Background: Proteolytic processing is involved in apoptosome-mediated caspase-9 activation, but its role is unknown.
Results: Processed caspase-9 has much higher activity than its unprocessed zymogen in the presence of the apoptosome.
Conclusion: Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation.
Significance: Our study challenges the common view that proteolytic processing is unnecessary for caspase-9 activation.
Keywords: Apoptosis, Caspase, Cell Biology, Cell Death, Enzyme Mechanisms, Apaf-1
Abstract
Maturation of the single-chain caspase-9 zymogen through autoproteolytic processing is mediated by the Apaf-1 apoptosome at the onset of apoptosis. Processed caspase-9 and the apoptosome form a holoenzyme with robust proteolytic activity that is 2–3 orders of magnitude higher than that of free processed caspase-9. An unresolved important question is the role of caspase-9 processing, with some experimental data suggesting its dispensability. In this study, we demonstrate that, in contrast to wild-type caspase-9, the unprocessed single-chain caspase-9 triple mutant E306A/D315A/D330A (Casp9-TM) could no longer be adequately activated by the apoptosome. Compared with the protease activity of wild-type caspase-9, that of Casp9-TM in the presence of the apoptosome was drastically reduced. The crippled protease activity of Casp9-TM in the presence of the apoptosome is likely attributable to a markedly reduced ability of Casp9-TM to form homodimers. These data identify an essential role for the autoproteolytic processing of caspase-9 in its activation.
Introduction
Programmed cell death, also known as apoptosis, is central to the development and tissue homeostasis of metazoans (1, 2). The extrinsic apoptosis pathway is triggered by extracellular cell death stimuli, such as death ligands TRAIL and TNF-α (3). In contrast, the intrinsic apoptosis pathway is initiated by a plethora of intracellular stress signals, including DNA damage, hypoxia, nutrition deprivation, and cytoskeleton and organelle disruption (1). Caspases, a family of cysteine proteases, are the executioners of apoptosis (4). Caspase-9 is the central enzyme controlling intrinsic apoptosis.
Caspases are divided into two classes: initiator caspase, such as mammalian caspase-2, -8, -9, and -10, and effector caspase, such as caspase-3, -6, and -7 (5). All caspases are synthesized as single-chain inactive zymogens and must be activated to facilitate cell death. An effector caspase (such as caspase-3) is activated by an initiator caspase (caspase-9) through an obligatory intrachain cleavage; the protease activity of a mature effector caspase is 4–5 orders of magnitude higher than that of its unprocessed zymogen. In contrast, an initiator caspase is activated by upstream multimeric complexes. Caspase-9 is activated by the apoptosome, whereas another initiator caspase, caspase-8, is activated by the death-inducing signaling complex (6).
In response to intracellular death stimuli, mitochondrial cytochrome c is released into the cytoplasm, where cytochrome c binds and activates Apaf-1 (7, 8). The Apaf-1·cytochrome c binary complex assembles into a heptameric apoptosome in the presence of ATP/dATP (9). Then, the apoptosome recruits the single-chain caspase-9 zymogen and facilitates its autocatalytic processing after Asp-315. Importantly, processed caspase-9 remains bound to the apoptosome as a holoenzyme, with a protease activity that is 2–3 orders of magnitude higher than that of free processed caspase-9 (10).
Because of its central role in apoptosis, caspase-9 has been the focus of intense investigation in the past 2 decades. Unlike effector caspases, for which intrachain cleavage is absolutely essential for activation, the activation of caspase-9 seemed to have little to do with autocatalytic processing (11–14). An uncleavable single-chain caspase-9 was shown to exhibit robust activity in the presence of the apoptosome; this activity was almost indistinguishable from that of processed WT caspase-9 (11, 13). In fact, it is generally believed by the cell death research community that, regardless of the intrachain cleavage, caspase-9 is activated to a similar extent by the apoptosome. In this study, we present compelling evidence that demonstrates an essential role for intrachain cleavage in caspase-9 activation.
EXPERIMENTAL PROCEDURES
Protein Preparation
Wild-type caspase-9, full-length Apaf-1, and procaspase-3(C163A) were expressed and purified as described (15). Apaf-1 residues 1–591 (Apaf-1(1–591)) was expressed in Escherichia coli and purified as described (15), with an additional step of gel filtration. The uncleavable caspase-9 triple mutant E306A/D315A/D330A (Casp9-TM) was cloned into vector pET-29b (Novagen) with a C-terminal hexahistidine tag and expressed and purified as WT caspase-9. The two-chain caspase-9 variants were generated by cloning the large subunit into vector pBB75 (Novagen) and the small subunit into vector pET-21b (Novagen) with a C-terminal hexahistidine tag. The two parts were coexpressed in E. coli BL21(DE3) cells and purified with the same procedure as used for WT caspase-9. The procaspase-3 zymogen was cloned into vector pET-21b with a hexahistidine tag at the C terminus, overexpressed in E. coli by induction with 0.2 mm isopropyl β-d-thiogalactopyranoside at 22 °C for 45 min, and then purified using nickel-nitrilotriacetic acid affinity columns (Qiagen). Equine heart cytochrome c and dATP were purchased from Sigma. The fluorogenic substrates Ac-LEHD-7-amino-4-trifluoromethylcoumarin (AFC)3 and Ac-DEVD-7-amino-4-methylcoumarin (AMC) were purchased from Enzo Life Sciences, Inc.
Caspase-9 Assay
The apoptosome was assembled by incubating full-length Apaf-1 and cytochrome c (molar ratio of 1:5) with 1 mm dATP or incubating only Apaf-1(1–591) with 1 mm dATP at 4 °C overnight. The reaction buffer used in the assays contained 100 mm KCl, 20 mm HEPES (pH 7.5), and 5 mm DTT. For the assays using Ac-LEHD-AFC as the substrate, WT or variant caspase-9 (0.2 μm) was incubated with the apoptosome (containing 0.4 μm Apaf-1) at 22 °C for 10 min, and Ac-LEHD-AFC was then added to a final concentration of 200 μm. The activity of caspase-9 was monitored using a fluorescence spectrophotometer (Hitachi F-4600), with excitation and emission wavelengths of 400 and 505 nm, respectively.
The activities of WT and variant caspase-9 with or without the apoptosome were also measured using procaspase-3(C163A) as the substrate. Procaspase-3(C163A) (40 μm) was incubated with caspase-9 (1 μm) and the apoptosome (containing 2 μm Apaf-1) for 1 h at 4 °C and for an additional 30 min at 22 °C. Reactions were stopped by the addition of 1 mm PMSF and immediately mixed with an equal volume of 2× SDS loading buffer. The samples were immediately heated at 96 °C for 5 min and separated on by 16% SDS-PAGE. The results were visualized by Coomassie Blue staining.
Gel Filtration Analysis
Superdex 200 HR 10/30 (GE Healthcare) was used in this study. The column was pre-equilibrated with 100 mm KCl, 20 mm HEPES (pH 7.5), and 5 mm DTT and calibrated with molecular weight standards (GE Healthcare). WT caspase-9 or its variants were injected into the column and eluted at a flow rate of 0.4 ml/min.
RESULTS
Essential Role for Intrachain Cleavage in Caspase-9 Activation
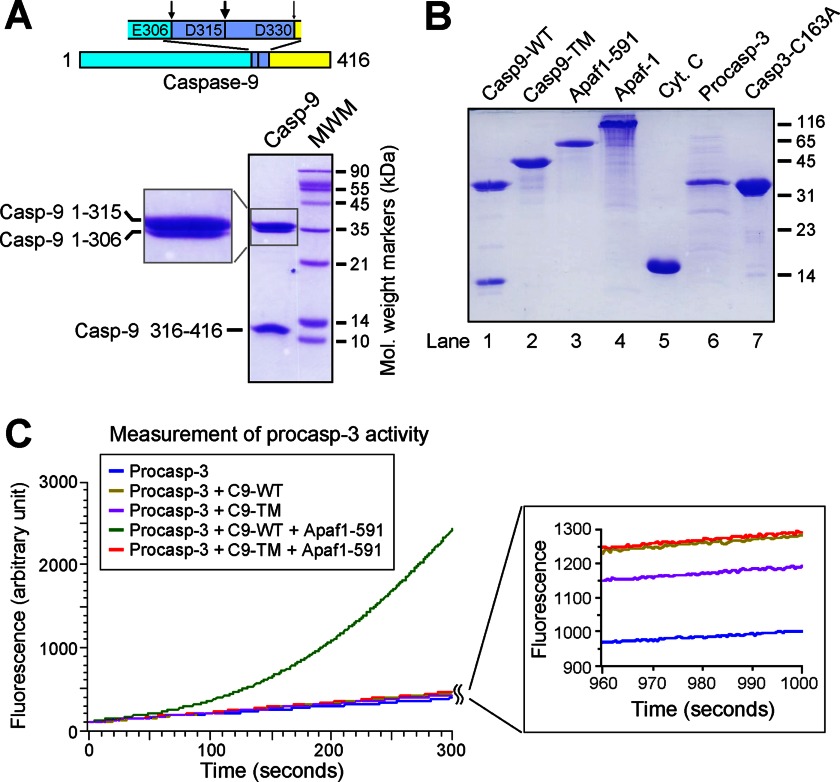
The caspase-9 zymogen is known to undergo intrachain cleavages at three sites (Fig. 1A, upper panel). Autocatalytic processing of caspase-9 occurs mainly after Asp-315 and, to a lesser extent, after Glu-306 (14, 16). In addition, cleavage after Asp-330 by activated caspase-3 in 293 cells or by autocatalytic processing in bacteria has also been reported (13, 16). Under our experimental conditions, WT caspase-9 was fully processed after Asp-315 and partially cleaved after Glu-306 when overexpressed in E. coli, resulting in two large subunits (residues 1–315 and 1–306) and one small subunit (residues 316–416) (Fig. 1A, lower panel). The quantity of fragment 1–306 was ∼10% of that of fragment 1–315. To ensure the generation of uncleavable single-chain caspase-9, we mutated all three potential cleavage sites. The resulting caspase-9 triple mutant E306A/D315A/D330A (Casp9-TM) was overexpressed as a single-chain protein and purified to homogeneity (Fig. 1B, lane 2). To reconstitute an in vitro caspase-9 activity assay, we also expressed and purified full-length Apaf-1, WD40-deleted Apaf-1 (Apaf-1(1–591)), and the full-length caspase-3 zymogen (residues 1–277) (Fig. 1B).
FIGURE 1.
Activation of the caspase-3 zymogen is greatly facilitated by WT caspase-9, but not uncleavable single-chain caspase-9, in the presence of the apoptosome. A, WT caspase-9 is autoproteolytically processed after Asp-315, with two additional minor processing sites after Glu-306 and Asp-330. The upper panel shows the three sites. The lower panel shows purified WT caspase-9 on an SDS-polyacrylamide gel stained with Coomassie Blue. B, recombinant proteins used in this study. Shown is an SDS-polyacrylamide gel stained with Coomassie Blue. Cyt. C, cytochrome c. C, the caspase-3 zymogen cannot be adequately activated by uncleavable single-chain caspase-9 in the presence of Apaf-1(1–591) and dATP. Apaf-1(1–591) is known to form a functional apoptosome in the presence of dATP (15). Shown is a time course of caspase-3-mediated cleavage of the substrate peptide Ac-DEVD-AMC. Only WT caspase-9 (C9) activated the caspase-3 zymogen in the presence of Apaf-1(1–591) and dATP.
Next, we examined the ability of caspase-9 to cleave and active its substrate protein, the caspase-3 zymogen. As anticipated, the caspase-3 zymogen exhibited a basal level of protease activity toward the fluorogenic caspase-3-specific substrate Ac-DEVD-AMC (Fig. 1C, blue line). WT caspase-9 alone failed to activate the caspase-3 zymogen to any significant extent (Fig. 1C, gold line) due to the relatively inactive nature of free caspase-9. Upon incubation with Apaf-1(1–591) and dATP, WT caspase-9 rapidly converted the caspase-3 zymogen into active caspase-3, as shown by the robust activity toward the substrate (Fig. 1C, green line). Apaf-1(1–591) is known to form a mini-apoptosome in the presence of dATP, which retains a similar ability to recruit and activate caspase-9 compared with the apoptosome (15). In sharp contrast to WT caspase-9, Casp9-TM failed to activate the caspase-3 zymogen in the presence of Apaf-1(1–591) and dATP (Fig. 1C, red line). This analysis strongly suggests that single-chain caspase-9 may have lost the ability to be activated by the apoptosome.
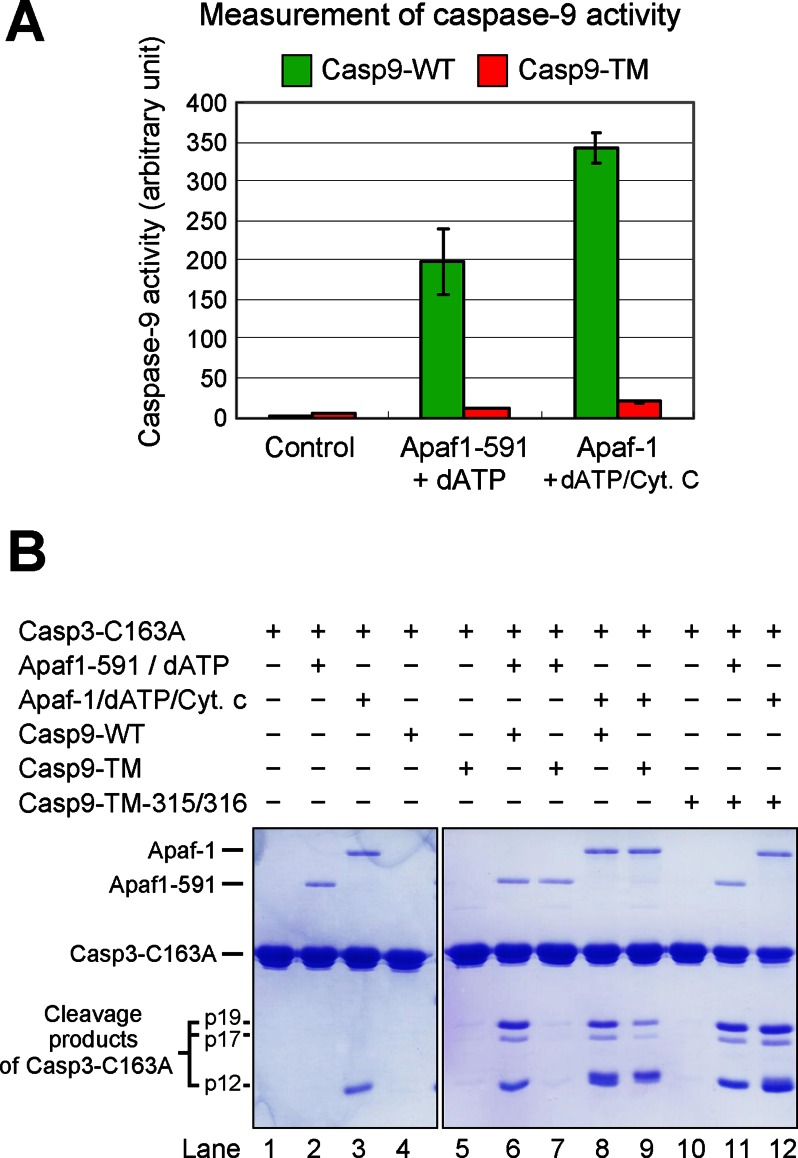
In these experiments, cleavage of caspase-3 substrate represents an indirect readout of the status of caspase-9 activation. To directly measure the protease activity of caspase-9, we employed the caspase-9 fluorogenic substrate Ac-LEHD-AFC. The results confirmed our preliminary analysis (Fig. 2A). Whereas processed WT caspase-9 was strongly activated by Apaf-1(1–591) in the presence of dATP, the protease activity of Casp9-TM was only marginally increased under similar conditions. Nearly identical results were obtained when Apaf-1(1–591) was replaced by full-length Apaf-1 and cytochrome c (Fig. 2A). In both cases, the protease activity of single-chain Casp9-TM represented ∼5% of that of processed WT caspase-9.
FIGURE 2.
WT caspase-9, but not Casp9-TM, is appropriately activated by the apoptosome. A, direct measurement of caspase-9 activity using the caspase-9-specific peptide substrate Ac-LEHD-AFC. The protease activity of WT caspase-9, but not uncleavable Casp9-TM, was markedly stimulated by the apoptosome formed by either Apaf-1(1–591) (Apaf1-591) or full-length Apaf-1. B, direct measurement of caspase-9 activity using caspase-3(C163A) as the substrate. Shown is an SDS-polyacrylamide gel stained with Coomassie Blue. Although the substrate protein was also cleaved by Casp9-TM in the presence of Apaf-1/cytochrome c (Cyt. c)/dATP (lane 9), the extent of cleavage was much smaller compared with WT caspase-9 (lane 8). Because the position of cytochrome c on the SDS-polyacrylamide gel nearly coincides with that of the p12 cleavage fragment, the majority of the lower band in lane 9 was contributed by cytochrome c.
In all experiments described above, fluorogenic peptides were used as the substrate for caspase-9 or caspase-3. To further corroborate our findings, we examined the cleavage of the caspase-3 zymogen, the biological substrate of caspase-9. To avoid self-cleavage, we generated the catalytic mutant C163A and purified the resulting caspase-3(C163A) to homogeneity (Fig. 1B, lane 7). Neither WT nor triple-mutant caspase-9 exhibited detectable protease activity toward caspase-3(C163A) (Fig. 2B, lanes 4 and 5). In the presence of Apaf-1(1–591) and dATP, WT caspase-9, but not Casp9-TM, was able to cleave the substrate protein caspase-3(C163A) into large subunits (p19 and p17) and a small subunit (p12) (Fig. 2B, lanes 6 and 7). In the presence of full-length Apaf-1, cytochrome c, and dATP, Casp9-TM also cleaved caspase-3(C163A), but the activity was markedly reduced compared with that of WT caspase-9 (Fig. 2B, lanes 8 and 9). Together, our experimental evidence demonstrates that the intrachain cleavage of caspase-9 is essential for its appropriate activation by the apoptosome.
Role of Different Intrachain Cleavages in Caspase-9
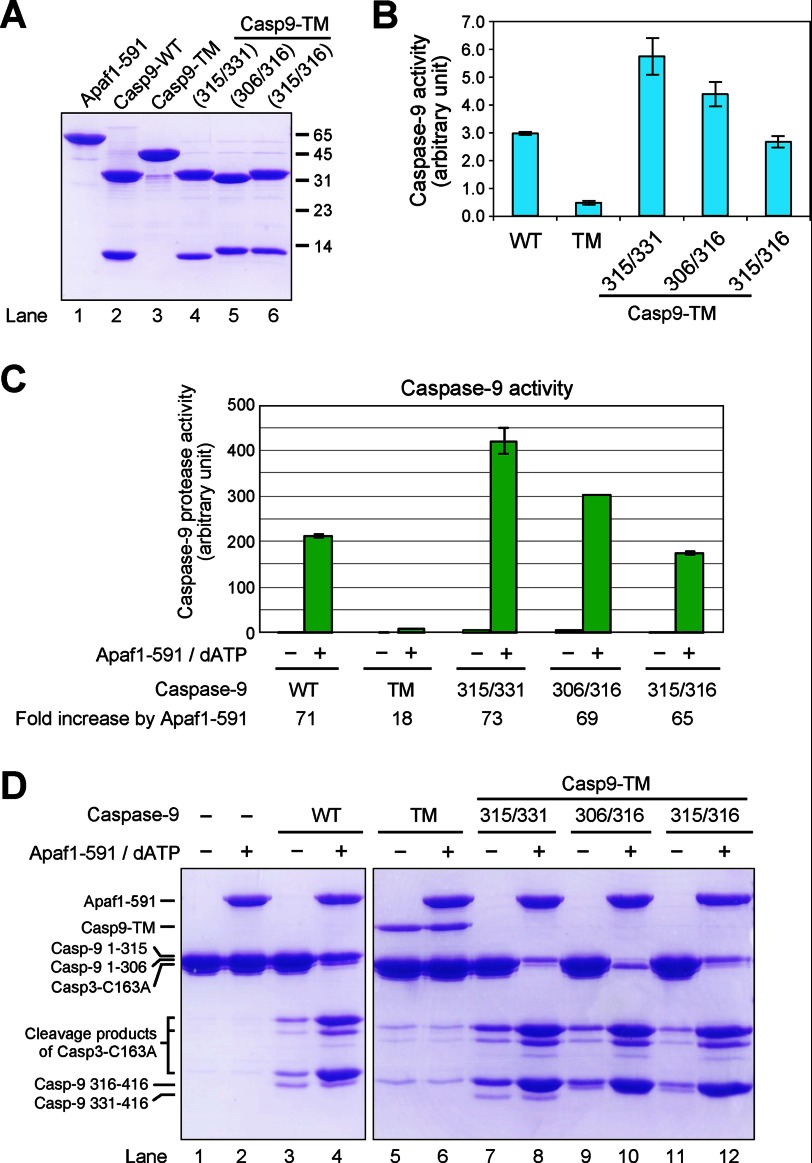
Because caspase-9 can be processed at three sites, we investigated whether different sites of processing affect the ability of caspase-9 to be activated by the apoptosome. To this end, we generated three caspase-9 variants, each containing one large subunit and one small subunit. The first variant, corresponding to the main product of caspase-9 activation cleavage, included residues 1–315 and 316–416 as the large and small subunits, respectively. To avoid secondary cleavages and to facilitate comparison with Casp9-TM, this variant contained three missense mutations, E306A, D315A, and D330A. This variant is hereafter referred to as Casp9-TM-315/316. The other two variants, referred to as Casp9-TM-315/331 and Casp9-TM-306/316, contained fragments 1–315 and 331–416 and fragments 1–306 and 316–416, respectively. They correspond to the two minor species following caspase-9 activation. Similar to Casp9-TM-315/316, the three residues Glu-306, Asp-315, and Asp-330 were all replaced with Ala to facilitate comparison with Casp9-TM. These three caspase-9 variants were individually purified to homogeneity (Fig. 3A).
FIGURE 3.
Two-chain caspase-9 variants are appropriately activated by the apoptosome. A, the three variants of two-chain caspase-9 were individually purified to homogeneity. Shown is an SDS-polyacrylamide gel stained with Coomassie Blue. Casp9–315/331 refers to residues 1–315 and 331–416. The other two variants are similarly defined. To ensure that no additional cleavages would occur in these variants, we introduced the missense mutations E306A, D315A, and D330A into the variants where applicable. B, measurement of caspase-9 activity in the absence of the apoptosome. The caspase-9-specific substrate Ac-LEHD-AFC was used in this assay. C, measurement of caspase-9 activity in the presence of Apaf-1(1–591) (Apaf1–591) and dATP. D, direct measurement of caspase-9 activity using caspase-3(C163A) as the substrate. Shown is a representative SDS-polyacrylamide gel stained with Coomassie Blue.
First, we used the fluorogenic substrate Ac-LEHD-AFC to examine the protease activity of these caspase-9 variants (Fig. 3B). In the absence of Apaf-1(1–591), the two-chain variant Casp9-TM-315/316 exhibited a similar level of protease activity compared with WT caspase-9 (Fig. 3B). The other two caspase-9 variants had slightly higher activities compared with WT caspase-9. At the protease concentrations used, the single-chain Casp9-TM variant displayed a lower protease activity compared with each of the three two-chain caspase-9 variants. In the presence of Apaf-1(1–591) and dATP, Casp9-TM-315/316 exhibited a drastically increased protease activity that was ∼65-fold higher compared with that of the free caspase-9 variant (Fig. 3C). The protease activities of WT caspase-9, Casp9-TM-315/331, and Casp9-TM-306/316 were increased by 71-, 73-, and 69-fold, respectively, in the presence of Apaf-1(1–591). Consequently, the protease activity of each of the three two-chain caspase-9 variants was either similar to or slightly higher than that of WT caspase-9. In sharp contrast, the protease activity of Casp9-TM, which was only 18-fold higher in the presence of Apaf-1(1–591) compared with its absence, was only 4.1, 3.2, and 2.3% of that of Casp9-TM-315/316, Casp9-TM-306/316, and Casp9-TM-315/331, respectively (Fig. 3C).
Next, we confirmed the results using the physiological substrate of caspase-9, caspase-3(C163A) (Fig. 3D). Both WT caspase-9 and Casp9-TM exhibited a low level of protease activity toward caspase-3(C163A) (Fig. 3D, lanes 3 and 5). In the presence of Apaf-1(1–591) and dATP, the protease activity of WT caspase-9, but not Casp9-TM, was stimulated to a markedly higher level (Fig. 3D, lanes 4 and 6). Similar to WT caspase-9 and different from Casp9-TM, each of the three caspase-9 variants exhibited a noticeably higher level of substrate cleavage in the presence of Apaf-1(1–591) and dATP (Fig. 3D, lanes 7–12). Together, our experimental evidence demonstrates that two-chain caspase-9, but not uncleavable single-chain Casp9-TM, can be adequately activated by the apoptosome. The first autoproteolytic cleavage (after Asp-315) was sufficient to confer upon caspase-9 the full ability to be activated by the apoptosome. Additional autocatalytic processing of caspase-9 does not appear to have a significant impact on this ability.
Mechanism of Compromised Ability of Casp9-TM to Be Activated by the Apoptosome
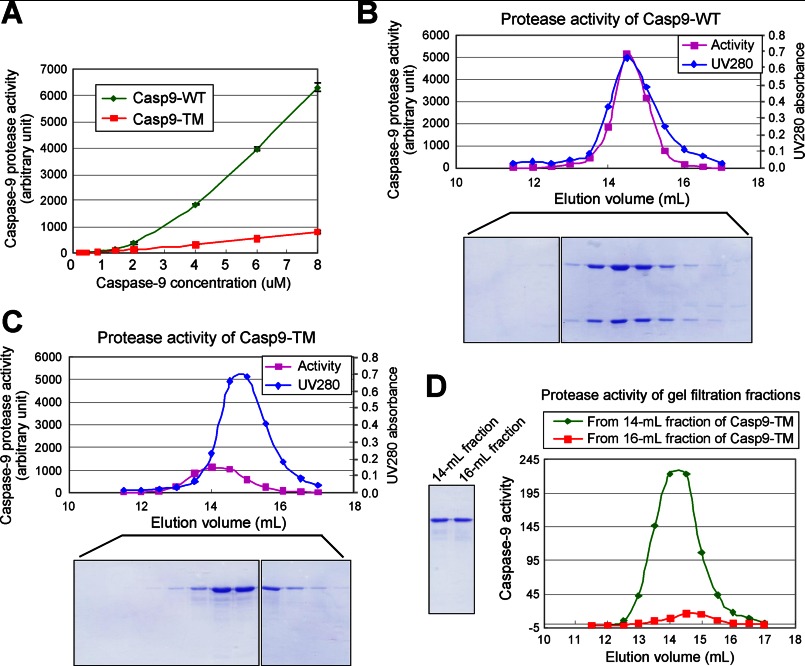
To investigate why single-chain Casp9-TM could no longer be fully activated by the apoptosome, we examined its catalytic activity at varying protease concentrations and compared the results with those obtained with two-chain WT caspase-9 (Fig. 4A). Strikingly, the catalytic activity of Casp9-TM was linearly proportional to the protease concentration over the entire range of 0.1–8 μm. In sharp contrast, the catalytic activity of WT caspase-9 increased nonlinearly and appeared to be second-order with respect to the protease concentration (Fig. 4A). In fact, the catalytic activity of Casp9-TM was higher than that of WT caspase-9 at low protease concentrations and fell below that of WT caspase-9 at 1 μm and above. At 8 μm, Casp9-TM exhibited a catalytic activity that was only 12% of that of WT caspase-9 (Fig. 4A).
FIGURE 4.
Uncleavable single-chain Casp9-TM is unable to undergo rapid equilibrium between inactive monomers and active homodimers. A, the catalytic activity of uncleavable single-chain Casp9-TM, but not WT caspase-9, is linearly proportional to the protease concentration. In contrast, the activity of WT caspase-9 appears to increase in a second-order relationship with respect to the protease concentration. B, the peak for the catalytic activity of WT caspase-9 coincides with that for the protease concentration upon gel filtration. C, the peak for the catalytic activity of single-chain Casp9-TM is ∼1.5 fractions earlier than that for the protease concentration. The elution volume for the peak activity of Casp9-TM corresponds to the approximate molecular weight of the caspase-9 homodimer. D, the elution volume for the peak activity of Casp9-TM remains unchanged during a second round of gel filtration.
This result is not surprising for WT caspase-9 because it is known to exist primarily as a catalytically inactive monomer in solution but has the ability to form a transient catalytically active homodimer. Formation of the WT caspase-9 homodimer should follow the mass equation, which explains the second-order relationship between catalytic activity and protease concentration. For Casp9-TM, however, the unanticipated result defies the mass equation. The linear relationship strongly suggests that a certain percentage of Casp9-TM is trapped in the catalytically active homodimeric form, and the catalytically inactive monomer is unable to form homodimers within the time frame of our experiments. To examine this notion, we subjected WT caspase-9 and Casp9-TM to gel filtration analysis and followed their catalytic activity (Fig. 4, B and C). The protease activity of WT caspase-9 coincided exactly with the protease concentrations (Fig. 4B), confirming the rapid equilibrium between the inactive monomer and the active homodimer. In contrast, the peak of the protease activity of Casp9-TM was shifted to earlier fractions with respect to the peak for Casp9-TM concentrations (Fig. 4C), supporting the notion that the protease activity is derived from the trapped homodimers. To further confirm this conclusion, we subjected the Casp9-TM fraction that contained the highest catalytic activity (the 14-ml fraction) to a second round of gel filtration analysis (Fig. 4D); the highest catalytic activity appeared exactly in the same fraction, the 14-ml fraction (Fig. 4D, green line), which corresponds to the molecular weight of a Casp9-TM homodimer. For a control, we also subjected the 16-ml fraction, which has approximately the same concentration of Casp9-TM as the 14-ml fraction, to gel filtration; the catalytic activity of this fraction was considerably lower than that of the 14-ml fraction (Fig. 4D, red line).
DISCUSSION
Autocatalytic processing of caspase-9 was initially reported to be unnecessary more than a decade ago (14). In that study, the cleavage of Ac-DEVD-AFC and Ac-DEVD-p-nitroanilide by caspase-3 was used as an indirect readout of caspase-9 activity in an in vitro reconstituted system involving cytosolic extract. The indirect activity measurement of caspase-3 may not faithfully represent the activation status of caspase-9 because a minute amount of active caspase-9 could be sufficient for full caspase-3 activation. This consideration, combined with the uncertain status of other activating factors from cytosolic extracts, suggests that the conclusion should be further examined. Following the initial report, two subsequent studies reported that the caspase-9 triple mutant had a similar ability to activated caspase-3 as WT caspase-9 (11, 13); in both studies, however, cleavage of the caspase-3-specific peptide substrate DEVD-AMC was measured as an indirect readout of caspase-9 activity. In a fourth study, the kcat and Km values for WT and single-chain CARD domain-deleted caspase-9, in the absence of the apoptosome, were determined to be similar using the caspase-9-specific substrate Ac-LEHD-AFC; this result led to the conclusion that the autocatalytic processing of caspase-9 was not only insufficient but also unnecessary for caspase-9 activation (12). As our results clearly show (Fig. 4A) that the activities of WT and single-chain caspase-9 have very different behavior in response to varying concentrations. The results derived from one concentration of caspase-9 may not faithfully represent its biochemical property.
In this study, we used highly purified biochemically homogeneous proteins to reconstitute a caspase-9 activity assay. Both the caspase-9-specific peptide substrate Ac-LEHD-AFC and the physiological substrate caspase-3(C163A) were used for direct measurement of caspase-9 protease activity. To avoid potential complication from minor secondary cleavages, all three specific two-chain caspase-9 variants also contained the relevant mutation E306A, D315A, or D330A. This experimental design helped validate the conclusion. Under the experimental conditions described, we saw clear and large differences between two-chain caspase-9 and uncleavable single-chain caspase-9 in terms of their ability to be activated by the apoptosome. This conclusion has been reproduced in a wide range of caspase-9 concentrations.
WT caspase-9 was once reported to exist in a kinetically trapped homodimeric state (12). Despite repeated attempts, we have never been able to detect such a state in our experimental system. Interestingly, uncleavable single-chain Casp9-TM appeared to have been partitioned into two states that barely exchanged with each other. Catalytically active homodimeric Casp9-TM could be readily isolated by gel filtration, and the protease activity still remained in the same fractions following a second round of gel filtration. A similar behavior was observed previously for another important initiator caspase, caspase-8 (17, 18).
Why is single-chain caspase-9 not fully activated by the apoptosome? We speculate that the L2 loop separating the large and small subunits of caspase-9 may play an important role in apoptosome-mediated activation. Prior to its proteolytic processing, the L2 loop may block formation of the caspase-9 homodimer. This hypothesis is consistent with the linear relationship between the protease activity and concentration of Casp9-TM, which argues that the fraction of dimerized Casp9-TM remains mostly unchanged with increasing concentrations. We further speculate that some amino acids in the L2 loop might interact with residues in the dimerization interface of caspase-9 and that autocatalytic processing after Asp-315 of caspase-9 likely weakens the interaction, thus allowing homodimerization.
Our study was performed exclusively using in vitro reconstituted systems, which include purified caspases and substrate proteins. The reliability of the in vitro assays employed in our study has been validated by a large number of previous studies (13, 18–23). Similar to our investigation, these studies employed in vitro assays to decipher the molecular mechanisms of caspase regulation in the execution of apoptosis. It is important to note that conclusions derived from such in vitro studies should be ideally verified by in vivo analysis.
This work was supported by Ministry of Science and Technology Grant 2009CB918801, National Natural Science Foundation of China Project 30888001, and the Beijing Municipal Commissions of Education, Science and Technology.
- AFC
- 7-amino-4-trifluoromethylcoumarin
- AMC
- 7-amino-4-methylcoumarin.
REFERENCES
- 1. Danial N. N., Korsmeyer S. J. (2004) Cell death: critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 2. Fuchs Y., Steller H. (2011) Programmed cell death in animal development and disease. Cell 147, 742–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lavrik I., Golks A., Krammer P. H. (2005) Death receptor signaling. J. Cell Sci. 118, 265–267 [DOI] [PubMed] [Google Scholar]
- 4. Riedl S. J., Shi Y. (2004) Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5, 897–907 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y. (2002) Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9, 459–470 [DOI] [PubMed] [Google Scholar]
- 6. Mace P. D., Riedl S. J. (2010) Molecular cell death platforms and assemblies. Curr. Opin. Cell Biol. 22, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 8. Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 [DOI] [PubMed] [Google Scholar]
- 9. Acehan D., Jiang X., Morgan D. G., Heuser J. E., Wang X., Akey C. W. (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9, 423–432 [DOI] [PubMed] [Google Scholar]
- 10. Rodriguez J., Lazebnik Y. (1999) Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13, 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) The Apaf-1·procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. (2001) Dimer formation drives the activation of the cell death protease caspase-9. Proc. Natl. Acad. Sci. U.S.A. 98, 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., Alnemri E. S. (2001) A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116 [DOI] [PubMed] [Google Scholar]
- 14. Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. (1999) Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 274, 8359–8362 [DOI] [PubMed] [Google Scholar]
- 15. Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933 [DOI] [PubMed] [Google Scholar]
- 16. Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1, 949–957 [DOI] [PubMed] [Google Scholar]
- 17. Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. (2003) A unified model for apical caspase activation. Mol. Cell 11, 529–541 [DOI] [PubMed] [Google Scholar]
- 18. Donepudi M., Mac Sweeney A., Briand C., Grütter M. G. (2003) Insights into the regulatory mechanism for caspase-8 activation. Mol. Cell 11, 543–549 [DOI] [PubMed] [Google Scholar]
- 19. Huang Y., Rich R. L., Myszka D. G., Wu H. (2003) Requirement of both the second and third BIR domains for the relief of X-linked inhibitor of apoptosis protein (XIAP)-mediated caspase inhibition by Smac. J. Biol. Chem. 278, 49517–49522 [DOI] [PubMed] [Google Scholar]
- 20. Zou H., Yang R., Hao J., Wang J., Sun C., Fesik S. W., Wu J. C., Tomaselli K. J., Armstrong R. C. (2003) Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J. Biol. Chem. 278, 8091–8098 [DOI] [PubMed] [Google Scholar]
- 21. Gao Z., Tian Y., Wang J., Yin Q., Wu H., Li Y. M., Jiang X. (2007) A dimeric Smac/Diablo peptide directly relieves caspase-3 inhibition by XIAP. Dynamic and cooperative regulation of XIAP by Smac/Diablo. J. Biol. Chem. 282, 30718–30727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burke S. P., Smith L., Smith J. B. (2010) cIAP1 cooperatively inhibits procaspase-3 activation by the caspase-9 apoptosome. J. Biol. Chem. 285, 30061–30068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keller N., Mareš J., Zerbe O., Grütter M. G. (2009) Structural and biochemical studies on procaspase-8: new insights on initiator caspase activation. Structure 17, 438–448 [DOI] [PubMed] [Google Scholar]