Background: The spatiotemporal dynamic behavior of Oct4 remains largely unknown.
Results: Oct4 is a nucleocytoplasmic shuttling protein, and Oct4 mutants with biased nucleocytoplasmic localization show limited potential for cellular reprogramming.
Conclusion: An appropriate nuclear retention of Oct4 is critical for cellular reprogramming but not for the self-renewal of ES cells.
Significance: Our findings will provide novel insight into the role of Oct4 during cellular reprogramming.
Keywords: Differentiation, Embryonic Stem Cell, Nuclear Transport, Reprogramming, Transcription Factors, Oct4, Nucleocytoplasmic Shuttling
Abstract
Oct4 is a member of the POU family of transcription factors and plays a critical role in both maintenance of the undifferentiated state of embryonic stem (ES) cells and in the reprogramming of somatic cells to induced pluripotent stem cells. Oct4 is imported into the nucleus where it functions as a transcription factor; however, the spatiotemporal dynamic behavior of Oct4 remains largely unknown. In the present study we show that Oct4 is a nucleocytoplasmic shuttling protein. Furthermore, although Oct4 mutants with altered nuclear import/export activity were able to maintain the self-renewal of ES cells, they displayed limited potential for cellular reprogramming. These results indicate that the intracellular localization of Oct4, which is dependent on nucleocytoplasmic shuttling, must be more strictly regulated for cellular reprogramming, suggesting that Oct4 plays differential roles in the self-renewal of ES cells and in somatic cell reprogramming.
Introduction
Oct4 is a homeodomain transcription factor that belongs to the POU (Pit-Oct-Unc) family (1–3) and is exclusively expressed in totipotent/pluripotent cell lineages, including oocytes, the inner cell mass, primitive ectoderm, primordial germ cells, and stem cell lines derived from the early embryo (4). It has been proposed that Oct4 regulates stem cell pluripotency and differentiation. Indeed, Oct4 is essential for various cellular processes associated with pluripotency, such as formation of the inner cell mass (5), the maintenance of embryonic stem (ES) cells (6), and induction of induced pluripotent cells (7, 8). In ES cells, Oct4 collaborates with Sox2 and Nanog to form a regulatory circuit that maintains ES-cell pluripotency (9, 10). Furthermore, several genomic profiling studies have revealed that Oct4 also maintains the pluripotency of ES cells by regulating genome-wide transcription either positively or negatively (11–14).
The expression level of Oct4 needs to be precisely controlled to sustain undifferentiated proliferation of ES cells (6). Indeed, a decrease in Oct4 expression to <50% of normal levels triggers cell differentiation toward the trophectoderm lineage, whereas a 50% increase in expression promotes cell differentiation into mesoderm or endoderm (6, 15). Although numerous studies have shown that the expression level of Oct4 is transcriptionally regulated, either positively or negatively, by various factors (16), the post-translational regulation of Oct4 is not well characterized. However, there is evidence that Oct4 is modified by ubiquitination (17), sumoylation (18), and phosphorylation (19–21).
The post-translational status of Oct4 is also regulated by its intracellular localization. After translation in the cytoplasm, Oct4 is imported into the nucleus via importin α, which binds to a conserved nuclear localization signal (NLS:RKRKR)3 at the N terminus of the homeobox domain (22–25). However, the dynamic behavior of Oct4 once imported into the nucleus is essentially unknown. In contrast, Oct6, another POU homeodomain transcription factor, is known to shuttle between the nucleus and cytoplasm (26). Thus, in the present study we analyzed the dynamic behavior of Oct4 and its role in the determination of cell fate. We demonstrated that Oct4 is a nucleocytoplasmic shuttling protein. Furthermore, we used Oct4 mutants with biased nucleocytoplasmic localization to show that the appropriate nuclear retention of Oct4 is critical for cellular reprogramming but not for the self-renewal of ES cells.
EXPERIMENTAL PROCEDURES
Cell Culture
NIH3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS). ZHBTc4 ES cells (6) were cultured on gelatin-coated dishes in DMEM supplemented with 10% FBS, 1 mm sodium pyruvate, 0.1 mm 2-mercaptoethanol, 0.1 mm nonessential amino acids, and 1000 units/ml murine leukemia inhibitory factor.
Plasmids
A retroviral vector expressing mutant Oct4 was generated by modifying the pMXs-Oct4 vector (constructed by the laboratory of Dr. Yamanaka; obtained from Addgene). First, the sequence at the C-terminal extremity of the Oct4 ORF was deleted, which resulted in the inclusion of a multiple cloning site containing the XhoI site at the C terminus of Oct4 (pMXs-Oct4*) and an additional 12 amino acids (LERPPAQWSTIK), with deletion of the final asparagine residue of endogenous Oct4. We subsequently inserted oligonucleotides coding for the NLS of the SV40 T-antigen (5′-TCGACAGTACTCCTCCAAAAAAGAAGAGAAAGGTAGAAGACC-3′, 5′-TCGAGGTCTTCTACCTTTCTCTTCTTTTTTGGAGGAGTACTG-3′; DSTPPKKKRKVEDL) or the nuclear export signal (NES) of HIV-1 Rev protein (5′-TCGACCTTCAGCTACCACCGCTTGAGAGACTTACTCTTGATTGTAACC-3′, 5′-TCGAGGTTACAATCAAGAGTAAGTCTCTCAAGCGGTGGTAGCTGAAGG-3′; DLQLPPLERLTLDCNL) into the XhoI site of pMXs-Oct4*. The Oct4-NES-NES mutant was also cloned by using the same oligonucleotides. The pMXs vector for the M10 NES mutant and Oct4 tandem alanine mutant (AAA) was generated by PCR-based mutagenesis. The Oct4-EGFP expression retrovirus (pMXs-Oct4-EGFP) was created by inserting the EGFP coding sequence into the XhoI site of pMXs-Oct4*, and the Oct4 (L260A,L262A)-EGFP mutant was generated by PCR-based mutagenesis using pMXs-Oct4-EGFP as a template. The pMXs vector for the Oct4 ΔPOU-EGFP mutant (lacking amino acids 150–188 of Oct4) was generated by PCR-based mutagenesis. A series of deletion mutants of Oct4 was generated by insertion of various Oct4 PCR fragments into the pEGFP-C1 expression vector (Clontech). In addition, pMXs-H2B-EGFP was generated by insertion of the H2B-EGFP fragment from pEGFP-N3-H2B into the multiple cloning site of the pMXs vector. To generate pGL4.14-Oct4, the E1B tata sequence was inserted into the XhoI-BglII site of the pGL4.14 vector (Promega), resulting in the production of pGL4.14-E1Btata. Subsequently, five repeats of the Oct4-binding site (5′-TATTTGCATATTTGCATATTTGCATATTTGCATATTTGCATAAGCT-3′, 5′-TATGCAAATATGCAAATATGCAAATATGCAAATATGCAAATAGTAC-3′) were inserted into the upstream KpnI-SacI site of pGL4.14-E1Btata. To generate pGL4.14-OS-nanog, 3 repeats of the Oct/Sox element (5′- TTTTGCATTACAATGTTTTGCATTACAATGTTTTGCATTACAATGAGCT-3′, 5′- CATTGTAATGCAAAACATTGTAATGCAAAACATTGTAATGCAAAAGTAC-3′) were inserted into the KpnI-SacI site of pGL4.14-E1Btata. The Tol2-based Oct4 expression vector (pT2AL200R175-CAGGS-Oct4) was generated by first producing pT2A-CMH, a Tol2 transposon-based vector containing a multiple cloning site, by modifying pT2AL200R175G (27). The coding region of Oct4 or mutant Oct4 was subsequently inserted into the multiple cloning site of pT2A-CMH.
Heterokaryon Assay
NIH3T3 cells were infected with the retroviral expression vector for 24 h. Subsequently, 3 × 105 HeLa cells and 3 × 105 infected NIH3T3 cells were mixed and plated onto coverslips (in a 6-well plate) and cultured for another 24 h. Co-cultures of the cells were preincubated with 100 μg/ml cycloheximide and with either leptomycin B (LMB; 10 nm) or vehicle (EtOH) for 60 min. Cell fusion was performed by inverting the coverslips onto a drop of prewarmed (37 °C) polyethylene glycol 8000, DMEM for 2 min. After washing the coverslips with PBS, fused cells were further cultured in the presence of 100 μg/ml cycloheximide and with either LMB (10 nm) or vehicle (EtOH) for 3 h. The cells were then fixed, mounted, and observed using a confocal microscope (LSM510).
Oct4 Rescue Experiments
For rescue experiments, 1 × 105 ZHBTc4 ES cells were co-transfected with 200 ng of pCAGGS-mT2TP, an expression plasmid containing the Tol2 transposase cDNA whose codons are optimized for mammals under control of the CAG promoter, and 200 ng of a Tol2 transposon-based pT2A-CMH vector expressing either wild-type or mutant Oct4. One-fifth of the total transfected cells was re-plated 48 h post-transfection and further cultured with ES medium containing 1 μg/ml doxycycline (DOX). After 7 days, colonies were stained for alkaline phosphatase activity (Sigma) or picked and expanded as clonal cell lines.
Fluorescence Loss in Photobleaching (FLIP)
For FLIP analysis, NIH3T3 cells grown on glass-bottom dishes were transfected with Oct4-EGFP, Oct4 Δ-POUs-EGFP, or H2B-EGFP expression plasmids for 24 h, and the cells were imaged at 37 °C with a laser-scanning LSM510 microscope (Carl Zeiss) using a 63 × 1.4 NA oil-immersion objective. A single Z-section image was obtained before and at specific time intervals after each bleaching, which was performed for 100 iterations in a circular region of 6-μm diameter within the cytoplasm using 100% power of an argon laser at 488 nm. Bleaching was repeated every 30 s during the period of the experiment (500 s). After background subtraction, the fluorescent intensities of the regions of interest (nuclei) were determined using LSM software and normalized to those of the pre-bleached images.
Fluorescence Recovery after Photobleaching (FRAP)
NIH3T3 cells expressing Oct4-EGFP Oct4 Δ-POUs-EGFP or H2B-EGFP expression vectors grown on glass-bottom dishes were observed using a laser-scanning LSM510 microscope (Carl Zeiss) at 37 °C. For FRAP analysis, the cells were photobleached using a 488-nm laser at 100% power for 20 iterations over a rectangular region (20 μm2) within the nucleus. A single Z-section image was obtained before and at specific time intervals after each bleaching. Images were recorded every 0.2 s for 30 s. After background subtraction, the fluorescent intensities of the regions of interest were determined using LSM software and normalized to those of the pre-bleached images.
Induction of Cellular Reprogramming
Mouse-induced pluripotent cells were generated essentially as described previously (28) using pMXs retroviruses expressing mouse Sox2, Klf4, and c-Myc (Addgene) together with wild-type Oct4 (Addgene) or a series of mutant Oct4 proteins. Briefly, Plat-E cells were transfected with the retroviral vectors using FuGENE 6 transfection reagent (Roche Applied Science). The medium was changed at 24 h post-transfection, and viral supernatants were harvested 48 h post-transfection, filtered through a 0.45-μm filter, and used to infect mouse embryonic fibroblasts (MEFs). Twenty-four hours post-infection, 3 × 103 cells were re-plated onto a 60-mm dish with mitomycin C-treated feeder cells, and the culture medium was replaced after 2 days with ES medium containing 20% knockout serum replacement (Invitrogen). The medium was changed every other day. Alkaline phosphatase staining was performed using the leukocyte alkaline phosphatase kit (Sigma).
Antibodies
The following primary antibodies were used: rabbit polyclonal anti-Nanog (Reprocell), mouse monoclonal anti-Oct4 (BD Biosciences), rabbit polyclonal anti-RanBP1 (Santa Cruz), and mouse monoclonal anti-GAPDH (Ambion).
Immunofluorescence Staining and Confocal Microscopy
Cells were grown on coverslips and fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. After permeabilization with 0.5% Triton X-100 in PBS for 5 min, the cells were incubated in blocking buffer (PBS containing 3% skim milk) for 30 min and subsequently incubated overnight with primary antibodies at 4 °C. After 4 washes with PBS, cells were incubated with secondary antibodies for 45 min. The cells were washed again with PBS, cell nuclei were stained with DAPI, and the coverslips were mounted with Vectashield (Vector Laboratories). Images were acquired using a confocal microscope (LSM 510 META, Carl Zeiss) equipped with a 63 × 1.4 NA oil objective lens (Carl Zeiss) and analyzed using Zeiss LSM software.
Immunoblotting Analysis
Cells were resuspended in a radioimmunoprecipitation assay buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors (leupeptin, aprotinin, and phenylmethylsulfonyl fluoride) and incubated on ice for 15 min. After centrifugation at 16,000 × g for 10 min at 4 °C, the supernatants were collected and used for further analysis. Protein extracts were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. After blocking with 3% skim-milk in TBST buffer (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 0.1% Tween 20) for 30 min at room temperature, the membrane was incubated overnight with primary antibodies at 4 °C. After incubation with secondary antibodies conjugated to horseradish peroxidase, bands were visualized using Pierce Western blotting substrate (Thermo Scientific).
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) and used for cDNA synthesis with the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science). PCR amplification was performed using rTaq DNA polymerase (Takara). The primer sequences are detailed in Table 1.
TABLE 1.
Oligonucleotides used for RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Cdx2 | AGGCTGAGCCATGAGGAGTA | CGAGGTCCATAATTCCACTCA |
| Fgf4 | GGGAGGCTACAGACAGCAAG | CTGTGAGCCACCAGACAGAA |
| Gapdh | GTGTTCCTACCCCCAATGTGT | ATTGTCATACCAGGAAATGAGCTT |
| Oct4 (3-UTR) | CAAGGCAAGGGAGGTAGACA | CAAAATGATGAGTGACAGACAGG |
| Sox2 | GAGTGGAAACTTTTGTCCGAGA | GAAGCGTGTACTTATCCTTCTTCAT |
| Nanog | AGGGTCTGCTACTGAGATGCTCTG | CAACCACTGGTTTTTCTGCCACCG |
| Hand1 | ATGAACCTCGTGGGCAGGTA | TCACTGGTTTAGCTCCAGCG |
RNA Interference
Cells were transfected with mouse Oct4 siRNA (5′-GUUCGAGUAUGGUUCUGUAdTdT-3′ and 5′-UACAGAACCAUACUCGAACdTdT-3′) or control non-targeting siRNA (5′-GGACAUGUAUUUUCAAACAdTdT-3′ and 5′-uguuugaaaauacauguccdTdT-3′) using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions (10 nm final concentration).
Statistical Analysis
Statistical analysis was carried out using the unpaired Student's t test. The p values ≤0.05 were considered to be a statistically significant difference (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Oct4 Is a Nucleocytoplasmic Shuttling Protein
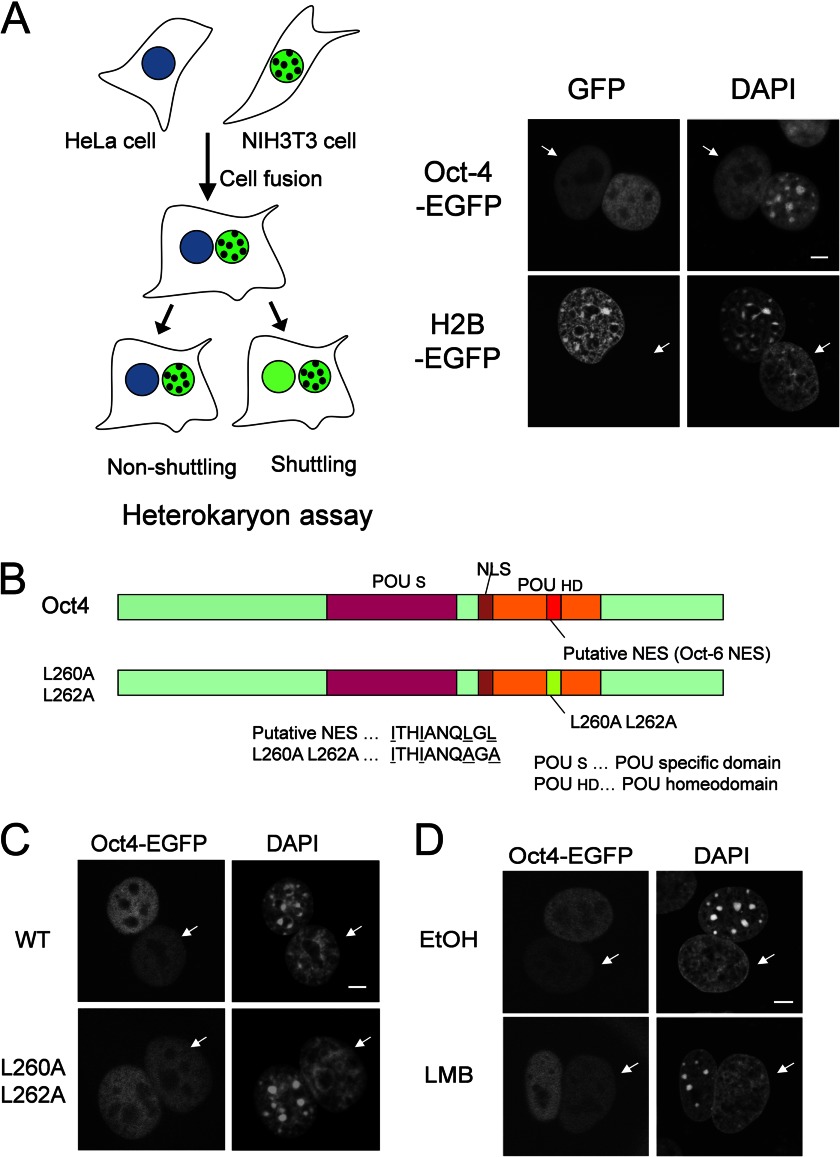
An interspecies heterokaryon assay was used to determine whether Oct4 shuttles between the nucleus and cytoplasm. NIH3T3 cells were retrovirally transduced with C-terminal EGFP-tagged Oct4 (Oct4-EGFP) and fused with HeLa cells to form heterokaryons in order to monitor the dynamic behavior of Oct4. DAPI staining allowed discrimination between NIH3T3 (punctate) and HeLa (homogenous) nuclei. As shown in Fig. 1A, Oct4-EGFP was detected in HeLa nuclei, indicating that Oct4-EGFP shuttles between the nucleus and the cytoplasm. In contrast, the control non-shuttling histone-H2B protein, also C-terminally tagged with EGFP, was not observed in HeLa nuclei of the heterokaryons.
FIGURE 1.
Oct4 is a Crm1/exportin 1-independent nucleocytoplasmic shuttling protein. A, mouse NIH3T3 cells were infected with retroviruses expressing Oct4-EGFP or histone H2B-EGFP. Heterokaryons were generated by fusion between retrovirus-infected NIH3T3 cells and human HeLa cells. Three hours after cell fusion, the cells were fixed, and fluorescence was analyzed. DAPI staining was used to distinguish between human and mouse nuclei within the heterokaryon. Arrows indicate the nuclei of HeLa cells. Mouse nuclei display a punctate staining pattern, whereas human nuclei are diffusely stained. Bar, 5 μm. B, shown is a schematic representation of wild-type and mutant (L260A,L262A) Oct4 proteins that contain double mutations within the putative NES sequence. The regions corresponding to the POU-specific domain (POUS), NLS, POU homeodomain (POUHD), and a putative NES are indicated. C, NIH3T3 cells were infected with retroviruses expressing either Oct4-EGFP or Oct4 (L260A,L262A)-EGFP, and heterokaryon assays were performed as described in A. Bar, 5 μm. D, shown is the effect of LMB on the shuttling of Oct4-EGFP. Heterokaryon assays were performed as described in A, with the exception that either LMB or EtOH were included. Arrows indicate the nuclei of HeLa cells. Bar, 5 μm.
Oct6, another member of POU transcription factor family, is exported from the nucleus by the nuclear export factor Crm1/exportin 1 (26). Thus, we subsequently examined whether Oct4 is also exported by Crm1. Oct4 harbors a putative leucine-rich NES, which is functional in Oct6 and is conserved among members of the POU transcription factor family (Fig. 1B). Therefore, we mutated two leucine residues to alanine (L260A and L262A) with the aim of disrupting the activity of the leucine-rich NES. However, the Oct4 (L260A,L262A) mutant retained the ability to shuttle between the nucleus and the cytoplasm (Fig. 1C). In addition, we confirmed these findings by using LMB, a specific inhibitor of Crm1, in the heterokaryon assay. As shown in Fig. 1D, LMB did not inhibit the shuttling of Oct4-EGFP. Thus, the nuclear export of Oct4 is not primarily mediated by Crm1.
Oct4 Is Exported from the Nucleus by Passive Diffusion
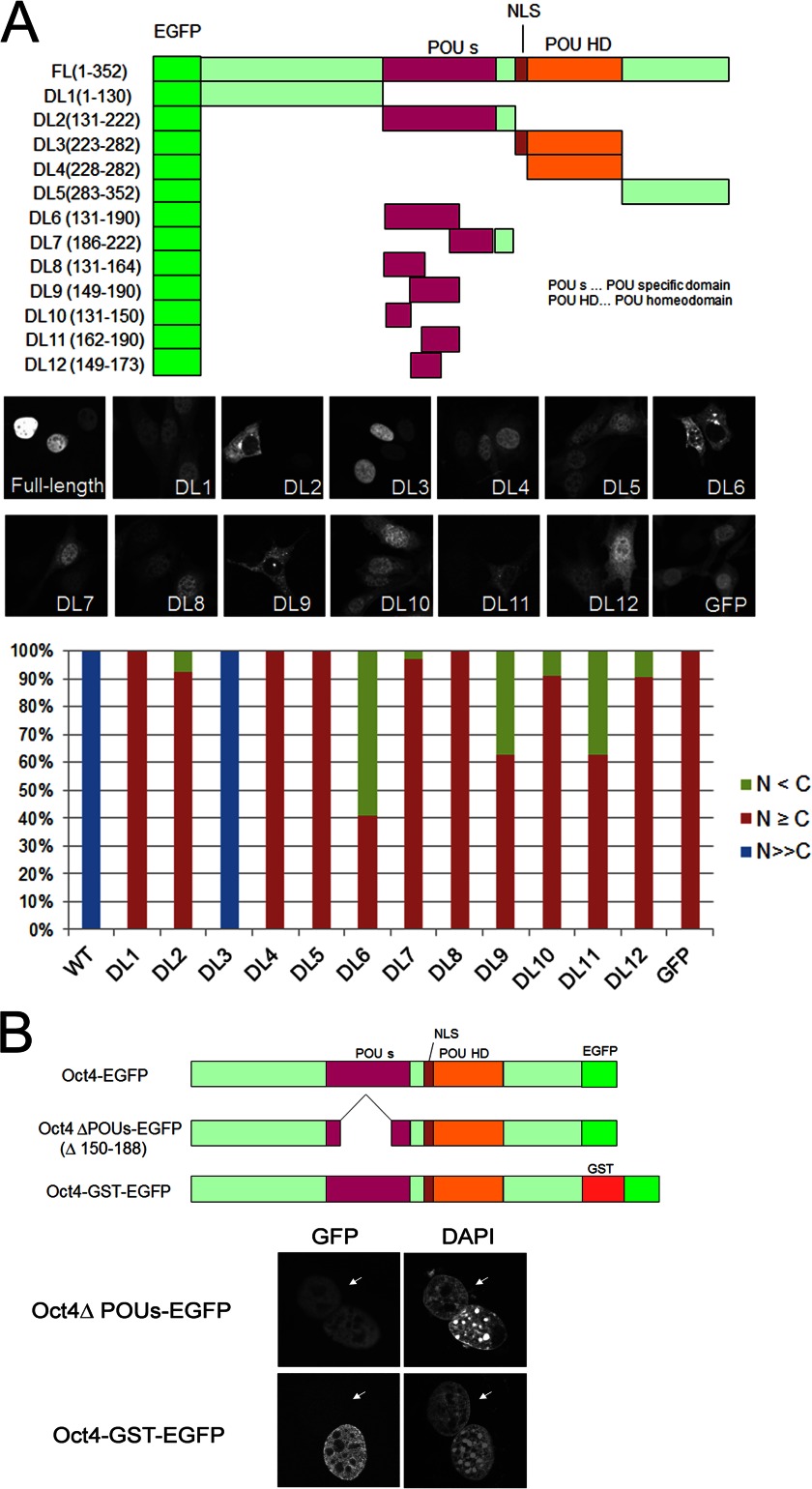
Oct4 consists of a POU domain, a DNA binding motif comprising a POU-specific domain and a POU homeodomain, and N- and C-terminal transactivation domains (29, 30). To determine whether Oct4 contains an NES, we constructed a series of deletion mutants that were N-terminally tagged with EGFP. As shown in Fig. 2A, Oct4 mutants displayed either diffuse distribution in both the nucleus and cytoplasm (DL1 and DL5) or exclusive nuclear localization (DL3). Moreover, deletion of the NLS present in DL3 (DL4) resulted in diffuse localization throughout the cell, which is consistent with a previous study (22). However, we noticed that a subpopulation of the cells expressing DL2 mutant, which contains the POU-specific domain, displayed predominant cytoplasmic localization. Therefore, additional deletion mutants of the POU-specific domain were generated to determine the relationship between the POU domain and cellular localization. As shown in Fig. 2A, predominant cytoplasmic localization was observed only for those containing the third helix of POU-specific domain (DL6, DL9, and DL11), which is also demonstrated by biochemical fractionation (data not shown), indicating that this portion of the POU-specific domain is essential for the cytoplasmic localization of Oct4.
FIGURE 2.
The POU-specific domain is not essential for Oct4 shuttling activity. A, shown is a schematic diagram and cellular localization of various Oct4 deletion mutants N-terminally tagged with EGFP. The regions corresponding to POUS, NLS, and POUHD are indicated. NIH3T3 cells were transfected with the constructs, and cells were fixed, permeabilized, and counterstained with DAPI at 24 h post-transfection. Subcellular localization of EGFP-Oct4 and its mutants was classified into three categories: nuclear (blue), nuclear dominant (red), and cytoplasmic dominant (green). B, shown is a schematic representation of the Oct4 ΔPOUs-EGFP and Oct4-GST-EGFP fusion proteins and heterokaryon assay results. NIH3T3 cells were infected with retroviruses expressing Oct4 ΔPOU-EGFP or Oct4-GST-EGFP, and heterokaryon assays were performed as described in Fig. 1. Arrows indicate the nuclei of HeLa cells.
To confirm whether the POU-specific domain of Oct4 plays a role in selective nuclear export, a deletion mutant (ΔPOU) was constructed that lacked the second and third helices of the POU-specific domain (amino acids 150–188). The resulting mutant protein was C-terminally fused to EGFP (ΔPOU Oct4-EGFP) and employed in the heterokaryon assay (Fig. 2B). However, the ΔPOU Oct4-EGFP mutant retained shuttling ability, suggesting that the Oct4 POU-specific domain may be important for cytoplasmic retention rather than selective nuclear export and that Oct4 may not possess an active nuclear export signal. Thus, efficient nuclear export of Oct4 is most likely to be mediated by passive diffusion. To confirm this hypothesis, an Oct4-GST-EGFP retroviral expression vector was generated. The predicted molecular mass of Oct4-GST-EGFP was more than 90 kDa, which is large enough to prevent its passive diffusion. As shown in Fig. 2B, the heterokaryon assay revealed that Oct4-GST-EGFP was retained in NIH3T3 cell nuclei, and it did not migrate into HeLa cell nuclei. Therefore, we concluded that the Oct4 protein is primarily exported via passive diffusion.
The Intranuclear Mobility of Oct4 Correlates with Its Export Efficiency
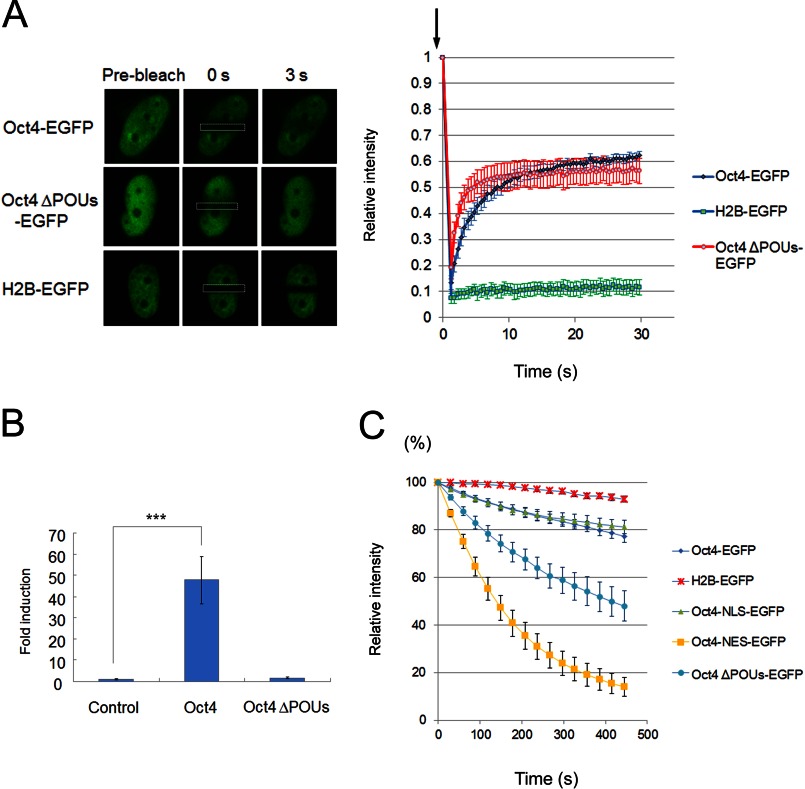
Because the nuclear export of Oct4 may be independent of export factors, it is possible that the intranuclear mobility of Oct4, which is dependent on its interaction with chromatin or nuclear protein complexes, affects Oct4 export efficiency. To examine this possibility, the dynamics of Oct4 in living cells were investigated in more detail. First, FRAP analysis was performed to examine the mobility of Oct4-EGFP within the nucleus. Consistent with a previous report (31), Oct4-EGFP displayed higher mobility compared with that of the control histone-H2B-EGFP protein (Fig. 3A). We also examined the mobility of ΔPOU Oct4-EGFP, which lacks the POU-specific domain required for DNA binding and transcriptional activity (Fig. 3B). FRAP analysis revealed that ΔPOU Oct4-EGFP showed a significant increase in the initial rate of fluorescence recovery compared with that of wild-type Oct4-EGFP (Fig. 3A), indicating that the ΔPOU Oct4 mutant is highly mobile within the nucleus.
FIGURE 3.
Intranuclear mobility of Oct4 is a key determinant of its export rate. A, FRAP analysis is shown. NIH3T3 cells were transfected with Oct4-EGFP, Oct4 ΔPOUs-EGFP, or H2B-EGFP, and FRAP experiments were performed with confocal laser scanning microscopy. The fluorescence intensity in the bleached region was measured and expressed as a relative ratio. Bars represent the means ± S.E. (n = 4–7). B, shown is a luciferase assay. NIH3T3 cells were transiently co-transfected with pRL-Renilla, the pGL4.14-Oct4 reporter plasmid, and the indicated Oct4-expressing plasmids. Results are presented as the firefly luciferase/Renilla luciferase ratio and normalized to the value obtained with the control (empty) vector. The results shown represent the mean ± S.D. of three independent experiments. *** indicates p < 0.001. C, shown is FLIP analysis. NIH3T3 cells were transfected with the indicated expression plasmids for 24 h, and the cytoplasm of cells expressing Oct4-EGFP, Oct4 ΔPOUs-EGFP, Oct4-NES-EGFP, Oct4-NLS-EGFP, or H2B-EGFP was irradiated every 30 s for ∼20 s with a laser. The fluorescence intensity in the nucleus was monitored after each bleaching and expressed as a relative ratio. Bars represent the means ± S.E. (n = 7–8).
The nucleocytoplasmic shuttling activity of these constructs was investigated in living cells by FLIP analysis. After repeated photobleaching in the cytoplasm, the nuclear fluorescence signal of wild-type Oct4-EGFP showed a slight but significant decrease compared with that of histone-H2B-EGFP (Fig. 3C). Furthermore, the ΔPOU Oct4-EGFP mutant displayed a considerable decrease in nuclear fluorescence intensity compared with wild-type Oct4-EGFP (Fig. 3C). Therefore, the intranuclear mobility of Oct4 correlated with protein export efficiency and is probably dependent on the binding of Oct4 to other nuclear proteins or chromatin. These results are also in good agreement with a previous report using a truncated mutant of Oct4 that lacked the C terminus, including the homeodomain (32).
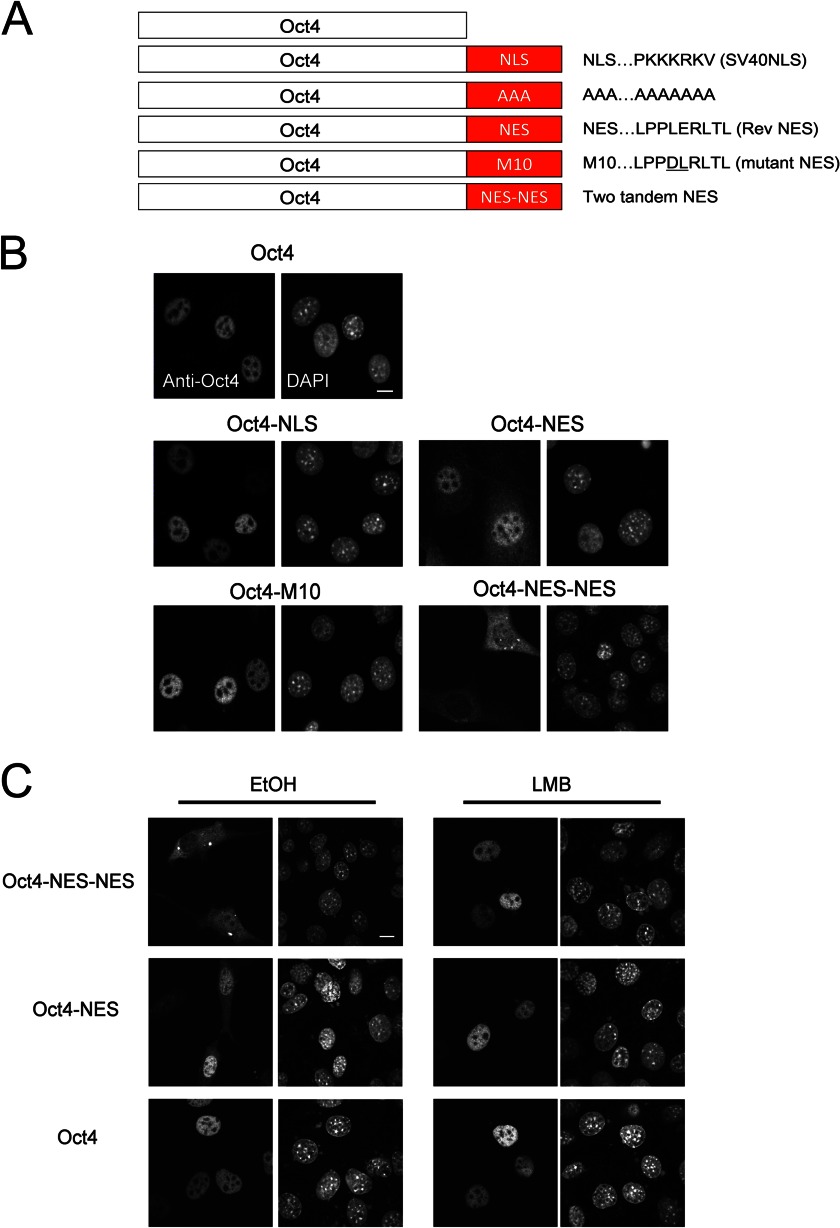
The Addition of Import/Export Signals Alters the Nucleocytoplasmic Localization of Oct4
We subsequently investigated whether the intracellular dynamic behavior of Oct4 affects its function. To disturb the spatiotemporal regulation of Oct4, a typical basic-type NLS derived from SV40 T antigen or a typical leucine-rich NES derived from the HIV Rev protein was fused to the C terminus of Oct4 to create Oct4-NLS or Oct4-NES mutants (Fig. 4A). In addition, constructs containing tandem alanine residues (AAA), an export-deficient NES mutant (M10), and two tandem leucine-rich NESs (NES-NES) were also generated. Confocal microscopy revealed that wild-type Oct4 was exclusively localized in the nucleus of NIH3T3 cells. In addition, Oct4-NLS and Oct4-M10 mutants displayed similar localization patterns to wild-type Oct4 (Fig. 4B). In contrast, Oct4-NES showed weak diffuse cytoplasmic localization as well as nuclear localization. FLIP analysis using EGFP fusion proteins revealed that the rate of nuclear export of Oct4-NES-EGFP was higher than that of ΔPOU Oct4-EGFP, a DNA binding-deficient mutant with a higher export rate than wild-type Oct4 (Fig. 3C). Furthermore, Oct4-NES-NES was either evenly distributed between the nucleus and cytoplasm or predominantly localized in the cytoplasm, indicating that its nuclear export efficiency is much higher than that of Oct4-NES. The addition of LMB to the culture medium led to the accumulation of cytoplasmic Oct4 mutant proteins in the nucleus (Fig. 4C), confirming that Oct4-NES and Oct4-NES-NES are first imported into the nucleus and then selectively exported by Crm1.
FIGURE 4.
Schematic representation and subcellular localization of various Oct4 mutants with biased intracellular localization. A, shown is a schematic representation of Oct4 mutants. NLS, nuclear localization signal of SV40 large T antigen; AAA, tandem alanine mutant; NES, nuclear export signal of human immunodeficiency virus Rev protein; M10, NES mutant; NES-NES, two tandem NES. B, shown is subcellular localization of Oct4 mutants. NIH3T3 cells were transfected with plasmids expressing Oct4 (WT or mutant) for 24 h. The cells were fixed and stained with anti-Oct4 antibody. DAPI was used to visualize nuclei. Bar, 10 μm. C, shown is the effect of LMB on the subcellular localization of Oct4 mutants. NIH3T3 cells were transfected with plasmids expressing Oct4 (WT or mutant) for 24 h and incubated in the presence of either EtOH (vehicle control) or LMB for 2 h. The cells were fixed and stained with anti-Oct4 antibody. DAPI was used to visualize nuclei. Bar, 10 μm.
Mutant Oct4 Proteins with Biased Nucleocytoplasmic Localization Maintain Self-renewal of ES Cells
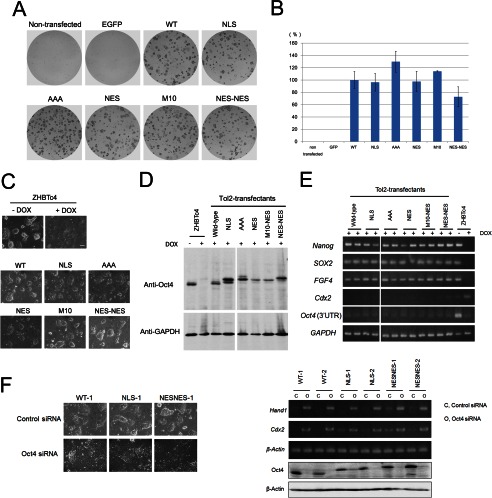
We examined whether the mutant Oct4 proteins can replace the function of wild-type Oct4 by using ZHBTc4 ES cells (6) that contain a tetracycline-regulated (Tet-off) wild-type Oct4 transgene and in which both alleles of endogenous Oct4 were inactivated by gene targeting. As a result, in the presence of tetracycline or DOX, the expression of Oct4 is repressed, and the cells differentiate toward trophectoderm lineages (6). We used the Tol2 transposon system (33) to stably introduce the Oct4 transgene into ZHBTc4 cells. In contrast to untransfected or control EGFP-expressing vector-transduced cells, stable expression of the wild-type Oct4 transgene enabled self-renewal of ZHBTc4 cells, which proliferated in an undifferentiated state (alkaline phosphatase positive) in the presence of DOX (Figs. 5, A and B). Unexpectedly, all of the Oct4 mutants retained the ability to form alkaline phosphatase-positive colonies in the absence of wild-type Oct4. Notably, the Oct4-NES-NES mutant, which was mainly localized in the cytoplasm, was also able to efficiently rescue the self-renewal of ZHBTc4 ES cells.
FIGURE 5.
Transgene expression of Oct4 mutants sustains the self-renewal of ES cells. Oct4 rescue experiments are shown. ZHBTc4 cells were transfected with a Tol2-based transposon expressing Oct4 transgenes. After 3 days, the cells were passaged in fresh media containing DOX to suppress tet-regulated wild-type Oct4. Eleven days after transfection, colonies were stained using an alkaline phosphatase staining kit. Representative images of the plates (A) and statistical analysis (B) are shown. Bars represent the mean ± S.D. of three independent experiments. C, shown is morphology of ZHBTc4 cells or isolated Tol2-Oct4-expressing ZHBTc4 clones cultured in the presence of DOX. Bar, 100 μm. D, shown are protein expression levels of wild-type or mutant Oct4 in isolated Tol2-Oct4-expressing ES clones. Cell lysates (20 μg) were used for Western blotting, and GAPDH was used as a loading control. E, shown are RT-PCR analyses of isolated ES cell clones. Total RNA was isolated from Tol2-Oct4-expressing ES clones (two clones each). Note that the primers specific for Oct4 (3′-UTR) detect expression of endogenous Oct4 but not Oct4 expression from transgenes. GAPDH was used as a loading control. F, shown is differentiation of Tol2-Oct4-expressing clones by Oct4 knockdown. Tol2-Oct4-expressing clones (WT, NLS, NES-NES) were transfected with siRNA against Oct4 or control siRNA. Left, shown are phase contrast images of clones at 4 days after transfection. Right top, shown is RT-PCR analysis of Cdx2, Hand1, and β-actin gene expression in control- or Oct4-siRNA-treated cell (4 days after transfection). Right bottom, shown is immunoblotting analysis of Oct4 and β-actin in control- or Oct4-siRNA-treated cell lysates (2 days after transfection).
Next, we isolated stable clones expressing Tol2-transposon-based mutant Oct4 in the presence of DOX. These clones were morphologically similar to the wild-type Oct4-expressing clone (Fig. 5C) and could be maintained for ≥10 passages. Immunoblotting analysis confirmed that the tetracycline-regulated wild-type Oct4 transgene was efficiently suppressed and that each cell expressed only Tol2 transposon-derived Oct4 (Fig. 5D). RT-PCR analysis (Fig. 5E) revealed that the clones expressing mutant Oct4 maintained the expression of pluripotency-associated genes, including Nanog, SOX2, and FGF4. Moreover, primer sets that amplify the 3′-UTR region of Oct4 were used to confirm that tetracycline-regulated wild-type Oct4 was efficiently suppressed in these clones in the presence of DOX. Furthermore, the expression of Cdx2, a trophectoderm marker, was also suppressed in these clones. Oct4 knockdown in those clones caused drastic morphological changes of ES cells concomitant with up-regulation of trophectoderm marker genes, Cdx2 and Hand1 (Fig. 5F), demonstrating that those clones at least possess differentiation potential into trophectoderm lineages. Confocal imaging using an anti-Oct4 antibody further confirmed the altered localization of Oct4-NES and Oct4-NES-NES. Moreover, in contrast to endogenous Nanog, the majority of Oct4-NES-NES was not accumulated in the nucleus (Fig. 6A). Thus, although Oct4-NES-NES enters the nucleus via its own NLS, it is rapidly and actively exported from the nucleus by the addition of two NES. Furthermore, the addition of LMB to the culture medium led to the accumulation of cytoplasmic Oct4-NES-NES proteins into the nucleus (Fig. 6B). Therefore, these results confirm that Oct4-NES-NES indeed exists at a low concentration in the nucleus due to its enhanced Crm1-mediated nuclear export yet is able to maintain the self-renewal of ES cells. This indicates that Oct4 maintains the pluripotent state of ES cells regardless of the length of time that Oct4 is present in the nucleus; transient localization in the nucleus is sufficient for Oct4 to maintain self-renewal of ES cells.
FIGURE 6.
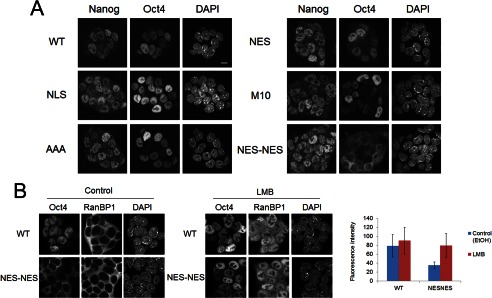
Analysis of subcellular localization of wild-type and mutant Oct4s. A, shown is immunocytochemical analysis of Tol2-Oct4-expressing ZHBTc4 clones. Cells were double-stained with anti-Nanog and anti-Oct4 antibodies. DAPI was used to visualize nuclei. Bar, 10 μm. B, shown is the effect of LMB on the subcellular localization of wild-type Oct4 and Oct4-NES-NES in stable ZHBTc4 clones. Cells were cultured in the presence of either EtOH (vehicle control) or LMB for 2 h, then fixed and stained with anti-Oct4 and anti-RanBP1 antibodies. The localization of endogenous RanBP1, an NES-containing protein known to shuttles between the nucleus and cytoplasm in a Crm1-dependent manner, was monitored to evaluate the effect of LMB. Mean fluorescence intensity of nuclear Oct4 staining for each sample is shown to the right. Data are the means ± S.D. The total number of analyzed nuclei (n) is as follows: WT, control (n = 89); WT, LMB (n = 119); NES-NES, control (n = 119); NES-NES, LMB (n = 112).
Mutant Oct4 Proteins with Biased Nucleocytoplasmic Localization Show Limited Potential for Cellular Reprogramming
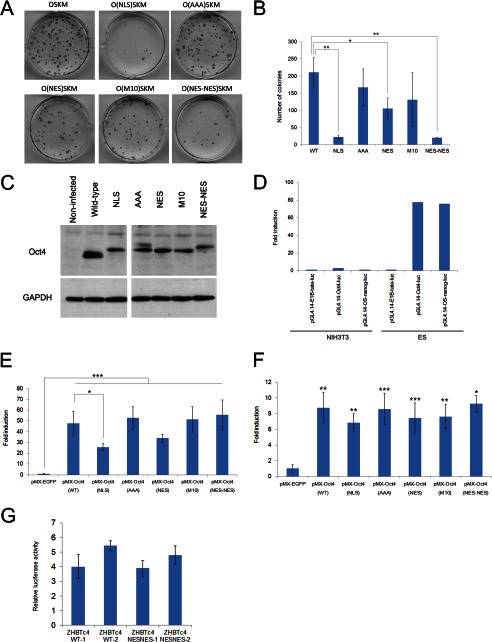
The ability of the aforementioned Oct4 mutants to reprogram fibroblasts into induced pluripotent cells was compared. To this aim, induced pluripotent cells were generated by transduction of MEFs with retroviral vectors expressing Sox2, Klf4, c-Myc, and either wild-type or a series of mutant Oct4 proteins. Immunoblotting analysis of infected MEF cell lysates revealed that comparable levels of expression were achieved for each mutant Oct4 protein (Fig. 7C). As shown in Figs. 7, A and B, the Oct4-NLS and Oct4-NES-NES mutants demonstrated a significantly reduced ability to reprogram cells to a pluripotent state compared with wild-type Oct4. Because the Oct4-NLS mutant contains an additional SV40 T-NLS, it is likely that Oct4-NLS is constitutively transported into the nucleus in an unregulated manner. These results suggest that both the appropriate nuclear entry and adequate intranuclear retention of Oct4 are crucial for efficient cellular reprogramming.
FIGURE 7.
Reprogramming efficiency and transcriptional activity of Oct4 mutants. A and B, reprogramming efficiency of Oct4 mutants is shown. 4F-infected MEFs were analyzed by alkaline phosphatase staining at 11 days post infection. Representative images of the plates (A) and statistical analysis (B) are shown. Bars represent the mean ± S.D. of three independent experiments. * and ** indicate p < 0.05 and p < 0.01, respectively. C, shown are protein expression levels of wild-type or mutant Oct4. MEFs were infected with retroviruses expressing Oct4 or Oct4 mutants. After 72 h, the cell lysates (10 μg) were used for Western blotting. GAPDH was used as a loading control. D, selective activation of reporter genes containing octamer or Oct4/Sox2 binding motifs in ES cells is shown. NIH3T3 cells or ES cells were co-transfected with a pRL-SV40-renilla luciferase plasmid and with either pGL4.14-E1Btata-luc, pGL4.14-Oct4-luc, or pGL4.14-OS-nanog-luc reporter plasmids. Luciferase assays were performed 24 h after transfection. Firefly luciferase activity was normalized to Renilla luciferase activity. E and F, shown is transcriptional activity of Oct4 (WT and mutant) for octamer (E) or Oct4/Sox2 binding motifs (F). NIH3T3 cells were co-transfected with the indicated plasmids expressing Oct4 (WT or mutant), Sox2 (for pGL4.14-OS-nanog-luc reporter assay only), a pRL-SV40-renilla luciferase plasmid, and either pGL4.14-Oct4-Luc or pGL4.14-OS-nanog-Luc reporter plasmids. Luciferase assays were performed as in D. Bars represent the mean ± S.D. of three independent experiments performed in duplicate. *, **, and *** indicate p < 0.05, p < 0.01, and p < 0.001, respectively. In F, the asterisks indicate significant differences from pMX-EGFP-transfected cells. G, Tol2-Oct4-expressing ZHBTc4 clones (WT-1, WT-2, NES-NES-1, or NES-NES-2) were co-transfected with a pRL-SV40-renilla luciferase plasmid and a pGL4.14-Oct4-luc reporter plasmid. Luciferase assays were performed as in D. Bars represent the mean ± S.D. of three independent experiments performed in duplicate.
Oct4-NES-NES Has Similar Transcriptional Activity to Wild-type Oct4
Finally, we performed a luciferase assay to determine the effect of Oct4 mutation on transcriptional activity. Reporter constructs were generated by inserting either five tandem copies of an octamer binding motif (pGL4.14-Oct4-luc) (34) or three tandem copies of an Oct4/Sox2 binding motif (pGL4.14-OS-nanog-luc) (35) upstream of the firefly luciferase gene containing the adenovirus E1B TATA-box minimal promoter (pGL4.14-E1B-tata-luc). As shown in Fig. 7D, both reporter genes were selectively activated in ES cells. We subsequently monitored the activation of pGL4.14-Oct4-luc by each Oct4 mutant in NIH3T3 cells. As shown in Fig. 7E, the transcriptional activities of Oct4-NLS was significantly reduced (about 50% compared with wild-type Oct4), which could explain the reduced ability of this mutant to reprogram somatic cells into induced pluripotent cells However, Oct4-NES-NES, which also demonstrated significantly reduced reprogramming efficiency (∼10% compared with wild-type Oct4), showed comparable transcriptional activity to wild-type Oct4. Therefore, these results indicate that transcriptional activity does not necessarily correlate with the ability of Oct4 mutants to reprogram cells.
To further examine the relationship between the transcriptional activity and reprogramming ability of Oct4 mutants, we subsequently employed the pGL4.14-OS-nanog-luc plasmid (Fig. 7F). Co-transfection of Sox2 with Oct4 resulted in about a 4-fold increase in luciferase activity, which is consistent with a previous report (Ref. 35 and data not shown). The transcriptional activities of Oct4 mutants, including Oct4-NES-NES, were comparable with those of wild-type Oct4. Finally, we compared the activity of pGL4.14-Oct4-luc in Tol2-Oct4 (wild type)- or Oct4-NES-NES-expressing ZHBTc4 clones. As shown in Fig. 7G, these clones showed a similar activation of a reporter. Thus, these results confirmed that cell reprogramming is not solely dependent on transcriptional activity of Oct4. Collectively, these findings indicate that post-translational, spatiotemporal regulation of Oct4 is critical for reprogramming cells.
DISCUSSION
Oct4 is functionally involved in two distinct processes: ES cell self-renewal and cellular reprogramming. However, it is currently unknown whether Oct4 functions in the same manner to mediate both activities. In this study we aimed to analyze the dynamic behavior of Oct4 and examine its role in the determination of cell fate. Oct4 mutants with biased nucleocytoplasmic distribution were able to maintain the self-renewal of ES cells; however, they displayed significantly reduced potential for cellular reprogramming compared with wild-type Oct4. Therefore, although the appropriate spatiotemporal regulation of Oct4 is essential for cellular reprogramming, it is not required for maintenance of the undifferentiated state of ES cells.
Notably, an Oct4-NES-NES mutant, which was primarily localized in the cytoplasm, was also capable of maintaining the undifferentiated state of ES cells. This suggests that despite its rapid export from the nucleus, Oct4 can still act as a transcription factor to drive genome-wide gene activation/repression necessary for self-renewal. Therefore, although it has previously been shown that the expression level of Oct4 is crucial for maintenance of the undifferentiated state of ES cells, our study suggests that the intranuclear level of Oct4 is not significant if continued nuclear import of Oct4 occurs. This is consistent with the recent finding that Oct4 kinetics, rather than absolute Oct4 expression levels, are critical for the determination of cell lineage patterning in vivo (32).
Oct4 associates with a number of different proteins (36–38) to form various functional complexes in the nucleus. We performed a luciferase assay with a reporter construct containing Oct4/Sox2 binding motifs to demonstrate that the Oct4-NES-NES mutant possessed comparable transcriptional activity to wild-type Oct4. Thus, even transient nuclear localization of a transcription factor complex containing Oct4 and Sox2 may ensure maintenance of the undifferentiated state of ES cells. On the other hand, it has been shown that chromatin remodeling and/or epigenetic changes are crucial for cellular reprogramming (39–47), which raises the possibility that Oct4-containing complexes required for chromatin remodeling and/or epigenetic changes may need to stay in the nucleus for a sufficient length of time to complete their function. Indeed, it was recently revealed that the majority of Oct4 binding sites during the initial phase of reprogramming are located in closed chromatin (48). Therefore, we propose that to induce cellular reprogramming, Oct4 must remain in the nucleus for a sufficient time period to access to its binding sites and allow recruitment of other factors required for chromatin remodeling and/or epigenetic modification.
However, we also found that the cellular reprogramming activity of Oct4-NLS was significantly reduced despite the fact that this mutant protein is constitutively localized in the nucleus. Because the Oct4-NLS mutant displayed reduced transcriptional activity, this reduction may significantly affect cellular reprogramming activity. Alternatively, because SV40 T-NLS possesses a strong import activity and is efficiently recognized by all members of the importin α family, regulated rather than constitutive nuclear import of Oct4 may be crucial to induce cellular reprogramming. Thus, the specific timing of Oct4 import into the nucleus may be important. On the other hand, SV40 T-NLS is composed of a stretch of basic amino acids that may cause nonspecific DNA binding in the nucleus. Therefore, it is possible that Oct4-NLS recruits its interacting partners to nonspecific regions of the genome, thereby inducing uncontrolled reactions that disturb cellular reprogramming.
We employed a heterokaryon assay to show that Oct4 shuttles between the nucleus and cytoplasm. How might Oct4 shuttle in vivo? Our results indicated that Oct4 is passively exported from the nucleus in a transport factor-independent manner and that Oct4 mobility within the nucleus parallels the export rate of nuclear Oct4. Thus, increased interactions between Oct4 and other nuclear proteins or DNA may retain Oct4 for longer within the nucleus. Indeed, we found that the Oct4 ΔPOU mutant that does not bind DNA showed an increased rate of nuclear export, which is in agreement with the previous report (32).
In summary, Oct4 shuttles between the nucleus and cytoplasm. To function as a transcription factor that maintains the undifferentiated state of ES cells, Oct4 only has to stay transiently in the nucleus. In contrast, to induce cellular reprogramming, Oct4 must stay in the nucleus for a sufficient length of time to induce essential transcription-independent reactions, which may involve chromatin remodeling and/or epigenetic changes. Thus, Oct4 plays distinct roles in the self-renewal of ES cells and in somatic cell reprogramming. Future studies focused on the characterization of cellular reprogramming processes induced by the Oct4-NES-NES mutant may shed light on how nuclear Oct4 retention functions to induce somatic cell reprogramming.
Acknowledgment
We are grateful to Dr. Hitoshi Niwa (RIKEN) for kindly providing ZHBTc4 ES cells.
This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Core Research for Evolutional Science and Technology (CREST) program of Japan Science and Technology Agency (JST).
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- LMB
- leptomycin B
- RanBP
- Ran-binding protein
- DOX
- doxycycline
- FLIP
- fluorescence loss in photobleaching
- FRAP
- fluorescence recovery after photobleaching
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472 [DOI] [PubMed] [Google Scholar]
- 2. Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M. (1990) A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692 [DOI] [PubMed] [Google Scholar]
- 3. Schöler H. R., Ruppert S., Suzuki N., Chowdhury K., Gruss P. (1990) New type of POU domain in germ line-specific protein Oct-4. Nature 344, 435–439 [DOI] [PubMed] [Google Scholar]
- 4. Ovitt C. E., Schöler H. R. (1998) The molecular biology of Oct-4 in the early mouse embryo. Mol. Hum. Reprod. 4, 1021–1031 [DOI] [PubMed] [Google Scholar]
- 5. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 6. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 8. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 9. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 12. Matoba R., Niwa H., Masui S., Ohtsuka S., Carter M. G., Sharov A. A., Ko M. S. (2006) Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE 1, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babaie Y., Herwig R., Greber B., Brink T. C., Wruck W., Groth D., Lehrach H., Burdon T., Adjaye J. (2007) Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells 25, 500–510 [DOI] [PubMed] [Google Scholar]
- 14. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimozaki K., Nakashima K., Niwa H., Taga T. (2003) Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development 130, 2505–2512 [DOI] [PubMed] [Google Scholar]
- 16. Pan G. J., Chang Z. Y., Schöler H. R., Pei D. (2002) Stem cell pluripotency and transcription factor Oct4. Cell Res. 12, 321–329 [DOI] [PubMed] [Google Scholar]
- 17. Xu H. M., Liao B., Zhang Q. J., Wang B. B., Li H., Zhong X. M., Sheng H. Z., Zhao Y. X., Zhao Y. M., Jin Y. (2004) Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279, 23495–23503 [DOI] [PubMed] [Google Scholar]
- 18. Wei F., Schöler H. R., Atchison M. L. (2007) Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- 19. Saxe J. P., Tomilin A., Schöler H. R., Plath K., Huang J. (2009) Post-translational regulation of Oct4 transcriptional activity. PLoS ONE 4, e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swaney D. L., Wenger C. D., Thomson J. A., Coon J. J. (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brehm A., Ohbo K., Schöler H. (1997) The carboxyl-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol. Cell. Biol. 17, 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan G., Qin B., Liu N., Schöler H. R., Pei D. (2004) Identification of a nuclear localization signal in OCT4 and generation of a dominant negative mutant by its ablation. J. Biol. Chem. 279, 37013–37020 [DOI] [PubMed] [Google Scholar]
- 23. Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawa Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. (2007) Triggering neural differentiation of ES cells by subtype switching of importin-α. Nat. Cell Biol. 9, 72–79 [DOI] [PubMed] [Google Scholar]
- 24. Li X., Sun L., Jin Y. (2008) Identification of karyopherin-α2 as an Oct4 associated protein. J. Genet. Genomics 35, 723–728 [DOI] [PubMed] [Google Scholar]
- 25. Young J. C., Major A. T., Miyamoto Y., Loveland K. L., Jans D. A. (2011) Distinct effects of importin α2 and α4 on Oct3/4 localization and expression in mouse embryonic stem cells. FASEB J. 25, 3958–3965 [DOI] [PubMed] [Google Scholar]
- 26. Baranek C., Sock E., Wegner M. (2005) The POU protein Oct-6 is a nucleocytoplasmic shuttling protein. Nucleic Acids Res. 33, 6277–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Urasaki A., Morvan G., Kawakami K. (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okada M., Oka M., Yoneda Y. (2010) Effective culture conditions for the induction of pluripotent stem cells. Biochim. Biophys. Acta 1800, 956–963 [DOI] [PubMed] [Google Scholar]
- 29. Herr W., Cleary M. A. (1995) The POU domain. Versatility in transcriptional regulation by a flexible two-in-one DNA binding domain. Genes Dev. 9, 1679–1693 [DOI] [PubMed] [Google Scholar]
- 30. Niwa H. (2001) Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 26, 137–148 [DOI] [PubMed] [Google Scholar]
- 31. van den Boom V., Kooistra S. M., Boesjes M., Geverts B., Houtsmuller A. B., Monzen K., Komuro I., Essers J., Drenth-Diephuis L. J., Eggen B. J. (2007) UTF1 is a chromatin-associated protein involved in ES cell differentiation. J. Cell Biol. 178, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Plachta N., Bollenbach T., Pease S., Fraser S. E., Pantazis P. (2011) Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 13, 117–123 [DOI] [PubMed] [Google Scholar]
- 33. Kawakami K., Noda T. (2004) Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics 166, 895–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun C., Nakatake Y., Akagi T., Ura H., Matsuda T., Nishiyama A., Koide H., Ko M. S., Niwa H., Yokota T. (2009) Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol. Cell. Biol. 29, 4574–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 37. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) An expanded Oct4 interaction network. Implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Plath K., Hochedlinger K. (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 [DOI] [PubMed] [Google Scholar]
- 40. Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. (2008) A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528 [DOI] [PubMed] [Google Scholar]
- 43. Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. (2011) Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang G., Taranova O., Xia K., Zhang Y. (2010) Butyrate promotes induced pluripotent stem cell generation. J. Biol. Chem. 285, 25516–25521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mali P., Chou B. K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R. A., Ohm J. E., Yu W., Baylin S. B., Yusa K., Bradley A., Meyers D. J., Mukherjee C., Cole P. A., Cheng L. (2010) Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singhal N., Graumann J., Wu G., Araúzo-Bravo M. J., Han D. W., Greber B., Gentile L., Mann M., Schöler H. R. (2010) Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943–955 [DOI] [PubMed] [Google Scholar]
- 47. Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., Wang J., Rendl M., Bernstein E., Schaniel C., Lemischka I. R. (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soufi A., Donahue G., Zaret K. S. (2012) Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]