6R over a time period of 24 h was determined via ELISA. ELISA data are the mean ± S.D. from three independent experiments.
Larger deletions within the IL-6R stalk, which have been shown to drastically diminish induced IL-6R shedding, also strongly blocked constitutive shedding. Deletion of 30 amino acids (ΔA333_V362) resulted in 17.6 ± 2.6% constitutive shedding compared with wild-type (Fig. 5B and Table 2), which was in the same range as induced ADAM10 shedding (13.3 ± 4.1%). Deletion of 10 additional amino acids (ΔA323_V362), which showed the strongest reduction in induced shedding (0.3 ± 0.08%), also revealed the lowest amount of constitutive shedding (3.1 ± 0.01%; Fig. 5B and Table 2). Overall, constitutive shedding was reduced in all IL-6R variants studied, which is in line with the observations made in the experiments with induced IL-6R shedding.
At Least a 22-Amino Acid-long Stalk Region Is Needed for IL-6 Classic Signaling
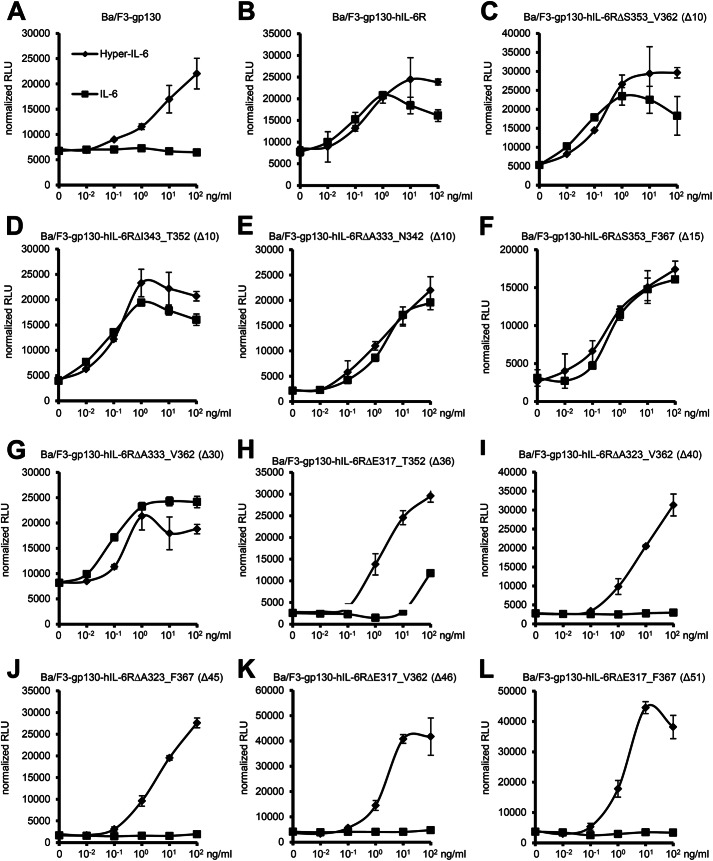
Besides its role in ectodomain shedding, the stalk region of the IL-6R is also suggested to position the domains D1-D3 of the IL-6R the correct distance from the plasma membrane to facilitate complex formation of IL-6·IL-6R with two gp130 molecules via site II and site III of IL-6 and CBM and D1 of gp130 (23). To test this hypothesis, we analyzed biological activity of all IL-6R stalk region deletion variants with respect to IL-6-induced cellular proliferation (see Fig. 2 for an overview). For this approach we again chose the Ba/F3-gp130-hIL-6R cells. Ba/F3 cells grow only in dependence of IL-3, but stable transduction with gp130 makes them responsive to IL-6/sIL-6R or Hyper IL-6, a designer cytokine where IL-6 is fused to the extracellular domains of the IL-6R. Ba/F3-gp130 cells do not express endogenous IL-6R (Fig. 2E) and, therefore, do not proliferate in the presence of IL-6 (Fig. 6A). As shown previously (7), expression of wild-type IL-6R renders Ba/F3-gp130 cells responsive to IL-6 (Fig. 6B). Importantly, cellular proliferation of Ba/F3-gp130-IL-6R was comparable irrespective of stimulation with IL-6 (via membrane-bound IL-6R) or Hyper-IL-6 (via soluble IL-6R) (Fig. 6B). Because all generated cell lines are independent clones and, therefore, might react quantitatively slightly different on IL-6, we used Hyper-IL-6-stimulated cellular proliferation as an internal cell clone-specific proliferation standard to judge IL-6-induced proliferation. Like in HEK293 cells, all our variants except IL-6RΔE317_F367 were expressed equally well on the cell surface of Ba/F3-gp130 cells as analyzed by flow cytometry (Fig. 2D). Truncation of the stalk region by 10 amino acids (hIL-6RΔS353_V362, ΔI343_T352, and ΔA333_N342) did not alter the responsiveness of the cells toward stimulation by IL-6, as the cells proliferated in a concentration-dependent manner (Fig. 6, C–E). The same was also true for deletions of 15 (ΔS353_F367; Fig. 6F) and 30 amino acids (ΔA333_V362; Fig. 6G). Interestingly, shortening of the stalk by 36 amino acids (ΔE317_T352; Fig. 6H) strongly reduced the IL-6 induced proliferation of Ba/F3-gp130 cells. Only high concentrations of IL-6 (100 ng/ml) resulted in weak cellular proliferation. Importantly, proliferation in response to Hyper-IL-6 was not impaired. In line with this finding, the remaining four deletion variants IL-6RΔA323_V362 (Fig. 6I), IL-6RΔA323_F367 (Fig. 6J), and IL-6RΔE317_V362 (Fig. 6K) or IL-6RΔE317_F367 (Fig. 6L) with 40, 45, 46, and 51 amino acid deletions, respectively, did not proliferate in response to IL-6 but proliferated normally when stimulated with Hyper-IL-6. In conclusion, we show that the length of the hIL-6R stalk is critical for IL-6 signal transduction and that between 16 to 22 amino acids are necessary for IL-6 classic signaling.
FIGURE 6.
Influence of alterations of the stalk region on IL-6 dependent signal transduction. A, equal numbers of Ba/F3-gp130 cells were cultured for 2 days with increasing amounts of Hyper-IL-6 (0- 102 ng/ml) or human IL-6 (0- 102 ng/ml). RLU, relative light units. B–L, equal numbers of stably transduced Ba/F3-gp130 cells were cultured for 2 days with increasing amounts of Hyper-IL-6 (0–102 ng/ml) or human IL-6 (0–102 ng/ml). The IL-6R variant is given above the respective diagram. Cellular proliferation was quantified as indicated under “Experimental Procedures.” One representative experiment of two or three performed is shown.
DISCUSSION
There are three major findings in this study. First, our data indicate that the target sequences for ADAM17 and the overall structure of the IL-6R stalk region contribute to efficient IL-6R shedding. Second, constitutive IL-6R shedding is also greatly diminished by deletions within the stalk region. Third, 22 amino acids are the minimal length of the stalk region needed for IL-6 classic signaling.
In IL-6 biology, signaling might be induced by IL-6 classic signaling via the membrane-bound IL-6R and by IL-6 trans-signaling via the soluble IL-6R. The membrane-bound IL-6R consists of three extracellular domains needed for efficient transport to the plasma membrane (D1) and binding of IL-6 (D2 and D3) (24). However, little is known about the functional role of the 52-amino acid-residue-long stalk region. Generation of soluble IL-6R by ectodomain shedding is mainly mediated by ADAM10 and ADAM17 (11–13) and the ADAM17 cleavage site of the IL-6R has been determined to be located within the stalk region between Gln-357 and Asp-358, in close proximity of the plasma membrane. In contrast, the cleavage site of ADAM10 has not been characterized so far. Genetic deletion of either ADAM10 or ADAM17 results in embryonic lethality (25, 26). This underlines the physiological importance of the two proteases. ADAM10 and ADAM17 show a partly overlapping, partly distinct spectrum of more than 100 substrates (27), for which practicably no consensus cleavage sequence exists (28). Inhibition of one or both proteases is considered as a therapeutic option in several diseases (29–31). However, how ADAM10 and ADAM17 recognize and selectively cleave their substrates is unknown, and whether all of them and/or under which conditions they are proteolytically shed in vivo is also not known. Therefore, addressing the cleavage of a single substrate in vivo is experimentally challenging. Nevertheless, exchange of small regions surrounding the ADAM cleavage sites between substrates and non-substrates has been shown to render proteolysis-resistant proteins susceptible to ADAM-mediated proteolysis and vice versa, including L-selectin and TNFα (32, 33). Genetically modified mice encoding an ADAM17-shedding-resistant L-selectin variant showed drastically reduced serum levels of soluble L-selectin and increased cell surface expression (34, 35). Genetic alteration of the ADAM cleavage site of a single substrate seems to be a promising strategy for in vivo analysis. Furthermore, the three-dimensional conformation of the substrate plays a role for ADAM-mediated shedding (13, 32, 33), which is recognized by the membrane-proximal domain of ADAM17 (36).
Here we have generated IL-6R deletion variants of the stalk region resulting in mild to severe ADAM10 and/or ADAM17 shedding defects and with full, reduced, or completely abrogated biological activity. In opposite to wild-type IL-6R, which is shed by ADAM10 and ADAM17, the IL-6RΔS353_V362 variant is not shed by ADAM17, indicating that ADAM10 and ADAM17 use different cleavage sites or have different structural requirements for substrate recognition and cleavage. However, the ADAM10 cleavage of IL-6RΔS353_V362 was largely absent in murine Ba/F3-gp130 cells. We have previously shown that human and murine ADAM17 differently shed murine IL-6R (13), and we now observed a similar effect concerning human IL-6R and human and murine ADAM10. Therefore, it seems to be important to perform ectodomain shedding experiments with protease and substrate from the same species.
IL-6RΔS353_F367 exhibits greatly reduced shedding by both ADAM10 and ADAM17, suggesting that the ADAM10 cleavage site is located between Ser-353 and Phe-367 but not between Ser-353 and Val-362. Finally, the 30-amino acid deletion variant IL-6RΔA333_V362 is not shed by ADAM17 and only minimally by ADAM10. Importantly, all three variants retained full biological activity with respect to induction of IL-6 classic signaling. Deletion of 36 or more amino acids of the stalk region results in even stronger shedding deficits of the IL-6R variants; however, these variants are biologically inactive or in the case of IL-6RΔE317_F367 are not efficiently transported to the plasma membrane. Additionally, we observed reduced constitutive shedding for all variants analyzed. IL-6RΔS353_V362 and IL-6RΔS353_F367 exhibited a reduction of constitutive shedding of about 60 and 50% despite shedding of ADAM17, and ADAM10/ADAM17 was almost completely abrogated, respectively. For the 30-amino acid deletion variant IL-6RΔA333_V362, constitutive shedding was reduced by 80%, and the most drastic effect on constitutive shedding was observed for the 40-amino acid deletion variant IL-6RΔA323_V362 with 97% reduced constitutive shedding. These findings are in line with observations that constitutive shedding is only partially reduced by ADAM10 and/or ADAM17 inhibitors as well as in ADAM10- and/or ADAM17-deficient murine embryonic fibroblasts or in ADAM17 hypomorphic (13) or conditional ADAM10-deficient mice,3 indicating the involvement of additional proteases responsible for constitutive shedding of IL-6R.
Besides its function in ectodomain shedding, the stalk region plays an important role in IL-6 classic signaling. Because cytokine binding domains of the signal transducing gp130 receptors are separated from the plasma membrane by three FNIII domains, the IL-6R stalk region might be considered as a spacer to position the IL-6R CBMs (D2 and D3) in the correct distance from the cell membrane to present IL-6 to the D1 domain of one gp130 and to the CBM (D2 and D3) of the second gp130 molecule. IL-6·IL-6R binding is mediated via the CBM of IL-6R and site I of IL-6. Importantly, neither IL-6 nor the IL-6R alone has an affinity toward gp130; therefore, the chronology of complex formation is binding of IL-6 to IL-6R and subsequent complex formation of IL-6·IL-6R with gp130. Importantly, the stalk region is not needed for IL-6 trans-signaling via the soluble IL-6R (15). With respect to biological activity, deletion of 30 amino acids was compatible, but deletion of 36 or more amino acid residues was not compatible with IL-6 classic signaling, albeit the IL-6RΔE317_T352 variant appeared to retain minimal biological activity. Therefore, we conclude that a stalk length of more than 16 amino acids but certainly 22 amino acids is sufficient to facilitate IL-6 classic signaling. There are several structural possibilities of how gp130 can look in the IL-6·IL-6R /gp130 complex. One possibility is that the three juxtamembrane FNIII domains are simply positioned in an elongated fashion (Fig. 7A). However, the crystal structure of the entire ectodomain of gp130 in an unliganded form has revealed that the fibronectin type III domains 4 and 5 are not elongated but form an acute bend in the gp130 structure with an wide open “C” shape (Fig. 7B and Ref. 37). As mentioned before, the calculated size of the FNIII domains in a stretched conformation is 96 Å or about 83 Å if arranged in a bend conformation. 52 amino acids of the IL-6R stalk can span about 198 Å, allowing both conformations of the FNIII domains and even additional conformational changes after ligand binding. 16–22 amino acids, however, span between 60.8 and 83.6 Å (3.8 Å/amino acids), indicating that the three juxtamembrane fibronectin domains of gp130 are not necessarily elongated but somehow flexed to allow IL-6 classic signaling and thereby are perfectly compatible with the view that the stalk region is needed for correct positioning of IL-6. After ligand binding, these fibronectin type III domains, which separate the gp130 cytokine binding domains from the plasma membrane, are considered to reposition to the correct proximity and orientation to induce trans-phosphorylation of receptor-associated JAKs within the intracellular domain. Activation of JAKs lead to subsequent activation of IL-6 signal transduction. It is, however, not known how and if the orientation of the FNIII domains is altered upon ligand binding. However, because this would also be enabled by a much shorter stalk region, additional functions such as regulation of inducible and constitutive shedding are encoded by the stalk region.
FIGURE 7.
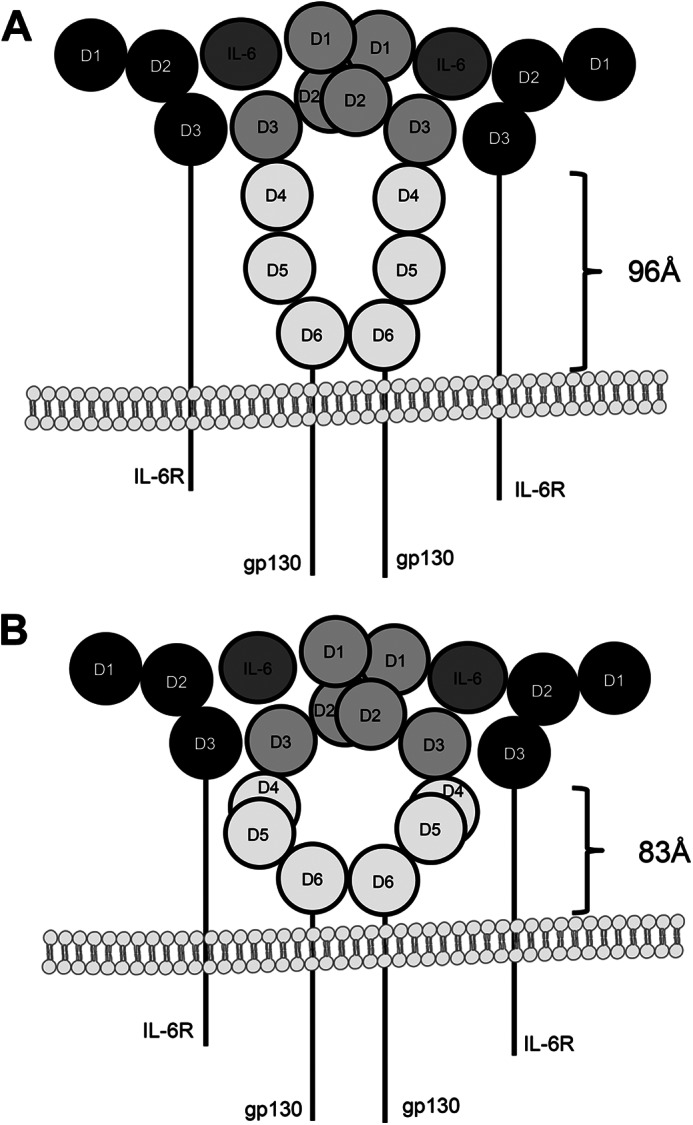
Schematic drawing of two possible gp130 conformations. A, the three juxtamembrane fibronectin type III domains (D4-D6) of gp130 are in an elongated fashion, which means that the stalk of the IL-6R must be able to span a distance of at least 96 Å. B, in contrast, the structure published by Xu et al. (37) shows that domains D4-D6 of gp130 are not necessarily elongated but are somehow flexed, which would allow IL-6 classic signaling with a shorter stalk of about 83 Å.
In conclusion, we provide evidence that a 15-amino acid sequence in direct proximity to the plasma membrane is absolutely required for PMA- and ionomycin-induced shedding of the IL-6R by ADAM proteases but only partially for constitutive shedding. Almost complete abrogation of constitutive shedding is then achieved by larger deletion variants, suggesting that additional proteases also contribute to constitutive shedding or that the three-dimensional structure of the IL-6R is also important for shedding. Moreover, we show that the stalk length of IL-6R is critical for IL-6 classic signaling as the cytokine binding module of the IL-6R has to be positioned in a correct way to enable IL-6·gp130 complex formation. Future experiments will show whether these findings can be transferred to the murine IL-6R. Shedding of murine IL-6R is further complicated as ADAM17 seems not to be the major sheddase of murine IL-6R (13, 38). In the future, the generation of an un-cleavable murine IL-6R variant will enable the detailed analysis of the membrane bound IL-6R for in vivo analysis.
J. Scheller, S. Rose-John, and P. Saftig, unpublished results.
- gp130
- glycoprotein 130 kDa
- CBM
- cytokine binding module
- ADAM
- a disintegrin and metalloprotease
- IL-6R
- interleukin-6 receptor
- sIL-6R
- soluble IL-6R
- hIL-6R
- human IL-6R
- PMA
- phorbol-12-myristate-13-acetate.
REFERENCES
- 1. Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. (2011) The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888 [DOI] [PubMed] [Google Scholar]
- 2. Garbers C., Hermanns H. M., Schaper F., Müller-Newen G., Grötzinger J., Rose-John S., Scheller J. (2012) Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 23, 85–97 [DOI] [PubMed] [Google Scholar]
- 3. Sprecher C. A., Grant F. J., Baumgartner J. W., Presnell S. R., Schrader S. K., Yamagiwa T., Whitmore T. E., O'Hara P. J., Foster D. F. (1998) Cloning and characterization of a novel class I cytokine receptor. Biochem. Biophys. Res. Commun. 246, 82–90 [DOI] [PubMed] [Google Scholar]
- 4. Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Hexameric structure and assembly of the interleukin-6/IL-6 α-receptor/gp130 complex. Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 5. Jostock T., Müllberg J., Ozbek S., Atreya R., Blinn G., Voltz N., Fischer M., Neurath M. F., Rose-John S. (2001) Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur. J. Biochem. 268, 160–167 [DOI] [PubMed] [Google Scholar]
- 6. Jones S. A., Scheller J., Rose-John S. (2011) Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 121, 3375–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garbers C., Thaiss W., Jones G. W., Waetzig G. H., Lorenzen I., Guilhot F., Lissilaa R., Ferlin W. G., Grötzinger J., Jones S. A., Rose-John S., Scheller J. (2011) Inhibition of classic signaling is a novel function of soluble glycoprotein 130 (sgp130), which is controlled by the ratio of interleukin 6 and soluble interleukin 6 receptor. J. Biol. Chem. 286, 42959–42970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garbers C., Spudy B., Aparicio-Siegmund S., Waetzig G. H., Sommer J., Hölscher C., Rose-John S., Grötzinger J., Lorenzen I., Scheller J. (2013) An interleukin-6 receptor-dependent molecular switch mediates signal transduction of the IL-27 cytokine subunit p28 (IL-30) via a gp130 protein receptor homodimer. J. Biol. Chem. 288, 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chalaris A., Garbers C., Rabe B., Rose-John S., Scheller J. (2011) The soluble Interleukin 6 receptor. Generation and role in inflammation and cancer. Eur J. Cell Biol. 90, 484–494 [DOI] [PubMed] [Google Scholar]
- 10. Lust J. A., Donovan K. A., Kline M. P., Greipp P. R., Kyle R. A., Maihle N. J. (1992) Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine 4, 96–100 [DOI] [PubMed] [Google Scholar]
- 11. Müllberg J., Schooltink H., Stoyan T., Günther M., Graeve L., Buse G., Mackiewicz A., Heinrich P. C., Rose-John S. (1993) The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 23, 473–480 [DOI] [PubMed] [Google Scholar]
- 12. Matthews V., Schuster B., Schütze S., Bussmeyer I., Ludwig A., Hundhausen C., Sadowski T., Saftig P., Hartmann D., Kallen K. J., Rose-John S. (2003) Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J. Biol. Chem. 278, 38829–38839 [DOI] [PubMed] [Google Scholar]
- 13. Garbers C., Jänner N., Chalaris A., Moss M. L., Floss D. M., Meyer D., Koch-Nolte F., Rose-John S., Scheller J. (2011) Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J. Biol. Chem. 286, 14804–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gearing A. J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A. H., Drummond A. H., Galloway W. A., Gilbert R., Gordon J. L. (1994) Processing of tumour necrosis factor-α precursor by metalloproteinases. Nature 370, 555–557 [DOI] [PubMed] [Google Scholar]
- 15. Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grötzinger J., Rose-John S. (1997) I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 142–145 [DOI] [PubMed] [Google Scholar]
- 16. Schroers A., Hecht O., Kallen K. J., Pachta M., Rose-John S., Grötzinger J. (2005) Dynamics of the gp130 cytokine complex. A model for assembly on the cellular membrane. Protein Sci 14, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mackiewicz A., Schooltink H., Heinrich P. C., Rose-John S. (1992) Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J. Immunol. 149, 2021–2027 [PubMed] [Google Scholar]
- 18. Chalaris A., Rabe B., Paliga K., Lange H., Laskay T., Fielding C. A., Jones S. A., Rose-John S., Scheller J. (2007) Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood 110, 1748–1755 [DOI] [PubMed] [Google Scholar]
- 19. Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K.-J., Rose-John S., Ludwig A. (2003) The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 20. Ludwig A., Hundhausen C., Lambert M. H., Broadway N., Andrews R. C., Bickett D. M., Leesnitzer M. A., Becherer J. D. (2005) Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb. Chem. High Throughput Screen 8, 161–171 [DOI] [PubMed] [Google Scholar]
- 21. Ketteler R., Glaser S., Sandra O., Martens U. M., Klingmüller U. (2002) Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 9, 477–487 [DOI] [PubMed] [Google Scholar]
- 22. Müllberg J., Oberthür W., Lottspeich F., Mehl E., Dittrich E., Graeve L., Heinrich P. C., Rose-John S. (1994) The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J. Immunol. 152, 4958–4968 [PubMed] [Google Scholar]
- 23. Grötzinger J., Kurapkat G., Wollmer A., Kalai M., Rose-John S. (1997) The family of the IL-6-type cytokines. Specificity and promiscuity of the receptor complexes. Proteins 27, 96–109 [PubMed] [Google Scholar]
- 24. Vollmer P., Oppmann B., Voltz N., Fischer M., Rose-John S. (1999) A role for the immunoglobulin-like domain of the human IL-6 receptor. Intracellular protein transport and shedding. Eur. J. Biochem. 263, 438–446 [DOI] [PubMed] [Google Scholar]
- 25. Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 26. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 27. Scheller J., Chalaris A., Garbers C., Rose-John S. (2011) ADAM17. A molecular switch to control inflammation and tissue regeneration. Trends Immunol. 32, 380–387 [DOI] [PubMed] [Google Scholar]
- 28. Caescu C. I., Jeschke G. R., Turk B. E. (2009) Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem. J. 424, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saftig P., Reiss K. (2011) The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17. Novel drug targets with therapeutic potential? Eur J. Cell Biol. 90, 527–535 [DOI] [PubMed] [Google Scholar]
- 30. Moss M. L., Sklair-Tavron L., Nudelman R. (2008) Drug insight. Tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 4, 300–309 [DOI] [PubMed] [Google Scholar]
- 31. Kenny P. (2007) TACE. A new target in epidermal growth factor receptor dependent tumors. Differentiation 75, 800–808 [DOI] [PubMed] [Google Scholar]
- 32. Althoff K., Müllberg J., Aasland D., Voltz N., Kallen K., Grötzinger J., Rose-John S. (2001) Recognition sequences and structural elements contribute to shedding susceptibility of membrane proteins. Biochem. J. 353, 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Althoff K., Reddy P., Voltz N., Rose-John S., Müllberg J. (2000) Shedding of interleukin-6 receptor and tumor necrosis factor α. Contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur. J. Biochem. 267, 2624–2631 [DOI] [PubMed] [Google Scholar]
- 34. Galkina E., Tanousis K., Preece G., Tolaini M., Kioussis D., Florey O., Haskard D. O., Tedder T. F., Ager A. (2003) L-selectin shedding does not regulate constitutive T cell trafficking but controls the migration pathways of antigen-activated T lymphocytes. J. Exp. Med. 198, 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venturi G. M., Tu L., Kadono T., Khan A. I., Fujimoto Y., Oshel P., Bock C. B., Miller A. S., Albrecht R. M., Kubes P., Steeber D. A., Tedder T. F. (2003) Leukocyte migration is regulated by L-selectin endoproteolytic release. Immunity 19, 713–724 [DOI] [PubMed] [Google Scholar]
- 36. Lorenzen I., Lokau J., Düsterhöft S., Trad A., Garbers C., Scheller J., Rose-John S., Grötzinger J. (2012) The membrane-proximal domain of A disintegrin and metalloprotease 17 (ADAM17) is responsible for recognition of the interleukin-6 receptor and interleukin-1 receptor II. FEBS Lett. 586, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 37. Xu Y., Kershaw N. J., Luo C. S., Soo P., Pocock M. J., Czabotar P. E., Hilton D. J., Nicola N. A., Garrett T. P., Zhang J.-G. (2010) Crystal structure of the entire ectodomain of gp130. Insights into the molecular assembly of the tall cytokine receptor complexes. J. Biol. Chem. 285, 21214–21218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arndt P. G., Strahan B., Wang Y., Long C., Horiuchi K., Walcheck B. (2011) Leukocyte ADAM17 regulates acute pulmonary inflammation. PLoS One 6, e19938. [DOI] [PMC free article] [PubMed] [Google Scholar]