Background: Expansion of CAG/CTG repeats causes familial neurological disorders, but the molecular basis is unknown.
Results: DNA polymerases β and δ effectively incorporate nucleotides to a CAG/CTG hairpin primer, leading to hairpin retention.
Conclusion: Coordinated actions by polymerases β and δ on hairpin primers during DNA synthesis promotes CAG/CTG repeat expansions.
Significance: The work discovers a novel mechanism for CAG/CTG repeat expansions.
Keywords: DNA Enzymes, DNA Polymerase, DNA Repair, Genomic Instability, Nucleotide Repeat Disease
Abstract
Expansion of CAG/CTG trinucleotide repeats causes certain familial neurological disorders. Hairpin formation in the nascent strand during DNA synthesis is considered a major path for CAG/CTG repeat expansion. However, the underlying mechanism is unclear. We show here that removal or retention of a nascent strand hairpin during DNA synthesis depends on hairpin structures and types of DNA polymerases. Polymerase (pol) δ alone removes the 3′-slipped hairpin using its 3′-5′ proofreading activity when the hairpin contains no immediate 3′ complementary sequences. However, in the presence of pol β, pol δ preferentially facilitates hairpin retention regardless of hairpin structures. In this reaction, pol β incorporates several nucleotides to the hairpin 3′-end, which serves as an effective primer for the continuous DNA synthesis by pol δ, thereby leading to hairpin retention and repeat expansion. These findings strongly suggest that coordinated processing of 3′-slipped (CAG)n/(CTG)n hairpins by polymerases δ and β on during DNA synthesis induces CAG/CTG repeat expansions.
Introduction
Expansion of CAG/CTG trinucleotide repeat (TNR)5 sequences causes at least 15 neurological and neurodegenerative disorders, including Huntington disease and myotonic dystrophy (1–4). They can be located in either coding or noncoding regions of a gene. Once the expansion exceeds a threshold, it inactivates the expression and function of the affected genes, leading to the onset of disease (5). However, the mechanisms that promote TNR expansion are not fully understood.
Hairpin formation and subsequent retention within CAG/CTG repeats in the nascent strand during DNA synthesis are considered a major path for repeat expansions (4). Numerous in vitro studies have revealed that CAG/CTG repeats can form a stable hairpin structures when there are more than two repeat units (6–9). A recent study by Liu et al. (10) demonstrated that the CAG/CTG hairpin formation indeed occurs in vivo, in a manner dependent on DNA replication. Increasing evidence suggests that hairpin formation occurs via DNA strand slippage during DNA metabolic processes that introduce DNA strand breaks within or near the repeat region (3, 4, 11). These processes include DNA replication (10, 12, 13), repair (2, 14, 15) and recombination (16, 17). In addition to DNA strand breaks, which provide opportunities for strand slippage, these processes share a common feature in DNA synthesis, a reaction catalyzed by DNA polymerases. Taken together, these studies suggest that the disease-causing CAG/CTG repeat expansion is likely originated from hairpin formations in the nick-residing nascent strand and subsequent error-prone DNA synthesis by DNA polymerases. However, little is known about the role of DNA polymerases in TNR expansions.
At least 15 mammalian DNA polymerases have been identified (18). These polymerases play distinct roles in genome maintenance, with a few of them functioning in the replication of the genome and the majority of them participating in DNA repair and translesion DNA synthesis (TLS). Replicative DNA polymerases, e.g. polymerase (pol) δ and pol ϵ, possess a 3′-5′ proofreading exonuclease activity and are essentially error-free. In contrast, DNA polymerases involved in TLS contain no proofreading activity and are highly mutagenic (18). Despite their distinct roles in DNA metabolic processes, recent evidence suggests that a transient switching between TLS polymerases and replicative polymerases occurs to deal with bulky DNA lesions during DNA synthesis (19). When a bulky DNA lesion blocks DNA synthesis by a replicative DNA polymerase, the replicative polymerase is replaced by a low fidelity TLS polymerase to bypass the lesion, followed by switching back to the replicative polymerase to resume the high-fidelity DNA synthesis. The formation of a CAG/CTG hairpin in the nascent strand would provide replicative and/or repair DNA polymerases with a huge lesion in their primer. How DNA polymerases handle a DNA hairpin in the nascent strand is unknown.
In this study, we constructed a series of (CAG)n or (CTG)n hairpin substrates that simulate the hairpin structures in the nascent strand during DNA synthesis, and examined HeLa nuclear extracts and several DNA polymerases for their ability to process these hairpin structures. We demonstrate here that pol δ can either retain or remove a primer hairpin, depending on the hairpin's 3′-end sequences/structures, and that pol β is capable of adding nucleotides to the hairpin primer to generate a hairpin structure with a 3′ complementary sequence. Surprisingly, in reactions containing both pol β and pol δ, the hairpin retention product is several folds more than that of reactions containing pol β or pol δ alone. This synergistic stimulation suggests a concerted cooperation between pol β and pol δ, leading a hairpin retention and TNR expansion.
EXPERIMENTAL PROCEDURES
Cell Culture and Nuclear Extract Preparations
HeLa S3 cells were cultured at 37 °C in a 5% CO2 atmosphere to a density of 5 × 105 cells/ml in RPMI 1640 supplemented with 5% fetal bovine serum. The nuclear extract was prepared as described (20).
Protein Preparations
Pol δ, replication factor C (RFC) and proliferating cell nuclear antigen (PCNA) were purified as described (21). Pol δ and RFC were expressed in High Five insect cells using the baculovirus system, and PCNA was expressed in Escherichia coli. The pol δ mutant defective in the 3′-5′ proofreading nuclease activity was similarly prepared by replacing wild type p125 catalytic subunit with the p125 mutant containing the D402A mutation. The p125 (wild-type) cDNA (22), a gift from Dr. Yoshihiro Matsumoto (Fox Chase Cancer Center), was modified to D402A by QuikChange II Site-directed mutagenesis kit (Agilent Technologies) and subcloned into a pFastBac vector for expression in insect cells. cDNAs of human pol β, pol κ, pol μ, and pol η were cloned into the pFastBac-HTb vector and expressed in insect High Five cells using the Bac-to-Bac expression system (Invitrogen, Carlsbad, CA). The expression of all proteins was confirmed by Western blot analysis. Protein concentration was determined by the Coomassie (Bradford) Protein Assay Reagent (Pierce). The purified proteins were stored in aliquots at −80 °C. T7 endonuclease I and BbvI were purchased from New England Biolabs.
DNA Hairpin Substrate Preparation
To produce DNA hairpin substrates that mimic 3′ CAG/CTG slippage during DNA replication or repair, oligonucleotides containing 15 CTG and 15 CAG repeats were annealed with ssDNA of M13mp18-(CAG)10 and M13mp18-(CTG)10 (23, 24) to form a CTG and a CAG hairpin substrates, respectively (Fig. 1). To prevent nuclease degradation, the first four 5′ bases of the oligonucleotide are phosphothioated bases. The presence of the hairpin in these substrates was verified by T7 endonuclease I and BbvI as described (25).
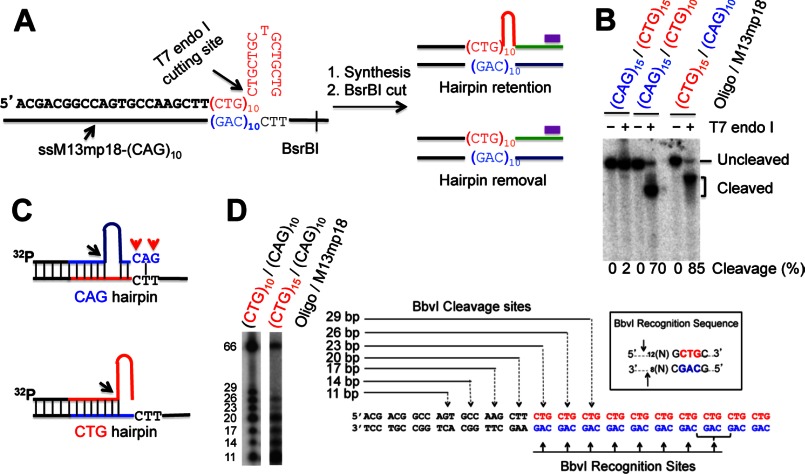
FIGURE 1.
DNA substrate and hairpin retention/removal assay. A, diagram of hairpin removal/retention assay by Southern blot analysis. The purple bar shows the 32P-labeled oligonucleotide probe, which specifically anneals to the newly synthesized strand near the BsrBI site. The complete primer sequence of a CTG hairpin substrate used in this study is also shown. B, substrate characterization. Each DNA substrate, which was 5′-32P-labeled in the primer strand, was digested with T7 endonuclease I, and the products were analyzed by denaturing polyacrylamide gel electrophoresis. The cleavage products were quantified. C, proposed structures for the CAG (upper) and CTG (lower) hairpin substrates. Black arrows point to the T7 cleavage sites, and red arrows indicate mismatches. The blue and red types/lines represent CAG and CTG repeats, respectively. D, digestion of a CTG hairpin by BbvI. The left panel shows the BbvI cleavage products of a (CTG)10/(CAG)10 homoduplex and a (CTG)15/(CAG)10 heteroduplex on a 15% denaturing polyacrylamide gel, and the right panel diagrams the recognition sites and the predicted cleavage sites of the (CTG)10/(CAG)10 homoduplex by BbvI.
Hairpin Removal/Retention Assay
Unless otherwise mentioned, hairpin removal/retention was assayed by Southern blot analysis. Hairpin substrates were incubated with HeLa nuclear extracts or purified polymerase (pol β or pol δ) systems for DNA synthesis at 37 °C for 15 min in a 40-μl reaction containing 110 mm KCl, 20 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 1 mm glutathione, 1.5 mm ATP, 0.1 mm of each dNTP, and 0.05 mg/ml BSA at 37 °C for 30 min. The reaction was terminated by incubating with 60 μl of proteinase K solution containing 0.67% (v/v) of SDS, 2.5 mm EDTA, and 20 mg/ml proteinase K. After phenol extraction and ethanol precipitation, the DNA sample was digested with 0.3 units of BsrBI (New England Biolab) and resolved in a 6% polyacrylamide denaturing gel, followed by Southern blotting analysis using a 32P end-labeled probe specifically annealing to the newly synthesized strand as described (23). The products were detected by a phosphorimager.
Hairpin Primer Extension Assay
Unless otherwise mentioned, hairpin primer extension was assayed by Southern blot hybridization. Oligonucleotides containing 15 CTG and 15 CAG repeats were annealed with ssDNA of M13mp18-(CAG)10 and M13mp18-(CTG)10 (23, 24) to form a CTG and a CAG hairpin substrates, respectively (Fig. 1). Individual hairpin substrates were incubated with HeLa nuclear extracts or purified polymerase (pol β or pol δ) systems for DNA synthesis at 37 °C for 15 min in a 40-μl reaction containing 110 mm KCl, 20 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 1.5 mm ATP, 0.1 mm of various combinations of dNTP and 0.05 mg/ml BSA. In the purified systems, each reaction also contained RFC (110 fmol) and PCNA (2 pmol) in addition to the indicated polymerase (600 fmol pol δ/D402A or 130 fmol pol β). The resulting products were digested with BsrBI before electrophoresis through a 10% denaturing polyacrylamide gel. The DNA molecules were subjected to Southern blot analysis using a 32P-labeled probe as described (23).
RESULTS
Polymerase β Promotes CAG/CTG Repeat Expansion in Nuclear Extract-catalyzed DNA Synthesis
To explore how human cells process a CAG or CTG hairpin formed in the nascent strand at the site of DNA synthesis, a CTG or CAG hairpin substrate that mimics the nascent strand hairpin formation was constructed (see Fig. 1A). To confirm the presence of a CTG or CAG hairpin at the 3′-end of the primer strand, the substrates were 32P-labeled at the 5′-end of the primer strand and digested with T7 endonuclease I and BbvI as described previously (25). As shown in Fig. 1B, at least 70% of the hairpin could be cleaved by T7 endonuclease I in each case, indicative of a hairpin formation in the primer strand. Interestingly, the cleavage product of the CTG substrate is bigger than that of the CAG substrate in size. These results suggest that the CTG hairpin is formed near the 3′-end while the CAG hairpin is closer (relative to the CTG hairpin) to the 5′-end, which likely contains a 3′ tail with mismatches (see Fig. 1C, also see Fig. 6 and related text for explanation). Fig. 1D shows the cleavage products of the CTG hairpin substrate by BbvI. The lack of the 29-bp product indicates that the BbvI recognition sequence at the 3′-end of the primer strand is interrupted by the CTG hairpin formation, i.e. the CTG hairpin was located at the very 3′-end of the primer (Fig. 1D).
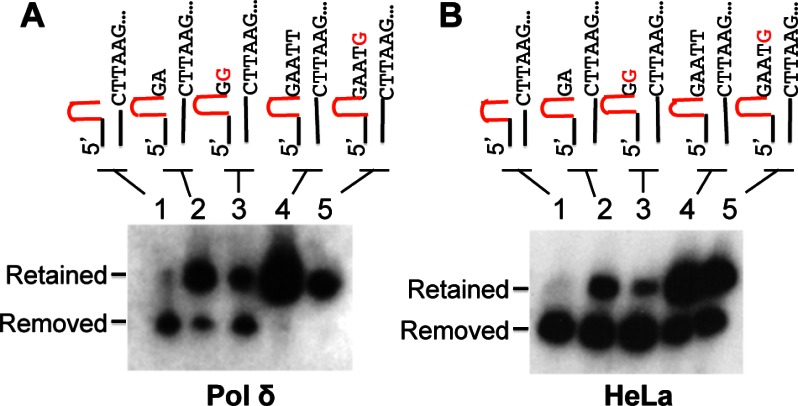
FIGURE 6.
Hairpin removal or retention activity of pol δ depends on the immediate 3′ sequence of the hairpin. A, processing of various hairpin substrates with or without a 3′ tail by the pol δ system that contains pol δ, RFC, and PCNA. B, processing of various hairpin substrates by HeLa nuclear extract. The reaction products were analyzed by Southern blot hybridization.
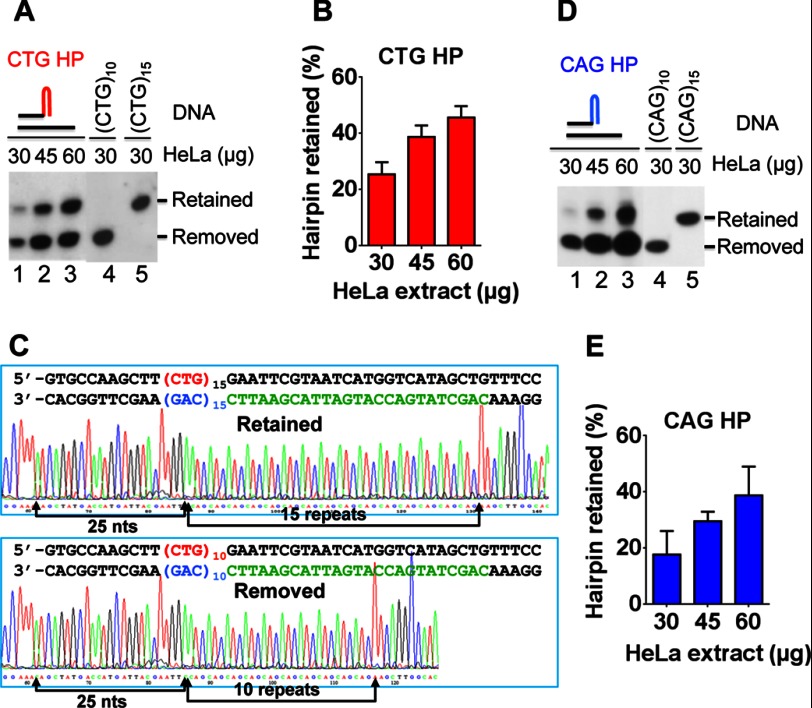
To determine how 3′-slipped hairpins are processed in human cells, the CAG and CTG hairpin substrates were incubated with HeLa nuclear extracts under conditions supporting DNA synthesis. The reaction products were subjected to Southern blot analysis using a probe (Fig. 1A, purple bar) specifically recognizing the downstream sequence (near the BsrBI site) of the newly synthesized strand. Thus, we can monitor whether the hairpin is removed or retained based on their relative mobility on denaturing gel electrophoresis. Incubation of the CTG hairpin substrate with HeLa nuclear extracts yielded two products with different molecular sizes (Fig. 2A, lanes 1–3), and the percentage of the larger product (upper band) increased proportionally to the increasing amount of HeLa nuclear extracts (Fig. 2B). Because the upper band and the lower band migrated in the positions corresponding to the molecular markers derived from the same primer extension assay using the bold typed sequence (Fig. 1A) as a primer and ssM13mp18-(CAG)15 and ssM13mp18-(CAG)10 as a template, respectively (Fig. 2A, lanes 4 and 5), we deduced that the upper and lower bands are the hairpin-retained and hairpin-removed products, respectively. To confirm this prediction, these bands were recovered from the gel and PCR-amplified, followed by DNA sequencing analysis. The results indeed revealed that the upper band contained 15 repeats (Fig. 2C, upper panel) and the lower band contained 10 repeats (Fig. 2C, lower panel). Similar results were also observed for the CAG hairpin substrate (Fig. 2, D and E). Given that the hairpin is located near the 3′-end of the primer (Fig. 1) and that expansions require DNA synthesis, the observed hairpin retention is likely due to direct incorporations of nucleotides to the 3′-end of the hairpin by a DNA polymerase.
FIGURE 2.
HeLa nuclear extract conducts CAG/CTG hairpin expansions during DNA synthesis. Hairpin primer extension assays were performed by incubating a CAG or CTG hairpin substrate with the indicated amount of HeLa nuclear extracts at 37 °C for 15 min, under the condition that supports for DNA synthesis. The resulting products were examined by Southern blot analysis. A, Southern blot analyses showing a (CTG)5 hairpin retention/removal in HeLa nuclear extracts. B, quantification of the CTG hairpin-retained product in the individual reactions shown in A. The data were from three independent experiments, and the error bar indicates the S.D. C, DNA sequence of the hairpin-retained product (top) and the hairpin-removed product (bottom) shown in A, which contains 15 and 10 CTG repeats, respectively. D, Southern blot analyses showing (CAG)5 hairpin retention/removal in HeLa nuclear extracts. E, quantification of the CAG hairpin-retained product in the individual reactions shown in D. The data were from three independent experiments, and the error bar represents S.D. Molecular markers (CAG)10, (CAG)15, (CTG)10, and (CTG)15 were produced by the pol δ system using the 21-mer bold black sequence shown in Fig. 1A as a primer and ssM13mp18-(CTG)10, ssM13mp18-(CTG)15, ssM13mp18-(CAG)10, and ssM13mp18-(CAG)15 (23, 24) as a template, respectively.
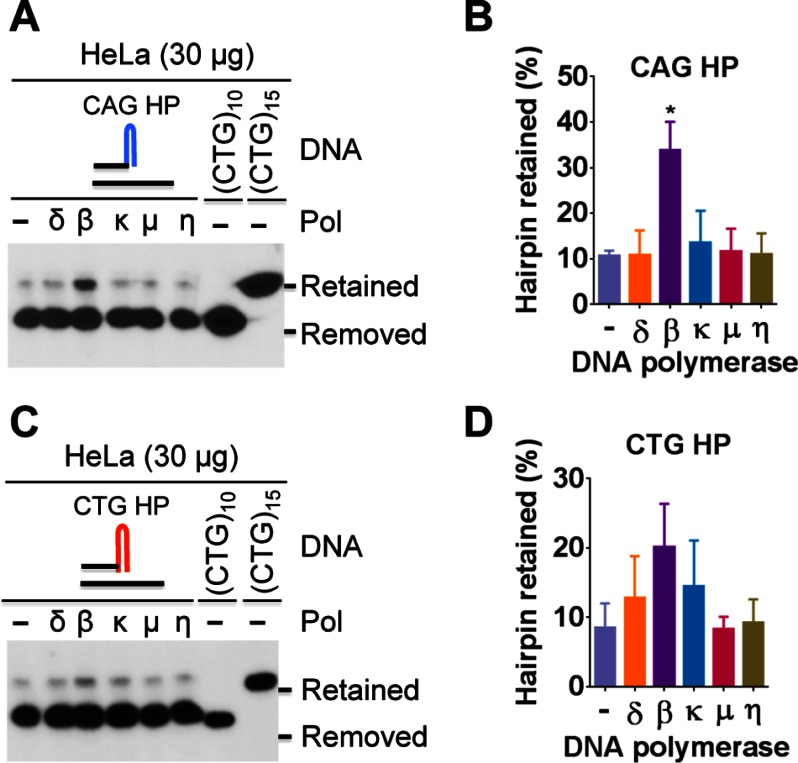
To test this possibility, DNA polymerase δ, β, κ, μ, or η was added to the hairpin primer extension reactions in the presence of 30 μg of HeLa extracts, which generates limited amounts of the hairpin-retained products (Fig. 2, A and D). Interestingly, among five DNA polymerases tested, pol β showed the strongest stimulation of hairpin retention for both the CAG (Fig. 3, A and B) and CTG (Fig. 3, C and D) hairpin substrates. These results suggest that pol β is a prominent candidate promoting CAG/CTG expansions during DNA synthesis. It is also noted that more hairpin-retained products were produced with the CAG substrate than with the CTG substrate (compare the two pol β reactions). This may be related to a more internal hairpin for the CAG structure as proposed in Fig. 1C (see more details below).
FIGURE 3.
Pol β enhances hairpin retention in HeLa nuclear extract. Hairpin primer extension assays were conducted essentially as described in the Fig. 2 legend, except using 30 μg of HeLa nuclear extracts in the presence or absence of a DNA polymerase (70 fmol), as indicated. A, Southern blot analysis showing that pol β promotes (CAG)5 hairpin retention. B, percentage of the CAG hairpin-retained species in each reaction. C, Southern blot analysis showing that pol β promotes (CTG)5 hairpin retention. D, percentage of the CTG hairpin-retained species in each reaction. The data shown in panels B and D were derived from three independent assays, and the error bars represent S.D. Statistical significance (p < 0.05) was determined by the One-way ANOVA test. Molecular markers were generated as described in the Fig. 2 legends.
Repeat Expansion Involves Concerted Actions of Pol β and Pol δ
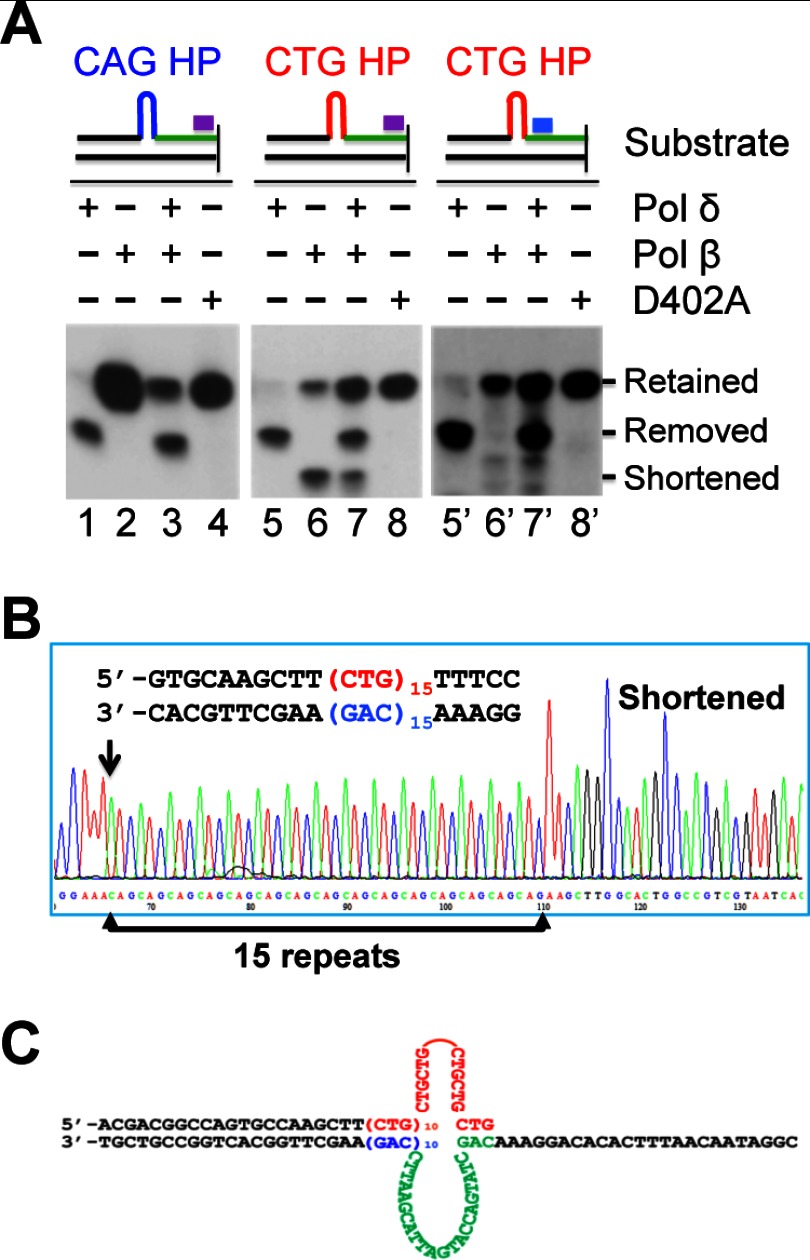
To determine if pol β is responsible for CAG/CTG repeat expansion during DNA synthesis, the hairpin-primer extension reactions were conducted in a purified protein system that contained replication factor C (RFC), proliferating cell nuclear antigen (PCNA), and pol β and/or pol δ. As shown in Fig. 4A, pol δ alone produced mainly the hairpin removal product for both the CAG (lane 1) and CTG (lane 5) hairpin substrates, suggesting that pol δ acts to remove CAG and CTG hairpins in the primer. When the primer extension reaction was carried out by the pol β-containing system, we observed only hairpin retention for the CAG hairpin substrate (Fig. 4A, lane 2), but two species for the CTG hairpin substrate, with one being a shortened product (see below for detail) and the other being the hairpin-retained species (Fig. 4A, lane 6). Processing of the same CTG substrate by a joint effort of pol β and pol δ produced three products, i.e. hairpin-retained, hairpin-removed, and shortened species (Fig. 4A, lane 7). Interestingly, in contrast to the products generated by pol β alone, the hairpin-retained species became the major product (Fig. 4A, compare products in lane 7 with lane 6). This phenomenon appears to be specific for the CTG hairpin, as about equal amounts of hairpin-retained and hairpin-removed products were observed when the CAG hairpin substrate was processed by both polymerases (Fig. 4A, lane 3). These results suggest that while pol β and pol δ can process a CAG hairpin primer independently, with pol β specifically catalyzing hairpin-retention and pol δ specifically catalyzing hairpin-removal, these two polymerases together synergistically promote CTG hairpin retention, i.e. CTG repeat expansion in the nascent DNA strand during DNA synthesis.
FIGURE 4.
Repeat expansion involves concerted actions of pol β and pol δ. Hairpin retention/removal assays were performed in reactions containing a purified polymerase system, as indicated, in the presence of PCNA and RFC. D402A is a mutant pol δ defective in 3′-5′ proofreading activity. Products were digested with BsrBI and analyzed by Southern blot analysis using a 32P-labeled probe (purple or blue bar) at the indicated location. The blue and red types/lines represent CAG and CTG repeats, respectively. A, Southern blot analyses showing hairpin retention and removal by pol β and/or pol δ. B, DNA sequence of the shortened band shown in A. C, proposed substrate structure that leads to the shortened DNA synthesis product. The arrow in B indicates the location where the 25-nt green fragment in C (also see Fig. 2C) is deleted in the shortened product.
As described above, a product smaller than the CTG hairpin-removed product was seen in the pol β-catalyzed reaction (Fig. 4A, lanes 6 and 7). To determine the nature of the shortened band, we performed DNA sequencing analysis, and found that the shortened band still contained 15 copies of the CTG/CAG repeat (Fig. 4B), i.e. the (CTG)5 hairpin was retained. However, in comparison with the sequence of the hairpin-retained band or the hairpin-removed band (Fig. 2C), the shortened band contained a deletion of 25 nucleotides (the green-typed and bracketed sequences shown in Fig. 2C) immediately 5′ to the CAG repeats (indicated by a black arrow in Fig. 4B) in the template strand or immediately 3′ to the CTG repeats in the primer strand. We then analyzed the sequence composition of the template strand and found that the missing 25-nucleotides could form a loop structure as shown in Fig. 4C, when the CAG sequence that is 22-nts 5′ to the (CAG)10 repeats in the template strand pairs with the 3′-end CTG sequence in the primer strand, which leads to the formation of a cruciform structure. In this case, the loop sequence would not be used as a template for DNA synthesis, thereby resulting in the shortened product. Thus, the shortened band is 25-nt and 10-nt smaller than the hairpin-retained and hairpin-removed products, respectively. Our Southern hybridization analysis using the loop sequencing as a probe could detect both the hairpin-retained and hairpin-removed bands, but not the shortened band (Fig. 4A, lanes 6′ and 7′), further confirming the proposed cruciform structure of the substrate (Fig. 4C). Given that this shorter product was not observed in the reaction with HeLa nuclear extract (Fig. 2, B and D) and pol δ (Fig. 4), it is likely that a bubble structure in the immediate template sequence is either inhibitory (to pol δ) or is repaired by the DNA hairpin repair pathway in extracts (23, 24, 26, 27). These data suggest that pol β conducts DNA synthesis in the presence of unusual structures in both template and primer strands. It is also possible that pol β binding may have specifically induced the formation of the cruciform structure.
Polymerase δ Removes (CAG)n/(CTG)n Hairpins via Its Proofreading Exonuclease Activity
Because pol δ possesses an intrinsic 3′-5′ proofreading nuclease activity (28–32), we propose that the proofreading activity is responsible for the CAG- and CTG-hairpin removal. This idea was first tested using a pol δ mutant that contains a D to A substitution at residue 402 (D402A) of the p125 subunit. The substitution inactivates the exonuclease activity but not the polymerase activity of pol δ (22). Unlike the reaction with pol δ, which mainly produced the hairpin-removed product, the reaction with pol δ(D402A) generated only the hairpin-retained species (Fig. 4A, lanes 4 and 8). We then performed the same experiment using a CTG hairpin substrate containing four phosphothiolated bases at the 3′-end of the hairpin, which prevent nuclease degradations from 3′-5′ orientations (29). We did not observe any hairpin retention and removal products (data not shown), indicative of the blockage of both the exonuclease and polymerase activities of pol δ. Taken together, our data show that the pol δ 3′-5′ proofreading nuclease activity is responsible for the hairpin removal.
We also tested pol δ for its ability to process (CAG)10 and (CTG)10 hairpins located at the 3′-end of a primer, and found that the polymerase can efficiently remove a (CAG)10 or (CTG)10 hairpin (data not shown). Taken together, the data presented here strongly suggest that the replicative DNA polymerase pol δ plays an important role in maintaining trinucleotide repeat stability by removing CAG and CTG repeat hairpins formed in the primer strand at the replication fork.
Pol β Initiates DNA Synthesis Regardless of Hairpin and Bubble Structures in the Primer and Template Strands
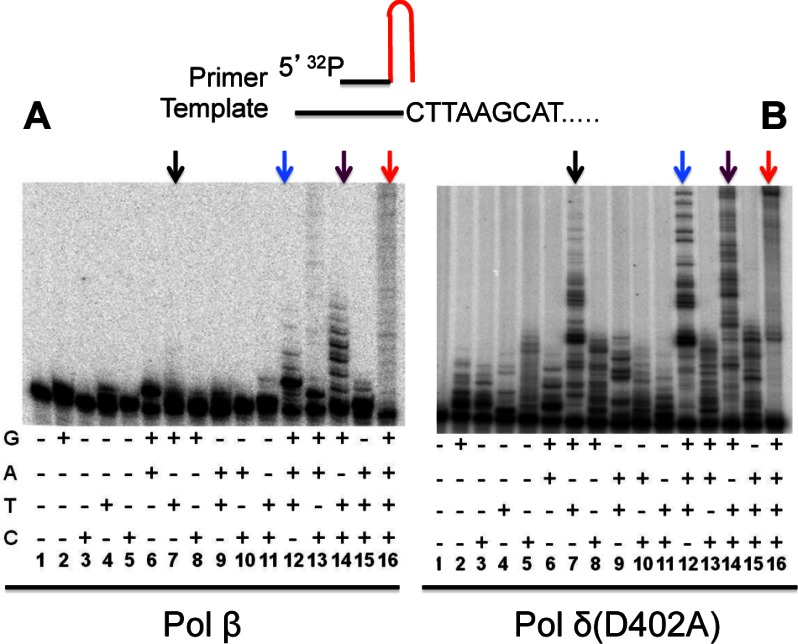
The mechanism by which pol δ and pol β synergistically promote CTG hairpin retention at the replication fork was investigated. We hypothesize that pol β initiates the hairpin retention by adding several nucleotides to the CTG hairpin primer and the resulting product can be effectively used as a primer for DNA synthesis by pol δ. To determine if pol β is capable of initiating DNA synthesis using a CTG hairpin primer, 5′-32P-labeled CTG hairpin substrate (see Fig. 1C) was incubated with pol β in the presence of different combinations of deoxynucleotide triphosphates (dNTPs). The resulting products were analyzed by polyacrylamide gel electrophoresis. The results showed that pol β could effectively incorporate correct or sometimes incorrect dNTPs at the 3′-end of the hairpin, depending on the availability of nucleotides (Fig. 5A). However, the enzyme has a limited processitivity, as judged by the limited extensive incorporations when three or all four dNTPs were provided (Fig. 5A, lanes 12–16). Nevertheless, these results revealed that despite limited processitivity, pol β can initiate DNA synthesis using the CTG hairpin as a primer.
FIGURE 5.
Pol β efficiently utilizes a hairpin primer for DNA synthesis. Incorporation of nucleotides to a CTG hairpin by pol β (A) and pol δ(D402A) (B), respectively. 5′ 32P-labeled CTG (top) was incubated with pol β or pol δ(D402A) in the presence of different combinations of dNTPs, as indicated. Products were resolved in 10% denaturing polyacrylamide gels, and detected by a phosphorimager. Colored arrows emphasize the difference in processitivity/faithfulness between pol β or pol δ(D402A).
The observation of the exclusive hairpin-retained product in the pol δ(D402A)-catalyzed system indicates that the pol δ exonuclease mutant can utilize the hairpin structure as a primer. Indeed, we observed an extensive incorporation of dNTPs at the 3′-end of the hairpin by the proofreading-deficient pol δ (Fig. 5B), suggesting that the failure to promote hairpin retention by WT pol δ is due to its proofreading activity (see below for explanation). Under normal circumstances, a nucleotide mis-incorporation by pol δ triggers its proofreading activity for mismatch removal. Because CTG hairpin contains a T-T mismatch in every three base pairs, it is possible that pol δ proofreading activity concomitantly removes these T-T mismatches in the hairpin upon activation, resulting in hairpin-removed species.
In addition, we observed two other unique properties of pol δ and pol β. First, pol δ(D402A) is much more processive than pol β, as judged by the fact that there are more slowly migrating molecules in pol δ(D402A)-catalyzed reactions than those catalyzed by pol β (compare corresponding reactions with arrows between Fig. 5A and Fig. 5B). Secondly, pol δ(D402A) is less faithful than pol β, because the former enzyme incorporates more incorrect nucleotides into the elongation chain (Fig. 5B).
Hairpin 3′ Sequence Dictates Hairpin Removal or Retention by Pol δ
To determine the condition under which pol δ promotes CTG hairpin retention during DNA synthesis, we tested the pol δ ability to process CTG hairpin substrates that carry a different number of bases with or without a mismatch at the immediate 3′-end of the hairpin. These substrates mimic the hairpin products generated by pol β as described above or hairpins formed within TNR sequences in nascent DNA strand via strand slippage during DNA replication or repair. As expected, processing of the CTG hairpin with an immediate 3′ complementary sequence after the hairpin by pol δ generated the hairpin-removed major product and the hairpin-retained minor products (Fig. 6A, lane 1). Incubation of the same hairpin substrate with a perfectly paired 2-nucleotides at the 3′-end of the hairpin rendered the system to switch the ratio of these two products (Fig. 6A, lane 2), i.e. the major and minor products are now the hairpin-retained and hairpin-removed species, respectively. However, when there was a mismatch at the last place of the 2-nucleotide 3′ tail (Fig. 6A, lane 3), pol δ produced less hairpin-retained but more hairpin-removed products as compared with the same substrate without a mismatch at the 3′-end (Fig. 6A, compare lane 2 with lane 3). These results suggest that a 3′ mismatch near the hairpin triggers the hairpin removal by the 3′-5′ proofreading nuclease of pol δ. This provides partially explanation as to why pol δ is capable of limited hairpin removal for the CAG hairpin (Fig. 4A, lane 1), which likely contains a mismatched 3′ tail (Fig. 1C, upper). Interestingly, when the 3′ complementary sequence reaches 5-nucleotides (Fig. 6A, lanes 4 and 5) or more (data not shown), pol δ could only promote hairpin retention, regardless of the presence or absence of a mismatch at the 3′-end, indicating the mismatch location can determine whether pol δ carries out the hairpin removal or hairpin retention.
Similar analyses were performed using HeLa nuclear extracts (Fig. 6B). Compared with the products generated from the pol δ system, a striking difference is that HeLa extracts generated more hairpin-removed products. This is likely due to the endonucleolytic removal of the hairpin by the DNA hairpin repair system in HeLa extracts as described previously (23), particularly in reactions with the hairpin substrate containing a 5-nucleotide tail in which no hairpin-removed products were detected in the pol δ system. However, it is noted that not all retained hairpins were removed by the repair system as there is still significant amount of hairpin molecules that escape the repair in each reaction (Fig. 6B).
DISCUSSION
As a major contributor to CAG/CTG repeat expansion, hairpin formation within the repeats is associated with DNA replication (10, 12, 13) and repair (2, 14). Because DNA expansion requires DNA synthesis, DNA polymerases must play a major role in this process. However, little is known about the mechanism by which DNA polymerases promote CAG/CTG expansion. In this study, we provide strong evidence that DNA polymerases can remove or retain a CAG/CTG hairpin formed in the nascent DNA strand during DNA synthesis, depending on the hairpin structure and the DNA polymerases involved in the DNA synthesis reaction.
Several surprising findings were made in this study. Firstly, we show that pol δ is capable of removing the hairpin primer if the primer contains no complementary 3′ sequences after the hairpin (Fig. 4A). Secondly, among DNA polymerases tested, the polymerase involved in base excision repair, pol β, is the most active enzyme in promoting hairpin retention in nuclear extracts (Fig. 3). Thirdly, the pol β hairpin-retention activity, especially for a CTG hairpin, is greatly stimulated by pol δ (Fig. 4A), indicating that the CTG hairpin retention is catalyzed by a collaborative effort of these two DNA polymerases.
Although the mechanism by which pol δ and pol β collaborate to promote TNR expansion awaits further investigations, The results presented here suggest that the hairpin retention by pol β and pol δ may involve polymerase switching, a concept originally established for the template strand lesion bypass by TLS polymerases (19) and recently adapted to propose possible TNR expansion (2, 14). Like many TLS polymerases, pol β possesses a PCNA-interacting protein (PIP) motif and can interact with PCNA (33), an essential property for polymerases to participate in polymerase switching (19). As a key enzyme involved in DNA base excision repair, pol β has been implicated in CAG repeat expansion during the repair of oxidative damage that occurs within CAG repeats (15). Previous studies have also shown that pol β can bypass a number of different types of lesions in the template strand, including cisplatin adducts, cyclobutanol pyrimidine dimers, and 6–4 photoproducts (34). We demonstrate here that pol β can also “bypass” a (CAG)n/(CTG)n hairpin lesion, but the “bypass” in our study occurs in the primer strand, leading to expansion of the repeats. Taken together, these observations suggest that pol β can tolerate a variety of unusual DNA lesions/structures in both the template and primer strands during DNA synthesis.
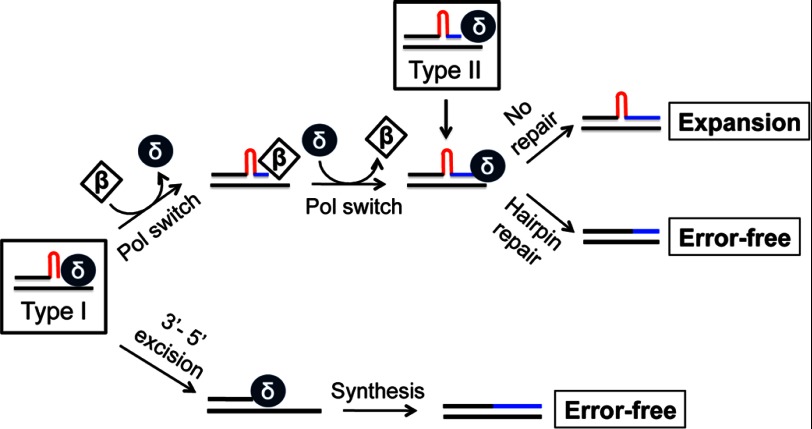
Fig. 7 shows a model describing the processing of CAG/CTG hairpins formed in the primer strand during DNA synthesis by pol β and pol δ. Two types of CAG/CTG hairpins can be formed via 3′ slippage in the nascent strand, one without a complementary 3′ tail (Type I) and the other with a 3′ tail (Type II). We hypothesize that the Type I hairpin primer can undergo both error-free synthesis if processed only by pol δ, but it undergoes error-prone (expansion) processing in the presence of both pol δ and pol β. In the latter case, pol β is recruited to the site for primer extension, which adds several nucleotides to the 3′-end of the hairpin, followed by re-recruitment of pol δ to resume the high-fidelity and highly processive DNA synthesis. It is the double-switch between pol δ and pol β that promotes hairpin preservation and repeat expansion. This model requires that (i) pol β is capable of adding nucleotides to the hairpin primer; and (ii) pol δ can use pol β-generated products for DNA synthesis, but not excision. Our experiments shown in Figs. 5 and 6 confirm that these are indeed the cases. For the Type II hairpin, which contains a complementary tail at the 3′-end, its processing by pol δ is essentially mutagenic (Fig. 7). This is because pol δ can efficiently incorporate nucleotides onto the 3′-end when a hairpin primer carries a complementary 3′ sequence with 2- or more nucleotides (Fig. 6A, lanes 4 and 5). If the retained hairpin is not removed by the DNA hairpin repair pathway (23, 24, 26, 27, 35), it causes expansions. However, when the Type II hairpin contains a 3′ tail of 1- or 2-nucleotides with a mismatch (Fig. 6A, lane 3), pol δ can conduct error-free synthesis by excising both the mismatch and the hairpin using its 3′-5′ proofreading activity. Therefore, whether or not pol δ conducts error-free or error-prone synthesis on a CAG/CTG hairpin primer totally depends on the hairpin structure and the presence of other DNA polymerases, such as pol β.
FIGURE 7.
Proposed model for DNA polymerase-catalyzed hairpin removal and retention. The 3′ slippage within CAG/CTG repeats in the nascent DNA strand leads to the formation of two different types of hairpins, i.e. Type I and Type II (highlighted with open squares). Processing of Type II hairpin, which carries a 3′ tail immediately after the hairpin, by pol δ results in hairpin retention. In the case of Type I hairpin, although pol δ by itself cannot directly use the hairpin as a primer for DNA synthesis, the polymerase is capable of removing the hairpin structure via its 3′-5′ proofreading activity, leading to error-free processing of the repeats. However, the blockage of pol δ-catalyzed DNA synthesis by the hairpin provokes DNA polymerase switching, which recruits pol β to the site for translesion synthesis. Pol β then incorporates several nucleotides to the 3′-end of the hairpin and the resulting Type II hairpin serves as an effective primer for continuous DNA synthesis by pol δ after a second polymerase switching. These coordinated actions result in hairpin retention and repeat expansion.
It is worth mentioning that despite that pol β exhibits the most potent hairpin-retention activity among polymerases examined, several TLS polymerases tested appear to display some weaker hairpin-retention activities (Fig. 3). In addition, we have not tested many other TLS polymerases, which may also be capable of extending the hairpin primer. Thus, the propensity for a 3′ slipped hairpin to retain during DNA synthesis in replication and repair is very high, especially when one or more such polymerases are overexpressed. Therefore, the 3′ slippage-formed hairpin and its subsequent processing by pol β (or a TLS polymerase) and pol δ may represent a major source of CAG/CTG repeat expansion.
In summary, we have identified in this study a novel mechanism for CAG/CTG repeat expansion, which likely involves a hairpin formation within the repeat units via 3′ slippage in the nascent DNA strand, DNA polymerase switching from pol δ to pol β for “translesion” synthesis and polymerase re-switching from pol β to pol δ for high-fidelity synthesis. The error-prone DNA synthesis preserves CAG/CTG hairpins, leading to CAG/CTG repeat expansions. Our in vitro system, which has implicated several key proteins in CAG/CTG repeat expansions, provides a new direction for studying the disease-causing TNR expansions in vivo.
Acknowledgments
We thank Dr. Charles Ensor for helpful comments and Dr. Yoshihiro Matsumoto for the plasmid expressing the pol δ p125 subunit.
The work was supported, in whole or in part, by National Institutes of Health Grant GM089684 (to G.-M. L.) and a China Scholarship Council Grant (Contract 2010601083) (to J. G.).
- TNR
- trinucleotide repeat
- TLS
- translesion synthesis
- PCNA
- proliferation cellular nuclear antigen
- RFC
- replication factor C
- pol δ
- DNA polymerase δ
- pol β
- DNA polymerase β
- dNTP
- deoxynucleotide triphosphate.
REFERENCES
- 1. Lopez Castel A., Cleary J. D., Pearson C. E. (2010) Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11, 165–170 [DOI] [PubMed] [Google Scholar]
- 2. McMurray C. T. (2010) Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11, 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pearson C. E., Nichol Edamura K., Cleary J. D. (2005) Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 [DOI] [PubMed] [Google Scholar]
- 4. Mirkin S. M. (2007) Expandable DNA repeats and human disease. Nature 447, 932–940 [DOI] [PubMed] [Google Scholar]
- 5. Li X. J., Li S. (2011) Proteasomal dysfunction in aging and Huntington disease. Neurobiol. Dis. 43, 4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith G. K., Jie J., Fox G. E., Gao X. (1995) DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 23, 4303–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gacy A. M., Goellner G., Jurani N., Macura S., McMurray C. T. (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81, 533–540 [DOI] [PubMed] [Google Scholar]
- 8. Moore H., Greenwell P. W., Liu C. P., Arnheim N., Petes T. D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. U.S.A. 96, 1504–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petruska J., Arnheim N., Goodman M. F. (1996) Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 24, 1992–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu G., Chen X., Bissler J. J., Sinden R. R., Leffak M. (2010) Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 6, 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petruska J., Hartenstine M. J., Goodman M. F. (1998) Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem. 273, 5204–5210 [DOI] [PubMed] [Google Scholar]
- 12. Pelletier R., Farrell B. T., Miret J. J., Lahue R. S. (2005) Mechanistic features of CAG*CTG repeat contractions in cultured cells revealed by a novel genetic assay. Nucleic Acids Res. 33, 5667–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krasilnikova M. M., Mirkin S. M. (2004) Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 24, 2286–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daee D. L., Mertz T., Lahue R. S. (2007) Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell Biol. 27, 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovtun I. V., Liu Y., Bjoras M., Klungland A., Wilson S. H., McMurray C. T. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kerrest A., Anand R. P., Sundararajan R., Bermejo R., Liberi G., Dujon B., Freudenreich C. H., Richard G. F. (2009) SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat. Struct. Mol. Biol. 16, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Usdin K., Grabczyk E. (2000) DNA repeat expansions and human disease. Cell Mol. Life Sci. 57, 914–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lange S. S., Takata K., Wood R. D. (2011) DNA polymerases and cancer. Nature Reviews. Cancer 11, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lehmann A. R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J. F., Kannouche P. L., Green C. M. (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 20. Holmes J., Jr., Clark S., Modrich P. (1990) Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl. Acad. Sci. U.S.A. 87, 5837–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Reconstitution of 5'-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 22. Fazlieva R., Spittle C. S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. (2009) Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 37, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou C., Chan N. L., Gu L., Li G. M. (2009) Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nat. Struct. Mol. Biol. 16, 869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang T., Huang J., Gu L., Li G. M. (2012) In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA Repair 11, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pearson C. E., Tam M., Wang Y. H., Montgomery S. E., Dar A. C., Cleary J. D., Nichol K. (2002) Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 30, 4534–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou C., Zhang T., Tian L., Huang J., Gu L., Li G. M. (2011) The Role of XPG in Processing (CAG)n/(CTG)n DNA Hairpins. Cell Biosci. 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panigrahi G. B., Lau R., Montgomery S. E., Leonard M. R., Pearson C. E. (2005) Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 12, 654–662 [DOI] [PubMed] [Google Scholar]
- 28. Byrnes J. J., Downey K. M., Black V. L., So A. G. (1976) A new mammalian DNA polymerase with 3' to 5' exonuclease activity: DNA polymerase δ. Biochemistry 15, 2817–2823 [DOI] [PubMed] [Google Scholar]
- 29. de Noronha C. M., Mullins J. I. (1992) Amplimers with 3'-terminal phosphorothioate linkages resist degradation by vent polymerase and reduce Taq polymerase mispriming. PCR Methods Appl 2, 131–136 [DOI] [PubMed] [Google Scholar]
- 30. Tanabe K., Bohn E. W., Wilson S. H. (1979) Steady-state kinetics of mouse DNA polymerase β. Biochemistry 18, 3401–3406 [DOI] [PubMed] [Google Scholar]
- 31. Wang T. S., Korn D. (1980) Reactivity of KB cell deoxyribonucleic acid polymerases α and β with nicked and gapped deoxyribonucleic acid. Biochemistry 19, 1782–1790 [DOI] [PubMed] [Google Scholar]
- 32. Kornberg A., Baker T. A. (1992) DNA Replication, W.H. Freeman and Company, New York [Google Scholar]
- 33. Kedar P. S., Kim S. J., Robertson A., Hou E., Prasad R., Horton J. K., Wilson S. H. (2002) Direct interaction between mammalian DNA polymerase β and proliferating cell nuclear antigen. J. Biol. Chem. 277, 31115–31123 [DOI] [PubMed] [Google Scholar]
- 34. Yamtich J., Sweasy J. B. (2010) DNA polymerase family X: function, structure, and cellular roles. Biochim. Biophys. Acta 1804, 1136–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan N. L., Hou C., Zhang T., Yuan F., Machwe A., Huang J., Orren D. K., Gu L., Li G. M. (2012) The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)(n)/(CTG)(n) hairpins. J. Biol. Chem. 287, 30151–30156 [DOI] [PMC free article] [PubMed] [Google Scholar]