Background: Cancer metastasis is a major hurdle in cancer therapy and needs identification of novel targets for drug designing.
Results: hnRNP-K is highly expressed in cancer cells and regulates extracellular matrix, cell motility, and angiogenesis pathways.
Conclusion: hnRNP-K is a potential target for metastasis therapy.
Significance: hnRNP-K expression level may serve as a marker of metastatic cancers, and hnRNP-K-inhibiting drugs could be candidate anti-cancer and anti-metastasis reagents.
Keywords: Angiogenesis, Cancer Biology, Cancer Tumor Promoter, Cell Growth, Cell Invasion, Cell Migration, Cell Proliferation, Cell Signaling, Tumor Metastases, hnRNP-K
Abstract
Cancer is a leading cause of death and still awaits effective therapies. Rapid industrialization has contributed to increase in incidence of cancer. One of the reasons why most of the cancers fail therapy is due to their metastatic property. Hence identification of factors leading to metastasis is highly important to design effective and novel anti-cancer therapeutics. In our earlier study (Inoue, A., Sawata, S. Y., Taira, K., and Wadhwa, R. (2007) Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl. Acad. Sci. U.S.A. 104, 8983–8988), we had reported that the involvement of heterogeneous nuclear ribonucleoprotein K (hnRNP-K) in metastasis. Here, we established hnRNP-K-overexpressing and -underexpressing derivative cell lines and examined their proliferation and metastatic properties in vitro and in vivo. Whereas hnRNP-K compromised cells showed delayed tumor growth, its overexpression resulted in enhanced malignancy and metastasis. Molecular basis of the hnRNP-K induced malignant and metastatic phenotypes was dissected by cDNA microarray and pathway analyses. We found that the hnRNP-K regulates extracellular matrix, cell motility, and angiogenesis pathways. Involvement of the selected genes (Cck, Mmp-3, Ptgs2, and Ctgf) and pathways was validated by gene-specific expression analysis. Our results demonstrated that the hnRNP-K is a potential target for metastasis therapy.
Introduction
Heterogeneous nuclear ribonucleoprotein K (hnRNP-K)3 is a 66-kDa unique RNA- and DNA-binding component of the ribonucleoprotein particles. It is a multifunctional signaling protein localized in the nucleus, cytoplasm, and mitochondria and is involved in the regulation of structural organization of the chromatin, transcription, pre-mRNA processing, splicing, mature mRNA transport to the cytoplasm, translation, nuclear transport, signal transduction, and DNA repair (1–8). Several studies have demonstrated its role as a docking platform to integrate cross-talk between nucleic acids and kinases that mediate signal transduction (9). Several proteins, including hepatitis C virus (HCV) core protein C, herpes simplex virus-1 (HSV-1) immediate early protein IE63, casein kinase 2, and p32 Y-box-binding protein, N-WASP, RNA binding motif protein-42, and androgen receptor have been shown to interact with hnRNP-K (10–17). Dividing cells have a higher level of hnRNP-K expression, suggesting its proproliferation function (18–20). Consistently, a variety of cancers, including breast carcinoma (21), esophageal squamous cell carcinoma (22), head-and-neck/oral squamous cell carcinomas (23, 24), lung adenocarcinoma (25, 26), pancreatic carcinoma (27), melanoma (28), colorectal carcinoma (29), and chronic myeloid leukemia (30) were found to be enriched in hnRNP-K expression. It was shown to regulate the anti-apoptotic protein FLICE-like inhibitory protein (FLIP) and VEGF in cancer cells (31, 32). The functional significance and molecular mechanism of the role of hnRNP-K in cancer cells yet remain unclear.
Using the loss-of-function screening system achieved by intracellular expression of single domain antibodies in human metastatic cells, we had previously identified the involvement of hnRNP-K in in vitro migration (33) of human cancer cells. To dissect its functional significance in metastasis, we established hnRNP-K-overexpressing and compromised derivative cell lines of NIH 3T3 (mouse immortal cells) and HT1080 (human fibrosarcoma). In vitro and in vivo phenotype and molecular analyses demonstrated that the hnRNP-K promotes metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Transfections
Mouse (NIH 3T3, immortal fibroblasts) and human (HT1080, fibrosarcoma) cells were obtained from Japanese Collection of Research Bioresources and human osteosarcoma (U2OS) from the ATCC and cultured in DMEM supplemented with 10% fetal bovine serum at 37 °C, in an atmosphere of 5% CO2 and 95% air in a humidified incubator. hnRNP-K cDNA was cloned into the NotI and XhoI site of pCMV-Tag1 vector. An expression vector encoding intracellular antibody to hnRNP-K (iAb-hnRNP-K) was generated as described previously (33). hnRNP-K expression plasmids were transfected into the non-malignant model cell lines NIH 3T3 and U2OS. The malignant model cell line, HT1080, was transfected with iAb-hnRNP-K vector using FuGENE 6 reagent (Roche Applied Science). Typically, 6 μg of plasmid DNA was used to transfect cells in 10-cm dishes at ∼70–80% confluency. Transfected cells were selected in a medium supplemented with G418 (500 μg/ml). hnRNP-K expression was examined in individual clones (10 clones for each cell line) by Western blotting and immunostaining with anti-hnRNP-K antibody. The clones with high level of expression were selected and maintained in the presence of G418 (300 μg/ml) for further analyses.
Gene Expression Analysis
Cells transfected with hnRNP-K and iAb-hnRNP-K expression plasmids were lysed in radioimmune precipitation assay buffer (Thermo Scientific). Aliquots of 20 μg of total protein were resolved on SDS-PAGE and examined for the expression of hnRNP-K and its downstream effectors by Western blotting as described previously (33), and antibodies such as anti-COX-2, anti-CCK, anti-CTGF, anti-VEGF, and anti-matrix metallopeptidase-3 (MMP) (Santa Cruz Biotechnology). Anti-actin antibody (Chemicon) was used as an internal control.
For immunostaining, cells (× 104) were plated on a glass coverslip placed in a 12-well culture dish. After 24 h, when cells had attached to the surface and spread well, they were washed with cold PBS three times and then fixed with prechilled methanol/acetone (1:1) mixture for 5 min. Fixed cells were washed twice with PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and blocked with 0.2% BSA/PBS for 10 min. They were incubated with anti-hnRNP-K antibody (ImmuQuest) for 1 h at room temperature, washed three times with 0.2% Triton X-100 in PBS, and then incubated with Alexa Fluor 594-conjugated goat anti-mouse (Molecular Probes) secondary antibodies. After extensive washings with 0.2% Triton X-100 in PBS, cells were examined on a Carl Zeiss microscope (Axiovert 200 m).
In Vitro and in Vivo Proliferation and Malignant and Metastasis Assays
hnRNP-K overexpressing non-malignant (NIH 3T3) and hnRNP-K compromised malignant cells (HT1080) cells were analyzed for their proliferation rate, colony-forming efficiency, chemotaxis, and invasion assays.
For proliferation rate, equal number of control and transfected cells were plated in 24-well plate. After 48 h, cells were trypsinized, and an aliquot (20 μl) was mixed with an equal volume of 0.4% trypan blue solution. After 5 min of incubation, number of viable (unstained) and dead (stained) cells was counted either by hemocytometer in a quadrant or Vi-CELL viability analyzer (Beckman Coulter).
For colony-forming assays, 500 cells were plated in a six-well plate and left to form colonies for the next 10–15 days with a regular change of medium on every third day. Colonies were fixed in methanol, stained with 0.1% crystal violet solution, photographed, and counted.
For chemotaxis assays, cells at 60–70% confluency were washed with cold PBS, trypsinized, and resuspended in DMEM supplemented with 0.5% bovine serum albumin (Sigma) at 5 × 104 cells/ml. 2.5 × 104 cells were plated in BioCoatTM MatrigelTM Invasion Chambers (8-mm pore, BD Biosciences), and the invasion assay was performed following the manufacturer's instructions. Cells that moved through the chamber were counted under a microscope.
For in vitro cell invasion assays, cells were grown in a monolayer. A wound was made in the monolayer of cells by scratching the cells in a line with a 200-μl pipette tip. Cells were washed a few times with PBS to remove cell debris, and a fresh medium was added. The time of scratching wound was designated as time 0. Cells were allowed to proliferate and migrate into the wound during the next 24 h and recorded under a phase contrast microscope with a 10× phase objective. Migration capacity was quantitated by measuring the percent of open area in 6–10 randomly captured images.
In vivo malignant and metastasis assays were performed using nude mice subcutaneous and tail vein xenograft models, respectively. Balb/c nude mice (4 weeks old, female) were bought from Nihon Clea (Tokyo, Japan). Animals used for experimentation received humane care. Cells were injected into the nude mice subcutaneously (1 × 107 suspended in 0.2 ml of growth medium) and through tail vein (1 × 106 suspended in 0.2 ml of growth medium) injection. Tumor formation and mice health (body weight) was monitored every alternate day. The experiment was repeated twice, using three mice for each injection. Volume of the subcutaneous tumors was calculated as V = L × W2/2, where L was length and W was the width of the tumor, respectively. For metastasis assay, the recipient mice were sacrificed after 20 days of tail vein injection, and their lungs were evaluated for the presence of tumors. This study was carried out in strict accordance with the recommendations in the Animal Experiment Committee, Safety and Environment Management Division, National Institute of Advanced Industrial Science & Technology (AIST), Tsukuba, Japan. The project was approved by the AIST Committee on the Ethics of Animal Experiments.
Microarray Assay and Gene Expression Analysis
Control and hnRNP-K overexpressing NIH 3T3 cells were grown in a monolayer prior to RNA isolation. Total RNA was isolated using RNeasy Mini kit (Qiagen). Single-strand DNA was synthesized using Ambion WT expression kit (Applied Biosystems, Carlsbad, CA). Biotinylated single-strand DNA was prepared using GeneChip DNA Labeling Reagent (Affymetrix, Santa Clara, CA). The single-strand DNA was hybridized with DNA probes on a GeneChip Mouse Gene 1.0 ST array (Affymetrix). The raw fluorescence intensity of each probe was quantified using GeneChip Command console software (Affymetrix). Data obtained from the control and hnRNP-K overexpressing cells were normalized to the individual median of fluorescent intensity using GeneSpringGX software (Agilent Technologies, Santa Clara, CA). Up-regulated genes in normalized sample compared with normalized control were selected, and similar entities were found using GeneSpringGX software.
Bioinformatics and Pathway Analysis
Pathway analysis and biological process association were mapped using PANTHER expression data analysis (34, 35). Conceptually, it is a simple binomial test to compare classifications of multiple clusters of the selected list with a reference list (NCBI, Homo sapiens genes) to statistically determine over- or under-representation of PANTHER classification categories. Biological process result was selected based on over-representation value and p value < 0.5. Protein-protein interactions for target genes were performed with Osprey based on the General Respository for Interaction Data sets (The GRID) and Gene Onthology (GO) (36).
Expression Analysis of hnRNP-K-regulated Genes
RT-PCR was employed with the total RNA isolated with the RNeasy Mini kit (Qiagen) as mentioned above. First-strand cDNA was synthesized using 1 μg of RNA with oligo(dT) as the primer with MMLV reverse transcriptase. PCR was performed using specific primers listed as follows. PCR product (20 μl) was resolved on a 1% agarose gel, stained with GelRed (Biotium), visualized, and photographed under UV light (Bio-Rad). Primers were as follows: Ptgs2, 5′-ACACACTCTATCACTGGCACC-3′ (forward) and 5′-TTCAGGGAGAAGCGTTTGC-3′ (reverse); Cck, 5′-TGCAGCCTTCTCCGTGGAACTCG-3′ (forward) and 5′-TCCTCATTCCACCTCCTCCAAGCAGGG-3′ (reverse); Mmp3, 5′-TGTACCAGTCTACAAGTCCTCCA-3′ (forward) and 5′-CTGCGAAGATCCACTGAAGAAGTAG-3′ (reverse); Ctgf, 5′-CATGCTTGCAGACAGACCTG-3′ (forward) and 5′-GTCTAACAGACAAGGCTCTGACTC-3′ (reverse); Mmp10, 5′-AGTTGCTCCTGCATGTTCTG-3′ (forward) and 5′-TGCATCCTCTCACCTACTGC-3′ (reverse); Tmsb4x, 5′-GGCTGTCCACTGGTCTGAAA-3′ (forward) and 5′-GAACCACATCGATGGCGGAA-3′ (reverse); Itga6, 5′-TGGTCTTACAAATGATGCCTTGTT-3′ (forward) and 5′-GGCTGTGTCCTTTAAAGCCAGT-3′ (reverse); Lilrb4, 5′-AATAAAGCCAAATCCTCCTCAA-3′ (forward) and 5′-CATTGATTAGTGTGCCCTGG-3′ (reverse); Rasa1, 5′-TCAACTGACAAGGAAACGCA-3′ (forward) and 5′-TCTTGGACAATTCCAACAAAAT-3′ (reverse); and Sdpr, 5′-TGAGGTGAATAATGGGTGTTGA-3′ (forward) and 5′-TTATTTAATCTTCTTTCACACAGGC-3′ (reverse).
RESULTS
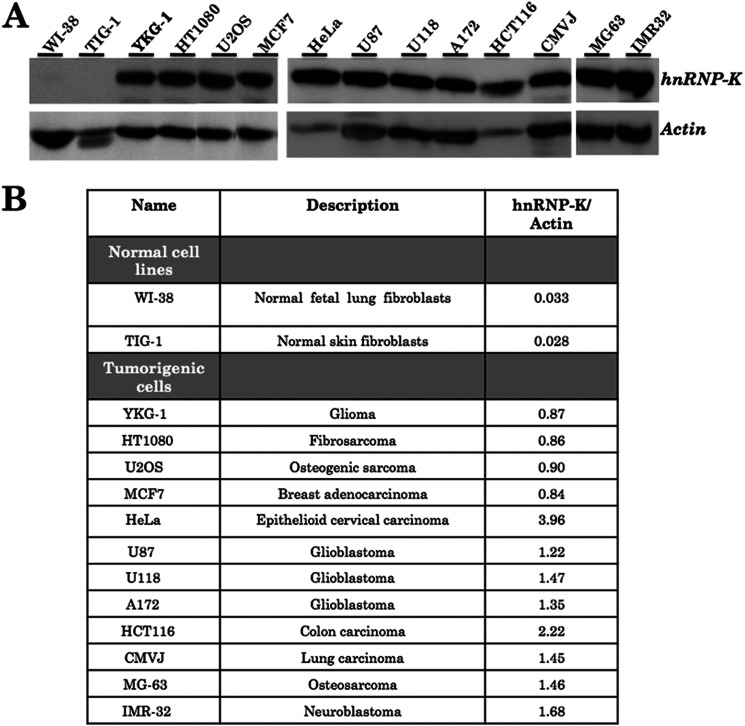
We had previously identified the role of hnRNP-K in in vitro migration of human cancer cells (33). To address the role of hnRNP-K in human carcinogenesis, we first examined its expression level in normal and cancer cells. As shown in Fig. 1, cancer cells showed many fold higher level of expression as compared with the control.
FIGURE 1.
hnRNP-K expression level in human normal and cancer cells. Cancer cells showed higher level of expression as compared with the normal (WI-38 and TIG-1) cells as seen by Western blotting (A). Quantitation of the signal normalized against internal control, actin, is shown in B.
hnRNP-K Intracellular Antibody Causes Tumor Suppression and Loss of Metastasis In Vivo
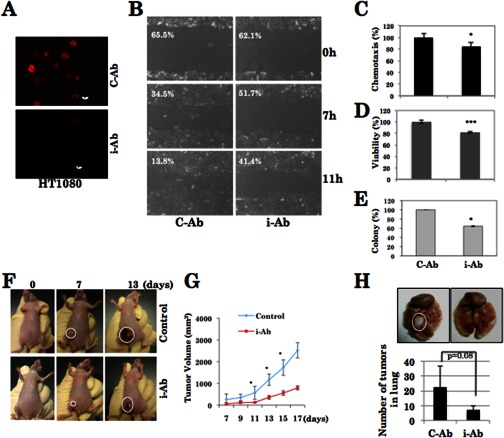
HT1080 cells transfected with hnRNP-K iAb expression vector were examined for hnRNP-K expression by immunostaining with specific antibody (Fig. 2A). hnRNP-K compromised cells were examined for their proliferation rate, colony-forming efficiency and invasion in vitro and in vivo. As shown, hnRNP-K-compromised cells showed delayed motility in scratch wound assays (Fig. 2B). Similar results were obtained in a chemotaxis assay. Quantitation from the three independent experiments showed that the hnRNP-K-compromised cells had 30% less chemotaxis (Fig. 2C), 22% less viability (Fig. 2D), and 40% less colony-forming efficiency (Fig. 2E). We next investigated in vivo tumor formation and metastasis of these cells in nude mice. As compared with the control cells, hnRNP-K compromised cells showed tumor suppression (Fig. 2, F and G), and most noticeably, the metastasis to lung was distinctly diminished (Fig. 2H). Whereas control cells formed big tumors in the lung, the hnRNP-K compromised cells showed either very small or no lung tumors in three independent experiments, suggesting that the hnRNP-K significantly contributes to the in vivo metastasis of cancer cells. Consistent with the decrease in metastatic capacity of iAb-expressing cells, mice injected with these cells looked healthier than the control mice. These mice maintained their body weight and showed increased survival rate as compared with the control cells (data not shown).
FIGURE 2.
hnRNP-K silencing by its iAb reduces its malignant properties in human fibrosarcoma (HT1080). HT1080 cells transfected with hnRNP-K iAb show reduced expression by immunostaining (scale bar, 10 μm; A). hnRNP-K iAb-transfected cells showed reduced mobility in scratch wound (B) and chemotaxis assays (C). These cells also showed moderate reduction in cell viability (D) and colony-forming capacity (E). In vivo tumor formation in nude mice by injecting HT1080 cells and the tumor suppression by iAb transfection are shown (F and G). iAb-transfected cells reduced the metastasis in lung (H). Statistical significance as determined by Student's t test was as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. C-Ab, control antibody, VHH (single heavy chain small antigen-binding fragment)-sfi expressing vector (pcDNA 3.1-Hygro) (33).
hnRNP-K Overexpression Contributes to the Malignant Properties of Cancer Cells
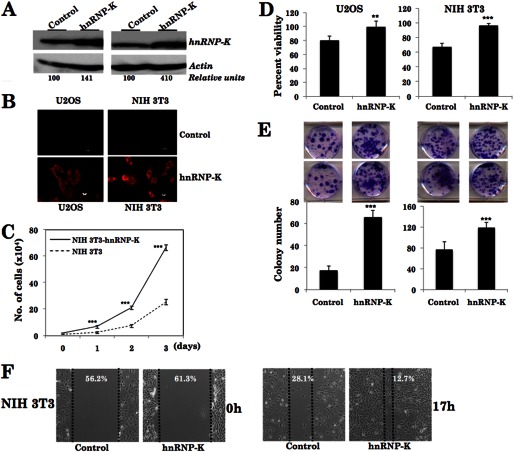
To address the functional significance of hnRNP-K in malignant and metastasis phenotype of cancer cells, we generated hnRNP-K-overexpressing derivatives of mouse immortal (NIH 3T3) and human osteosarcoma (U2OS) cells that are non-malignant and non-metastatic in the nude mice model. Derivative cells were first analyzed for hnRNP-K expression by Western blotting and immunostaining (Fig. 3, A and B) and then subjected to proliferation, viability, colony-forming, and invasion assays. Growth curves of control and hnRNP-K-overexpressing cells revealed higher proliferation rate of the latter (Fig. 3C and data not shown). Furthermore, hnRNP-K-overexpressing, both NIH 3T3 and U2OS, cells showed higher viability, colony-forming efficacy, and invasion as compared with the control parent cells (Fig. 3, D–F, and data not shown). In the in vivo nude mice model, control NIH 3T3 cells neither formed tumors in subcutaneous xenografts nor showed any metastasis in lung in tail vein injection model (Fig. 4, A and C). hnRNP-K-overexpressing NIH 3T3 cells formed subcutaneous tumors and showed metastatic tumors in the lungs (Fig. 4C). Consistent with these results, Western blotting of lung tissues, isolated from mice injected with either control or hnRNP-K-overexpressing NIH 3T3 cells, revealed a higher level of expression of MMP-3 in the latter (Fig. 4D). Furthermore, we found that the hnRNP-K-overexpressing NIH 3T3 cells have a higher level of expression of VEGF, an established marker of angiogenesis (Fig. 4, E and F).
FIGURE 3.
hnRNP-K overexpression in NIH 3T3 cells increased their viability, colony-forming efficiency, and motility. Expression level of hnRNP-K, in control and its overexpressing NIH 3T3 derivatives, is shown by Western blotting (A) and immunostaining (B). hnRNP-K-overexpressing cells showed higher proliferation (C) viability (D), colony-forming efficiency (E), and motility in wound-scratch assays (F). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student's t test).
FIGURE 4.
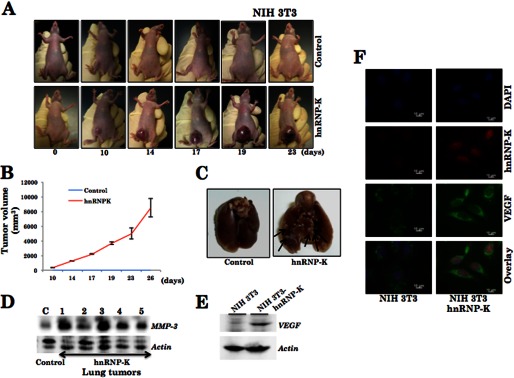
hnRNP-K overexpression induced malignant transformation of NIH 3T3 cells. hnRNP-K-overexpressing cells when xenografted in nude mice lead to tumor formation (A). B, average volume of hnRNP-K induced tumors from six mice is shown. Overexpression of hnRNP-K in NIH 3T3 cells resulted in lung metastasis (C). D, Western blot indicates a high level of expression of MMP-3 in lung tumor tissue extracts. A high level of expression VEGF (an angiogenesis marker) was detected in hnRNP-K-overexpressing NIH 3T3 cells by Western blotting as well as immunostaining (F). Actin was used as a loading control.
hnRNP-K-regulated Extracellular Matrix and Cell Motility Genes
To investigate the molecular mechanism of contribution of hnRNP-K in cancer cell malignancy and metastasis, we performed cDNA array analysis of control and malignantly transformed NIH 3T3 cells. As shown in Fig. 5, hnRNP-K-overexpressing malignant derivatives showed either up- (Table 1) or down-regulation (Table 2) of several genes. The most significant (∼5-fold change in the expression level) of these genes, as identified by GeneSpringGX software, are shown in Table 1. Among these, matrix metallopeptidases (Mmp-3 and Mmp-10) showed the highest up-regulation (6.35- and 4-fold, respectively) followed by connective tissue growth factor, thymosin, integrin, and Ras-p21 (∼3-fold increase). We also applied bioinformatics tools on the gene targets (Table 1 and Fig. 5) selected from the array analysis. The top pathways selected were plasminogen cascade, integrin and inflammation signaling along with biological processes, including cell communication and signal transduction (Fig. 5, B and C). The selected target genes could be grouped into six clusters. These genes indicated that the hnRNP-K is a multifunctional protein, and is involved in pathways regulating transcription, signal transduction, and extracellular matrix. For example, the Sdpr cluster signified up-regulation of genes involved in transcription, the Il2rg cluster signified proproliferation. The Kng1-Ctgf cluster signified anti-apoptosis and angiogenesis signaling, Mmp3-Mmp10-Col1a1 and Tmsb4x gene clusters signified extracellular matrix signaling involved in cell motility. Ptgs2 compose cluster for cell growth control (Fig. 5D). Of note, the analyses indicated that 32% of genes involved in extracellular matrix were regulated by hnRNP-K, suggesting the major contribution of extracellular matrix in hnRNP-K-mediated malignancy and metastasis (Fig. 5E).
FIGURE 5.
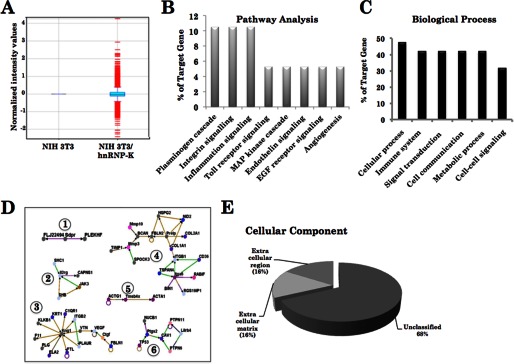
cDNA array of control and hnRNP-K-overexpressing NIH 3T3 cells. hnRNP-K overexpression showed up- and down-regulation of several genes (A). The pathway analysis on the up- and down-regulated genes (as shown in Tables 1 and 2) revealed that the hnRNP-K regulates pathways and biological processes as shown (B–E).
TABLE 1.
The top 10 most up-regulated genes induced by overexpressing hnRNP-K in NIH 3T3 cells
| Gene | Fold change | Biological progress |
|---|---|---|
| Mmp3 | 6.35 | Collagen catabolic process; metabolic process; proteolysis |
| Mmp10 | 4.00 | Collagen catabolic process; metabolic process; proteolysis |
| Ctgf | 3.62 | Angiogenesis; cell adhesion; cell differentiation; cell migration; cell-matrix adhesion; fibroblast growth factor receptor signaling pathway; integrin-mediated signaling pathway; lung development |
| Tmsb4x | 3.51 | Actin cytoskeleton organization; cytoskeleton organization; regulation of cell migration; sequestering of actin monomers |
| Itga6 | 3.32 | Cell adhesion molecule activity; up regulation of cell-cell adhesion |
| Lilrb4 | 3.23 | Immune response |
| Rasa1 | 3.19 | Vasculogenesis; embryonic development; negative regulation of neuron apoptosis |
| Cck | 2.91 | Axonogenesis; eating behavior; neuronal migration |
| Sdpr | 2.89 | Up-regulation of transcription from RNA polymerase II promoter |
| Ptgs2 | 1.73 | Prostaglandin biosynthetic process; cell motility; cyclooxygenase pathway; regulation of inflammatory response |
TABLE 2.
The top seven most down-regulated genes induced by overexpressing hnRNP-K in NIH 3T3 cells
| Gene | Fold change | Biological progress |
|---|---|---|
| Col12a1 | 6.99 | Cell adhesion |
| Prelp | 5.14 | Cell aging |
| Igsf10 | 5.03 | Ossification; protein amino acid phosphorylation; vascular endothelial growth factor receptor signaling pathway |
| Slc47a1 | 4.56 | Multidrug transport; transmembrane transport; transport monomers |
| Il2rg | 3.87 | Positive regulation of B cell differentiation; positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T celldifferentiation; positive regulation of T cell differentiation in the thymus |
| Tcn2 | 3.30 | Cobalamin transport; cobalt ion transport; ion transport; transport |
| Kng1 | 2.97 | Blood coagulation; inflammatory response; regulation of blood vessel size; vasodilation |
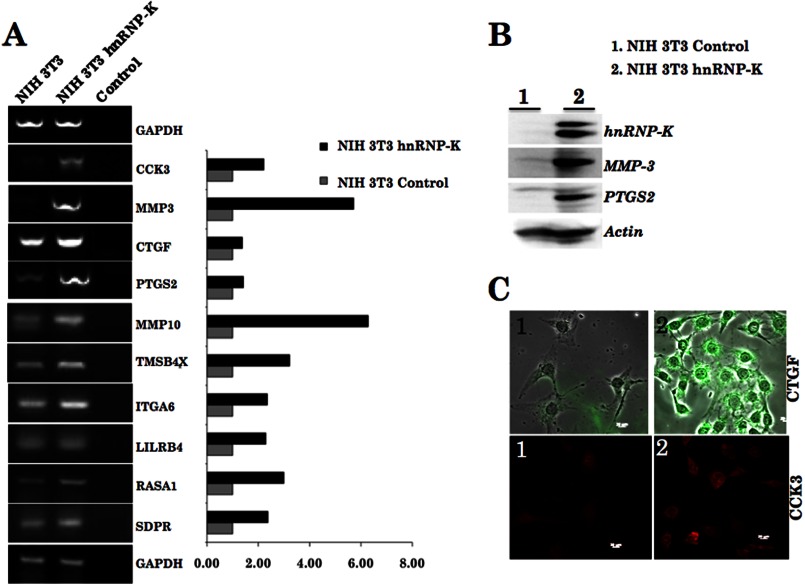
We next validated the outcome of cDNA array and bioinformatics by gene-specific RT-PCR analysis. As shown, hnRNP-K-overexpressing NIH 3T3 cells showed up-regulation of all of the 10 identified genes (Fig. 6A). Mmp-3 and Mmp-10 showed highest up-regulation followed by Tmsb4x, Rasa1, Cck, Lilrb4, and Itga6. We also examined the protein expression level in control and hnRNP-K-overexpressing malignant cells and found that the selected genes such as Ptgs2, Mmp-3, Ctgf, and Cck were enriched in the latter (Fig. 6, B and C). Of note, as described above, tumors formed by hnRNP-K-overexpressing NIH 3T3 cells revealed a high level of MMP-3 expression as compared with the control (Fig. 4D).
FIGURE 6.
Validation of the hnRNP-K-regulated gene targets identified by cDNA array. Gene-specific RT-PCR in control and hnRNP-K-overexpressing cells confirmed the up-regulation of the downstream genes as shown (A). hnRNP-K-overexpressing cells showed higher expression level of MMP-3 and PTGS2 (Western blotting, B), CTGF, and CCK3 (immunostaining, C).
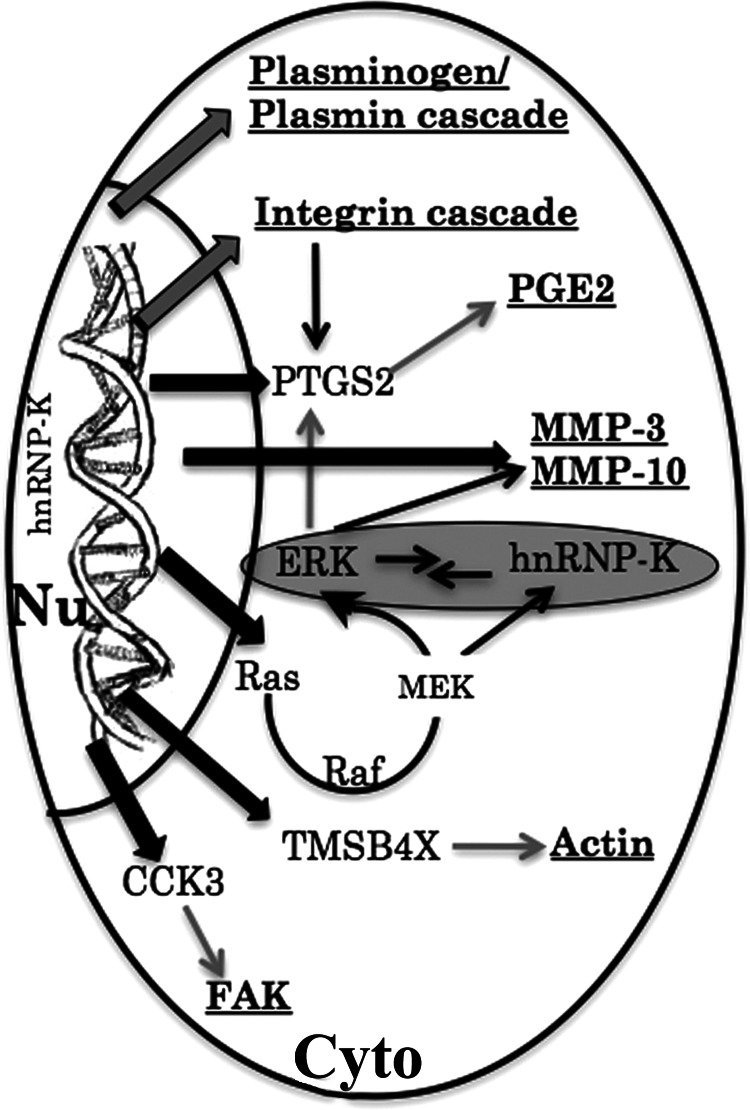
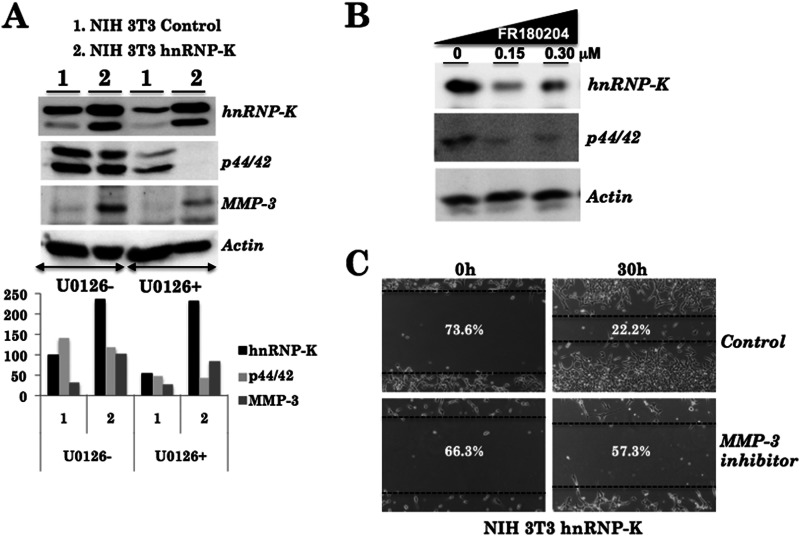
Based on these data, we predicted that hnRNP-K might regulate cell migration in two ways. In the nucleus, it may cause transcriptional up-regulation of Ptgs2, Mmp-3, Mmp-10, Tmsb4x, Cck, and Ras (Fig. 6A). Activated Ras and MEK stabilize hnRNP-K in the cytoplasm either directly or through the ERK activation. In the cytoplasm, it might activate ERK in a regulatory loop that may in turn regulate cell migration by the MMP-3/MMP-10 pathway (Fig. 7). In addition to these molecular events, hnRNP-K was also predicted to affect plasmin and integrin cascades that affect the PTGS2 signaling and cell migration (Fig. 7). According to this hypothesis, hnRNP-K was predicted to affect MMP-3/MMP-10 and cell migration through ERK signaling. We investigated this hypothesis by adopting the MAPK/ERK kinase inhibitor approach. As shown in Fig. 8A, control and hnRNP-K-overexpressing cells were treated with U0126 (1,4-diamino-2, 3-dicyano-1, 4-bis[2-aminophenylthio]butadiene), a highly selective inhibitor of MEK-1/MEK-2. As shown in Fig. 8A, U0126 caused inhibition of ERKp44/42, both in the control and hnRNP-K overexpressing NIH 3T3 cells. Of note, hnRNP-K-induced up-regulation of MMP-3 was compromised by treatment with U0126 in the hnRNP-K-overexpressing cells. The result endorses that the hnRNP-K up-regulates MMP-3 expression by activation of ERK kinase. Furthermore, treatment of cells with specific inhibitor of ERK1 and ERK2 (FR180204) led to reduced levels of hnRNP-K (Fig. 8B). hnRNP-K-overexpressing cells showed compromised cell migration when treated with MMP-3 inhibitor (MMP-3 inhibitor, Santa Cruz Biotechnology) (Fig. 7C), suggesting that hnRNP-K-induced cell migration occurs through ERK-MMP-3 activation.
FIGURE 7.
Schematic model showing the role of hnRNP-K in regulation of cell migration through PTGS2, CCK3, RAS, ERK, and MMP-3.
FIGURE 8.
Involvement of MEK and ERK in hnRNP-K-induced cell migration. Cells treated with MEK1/MEK2 inhibitor, U0126, showed decrease in ERK (p44/42). Increase in MMP-3 in hnRNP-K-overexpressing cells was compromised by treatment with the inhibitor (A). ERK1/ERK2 inhibitor FR180204 resulted in decreased level of hnRNP-K confirming cross-talk between MEK, ERK, and hnRNP-K as proposed in the model (Fig. 7). Treatment of hnRNP-K-overexpressing cells with MMP-3 inhibitor resulted in their decreased invasion capacity.
DISCUSSION
hnRNP-K-overexpressing cells showed enhanced malignant and metastatic properties. Molecular analysis of the hnRNP-K-overexpressing NIH 3T3 derivatives revealed that it regulates a variety of biological cascades of which cell adhesion and invasion signaling pathways that regulate extracellular matrix, cell motility, and angiogenesis were most prominent (Table 1 and Fig. 6). Bioinformatics analysis on the up-regulated genes revealed that the plasminogen and integrin signaling were the most up-regulated pathways in hnRNP-K-overexpressing cells. It has been established that the activation of plasminogen cascade is required for the movement of tumor cells through an extracellular matrix, an essential step in metastatic spread of cancer from the primary tumor to a secondary remote site. Tumor cells induce breakdown of the extracellular matrix by a plasminogen/plasmin and matrix metalloproteinase system. Integrins have been shown to regulate adhesion and invasiveness of tumor cells and stimulate endothelial cells for angiogenesis. It is regulated by a cross-talk between the integrin family and other signaling proteins. Integrin α3β1 was shown to up-regulate COX-2/PTGS in tumor cells (37). COX-2/PTGS expression is frequently enriched in premalignant, malignant, and metastatic tumors (38). It was also shown that COX-2 induces prostaglandin E2, which has a role in tumor cell invasion and cross-talk to endothelial cells for angiogenesis. COX-2 was also shown to be regulated by Akt and ERK1/2 pathways (39). Furthermore, COX-2 inhibition caused tumor suppression (40). The fact that the overexpression of hnRNP-K resulted in up-regulation of Cox-2 RNA as well as protein suggesting that it might regulate Cox-2 at the transcriptional level. A recent study showed that hnRNP-K binds to the Cox-2 promoter as well as the UTR and regulates its stability (41). We also found that cholecystokinin (CCK3) was up-regulated in hnRNP-K-overexpressing malignant and metastatic cells. CCK3 has been shown to have role in the development of gastric, pancreatic, and colon cancers (42). It has been reported to activate focal adhesion kinase, involved in the progression of malignancies, invasion, and lymph node metastasis (43). These data revealed that the overexpression of hnRNP-K promoted metastasis by induction of signaling cascades involved in extracellular matrix, cell adhesion, invasion, and angiogenesis. Tmsb4x, an actin monomer-binding protein involved in cell migration and angiogenesis, was shown to be associated with tumor development (44, 45).
Matrix metalloproteinases MMP-3 and MMP-10 showed the highest up-regulation in hnRNP-K-overexpressing cells. Role of matrix metalloproteinases has been established in tumor metastasis. They, along with plasminogen and integrin family proteins, regulate degradation of extracellular matrix, invasion of cells through the matrix, and cross-talk with endothelial cells to generate new vessels for blood supply. Matrix metalloproteinases are known to be regulated by the RAS-MEK-ERK cascade. Because RAS also gets up-regulated in hnRNP-K-overexpressing cells, we examined the role of the RAS-MEK-ERK cascade in these cells by employing specific inhibitor of MEK1 and MEK2. As expected and shown in Fig. 8A, U0126 caused a reduction in the phosphorylation of ERK1 and ERK2. Similar data were obtained by treating the cells with another specific inhibitor of ERK1 and ERK2, FR180204 (Fig. 8B), further endorsing the involvement of the RAS-MEK-ERK cascade. Furthermore, NIH 3T3 cells treated with U0126 lost hnRNP-K-induced up-regulation of MMP-3, demonstrating that the hnRNP-K-regulated cell migration is by the Ras/MEK/ERK-MMP-3 pathway. Mikula et al. (46) used chromatin immunoprecipitations to study co-recruitment of hnRNP-K and ERK cascade activity along with the Egr-1 gene and found that the spatiotemporal binding patterns of ERK cascade transducers (GRB2, SOS, B-Raf, MEK, and ERK) at the EGR-1 locus resemble both hnRNP-K and RNA polymerase II. Furthermore, knockdown of hnRNP-K by siRNA decreased the level of active MEK and ERK suggesting that hnRNP-K regulates MEK and ERK as seen in the present study. Taken together, in continuation to our first identification of hnRNP-K as a gene involved in cell migration in intracellular antibody screening system, we, in the present study, demonstrate that the hnRNP-K regulates metastasis in vivo by regulation of extracellular matrix components through the ERK signaling pathway. Thus the inhibitors of hnRNP-K such as siRNA, intracellular antibody or the small molecules might be useful for treatment of metastatic cancers.
Acknowledgment
We thank Masumi Maruyama Takano for help in the cDNA array data analysis.
Footnotes
- hnRNP-K
- heterogeneous nuclear ribonucleoprotein K
- iAb
- intracellular antibody
- MMP
- matrix metallopeptidase
- CCK
- cholecystokinin
- PTGS
- prostaglandin-endoperoxide synthase
- CTGF
- connective tissue growth factor
- sdpr
- serum deprivation response.
REFERENCES
- 1. Matunis M. J., Michael W. M., Dreyfuss G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell Biol. 12, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan J. Y., Huang S. M., Liu S. T., Huang C. H. (2008) The transactivation domain of heterogeneous nuclear ribonucleoprotein K overlaps its nuclear shuttling domain. Int. J. Biochem. Cell Biol. 40, 2078–2089 [DOI] [PubMed] [Google Scholar]
- 3. Lee M. H., Mori S., Raychaudhuri P. (1996) Trans-activation by the hnRNP K protein involves an increase in RNA synthesis from the reporter genes. J. Biol. Chem. 271, 3420–3427 [DOI] [PubMed] [Google Scholar]
- 4. Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. (1996) Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16, 2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ostrowski J., Wyrwicz L., Rychlewski L., Bomsztyk K. (2002) Heterogeneous nuclear ribonucleoprotein K protein associates with multiple mitochondrial transcripts within the organelle. J. Biol. Chem. 277, 6303–6310 [DOI] [PubMed] [Google Scholar]
- 6. Denisenko O., Bomsztyk K. (2002) Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol. Cell. Biol. 22, 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moumen A., Masterson P., O'Connor M. J., Jackson S. P. (2005) hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y., Zhou X., Liu N., Wang C., Zhang L., Mo W., Hu G. (2008) Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 582, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 9. Bomsztyk K., Denisenko O., Ostrowski J. (2004) hnRNP K: one protein multiple processes. Bioessays 26, 629–638 [DOI] [PubMed] [Google Scholar]
- 10. Hsieh T. Y., Matsumoto M., Chou H. C., Schneider R., Hwang S. B., Lee A. S., Lai M. M. (1998) Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 273, 17651–17659 [DOI] [PubMed] [Google Scholar]
- 11. Miau L. H., Chang C. J., Shen B. J., Tsai W. H., Lee S. C. (1998) Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPβ-mediated gene activation. J. Biol. Chem. 273, 10784–10791 [DOI] [PubMed] [Google Scholar]
- 12. Wadd S., Bryant H., Filhol O., Scott J. E., Hsieh T. Y., Everett R. D., Clements J. B. (1999) The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 274, 28991–28998 [DOI] [PubMed] [Google Scholar]
- 13. Bryant H. E., Matthews D. A., Wadd S., Scott J. E., Kean J., Graham S., Russell W. C., Clements J. B. (2000) Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J. Virol. 74, 11322–11328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shnyreva M., Schullery D. S., Suzuki H., Higaki Y., Bomsztyk K. (2000) Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J. Biol. Chem. 275, 15498–15503 [DOI] [PubMed] [Google Scholar]
- 15. Yoo Y., Wu X., Egile C., Li R., Guan J. L. (2006) Interaction of N-WASP with hnRNPK and its role in filopodia formation and cell spreading. J. Biol. Chem. 281, 15352–15360 [DOI] [PubMed] [Google Scholar]
- 16. Fukuda T., Naiki T., Saito M., Irie K. (2009) hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells 14, 113–128 [DOI] [PubMed] [Google Scholar]
- 17. Mukhopadhyay N. K., Kim J., Cinar B., Ramachandran A., Hager M. H., Di Vizio D., Adam R. M., Rubin M. A., Raychaudhuri P., De Benedetti A., Freeman M. R. (2009) Heterogeneous nuclear ribonucleoprotein K is a novel regulator of androgen receptor translation. Cancer Res. 69, 2210–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ostrowski J., Bomsztyk K. (2003) Nuclear shift of hnRNP K protein in neoplasms and other states of enhanced cell proliferation. Br. J. Cancer 89, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Backe P. H., Messias A. C., Ravelli R. B., Sattler M., Cusack S. (2005) X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 20. Laury-Kleintop L. D., Tresini M., Hammond O. (2005) Compartmentalization of hnRNP-K during cell cycle progression and its interaction with calponin in the cytoplasm. J. Cell. Biochem. 95, 1042–1056 [DOI] [PubMed] [Google Scholar]
- 21. Mandal M, Vadlamudi R, Nguyen D, Wang RA, Costa L, Bagheri-Yarmand R, Mendelsohn J, Kumar R. (2001) Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J. Biol. Chem. 276, 9699–9704 [DOI] [PubMed] [Google Scholar]
- 22. Hatakeyama H., Kondo T., Fujii K., Nakanishi Y., Kato H., Fukuda S., Hirohashi S. (2006) Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics 6, 6300–6316 [DOI] [PubMed] [Google Scholar]
- 23. Roychoudhury P., Chaudhuri K. (2007) Evidence for heterogeneous nuclear ribonucleoprotein K overexpression in oral squamous cell carcinoma. Br. J. Cancer 97, 574–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matta A., Tripathi S. C., DeSouza L. V., Grigull J., Kaur J., Chauhan S. S., Srivastava A., Thakar A., Shukla N. K., Duggal R., DattaGupta S., Ralhan R., Michael Siu K. W. (2009) Heterogeneous ribonucleoprotein K is a marker of oral leukoplakia and correlates with poor prognosis of squamous cell carcinoma. Int. J. Cancer 125, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 25. Chen Y., Li W., Zhang S. (2008) hnRNP K expression and its clinical significance in human lung cancer tissues. Zhongguo Fei Ai Za Zhi 11, 241–245 [DOI] [PubMed] [Google Scholar]
- 26. Tang F. M., Li W. M., Chen Y., Wang D. M., Han J. (2008) Expression of hnRNP K in lung adenocarcinoma cells. Sichuan Da Xue Xue Bao Yi Xue Ban 39, 823–826 [PubMed] [Google Scholar]
- 27. Zhou R., Shanas R., Nelson M. A., Bhattacharyya A., Shi J. (2010) Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int. J. Cancer 126, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wen F., Shen A., Shanas R., Bhattacharyya A., Lian F., Hostetter G., Shi J. (2010) Higher expression of the heterogeneous nuclear ribonucleoprotein k in melanoma. Ann. Surg. Oncol. 17, 2619–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang F., Zhang P., Shi C., Yang Y., Qin H. (2011) Immunohistochemical detection of HSP27 and hnRNP K as prognostic and predictive biomarkers for colorectal cancer. Med. Oncol. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 30. Du Q., Wang L., Zhu H., Zhang S., Xu L., Zheng W., Liu X. (2010) The role of heterogeneous nuclear ribonucleoprotein K in the progression of chronic myeloid leukemia. Med. Oncol. 27, 673–679 [DOI] [PubMed] [Google Scholar]
- 31. Chen L. C., Chung I. C., Hsueh C., Tsang N. M., Chi L. M., Liang Y., Chen C. C., Wang L. J., Chang Y. S. (2010) The antiapoptotic protein, FLIP, is regulated by heterogeneous nuclear ribonucleoprotein K and correlates with poor overall survival of nasopharyngeal carcinoma patients. Cell Death Differ. 17, 1463–1473 [DOI] [PubMed] [Google Scholar]
- 32. Feliers D., Lee M. J., Ghosh-Choudhury G., Bomsztyk K., Kasinath B. S. (2007) Heterogeneous nuclear ribonucleoprotein K contributes to angiotensin II stimulation of vascular endothelial growth factor mRNA translation. Am. J. Physiol. Renal Physiol. 293, F607–615 [DOI] [PubMed] [Google Scholar]
- 33. Inoue A., Sawata S. Y., Taira K., Wadhwa R. (2007) Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl. Acad. Sci. U.S.A. 104, 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mi H., Guo N., Kejariwal A., Thomas P. D. (2007) PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35, D247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomas P. D., Kejariwal A., Guo N., Mi H., Campbell M. J., Muruganujan A., Lazareva-Ulitsky B. (2006) Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Breitkreutz B. J., Stark C., Tyers M. (2003) The GRID: the General Repository for Interaction Datasets. Genome Biol. 4, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mitchell K., Svenson K. B., Longmate W. M., Gkirtzimanaki K., Sadej R., Wang X., Zhao J., Eliopoulos A. G., (2010) Berditchevski, F., and Dipersio, C. M. Suppression of integrin α3β1 in breast cancer cells reduces cyclooxygenase-2 gene expression and inhibits tumorigenesis, invasion, and cross-talk to endothelial cells. Cancer Res. 70, 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Niki T., Kohno T., Iba S., Moriya Y., Takahashi Y., Saito M., Maeshima A., Yamada T., Matsuno Y., Fukayama M., Yokota J., Hirohashi S. (2002) Frequent co-localization of Cox-2 and laminin-5 γ2 chain at the invasive front of early-stage lung adenocarcinomas. Am. J. Pathol. 160, 1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai T. Y., Chen L. M., Lin J. Y., Tzang B. S., Lin J. A., Tsai C. H., Lin Y. M., Huang C. Y., Liu C. J., Hsu H. H. (2010) 17β-estradiol inhibits prostaglandin E2-induced COX-2 expressions and cell migration by suppressing Akt and ERK1/2 signaling pathways in human LoVo colon cancer cells. Mol. Cell. Biochem. 342, 63–70 [DOI] [PubMed] [Google Scholar]
- 40. Fang W., Han A., Bi X., Xiong B., Yang W. (2010) Tumor inhibition by sodium selenite is associated with activation of c-Jun NH2-terminal kinase 1 and suppression of β-catenin signaling. Int. J. Cancer 127, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shanmugam N., Reddy M. A., Natarajan R. (2008) Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 283, 36221–36233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo Y. S., Townsend C. M., Jr. (2000) Roles of gastrointestinal hormones in pancreatic cancer. J. Hepatobiliary. Pancreat. Surg. 7, 276–285 [DOI] [PubMed] [Google Scholar]
- 43. Yu H. G., Tong S. L., Ding Y. M., Ding J., Fang X. M., Zhang X. F., Liu Z. J., Zhou Y. H., Liu Q. S., Luo H. S., Yu J. P. (2006) Enhanced expression of cholecystokinin-2 receptor promotes the progression of colon cancer through activation of focal adhesion kinase. Int. J. Cancer 119, 2724–2732 [DOI] [PubMed] [Google Scholar]
- 44. Györffy B., Dietel M., Fekete T., Lage H. (2008) A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int. J. Gynecol. Cancer 18, 1215–1233 [DOI] [PubMed] [Google Scholar]
- 45. Olbryt M., Jarzab M., Jazowiecka-Rakus J., Simek K., Szala S., Sochanik A. (2006) Gene expression profile of B 16(F10) murine melanoma cells exposed to hypoxic conditions in vitro. Gene Expr. 13, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mikula M., Bomsztyk K. (2011) Direct recruitment of ERK cascade components to inducible genes is regulated by heterogeneous nuclear ribonucleoprotein (hnRNP) K. J. Biol. Chem. 286, 9763–9775 [DOI] [PMC free article] [PubMed] [Google Scholar]