Background: DnaJ is an Hsp40 molecular chaperone able to bind and remodel native substrates as RepE, the repressor/activator of plasmid F replication.
Results: We describe the structure of DnaJ bound to RepE and two mutants and the resulting protein conformational changes.
Conclusion: DnaJ conformational plasticity allows binding of different substrates.
Significance: We present the first structure of an Hsp40 chaperone bound to a client.
Keywords: Electron Microscopy (EM), Heat Shock Protein, Protein Complexes, Protein Conformation, Protein Structure, DnaJ, Hsp40, Chaperones, Protein Folding
Abstract
Hsp40 chaperones bind and transfer substrate proteins to Hsp70s and regulate their ATPase activity. The interaction of Hsp40s with native proteins modifies their structure and function. A good model for this function is DnaJ, the bacterial Hsp40 that interacts with RepE, the repressor/activator of plasmid F replication, and together with DnaK regulates its function. We characterize here the structure of the DnaJ-RepE complex by electron microscopy, the first described structure of a complex between an Hsp40 and a client protein. The comparison of the complexes of DnaJ with two RepE mutants reveals an intrinsic plasticity of the DnaJ dimer that allows the chaperone to adapt to different substrates. We also show that DnaJ induces conformational changes in dimeric RepE, which increase the intermonomeric distance and remodel both RepE domains enhancing its affinity for DNA.
Introduction
DnaJ from Escherichia coli belongs to the type I Hsp40 protein family, whose members mainly act as cochaperones of their Hsp70 partners to control protein homeostasis in the cell (1–3). As a cochaperone, DnaJ enhances the ATPase activity of DnaK, the major bacterial Hsp70, synergistically with substrates. This function relies on the highly conserved N-terminal domain of DnaJ, the J-domain, which interacts with the nucleotide-binding domain of DnaK (4). In addition to this cochaperone function, DnaJ and Ydj1p, its yeast homolog, can also behave as independent chaperones, exhibiting a “holdase” activity that allows their interaction with unfolded polypeptides preventing their aggregation (5, 6). Moreover, DnaJ also interacts with folded proteins such as σ32, the bacterial heat shock transcription factor, and members of the Rep protein family, altering their conformational and functional properties (7–10).
DnaJ consists of four domains: the N-terminal J-domain, a peptide stretch rich in G and F residues (G/F-domain), a Zn-binding domain (ZBD),5 and the C-terminal domain, which contains two distinct regions (CTDI and CTDII) (see Fig. 2A). Several reports have implicated the last three domains in substrate binding. We previously found that the G/F-domain is required to bind a folded substrate, RepE, whereas it is dispensable to bind unfolded polypeptides (10). The ZBD of DnaJ and other type I Hsp40s participates in binding of denatured substrates (6), but its role may be indirect because its mutation reduces the affinity of DnaJ for the substrates but does not completely abolish their binding (5, 11). Finally, the C-terminal domains of Hsp40s contain a peptide binding site (12, 13) and the dimerization domain. Substitution of the residues that form the binding site severely affects peptide binding, refolding of denatured substrates and cell viability (14, 15). On this basis, Hsp40 proteins may have a large and adaptable interaction surface formed by several domains to accommodate different protein substrates.
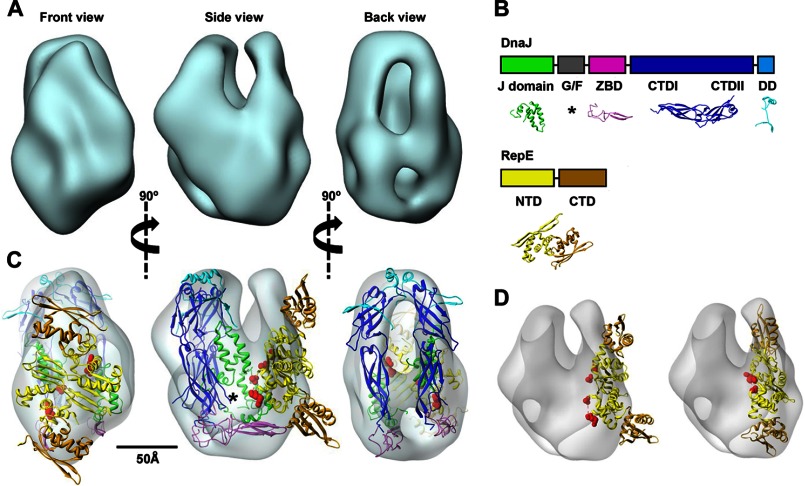
FIGURE 2.
Architecture of the DnaJ-RepE complex. A, three-dimensional reconstruction of the DnaJ-RepE complex shown in three different, orthogonal views. B, domain arrangement of DnaJ and RepE proteins and structures used in this study: J-domain (green; PDB code 1XBL), ZBD (magenta; PDB code 1EXK), CTDI and CTDII domains (blue; PDB code 1NLT), and dimerization domain (DD) (cyan; PDB code 1XAO). Disordered G/F-domain is marked by an asterisk. In the RepE monomer, NTD and CTD are shown in yellow and brown, respectively (extracted from PDB code 2Z9O). C, docking of the atomic model of DnaJ and the atomic structure of RepE into the corresponding masses of the three-dimensional reconstruction; color coding is the same as in B. The disordered G/F-domain is tentatively located in the empty region marked by an asterisk. Red spheres indicate residues Ile46 and Asp57, located, respectively, at the beginning and end of Lα2-β2. See supplemental Movie S1 for an easier interpretation of the docking. D, docking of the DNA-bound RepE structure (PDB code 2Z9O) into the DnaJ-RepE complex three-dimensional reconstruction before (left) and after (right) applying flexibility to the monomers. A 25° tilt of the RepE C-terminal domains substantially improves the quality of the docking.
To gain knowledge on the interaction of Hsp40s with protein substrates, we have characterized the binding of DnaJ to RepE, the replication initiation factor of plasmid mini-F (16). RepE dimers (repressors) contain an N-terminal domain responsible for the dimerization of the protein (NTD) and a CTD involved in DNA binding (17). Using electron microscopy (EM) and other biochemical and biophysical techniques, we show that DnaJ induces a conformational change in the RepE dimer that remodels both domains of the protomers, which in turn increases its affinity for DNA. We also show that DnaJ displays a conformational plasticity that may be important to adapt to different substrate proteins.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
RepE C-terminal deletion mutants were amplified by standard PCR techniques and cloned into pHAT vector (18), which contains and N-terminal His tag. RepEΔ51–55 was produced by an overlap extension method and cloned in pHAT. All deletion mutants, as well as His-tagged WT RepE, were expressed and purified as described (10). DnaJ, DnaK, WT RepE, and RepE54 were purified following published protocols (19–21).
Fluorescence Techniques
Fluorescence anisotropy isotherms were obtained as published (10). Fluorescence spectra of RepE and its variants were recorded in a Fluorolog spectrofluorometer (Jobin Yvon) with excitation wavelength set at 295 nm to avoid absorption of tyrosines and phenylalanines. Excitation and emission slits were 4 and 5 nm, respectively. The emission maximum wavelength was obtained form the first derivative of the spectra. Data were fitted to a quadratic equation modeling a single binding site. To form the complex, RepE proteins (1 μm) and DnaJ (0–40 μm) were previously incubated for 1 h at 25 °C in 20 mm Hepes, pH 7.4, 50 mm KCl, 5 mm DTT, 0.1 mm EDTA. In the fluorescence quenching experiments, the emission was measured at 345 nm upon excitation at 295 nm, using 2-nm slit widths. Acrylamide was gradually added to 10 μm RepE in the absence or presence of 10 μm DnaJ, and dilution effects were corrected.
Pulldown Assays
To determine DnaJ-RepE complex stoichiometry, 20 μm His-tagged RepE and increasing concentrations of DnaJ (0–40 μm) were mixed and incubated for 90 min at 25 °C in 20 mm Hepes/KOH, pH 7.4, 50 mm KCl, 0.5 mm MgCl2, 50 mm imidazole. Ni-nitrilotriacetic acid beads (Qiagen) were added and incubated for 90 min at 25 °C with gentle shaking. Supernatants were removed, and the beads were washed with buffer. Pellets and supernatants were analyzed by 12% SDS-PAGE. The amounts of DnaJ and RepE were measured by densitometry of each lane using an in gel reference of total input protein. To identify the binding site, pulldown experiments with His-tagged versions of RepE1–152, RepE1–144, RepE1–139, and RepEΔ51–55 were performed as described (10).
Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra were recorded at 25 °C in a Jasco J-810 spectropolarimeter (Jasco, Tokyo, Japan) equipped with a thermoelectric cell holder, using a 0.1-mm path length cuvette. Proteins were dissolved in 20 mm potassium phosphate, pH 7.0, 50 mm KCl, 5 mm MgCl2, at 50 μm for WT RepE and RepEΔ51–55, and 100 μm for the C-terminal deletion mutants of RepE. Near-UV CD spectra were obtained using a 1-cm path length cuvette. All RepE variants proteins were at 30 μm in the above buffer. DnaJ complexes were performed by incubating RepE or the corresponding mutants with 30 μm DnaJ for 1 h at 25 °C.
EM and Image Processing
Samples (either DnaJ, DnaJ-RepE, DnaJ-RepE1–144, or DnaJ-RepE54 complexes) were applied onto carbon-coated copper grids and stained with 2% uranyl acetate. Micrographs were taken under minimal dose conditions in a JEOL JEM1200EXII microscope operated at 100 kV and digitized in a Zeiss SCAI scanner with a sampling window corresponding to 2.33 Å/pixel. Individual particles were manually selected using XMIPP (22). Image classification was performed using a free pattern maximum-likelihood multireference refinement (23). Homogeneous populations were obtained and averaged for a final two-dimensional characterization.
For three-dimensional reconstructions, several volumes were prepared as starting templates for angular refinement in EMAN (24). Starting models were generated by either common lines or by using artificial noisy models and Gaussian blobs with the rough dimension of the proteins. The different strategies converged to similar solutions (data not shown). The resolution of the reconstructions was determined by the FSC 0.5 criterion for the Fourier shell correlation coefficient between two independent reconstructions. The density maps and atomic structures were visualized with UCSF Chimera (25). The atomic structures were manually fitted into the three-dimensional reconstructions, and the handedness providing the best fit was chosen to render the three-dimensional reconstruction.
Trypsin Partial Proteolysis
Complexes of DnaJ with RepE, RepEΔ51–55, and RepE1–152 were subjected to controlled trypsin degradation as described (10).
Cross-linking Experiments
RepE or RepEΔ51–55 (10 μm) was incubated with DnaJ (0–15 μm) for 1 h at 25 °C in 20 mm Hepes/KOH, pH 7.4, 50 mm KCl, 0.5 mm MgCl2. Bis(sulfosuccinimidyl) suberate (Pierce, Thermo Scientific) was added at 20 μm final concentration, and samples were incubated for 30 min. Reactions were stopped by the addition of 50 mm Tris, pH 7.5, and samples were analyzed by SDS-PAGE.
Gel Retardation Assays
A 180-bp DNA probe containing the inverted repeat from mini-F plasmid oriT was obtained by PCR using plasmid pDAG114 as a template. 50 nm DNA was incubated with 90 nm RepE or RepEΔ51–55 in the absence or presence of DnaJ (0.5 or 1 μm) for 30 min at 30 °C in 20 mm Tris/HCl, pH 7.5, 40 mm KCl, 40 mm NaCl, 0.1 mm EDTA, 1 mm DTT. Samples were loaded in a 10% polyacrylamide gel and run for 6 h at 40 V. Gels were stained with SYBR-safe (Invitrogen, Molecular Probes) and photographed in a UV-transilluminator.
DnaK ATPase Activity
The steady-state ATPase activity of DnaK in the presence of RepE (0–15 μm) was measured as described (20). DnaJ was added at 0.1 μm.
RESULTS
DnaJ Binds to the N-terminal Domain of RepE with a 1:1 Stoichiometry
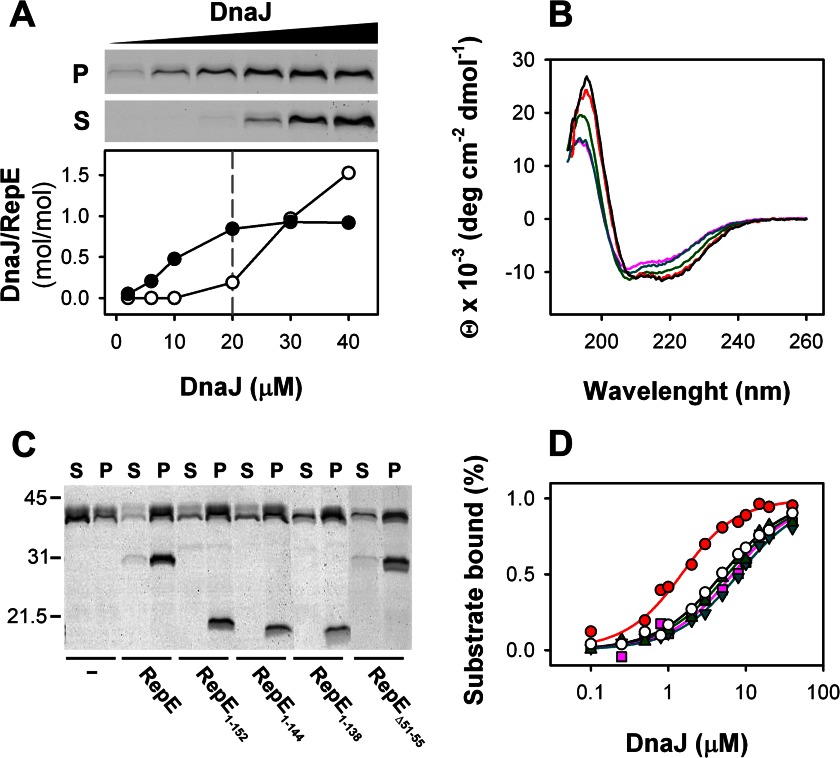
To obtain the stoichiometry of the DnaJ-RepE complex, a fixed amount of a His-tagged RepE was titrated with increasing DnaJ concentrations. The concentration of RepE (20 μm) was four times higher than the Kd value of 5 μm (10) to ensure an almost quantitative binding of substoichiometric amounts of DnaJ until saturation of the immobilized RepE. DnaJ-RepE complexes were preformed before adding nickel-nitrilotriacetic acid beads, and 50 mm imidazole was included in the buffer to minimize unspecific binding of DnaJ. It should be noted that approximately only 60% of total input RepE was recovered after washing the beads, very likely due to the high concentration of imidazole in the buffer. As shown in Fig. 1A, all DnaJ binds to RepE below 20 μm, whereas above this concentration DnaJ excess appears in the supernatant. The densitometry demonstrated that the stoichiometry of the DnaJ-RepE complex is 1:1, i.e. a DnaJ dimer binds one RepE dimer, as found for the homolog protein RepA (8).
FIGURE 1.
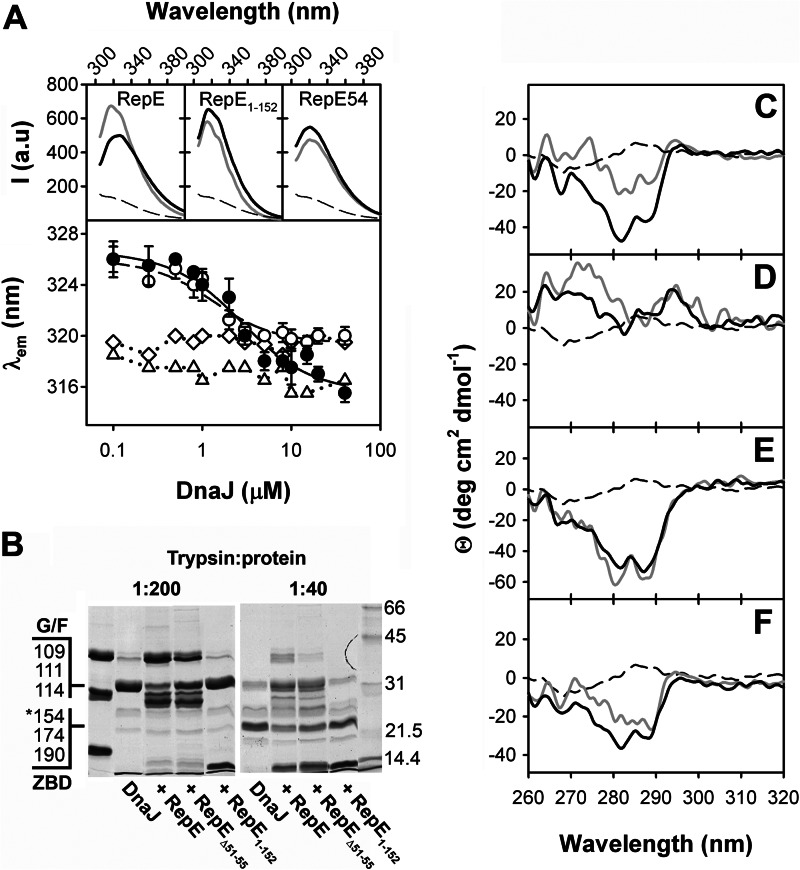
Stoichiometry of the DnaJ-RepE complex and mapping of RepE interaction domain. A, pulldown assays of increasing concentrations of DnaJ with N-His RepE. Upper panel, SDS-PAGE of DnaJ obtained in the pellet and supernatant, after washing and elution of nickel-nitrilotriacetic acid beads. Lower panel, ratio between DnaJ in pellets (filled circles) and supernatants (open circles) and RepE in pellets estimated by densitometry and plotted as a function of total DnaJ. B, far-UV CD spectra of RepE (black), RepE1–152 (green), RepE1–144 (magenta), RepE1–138 (cyan), and RepEΔ51–55 (red). C, pulldown of DnaJ with N-terminal His-tagged versions of RepE, the C-terminal domain deletion mutants, and RepEΔ51–55. Supernatants (S) and pellets (P) were analyzed separately in 12.5% SDS-PAGE. D, formation of complexes between DnaJ and WT RepE (open circles), RepE1–152 (green triangles), RepE1–144 (cyan inverted triangles), RepE1–138 (magenta squares), and RepEΔ51–55 (red circles) followed by fluorescence anisotropy measurements. Solid lines represent the best fits obtained with a single site binding model. Data points are the average of at least three independent experiments. Error bars are omitted for clarity.
To identify the domain where DnaJ binds, we constructed a RepE deletion mutant that lacks the C-terminal domain, RepE1–152. This mutant has a secondary structure compatible with a folded protein and similar to that of WT RepE, albeit with a slightly reduced helical content (Fig. 1B), as expected from the crystallographic structure of the protein. The structural integrity of RepE1–152 is also demonstrated by its ability to form dimers (26). Pulldown experiments (Fig. 1C) and fluorescence anisotropy binding isotherms (Fig. 1D and Table 1) show that RepE1–152 interacts with DnaJ with affinity similar to that of WT RepE. This result demonstrates that DnaJ binds to the N-terminal domain of RepE and that the contribution of the C-terminal domain to the overall complex stability is low. Regarding the binding site within the N-terminal domain, Wickner and colleagues identified helix α5 as the binding site for DnaJ in the homolog RepA (27). Besides this putative binding site, RepE might contain another DnaJ binding site in a disordered loop at the N-terminal domain (Lα2-β2) within residues 51–57 (G51TLQEHD57), according to the consensus binding sequence found for the yeast homolog Ydj1p (28). To test whether α5 and Lα2-β2 are involved in the formation of DnaJ-RepE complexes, we progressively eliminated helix α5 in two C-terminal deletion mutants, RepE1–144 and RepE1–139, and constructed a mutant in which Lα2-β2 was partially deleted, RepEΔ51–55. The far-UV CD spectra of RepE1–144 and RepE1–139 show a gradual reduction of α helical content due to deletion of α5, and the spectrum of RepEΔ51–55 is similar to that of the WT protein, demonstrating that the mutants are correctly folded (Fig. 1B). Our results show that these mutants interact with DnaJ (Fig. 1C), and whereas deletion of α5 does not modify the affinity of DnaJ for the protein, DnaJ binds RepEΔ51–55 with a slightly increased affinity (Fig. 1D and Table 1). Thus, the contribution of the predicted binding sites to complex formation is low. The discrepancy between RepA and RepE in the involvement of α5 in cochaperone binding might be due to the lower hydrophobicity of this structural element in RepE.
TABLE 1.
Dissociation constants and quenching Stern-Volmer constants for the complexes of DnaJ with WT RepE and the mutants used in this study
| Complex | Kda |
KSVb |
|
|---|---|---|---|
| −DnaJ | +DnaJ | ||
| μm | m−1 | m−1 | |
| RepE | 5.2 ± 0.8 | 6.8 | 2.3 |
| RepE1–152 | 4.6 ± 0.9 | 5.2 | 4.5 |
| RepE1–144 | 5.8 ± 0.7 | NDc | ND |
| RepE1–138 | 7.1 ± 0.9 | ND | ND |
| RepEΔ51–55 | 0.7 ± 0.1 | 7.2 | 5.8 |
| RepE54 | 7.7 ± 0.8 | 3.8 | 4.0 |
a Standard deviations are given.
b Quenching was linear within the acrylamide concentration range used, and KSV values were obtained the from the Stern-Volmer equation: F0/F = 1 + KSV[acrylamide].
c ND, not determined.
The Three-dimensional Reconstruction of DnaJ-RepE Complex
To generate a three-dimensional reconstruction by EM and image processing, the DnaJ-RepE complex was negatively stained, and 14,327 individual particles were selected, aligned, and classified into homogeneous classes as described under “Experimental Procedures.” The resolution of the model was estimated to be 19 Å (data not shown). The reconstructed complex has an elongated and symmetrical structure of 105 Å long and 85 Å diameter and consists of two masses running in parallel along the longitudinal axis and interacting at the central and bottom area (Fig. 2A, side view). One mass is compact and has a clear two-symmetrical fold (Fig. 2A, front view), whereas two rod-like masses, parallel to the longitudinal axis, are observed in the other (Fig. 2A, back view). Both masses show symmetrical features, in accordance with the described dimer:dimer stoichiometry.
We then resorted to the docking of the atomic structures of the two molecules (Fig. 2B) into the reconstructed volume. Despite the diversity of the Hsp40 protein family, the structural alignment of the ZBD and C-terminal domains of E. coli DnaJ, yeast Ydj1p and Sis1p, and human Hdj1 reveals a high degree of structural homology (data not shown). Thus, the atomic coordinates of the C-terminal domain of Ydj1 (PDB code 1NLT), the closest homolog of DnaJ, were used for the docking. The characteristic L-shape of the C-terminal domains and the bell-shaped structure adopted by Ydj1 dimers (29) enabled us to dock each monomer in the rod-like masses observed in the back view of the model (Fig. 2C and supplemental Movie S1). Fitting of domains CTDI and CTDII into these masses was very good, placing the CTDIIs in close proximity and leaving space at the top of the volume to dock the C-terminal dimerization domain (PDB code 1XAO), missing in the Ydj1 x-ray structure. The ZBDs could also be fitted in the bottom of the volume, but it required a rotation of these domains from their orientation in the Ydj1 modeled dimer (29). Next, the atomic structure of the RepE dimer bound to the operator DNA (PDB code 2Z9O) (17) was fitted to the assigned compact, symmetrical density (Fig. 2C, front view). Although the general shape of dimeric RepE fitted very well in this mass, the ends of the concave structure of DNA-bound RepE did not dock into the DnaJ-RepE three-dimensional envelope (Fig. 2C, side view). The quality of the docking was substantially improved when the RepE dimer structure was flattened by a ∼25° tilt of the RepE C-terminal domains (Fig. 2D). Thus, docking of RepE into the DnaJ-RepE complex suggests that binding to DnaJ induces a conformational change in RepE, particularly in its C-terminal domain.
Finally, we performed the docking of the two J-domains. It should be mentioned that the localization of these domains might be ambiguous due to the flexibility and the lack of a defined structure for the connecting G/F-domain. In the DnaJ-RepE complex, the volume obtained has enough space to place the J-domains in an area located between the DnaJ and RepE molecules (Fig. 2C, front and side views). Supporting previous results that discarded the involvement of the J-domain in the interaction with RepE (10), these domains barely contact the client in the two complexes. The fitting also leaves space in the proximity to the RepE dimer where the G/F-domain could be accommodated, in agreement with the requirement of this domain to interact with RepE (10).
Three-dimensional Reconstruction of the DnaJ-RepE1–144 Complex
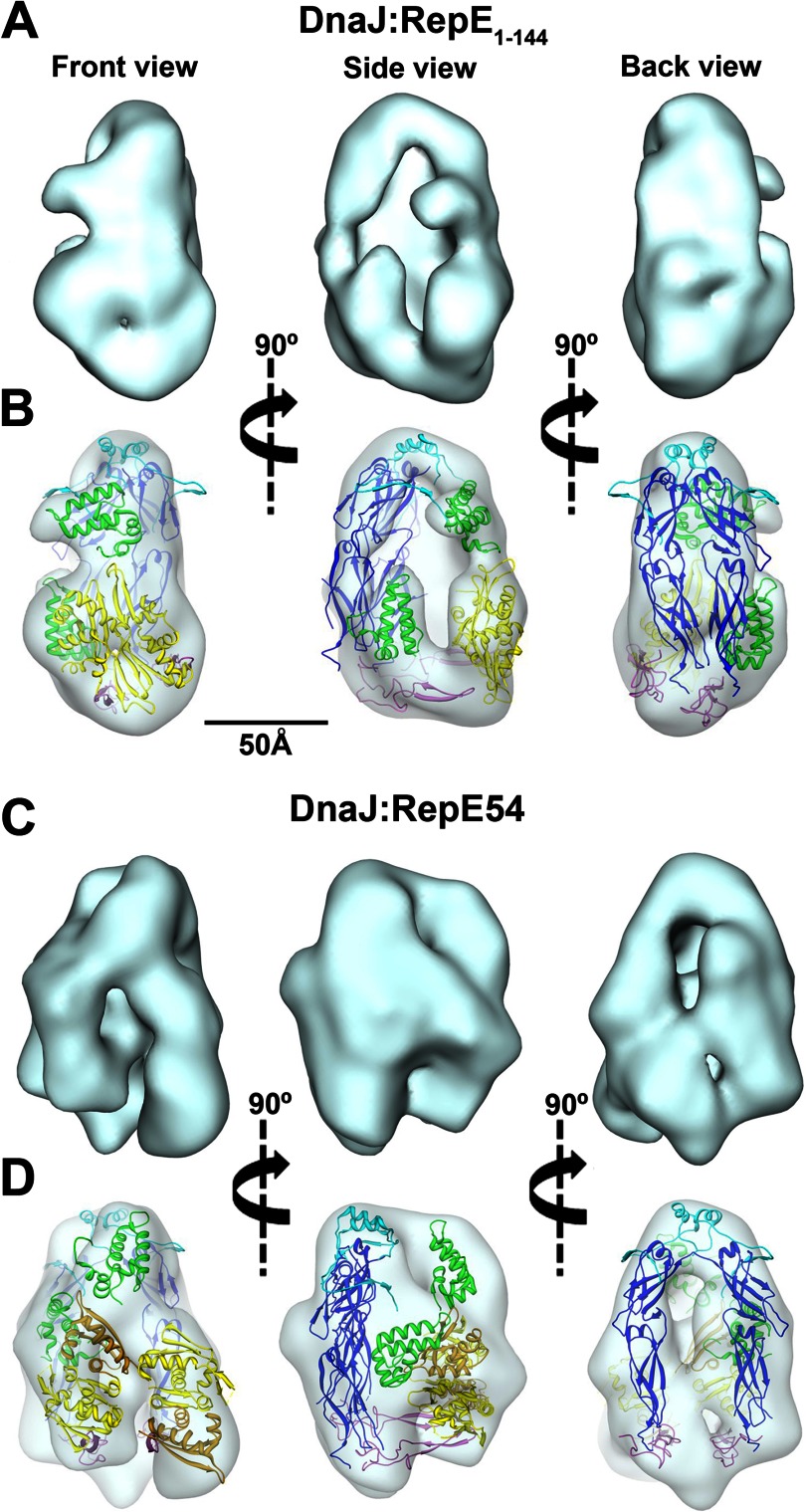
To confirm the localization of DnaJ and RepE in the three-dimensional volume, we performed a similar structural analysis with the deletion mutant RepE1–144 that lacks the C-terminal domain but maintains the dimeric structure. We selected 9,623 individual particles from negative stained micrographs and classified them into homogeneous classes to generate a final model with an estimated resolution of 20 Å (Fig. 3A). As in the complex with WT RepE, the structure of the DnaJ-RepE1–144 complex is elongated and has the same length (105 Å) but a shorter diameter (75 Å). The overall shape of the structure is less symmetrical, with a large mass running parallel to the longitudinal axis (Fig. 3A, back view) and a smaller, round mass, placed asymmetrically at the bottom half of the complex (Fig. 3A, front view). The three-dimensional reconstruction reveals two small lobules not observed before (Fig. 3A, side view), protruding from the longitudinal mass, and a large cavity in the center of the density. The smaller round mass observed in the front view, as well as the larger central cavity, can account for the lack of the C-terminal domain in RepE1–144. Thus, by comparison of the two reconstructions, we could assign the 2-fold symmetrical compact mass and the rod-like masses observed to RepE and DnaJ, respectively.
FIGURE 3.
Architecture of DnaJ-RepE1–144 and DnaJ-RepE54 complexes. A, three-dimensional reconstruction of the DnaJ-RepE1–144 complex. B, docking of the different domains of DnaJ and RepE1–144 into the three-dimensional reconstruction of the DnaJ-RepE1–144 complex. C, three-dimensional reconstruction of the DnaJ-RepE54 complex. D, docking of the different domains of DnaJ and RepE54 into the three-dimensional reconstruction of the DnaJ-RepE54 complex. The views shown are the same as in the DnaJ-RepE complex. Domain color coding is the same as in Fig. 2. Docking of the atomic structures can also be viewed in supplemental Movies S2 and S3.
We also carried out a docking of the atomic structures of the two molecules (DnaJ and RepE1–144) into the reconstructed volume. For that, we used the same strategy as for WT RepE, obtaining similar results (Fig. 3B and supplemental Movie S2). The J-domains can be easily located in two protrusions located in the upper and bottom part of the DnaJ-RepE1–144 complex (Fig. 3B, front and side views). Docking of domains CTDI and CTDII into the longitudinal mass in the three-dimensional volume obtained for RepE1–144 was very accurate (Fig. 3B, back view), but, interestingly, the aperture of the bell-shaped DnaJ dimer had to be reduced. The bottom of the envelope could fit the ZBDs as in the complex with WT RepE. The atomic structure of dimeric RepE encompassing residues 1–144 could also be accurately fitted to the round mass observed in the front view, although with a ∼15° rotation of the mass compared with that of WT RepE (compare front views in Figs. 2C and 3B), suggesting that DnaJ binds differently the smaller substrate. In contrast to the complex with WT RepE, a conformational rearrangement of the crystallographic structure was not required to dock RepE1–144 into the corresponding mass of the reconstructed volume (Fig. 3B, side view), which confirms the good docking of the RepE N-terminal domain into the corresponding mass of the DnaJ-RepE complex.
DnaJ Can Bind Two Monomers of RepE Separately
To further characterize the structural plasticity of the DnaJ-substrate complexes, we used the mutant RepE54, which carries the mutation R118P and is a monomer in solution (16). The three-dimensional reconstruction of this complex (Fig. 3C) reveals a structure similar to those previously shown, although less symmetrical, with two large masses running in parallel to the longitudinal axis (Fig. 3C, back view), and two other smaller masses with the same orientation, albeit slightly tilted in the other side of the molecule (Fig. 3C, front view). By comparison with the other two complexes, the symmetrical masses can be assigned to DnaJ and the two separated, tilted masses to the RepE54 monomers. The good docking of the atomic structure of RepE54 (PDB code 1REP) (30) into these regions shows that the two monomers bind to the two DnaJ monomers separately (Fig. 3D and supplemental Movie S3), although maintaining a similar orientation than the two WT RepE protomers bound to DnaJ (Fig. 2C, front view). This result confirms the structural plasticity in the interaction between DnaJ and its substrates, in particular RepE.
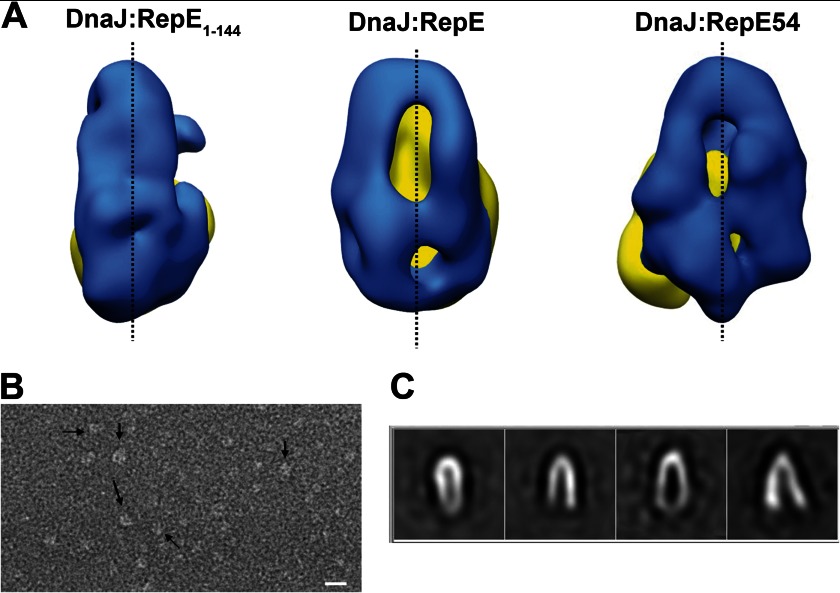
The Inherent Flexibility of DnaJ
One of the most remarkable differences between the three-dimensional reconstructions shown here is the arrangement of the DnaJ monomers, being close to each other in the case of the deletion mutant, separated in the case of the wild-type and more open in the case of the monomeric mutant (Fig. 4A). This speaks of an intrinsic flexibility of DnaJ dimers that might serve to accommodate different substrates. To explore the hypothesis, we performed an EM analysis of substrate-free DnaJ. 5,225 individual particles were selected and classified into homogeneous classes. All classes had a bell shape, in good agreement with the modeled Ydj1 dimer, but showed different conformational states represented by distinct classes with a variable degree of aperture of its two arms, ranging from 7° to 28° (Fig. 4C). The N terminus of the protein, comprising the J-domain, was not observed in our EM study very likely due to its small size and the variability imposed by the intrinsically unstructured G/F-rich domain. It is noticeable that the conformations of the DnaJ dimer observed in the three-dimensional reconstructions of DnaJ complexed with RepE, RepE1–144, and RepE54 resembled the conformation in some of the classes of substrate-free DnaJ. These results suggest that the DnaJ dimer is intrinsically flexible and may adapt its conformation to the size of the client protein.
FIGURE 4.
Flexibility of DnaJ dimer. A, back views of the three-dimensional reconstructions of DnaJ-RepE1–144 (left), DnaJ-RepE (center), and DnaJ-RepE54 (right) showing the different degrees of aperture of the DnaJ monomers. Blue color indicates the DnaJ monomers, and yellow the corresponding RepE molecules. B, electron microscopy field of substrate-free DnaJ dimers. C, reference-free two-dimensional averages obtained from the data set containing images of DnaJ dimers. The images reveal, albeit with a different degree of aperture, the bell shape of dimeric DnaJ.
DnaJ Remodels the Conformation of the Two Domains of RepE
We used fluorescence spectroscopy and near-UV CD to characterize the conformation of RepE when complexed to DnaJ. Both spectroscopic techniques may monitor specifically the conformation of RepE in the complex because DnaJ lacks Trp residues, whereas RepE has two (Trp114 and Trp187, in the N- and C-terminal domains, respectively). Binding to DnaJ promotes a blue shift of RepE fluorescence emission maximum (λem) from 326 to 316 nm (Fig. 5A), compatible with a reduction of the polarity surrounding the Trp residues. A similar effect was also observed for RepEΔ51–55, albeit the λem of the DnaJ-bound conformation of the mutant is centered at 320 nm. The experimental data could be fitted to a single-site binding model giving Kd values of 2.3 and 0.8 μm for RepE and RepEΔ51–55, respectively, similar to those found by anisotropy measurements. In contrast, RepE1–152 has a λem at 317 nm, corresponding to a highly hydrophobic environment, which was not significantly altered by DnaJ. Furthermore, the DnaJ-induced change in the λem is specific for dimeric RepE because it was not detected for the monomeric mutant RepE54, which has a λem at 320 nm regardless of the presence of DnaJ.
FIGURE 5.
DnaJ binding induces a conformational change in the C-terminal domain of RepE. A, upper panels, fluorescence emission spectra of RepE, RepE1–152, and RepE54 (1 μm) in the absence (black line) or the presence (gray line) of 10 μm DnaJ. The thin broken line represents the spectrum of 10 μm DnaJ. A, lower panel, emission maximum of RepE (filled circles), RepEΔ51–55 (open circles), RepE1–152 (triangles), and RepE54 (diamonds) plotted as a function of DnaJ concentration. Data were fitted to a quadratic equation modeling a single binding site (solid and broken line for WT and RepEΔ51–55, respectively). B, partial proteolysis of DnaJ (20 μm) complexed with WT RepE, RepE1–152, and RepEΔ51–55 (20 μm). The complexes were digested at a 1:200 trypsin:protein ratio (w/w) for 30 s and 1:40 ratio for 5 min. The first line contains the undigested proteins as reference. The main tryptic sites in the G/F-rich domain and ZBD identified by mass spectrometry (10) that give rise to C-terminal DnaJ fragments of molecular mass higher than 20 kDa are indicated. The fragment marked with an asterisk is produced by double digestion in the ZBD and C-terminal dimerization domain of DnaJ. C–F, near-UV CD spectra of RepE (C), RepE1–152 (D), RepE54 (E), and RepEΔ51–55 (F) free in solution (black lines) and complexed to DnaJ (gray lines). DnaJ spectrum is shown as a black broken line.
Interaction with DnaJ also promotes a reduction of RepE Trp solvent accessibility, reflected by a decrease of the KSV from 6.8 to 2.3 m−1 in fluorescence quenching experiments (Table 1). However, no change in the accessibility of RepE1–152 and RepE54 was observed in the presence of DnaJ, their KSV values being intermediate between those of free WT RepE and its complex with DnaJ. Interestingly, DnaJ only slightly reduced the KSV of RepEΔ51–55, suggesting that the mutant adopts a different conformation than RepE when complexed with the chaperone. The changes in fluorescence properties suggest that DnaJ induces a structural modification of RepE C-terminal domain and that Lα2-β2 in the N-terminal domain is necessary to fully promote the conformational change.
We performed partial proteolysis experiments (Fig. 5B) to study whether RepE1–152 and RepEΔ51–55 were able to protect the G/F-rich domain and ZBD of DnaJ from protease attack as WT RepE (10). It should be noted that the proteolytic pattern of RepE proteins is not modified by the interaction with DnaJ, as published (10). In contrast to WT RepE, RepE1–152 is unable to prevent proteolysis in these DnaJ domains. This might be caused by a lower steric hindrance due to the lack of RepE C-terminal domain. RepEΔ51–55 protects the G/F-rich domain and ZBD of DnaJ, albeit to a lower extent than WT RepE. This finding supports that Lα2-β2 could modulate a rearrangement of RepE C-terminal domain in the complex.
To further prove the conformational change in RepE complexed to DnaJ, we characterized the protein structure of the client protein by near-UV CD spectroscopy (Fig. 5, C–F). The CD spectrum of a protein in this spectral region mainly reflects the environment of aromatic amino acid side chains, its shape and intensity being modulated, among other factors, by the rigidity of the protein. Therefore changes in the CD spectrum of a protein are usually interpreted as modifications of its tertiary structure. RepE displays a negative signal with two minima centered at 283 and 290 nm, which arise from the absorption of tryptophan residues, whereas DnaJ does not show any significant absorption in the same spectral region (Fig. 5C). The spectra of isolated RepE54 and RepEΔ51–55 are similar to that of the WT protein, whereas RepE1–152 shows a weak absorption lacking the fine structure characteristic of WT RepE (Fig. 5, D–F). To estimate the spectral features of these RepE variants bound to DnaJ, the weak contribution of DnaJ was subtracted from the spectrum of the complex, assuming that the conformational changes would not significantly modify the chaperone spectrum in this spectral region. This assumption is reasonable because DnaJ does not contain Trp residues. The comparison of the spectra of DnaJ-bound states of RepE and the different variants with those of the free proteins reveals that (i) the DnaJ-induced change in WT RepE arises mainly from its C-terminal domain (Fig. 5, C and D); (ii) the spectrum of monomeric Rep54 does not change in the presence of DnaJ (Fig. 5E); and (iii) the intensity of the spectrum of RepEΔ51–55 complexed to DnaJ is reduced to a lesser extent than that of WT RepE (Fig. 5, C and F). Taken together, these experiments demonstrate that DnaJ modifies the conformation of RepE C-terminal domain, as suggested by the three-dimensional reconstruction of the DnaJ-RepE complex, and that the interaction with Lα2-β2 is essential to fully transmit this conformational change to the substrate protein.
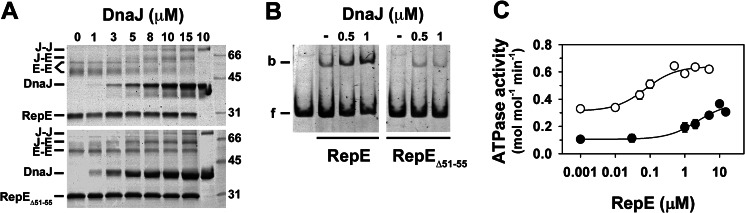
Then we studied how DnaJ affects the stability and biochemical function of RepE. Cross-linking of isolated RepE with bis(sulfosuccinimidyl) suberate produces two bands of apparent molecular size compatible with a dimer (Fig. 6A, upper panel). The different mobility of these adducts could be due to a variable number of cross-linked pairs between the monomers. In the presence of increasing amounts of DnaJ, formation of dimeric RepE adducts is progressively reduced and abolished at the highest DnaJ concentration, with the concomitant appearance of two new bands corresponding to the DnaJ-RepE complex and the DnaJ dimers. Similar results were obtained for the C-terminal deletion mutant RepE1–152 (data not shown). In contrast, the dimer adducts are still formed in the complex of RepEΔ51–55 with DnaJ (Fig. 6A, lower panel), thus discarding an impaired accessibility of bis(sulfosuccinimidyl) suberate to specific lysine residues at the dimerization interface. These results demonstrate that DnaJ increases the intermonomeric distance of RepE dimers, in an Lα2-β2-dependent manner.
FIGURE 6.
Functional consequences of the DnaJ-induced rearrangement of RepE. A, cross-linking of RepE (upper panel) or RepEΔ51–55 (lower panel) at increasing DnaJ concentrations. The last lane represents the cross-linking of DnaJ in the absence of substrate. B, binding of RepE (left) and RepEΔ51–55 (right) to DNA measured by gel retardation in the absence or presence of DnaJ. C, ATPase activity of DnaK at increasing RepE concentrations in the absence (filled circles) or presence (open circles) of DnaJ.
Next we addressed whether DnaJ modified the DNA binding properties of RepE. In agreement with published observations (9), saturating concentrations of DnaJ increase binding of RepE to a DNA probe containing the operator sequence of mini-F plasmid (Fig. 6B). However, the chaperone does not enhance to the same extent DNA binding of RepEΔ51–55, emphasizing the importance of Lα2-β2 as a regulatory element of DnaJ-induced conformational changes in RepE. Finally, DnaK also interacts with RepE and is essential to regulate its function (31, 32), thus we explored the effect of DnaJ in this interaction. As a bona fide substrate, RepE stimulates 3–4-fold the ATPase activity of DnaK, with a K0.5 of 2.5 μm (Fig. 6C). In the presence of DnaJ and the substrate, DnaK ATPase activity is synergistically stimulated and the K0.5 for RepE lowered to 70 nm. These results suggest that the DnaJ-induced conformational changes in RepE increase the affinity of DnaK for this client protein.
DISCUSSION
In this work, we have characterized the complex between DnaJ and RepE, the conformational changes that both proteins undergo upon complex formation, and the functional consequences of this interaction. The three-dimensional reconstruction of the DnaJ-RepE complex represents the first structure of an Hsp40 in its dimeric state complexed with a client protein. The model shows that DnaJ binds a RepE dimer through a large interaction surface in which the two DnaJ monomers are involved in substrate binding, in agreement with the inability of dimerization domain deletion mutants of DnaJ and Sis1 to bind and chaperone substrates (13, 33). The comparison of the complexes with WT RepE, a C-terminal deletion (RepE1–144), and a monomeric mutant (RepE54) speaks of a large structural plasticity of DnaJ that allows adaptation to different substrates. Interaction of DnaJ with RepE1–144 forces a closure of the bell-shaped DnaJ dimer, whereas binding to two separate monomeric RepE molecules induces a separation of the DnaJ monomers. In fact, the EM study of substrate-free DnaJ reveals an intrinsic variety of conformations of the DnaJ dimer with different intermonomeric angles. This suggests that the dimerization domain allows the rearrangement of the monomers to adjust the binding interface to substrates of different size.
Our data demonstrate that the interaction with DnaJ induces conformational changes that affect both domains of RepE. The DnaJ-bound conformation of WT RepE is characterized by (i) an increased intermonomeric distance, (ii) a reduced polarity and accessibility to polar quenchers of Trp187 located in the C-terminal domain, and (iii) a less rigid tertiary structure of the C-terminal domain that results in the reduction of its near-UV CD signal. This DnaJ-induced conformational changes lead to the “activation” of RepE that is reflected in an increased affinity for specific DNA probes. Analysis of the RepE deletion mutants demonstrates that the N-terminal domain is sufficient to form a stable complex, confirmed by the good docking of this domain in the reconstruction of the complex between DnaJ and RepE1–144. According to the three-dimensional reconstruction of the DnaJ-RepE complex and the conformational changes described above, the chaperone promotes a rearrangement at the level of the C-terminal domain of RepE distant from the binding site, as has been shown for other DnaJ substrates (7, 34). We demonstrate that the disordered loop encompassing residues 47–57 (Lα2-β2) at the N-terminal domain of RepE is essential to promote the conformational activation of the substrate. Partial deletion of this structural element hampers the full conformational rearrangement that DnaJ induces in RepE, although it does not significantly affect the stability of the complex. It is likely that DnaJ interacts with Lα2-β2 because this structural feature is in close proximity to DnaJ CTDI in the model obtained upon docking the crystallographic structures of the proteins into the three-dimensional envelope of the complex. The large interaction interface between DnaJ and RepE suggests the existence of interaction sites different from this loop, as it could be α5, within the N-terminal domain of the substrate protein, which would explain the null effect of its deletion in the affinity for the cochaperone.
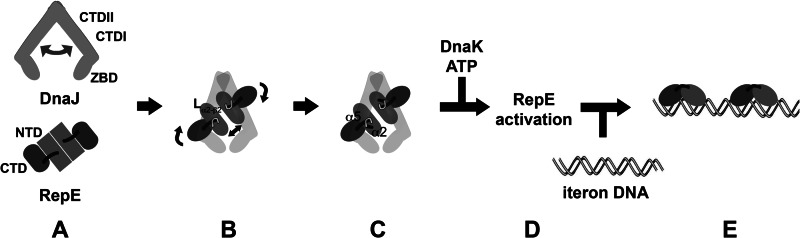
We propose a model in which DnaJ interacts mainly with the N-terminal domain of RepE (Fig. 7). In particular, the interaction with Lα2-β2 seems to be essential to modulate the conformation of RepE. Lα2-β2 connects strand β2 at the dimerization interface with helix α2, which establishes hydrophobic contacts with structural elements of the N-terminal domain such as the linker helix α5. Thus, the interaction with Lα2-β2 might both alter the intermonomeric distance and propagate the conformational change to the C-terminal domain of RepE through the linker, enhancing its affinity for DNA. This structural transition could also trigger the exposure of a predicted DnaK binding site in α2 (17), increasing its affinity for RepE and allowing the subsequent action of the chaperone to fully activate the client protein.
FIGURE 7.
Model for the interaction of DnaJ with RepE. A, the chaperone DnaJ functions as an adjustable dimer able to bind folded protein clients as RepE. B and C, DnaJ binds to the N-terminal, dimerization domain of RepE forming a stable complex. Complex formation induces conformational changes in RepE that affect the N- and C-terminal domains, forcing the flattening of the concave DNA-bound structure, and increases the intermonomeric distance. These structural rearrangements depend on the interaction of DnaJ with Lα2-β2 of RepE. D and E, the subsequent action of DnaK leads to full activation of RepE (35), promoting binding of RepE monomers to the iteron.
In summary, the results presented here put forward the ability of DnaJ to adjust its conformation to build a large binding interface made by both monomers and to adapt to different client proteins. In the absence of ATPase activity, DnaJ might rely on this large and variable binding interface to exert an independent chaperone activity on folded substrates as found here for RepE. Recently, different modes of substrate binding have also been found for DnaK (32), which indicates the conformational plasticity of this chaperone system.
Supplementary Material
Acknowledgment
We thank Dr. Jean-Yves Bouet for plasmid pDAG114.
This work was supported in part by Ministerio de Educación e Innovación Grants BFU2010-15443 (to A. M.) and BFU2010-15703 (to J. M. V.), University of Basque Country and Gobierno Vasco Grant IT-358-07 (to A. M.), and Comunidad Autónoma de Madrid Grant S2009MAT-1507 (to J. M. V.).

This article contains supplemental Movies S1–S3.
The three-dimensional reconstructions of the different complexes have been deposited in the EMDataBank with codes emd-2332 (DnaJ-RepE), emd-2333 (DnaJ-RepE1–144), and emd-2334 (DnaJ-RepE54).
- ZBD
- zinc-binding domain
- CTD
- C-terminal domain
- Kd
- dissociation constant
- KSV
- Stern-Volmer constant
- NTD
- N-terminal domain.
REFERENCES
- 1. Genevaux P., Georgopoulos C., Kelley W. L. (2007) The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 66, 840–857 [DOI] [PubMed] [Google Scholar]
- 2. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 3. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahmad A., Bhattacharya A., McDonald R. A., Cordes M., Ellington B., Bertelsen E. B., Zuiderweg E. R. (2011) Heat shock protein 70-kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. U.S.A. 108, 18966–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu Z., Cyr D. M. (1998) The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273, 5970–5978 [DOI] [PubMed] [Google Scholar]
- 6. Szabo A., Korszun R., Hartl F. U., Flanagan J. (1996) A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 15, 408–417 [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez F., Arsène-Ploetze F., Rist W., Rüdiger S., Schneider-Mergener J., Mayer M. P., Bukau B. (2008) Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol. Cell 32, 347–358 [DOI] [PubMed] [Google Scholar]
- 8. Wickner S. H. (1990) Three Escherichia coli heat shock proteins are required for P1 plasmid DNA replication: formation of an active complex between E. coli DnaJ protein and the P1 initiator protein. Proc. Natl. Acad. Sci. U.S.A. 87, 2690–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawasaki Y., Wada C., Yura T. (1992) Binding of RepE initiator protein to mini-F DNA origin (ori2): enhancing effects of RepE mutations and DnaJ heat shock protein. J. Biol. Chem. 267, 11520–11524 [PubMed] [Google Scholar]
- 10. Perales-Calvo J., Muga A., Moro F. (2010) Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. J. Biol. Chem. 285, 34231–34239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banecki B., Liberek K., Wall D., Wawrzynów A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. (1996) Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 271, 14840–14848 [DOI] [PubMed] [Google Scholar]
- 12. Li J., Qian X., Sha B. (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 13. Sha B., Lee S., Cyr D. M. (2000) The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure 8, 799–807 [DOI] [PubMed] [Google Scholar]
- 14. Li J., Sha B. (2005) Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem. J. 386, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S., Fan C. Y., Younger J. M., Ren H., Cyr D. M. (2002) Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J. Biol. Chem. 277, 21675–21682 [DOI] [PubMed] [Google Scholar]
- 16. Ishiai M., Wada C., Kawasaki Y., Yura T. (1994) Replication initiator protein RepE of mini-F plasmid: functional differentiation between monomers (initiator) and dimers (autogenous repressor). Proc. Natl. Acad. Sci. U.S.A. 91, 3839–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura A., Wada C., Miki K. (2007) Structural basis for regulation of bifunctional roles in replication initiator protein. Proc. Natl. Acad. Sci. U.S.A. 104, 18484–18489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]
- 19. Zylicz M., Yamamoto T., McKittrick N., Sell S., Georgopoulos C. (1985) Purification and properties of the DnaJ replication protein of Escherichia coli. J. Biol. Chem. 260, 7591–7598 [PubMed] [Google Scholar]
- 20. Moro F., Fernández V., Muga A. (2003) Interdomain interaction through helices A and B of DnaK peptide binding domain. FEBS Lett. 533, 119–123 [DOI] [PubMed] [Google Scholar]
- 21. Zzaman S., Abhyankar M. M., Bastia D. (2004) Reconstitution of F factor DNA replication in vitro with purified proteins. J. Biol. Chem. 279, 17404–17410 [DOI] [PubMed] [Google Scholar]
- 22. Sorzano C. O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J. R., Scheres S. H., Carazo J. M., Pascual-Montano A. (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 [DOI] [PubMed] [Google Scholar]
- 23. Scheres S. H., Valle M., Nuñez R., Sorzano C. O., Marabini R., Herman G. T., Carazo J. M. (2005) Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348, 139–149 [DOI] [PubMed] [Google Scholar]
- 24. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 25. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 26. Nakamura A., Komori H., Kobayashi G., Kita A., Wada C., Miki K. (2004) The N-terminal domain of the replication initiator protein RepE is a dimerization domain forming a stable dimer. Biochem. Biophys. Res. Commun. 315, 10–15 [DOI] [PubMed] [Google Scholar]
- 27. Kim S. Y., Sharma S., Hoskins J. R., Wickner S. (2002) Interaction of the DnaK and DnaJ chaperone system with a native substrate, P1 RepA. J. Biol. Chem. 277, 44778–44783 [DOI] [PubMed] [Google Scholar]
- 28. Kota P., Summers D. W., Ren H. Y., Cyr D. M., Dokholyan N. V. (2009) Identification of a consensus motif in substrates bound by a type I Hsp40. Proc. Natl. Acad. Sci. U.S.A. 106, 11073–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y., Li J., Jin Z., Fu Z., Sha B. (2005) The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J. Mol. Biol. 346, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 30. Komori H., Matsunaga F., Higuchi Y., Ishiai M., Wada C., Miki K. (1999) Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J. 18, 4597–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawasaki Y., Wada C., Yura T. (1990) Roles of Escherichia coli heat shock proteins DnaK, DnaJ, and GrpE in mini-F plasmid replication. Mol. Gen. Genet. 220, 277–282 [DOI] [PubMed] [Google Scholar]
- 32. Schlecht R., Erbse A. H., Bukau B., Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
- 33. Shi Y. Y., Hong X. G., Wang C. C. (2005) The C-terminal(331–376) sequence of Escherichia coli DnaJ is essential for dimerization and chaperone activity: a small angle x-ray scattering study in solution. J. Biol. Chem. 280, 22761–22768 [DOI] [PubMed] [Google Scholar]
- 34. Puvirajesinghe T. M., Elantak L., Lignon S., Franche N., Ilbert M., Ansaldi M. (2012) DnaJ (Hsp40 protein) binding to folded substrate impacts KplE1 prophage excision efficiency. J. Biol. Chem. 287, 14169–14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zzaman S., Bastia D. (2005) Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol. Cell 20, 833–843 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.