Background: Src homology 2 (SH2) domains are known to specifically bind to phosphotyrosine followed by a few amino acids.
Results: A novel interaction region was revealed by the solution structure of the C-terminal Src kinase SH2 domain in complex with the Csk-binding protein.
Conclusion: The novel interaction region was required for tumor suppression.
Significance: The structure sheds new light on the interaction mode of SH2 domains.
Keywords: NMR, Protein Structure, SH2 Domains, Src, Tumor, Tyrosine Protein Kinase (Tyrosine Kinase), C-terminal Src Kinase, Csk-binding protein
Abstract
Proteins with Src homology 2 (SH2) domains play major roles in tyrosine kinase signaling. Structures of many SH2 domains have been studied, and the regions involved in their interactions with ligands have been elucidated. However, these analyses have been performed using short peptides consisting of phosphotyrosine followed by a few amino acids, which are described as the canonical recognition sites. Here, we report the solution structure of the SH2 domain of C-terminal Src kinase (Csk) in complex with a longer phosphopeptide from the Csk-binding protein (Cbp). This structure, together with biochemical experiments, revealed the existence of a novel binding region in addition to the canonical phosphotyrosine 314-binding site of Cbp. Mutational analysis of this second region in cells showed that both canonical and novel binding sites are required for tumor suppression through the Cbp-Csk interaction. Furthermore, the data indicate an allosteric connection between Cbp binding and Csk activation that arises from residues in the βB/βC loop of the SH2 domain.
Introduction
Src homology 2 (SH2)3 domains are noncatalytic regions commonly observed in various types of signal transduction proteins. They function as modules that mediate the interaction between proteins by recognizing a phosphotyrosine (Tyr(P)) in the target proteins. Structural and quantitative binding analyses of many SH2 domains in complexes with related ligand peptides have shown that SH2 domains generally recognize Tyr(P) with three amino acid residues toward the C terminus (Tyr(P)-Xaa1-Xaa2-Xaa3) of the target ligands using two recognition pockets on the surface of the SH2 domains (1–4). One pocket recognizes Tyr(P) of the target primarily through electrostatic interactions and hydrogen bonds, whereas the other recognizes the remaining three amino acids (Xaa1-Xaa2-Xaa3) specifically through hydrophobic interactions. This specificity is considered to generate versatility in the interaction of SH2 domains (5, 6). However, it is still controversial whether the latter pocket alone is sufficient to determine the specificity of the associated interactions (4, 7).
The protein C-terminal Src kinase (Csk) includes SH3, SH2, and kinase domains. This kinase specifically phosphorylates a regulatory Tyr in the C-terminal tail of Src Tyr kinases (SFKs) (8, 9). This event leads to an intramolecular interaction between the Tyr(P)-containing tail and the SH2 domain of the phosphorylated SFK, shifting it to an inactive closed conformation. Thus, Csk negatively regulates the kinase activity of SFKs and plays an important role in physiological functions via signaling pathways for cell proliferation, differentiation, adhesion, and migration (10).
Although SFKs are anchored to membranes via their fatty-acylated N termini, Csk, which lacks such a fatty acylation site, exists in the cytoplasm (9). Thus, for Csk to efficiently access SFKs, Csk interacts with the Csk-binding protein (Cbp/PAG), which is localized to membrane microdomains enriched in cholesterol, glycosphingolipids, and lipid rafts, and it is subsequently recruited to the reaction space (11, 12). This interaction is known to occur between the SH2 domain of Csk and the SFK-phosphorylated Tyr-314 of Cbp, but it remains unknown whether any other region in Cbp is involved in the interaction (13). In addition, unlike SFKs, which conserve specific Tyr in their C-terminal tails for regulating activity, Csk lacks such a functional tail (9). Nevertheless, the crystal structure of Csk by itself exhibited the existence of two conformers corresponding to active and inactive forms (14). The mechanism of Csk activation is described in detail in a previous review (15). The SH2 domain appears to be required for stabilizing the active form of Csk through connection between the βB/βC loop (SH2) and the β3/αC loop (kinase). It was also reported that Csk activity is increased through interactions with phosphorylated Cbp or its peptides (13, 16–19). Therefore, it can be speculated that Cbp binding shifts the dynamic equilibrium of at least these two conformers to the more active form. However, the mechanism through which Cbp binding increases Csk activity remains unclear.
To elucidate these mechanisms, we first analyzed the interaction of Csk with various lengths of Tyr-314-containing regions of Cbp using gel filtration chromatography. On the basis of this biochemical result, we determined the tertiary solution structure of the complex of Csk-SH2 with a region of Cbp that contained both Tyr-296 and Tyr(P)-314 using liquid-state nuclear magnetic resonance (NMR) spectroscopy. We found that Csk recognizes not only the four canonical amino acids beginning with Tyr(P)-314 but also a region on the N-terminal side of Tyr(P)-314 that contains Tyr-296.
EXPERIMENTAL PROCEDURES
Preparation of Cbp Mutants
The DNA fragment for rat Cbp peptide (from 289 to 321; Cbp5) was amplified using the pGEX-6P-1 plasmid (GE Healthcare) containing the DNA fragment encoding the region from 195 to 328 of Cbp (Cbp3) by PCR using 5′-CGC GGA TCC AAG AGA TTT AGT TCC TTG TCA-3′ and 5′-GGC GAA TTC CTA TCC AGG CTT ATT CAC TGA AGA-3′ as primers (Fig. 1A). The amplified DNA fragment was cleaved with BamHI and EcoRI, and the resulting gene was inserted into the BamHI-EcoRI site of pGEX-6P-1. The pGEX-6P-1 plasmid containing the DNA fragment encoding the region from 302 to 321 of Cbp (Cbp6) or a mutant of Cbp5 (Y296F) was produced by the same method as described above using the following primers: 5′-CGC GGA TCC CCA ACT CTT ACA GAA GAG GAG-3′ and 5′-GGC GAA TTC CTA TCC AGG CTT ATT CAC TGA AGA-3′ for Cbp6 and 5′-CGC GGA TCC AAG AGA TTT AGT TCC TTG TCA TTC AAG TCT CGA-3′ and 5′-GGC GAA TTC CTA TCC AGG CTT ATT CAC TGA AGA-3′ for Cbp5-Y296F. The DNA fragment encoding the region from 312 to 321 of Cbp (Cbp7) was amplified by annealing 5′-GATCC ATG TAT TCT TCA GTG AAT AAG CCT GGA TAG G-3′ and 5′-ATTTC CTA TCC AGG CTT ATT CAC TGA AGA ATA CAT G-3′, and the amplified DNA fragment was inserted into the BamHI-EcoRI site of pGEX-6P-1. For a doubly phosphorylated Tyr(P)-296/Tyr(P)-314 peptide, a synthetic product (TORAY) was used.
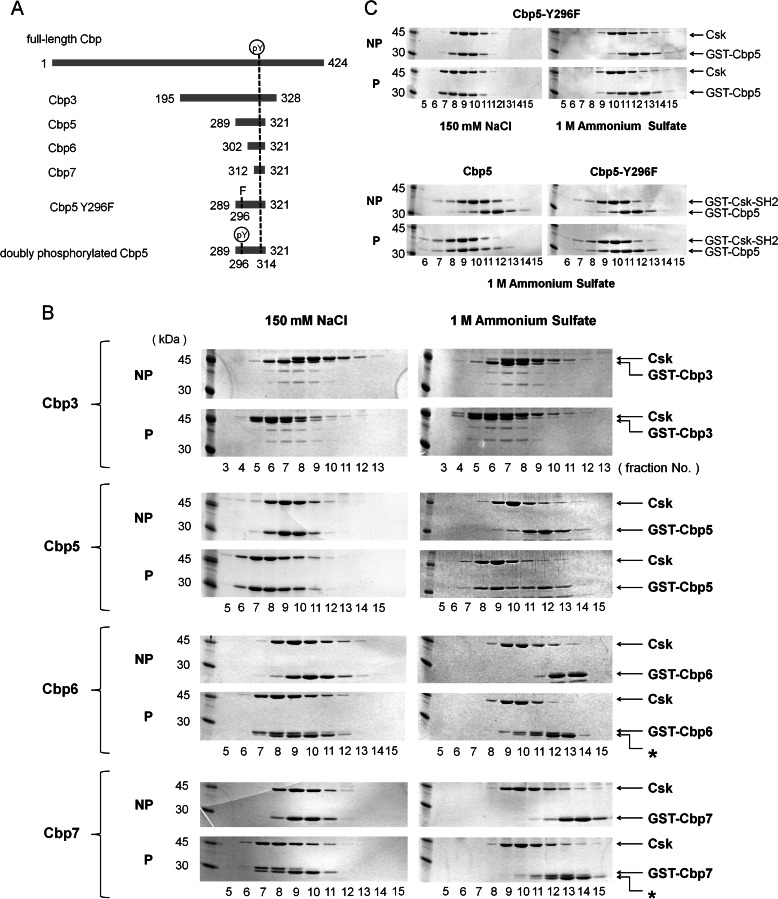
FIGURE 1.
Gel filtration assay using Cbp peptides of different lengths. A, schematic representation of intact Cbp and its various peptide fragments that appear in the text. The N- and C-terminal residue numbers are indicated on the left and right of each peptide. Tyr(P) represents the 296th or 314th phosphorylated Tyr. B, SDS-PAGE of gel-filtered fractions, including GST-fused Cbp3, Cbp5, Cbp6, or Cbp7, and Csk showing a secondary binding region between Lys-289 (N-terminal of Cbp5) and Ser-302 (N-terminal of Cbp6) on the N-terminal side of the Tyr(P) of Cbp. NP and P indicate the corresponding nonphosphorylated and phosphorylated Cbp peptides, respectively. Fractions simultaneously containing both the Cbp peptide and Csk indicate that the complex was formed in the corresponding condition. Asterisks indicate nonphosphorylated samples. C, SDS-PAGE of gel-filtered fractions, including GST-fused Cbp5-Y296F and Csk or GST-fused Csk-SH2, shows that Tyr-296 of Cbp is at least involved in the interaction with Csk or Csk-SH2. NP and P indicate the corresponding nonphosphorylated and phosphorylated Cbp peptides, respectively. All the gel filtration assays were performed in a buffer containing 150 mm NaCl (left) or 1 m (NH4)2SO4 (right).
Expression and Purification of Various Cbp Peptides
Escherichia coli BL21(DE3) cells were transformed with pGEX-6P-1 containing DNA encoding each of the Cbp peptides and grown in LB media. The cells were incubated at 25 °C with shaking at 240 rpm for 12 h. The expression of GST-fused Cbp peptides was induced by addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.1 mm when the absorbance at 600 nm was between 0.3 and 0.6. The cells were further incubated overnight, harvested by centrifugation, and stored at −80 °C. For purification, the cells were dissolved at 4 °C and disrupted by sonication in 100 mm Tris-HCl buffer (pH 7.4) containing 150 mm NaCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 1% Nonidet P-40 (lysis buffer). After centrifugation, the supernatant was applied to the GSTrap FF affinity column (GE Healthcare), and adsorbed proteins were eluted using lysis buffer (pH 9.0) containing 20 mm reduced glutathione. All purified GST-Cbp fusion proteins were phosphorylated using recombinant Fyn (Millipore) in 50 mm Tris-HCl buffer (pH 7.4) containing 3 mm MgCl2, 1 mm β-mercaptoethanol, and 4 mm ATP.
Expression and Purification of Csk
Full-length (amino acids 1–450) rat Csk was expressed using a baculovirus vector in Sf9 insect cells, as described previously (13). Cells were lysed using the above-mentioned lysis buffer containing EDTA-free protease inhibitor mixture (Nacalai Tesque) and disrupted using a Dounce homogenizer. The supernatant was collected by centrifugation and applied to the HiTrap Q HP anion exchange column (GE Healthcare) equilibrated with 50 mm Tris-HCl buffer (pH 8.0) containing 1 mm EDTA, 5% glycerol, 5 mm β-mercaptoethanol, and 0.02% octyl-d-glucoside (buffer A). The protein was eluted with a linear gradient of 75–300 mm NaCl. Protein-containing fractions were applied to the HiTrap SP HP cation exchange column (GE Healthcare) equilibrated with buffer A. The protein, eluted with a linear gradient of 50–300 mm NaCl, was applied to the Superdex 200 gel filtration column (GE Healthcare).
Expression and Purification of 13C- and 15N-Labeled Csk-SH2
To obtain 13C- and 15N-labeled proteins, E. coli Origami B(DE3) cells were transformed with pGEX-6P-1(GE Healthcare) containing the gene of Csk-SH2 and were grown in M9 minimal medium containing 1.5 g/liter 15NH4Cl and 2.0 g/liter d-[U-13C6]glucose, as nitrogen and carbon sources, respectively. For the expression of uniformly 15N-labeled proteins, d-[U-13C6]glucose was replaced with 4.0 g/liter d-glucose, and 0.1% glycerol was added to the minimal medium. Cells were incubated at 37 °C with shaking. The expression of proteins was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm when the absorbance of cells reached an A600 of 0.6. Bacteria were grown for an additional 3 h at 37 °C. Cells were collected by centrifugation and disrupted by sonication in 50 mm Tris-HCl buffer (pH 7.0) containing 400 mm NaCl, 0.1% Tween 20, 1 mm EDTA, and 1 ml of protease inhibitor mixture (Sigma). After centrifugation, the supernatant solution was passed through the DEAE-Sepharose anion exchange column (GE Healthcare) to remove contaminating nucleic acids. The eluate was applied to the GSTrap FF column (GE Healthcare), and the proteins were eluted with 50 mm Tris-HCl buffer (pH 8.0) containing 10 mm reduced glutathione. The eluate was dialyzed for a few hours at 4 °C against 2 liters of the sonication buffer. After addition of 20 μl of PreScission protease (2 units/μl; GE Healthcare) to the solution, it was further dialyzed over 17 h for cleavage of the fusion proteins. In some cases, GST was cut off by the same protease before elution of the fusion protein from the GSTrap FF column during overnight incubation at 4 °C. The proteins were concentrated using an Amicon ultra-4-centrifugal filter unit (Millipore; molecular cutoff, 3000) and were applied to the Superdex 75 gel filtration column (GE Healthcare) with a 20 mm sodium phosphate buffer solution (pH 6.0) containing 50 mm NaCl. The obtained protein (Csk-SH2) ranged from Met-80 to Met-173 with a tag derived from the expression vector (H2N-Gly-Pro-Leu-Gly-Ser-) attached to the N terminus. Protein concentration was estimated using absorbance at 280 nm (A280) with the calculated molar absorption coefficient of 16,000.
Expression and Purification of 13C- and 15N-Labeled Cbp Peptide
The Cbp5 peptide was expressed in E. coli BL21(DE3) cells (Takara) as a fusion protein with GST, which was labeled with 13C- and 15N-stable isotopes. The procedures for expression and purification were almost the same as those for Csk-SH2. After elution from the GSTrap FF column, the eluate was diluted with the same amount of 20 mm Tris-HCl buffer (pH 8.0). The solution was applied to the HiTrap DEAE FF anion exchange column (GE Healthcare), and the Cbp peptide was phosphorylated by addition of Fyn and ATP prior to removal of GST. A solution containing Csk-SH2 and Cbp5 was applied to the gel filtration column, and the fractions containing the complex were collected.
Protein Binding Assays Using Gel Filtration Chromatography
Each peptide was mixed with equimolar intact Csk or Csk-SH2 in 100 mm Tris-HCl buffer (pH 8.5) containing 150 mm NaCl or 1 m (NH4)2SO4, 5% glycerol, 5 mm β-mercaptoethanol, and 0.02% octyl-d-glucoside with or without ATP. The solution was applied to the Superdex 200 HR 10/30 (GE Healthcare) gel filtration column; A280 was monitored, and each fraction was analyzed by SDS-PAGE. The synthesized peptide and Csk were mixed in a molar ratio of 2:1, and the mixture was assayed as described above.
NMR Spectroscopy
All NMR spectra were acquired at 298 K, except the three-dimensional aromatic 13C-edited nuclear Overhauser effect spectroscopy (NOESY), which was performed at 288 K (20), using Bruker DRX-500 and DRX-600 instruments equipped with shielded triple-axis gradient triple-resonance probes, and DRX-800, AvanceII-800, and AvanceIII-950 instruments equipped with z axis gradient triple resonance cryogenic probes. For assignments of 1H, 13C, and 15N resonances, a series of two- and three-dimensional experiments were performed (21). Two-dimensional 1H-15N heteronuclear single- quantum correlation (HSQC), three-dimensional HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, HN(CA)CO, and HBHA(CBCACO)NH spectra were acquired for assignment of backbone signals. For assignment of the aliphatic side-chain signals, two-dimensional 1H-13C constant time HSQC, three-dimensional 15N-edited total correlation spectroscopy (TOCSY) with a mixing time of 79.4 ms, HCCH-TOCSY with a mixing time of 22.6 ms, and C(CO)NH and H(CCO)NH with a mixing time of 22.6 ms spectra were used. For assignment of the aromatic side-chain signals, two-dimensional 1H-1H double-quantum filtered correlation spectroscopy (22–24) and three-dimensional aromatic 13C-edited NOESY with a mixing time of 100 ms were used. The chemical shifts of the 1Hδ/ϵ spins in the aromatic residues were assigned by means of two-dimensional (Hβ)Cβ(CγCδ)Hδ and (Hβ)Cβ(CγCδCϵ)Hϵ experiments (25). For detection of intermolecular NOEs, a series of filter-related experiments were conducted, namely a 13C-filtered/13C-edited NOESY using 13C,15N-labeled Csk-SH2 complexed with nonlabeled Cbp5 peptide and a 13C-filtered/13C-edited NOESY and 13C,15N-filtered/15N-edited NOESY using 13C,15N-labeled Cbp5 peptide complexed with nonlabeled Csk-SH2 (26).
Structure Calculations
The NOE peaks were manually assigned using Sparky. Distance restraints were generated according to the assignment of the NOE cross-peaks, and pseudo-atom corrections were applied to the upper bound restraints involving methyl, methylene, and aromatic ring protons as described previously (27). Torsion angle restraints were derived using TALOS+ (28) with the assigned chemical shifts of 1Hα, 13Cα, 13Cβ, 13CO, and 15N, in reference to the x-ray structure of intact Csk (14). Hydrogen bond restraints, 2.5–3.3 Å for N-O pairs and 1.8–2.3 Å for H-O pairs, were added only to secondary structural regions as confirmed through the corresponding NOE patterns. One disulfide bond between Cys-122 and Cys-164 was confirmed through characteristic 13Cα and 13Cβ chemical shifts (29) and used as a restraint for structure calculations. Structure calculations with torsion angle dynamics were performed using CYANA-2.1 (30). A total of 100 structures were calculated with 40,000 steps, and after a root-mean-square deviation (r.m.s.d.) for the backbone atoms had reached 1.0 Å, the r−6 sum averaging method, originally implemented in the CYANA calculations, was applied instead of the pseudo-atom corrections. Finally, the 20 structures with the lowest target functions were selected. Molecular models were prepared using MOLMOL (31) and UCSF Chimera (32). Hydrogen bonds in the complex structure were defined using UCSF Chimera. An accessible surface area for each residue was calculated using MOLMOL. The electrostatic potential of the complex structure was calculated using Delphi (33). All the amino acid sequence alignments were performed using ClustalW, version 2.0 (34).
Relaxation Analysis
The 15N-spin longitudinal (R1) and transverse (R2) relaxation rates and {1H}–15N steady-state NOE values were acquired on Bruker DRX-500 and AvanceII-800 spectrometers (26). R1 relaxation delays were set at 5, 100, 200, 300, 400, 550, 700, 850, 1000, 1150, and 1300 ms, and R2 relaxation delays were set at 7.2, 36, 72, 108, 144, 180, 216, 252, 288, 324, and 360 ms in corresponding experiments. Single exponential curve fitting was performed using Sparky (35). The model-free analysis developed by Lipari and Szabo (36), with the isotropic rotational diffusion model assumed, was performed using Tensor2 (37). All NMR data were processed and analyzed using NMRPipe (38) and Sparky, respectively.
zz-Exchange Spectroscopy
Chemical exchange associated with the interaction was monitored using 15N-labeled Csk-SH2 mixed with each of the nonlabeled phosphopeptides, Cbp5, Cbp6, and Cbp7, at a molar ratio of 2:1 by two-dimensional 15N zz-exchange spectroscopy (39–41) on a Bruker Avance-III 950 spectrometer with a cryogenic TCI probe. The spectra were acquired with mixing times, τm, of 0, 20, 50, 100, 200, 350, 550, and 800 ms. For each time point, four peaks observed with intensities denoted by IAA(τm), IBB(τm), IBA(τm), and IAB(τm) are, respectively, governed by Equation 1,
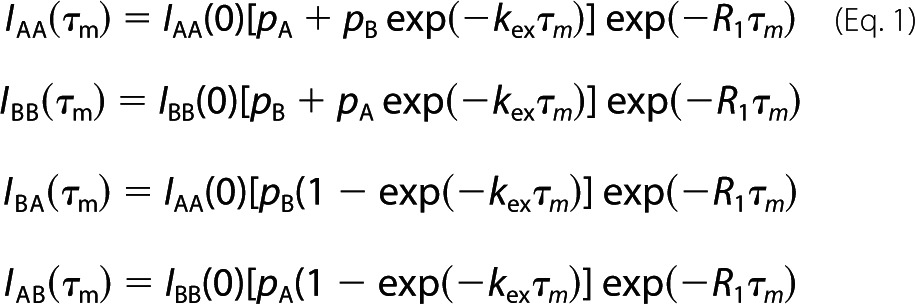 |
where pi represents the relative population for the site i; kex represents the sum of the forward, k1, and reverse, koff, kinetic rate constants for interconversion between the sites; and R1 represents the longitudinal relaxation rate of the 15N nucleus in the observed spin system. Note that k1 is an apparent pseudo-first-order rate constant that corresponds to the product of the second-order association rate constant, kon, and the concentration of free Cbp, i.e. k1 = kon[Cbp]free. The total cross-peak intensity (IBA(τm) + IAB(τm)) for each time point, τm, was fitted to the sum of the third and forth equalities given in Equation 1 to obtain kex and R1. pA and pB were estimated from the intensity ratio of the monomer and complex peaks in a two-dimensional 1H-15N HSQC spectrum. All NMR data were processed and analyzed using NMRPipe and Sparky, respectively.
Cells and Gene Transfer
A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Genes were transfected into the A549 cells using a previously described method (42). In brief, cDNAs encoding wild-type human Cbp and its mutant, in which Tyr-296 was substituted with Phe (Y296F) or both Tyr-296 and Tyr-314 were substituted with Phe (Y296F/Y314F), were subcloned into the retroviral vectors pCX4puro and pCX4bleo, respectively. All constructs were generated using a PCR-based procedure. Retroviral vectors were co-transfected into Plat-E with pGP+ pE-Ampho (Takara Bio, Japan), which is an amphotropic retrovirus-packaging construct. Transformed cell populations were selected with puromycin or bleomycin, and mixed cell populations were assayed.
Immunoprecipitation and Immunoblotting
Cells were lysed on ice in 20 mm Tris-HCl (pH 7.4) buffer containing 250 mm NaCl, 0.5 mm EDTA, 2% n-octyl-β-d-glucoside, 1% Nonidet P-40, 10% glycerol, 1 mm Na3VO4, 50 mm NaF, and protease inhibitor mixture and were clarified by centrifugation. Immunoprecipitation was performed as described previously (43). In brief, cell lysates were quantified; equal amounts of the total cell proteins were incubated with anti-Cbp antibody, and the immune complex was collected on protein G-Sepharose beads (GE Healthcare). The beads were washed three times in lysis buffer and boiled in 50 μl of the SDS sample buffer. Protein samples were separated by SDS-PAGE, transferred to PVDF membranes, immunoblotted with the indicated antibodies, and subjected to chemiluminescent detection (PerkinElmer Life Sciences). Anti-human Cbp antibody was generated by immunizing rabbits with a GST-Cbp (residues 331–430) fusion protein (44). The anti-Src antibody (Ab-1) was purchased from Calbiochem, anti-Src pY418 antibody from BIOSOURCE, anti-Csk antibody from Santa Cruz Biotechnology, and anti-Tyr(P) antibody (4G10) was from Upstate Biotechnology.
Soft Agar Colony Formation Assay
Single cell suspensions of 1 × 104 cells in 1.5 ml of DMEM containing 10% FBS and 0.36% agar were plated onto a layer of 2.5 ml of the same medium containing 0.7% agar in six-well culture dishes. Viable colonies were stained with 3-(4,5-dimethylthiazol-2–2-yl)-2,5-diphenyltetrazolium bromide 10 days after plating. Colonies larger than ∼0.1 mm in diameter were counted.
RESULTS
Gel Filtration Assay Using Cbp Peptides of Different Lengths
To identify the Cbp region responsible for the interaction with Csk, we assayed the interaction between each of the Cbp peptides of different lengths containing Tyr(P)-314 or Tyr-314 and intact Csk using gel filtration chromatography and SDS-PAGE. Deletion mutants of Cbp used for the assay, Cbp3 (195–328), Cbp5 (289–321), Cbp6 (302–321), and Cbp7 (312–321; Fig. 1A), were fused with GST and expressed in E. coli, and intact Csk was expressed in a baculovirus/insect Sf9 cell system. The GST-fused Cbp peptides were phosphorylated at Tyr-314 by addition of recombinant Fyn and ATP. We confirmed that this reaction did not phosphorylate Tyr-296 by a peptide mass fingerprinting method. Nonphosphorylated Cbp peptides were prepared as controls by mixing each peptide with Fyn in the absence of ATP.
Each of the GST-fused peptides, containing Tyr-314 or Tyr(P)-314, was mixed with Csk, and the solutions were applied to a gel filtration column. The eluted fractions were applied to SDS-PAGE to detect the contents. To evaluate the affinity qualitatively, we used two types of running buffers in the gel filtration assay, one containing 150 mm NaCl and the other containing 1 m (NH4)2SO4. The results showed that phosphorylated Cbp3 and Cbp5 eluted from the column as one peak and formed a complex with Csk at both salt concentrations, whereas the nonphosphorylated peptides eluted separately from Csk or later than the corresponding phosphorylated peptides without dominantly forming any complex with Csk (Fig. 1B). Moreover, the peak corresponding to Csk in the experiment using the phosphorylated peptides appeared earlier than that using the nonphosphorylated peptides, indicating that the former eluate contained Csk of high molecular weight, because of complex formation with the phosphorylated peptides (Fig. 1B). When Tyr-314 was not phosphorylated, none of the examined peptides dominantly formed a complex in either salt concentration. This indicates that the phosphorylation of Tyr-314 is at least necessary for interaction with Csk. Interestingly, shorter peptides lacking N-terminal regions, namely phosphorylated Cbp6 and Cbp7, formed a complex with Csk during gel filtration in the running buffer containing 150 mm NaCl but not in the buffer containing 1 m (NH4)2SO4 (Fig. 1B). Because the interaction involving the phosphate group of Tyr(P)-314 was weakened by a high salt concentration, it is expected that this interaction is electrostatic in nature. The differences observed in elusion patterns of Cbp5 and Cbp6 at high salt concentrations indicate that a region within Cbp5 but outside Cbp6 (i.e. 289–302) generates an additional interaction to Tyr(P)-314 that enables high affinity binding to Csk even at high salt concentrations.
Gel Filtration Assay Using Y296F Mutant and Phospho-Tyr-296
Here a question emerged as to which residues between 289 and 302 mediate the secondary interaction. We focused on Tyr-296 because it is a unique Tyr in this region, and we expected its phosphorylation to strengthen the interaction with Csk in combination with phosphorylation of the canonical Tyr-314. To study the possible secondary interaction region, we prepared a mutant of GST-fused Cbp5 peptide in which Tyr-296 was replaced with Phe, and we assayed its interaction with Csk or Csk-SH2 using gel filtration chromatography as described above. Although both interactions were maintained in 150 mm NaCl, they were broken in the presence of 1 m (NH4)2SO4 (Fig. 1C). As revealed by NMR spectroscopy (see below), replacement of Tyr-296 with Phe in Cbp5 weakened its interaction with Csk and Csk-SH2, probably because of the absence of a hydrogen bond between the hydroxyl group of Cbp Tyr-296 and the guanidinium group of Csk-SH2 Arg-107.
Overall Structure of the Complex of Csk-SH2 with Cbp5
The biochemical data described above, including gel filtration assays, indicate that Tyr(P)-314 interacts with Csk and that at least Tyr-296 contributes to binding affinity. To obtain detailed structural information about this interaction, we have determined the tertiary structure of the complex of Csk-SH2 with Cbp5 using multidimensional and multiresonance NMR spectroscopy. We confirmed that Csk-SH2 and phosphorylated Cbp5 (without GST) maintained a stable complex during gel filtration, whereas Csk-SH2 and nonphosphorylated Cbp5 did not by MALDI-TOF mass spectrometry. In analysis, we used not only isotope-labeled Csk-SH2 but also isotope-labeled Cbp5, which was overexpressed as a GST fusion protein in E. coli. Most of the phosphopeptides that have been used to date for structural analyses were not labeled because they have often been synthesized chemically as phosphorylated forms. However, with the isotopically labeled phosphopeptide of Cbp, we successfully obtained a high quality solution structure of the complex. The Cbp peptide was phosphorylated by Fyn and ATP during purification, and its purity was estimated to be >99% by mass spectrometry. The sequence of Cbp5 includes 33 amino acid residues, which was much longer in the N-terminal direction compared with the sequences of various other peptides that have been used for structural analyses of complexes with SH2 domains (Fig. 2A). This N-terminal sequence is not conserved in other ligands that interact with Csk-SH2 (Fig. 2B).
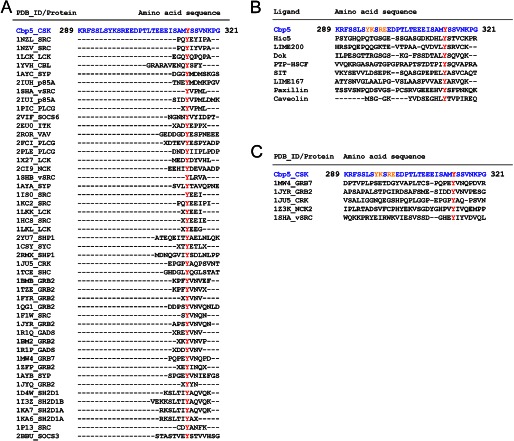
FIGURE 2.
Comparison of the amino acid sequence of Cbp5 with those of various peptide ligands associated with SH2 domains registered in PDB. The sequences were aligned using ClustalW2. The sequence of Cbp5 is shown in blue and the Tyr(P) residues in red. A, comparison of Cbp5 with other peptide ligands associated with different SH2 domains. B, comparison of Cbp5 with other peptide ligands that can interact with Csk-SH2. The secondary binding sites with Csk-SH2 are shown in orange. C, comparison of the N-terminal sequence from Tyr(P) of Cbp5 with the corresponding sequences of other peptide ligands associated with representative SH2 domains. The secondary binding sites with Csk-SH2 are shown in orange.
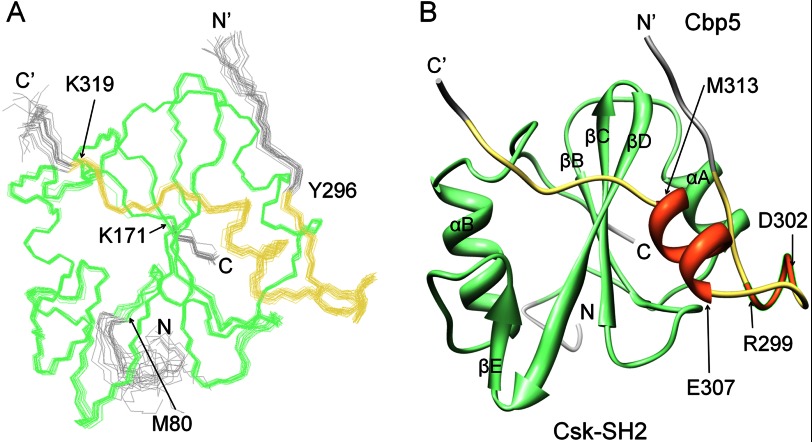
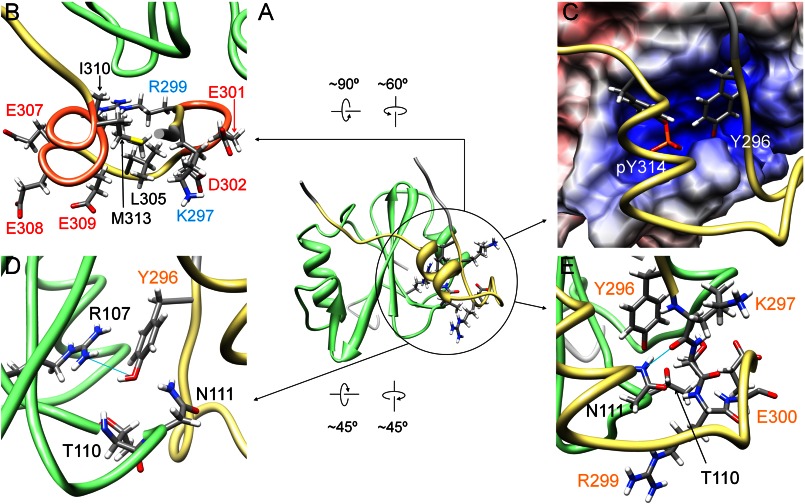
Fig. 3A shows an overlay of the final 20 structures, which exhibited the best target functions among 100 calculated structures (PDB code 2rsy). In Fig. 3B, a ribbon diagram of the representative structure with the minimum target function is presented. The coordinates of the backbone atoms of Cbp5 were well defined in the wide range between Tyr-296 and Lys-319 with respect to the coordinates of Csk-SH2. These gave an average r.m.s.d. value to the mean structure of 0.26 ± 0.04 Å for the backbone of Tyr-296–Lys-319 (Cbp) and Met-80–Lys-171 (Csk-SH2; Table 1). Bound Cbp5, regardless of its length, had a compact conformation comprising an α-helix between Glu-307 and Met-313 and a turn between Arg-299 and Asp-302 (Fig. 3B).
FIGURE 3.
Overall structure of the complex of Csk-SH2 with Cbp5. A, overlay of the best 20 structures of Csk-SH2 in complex with Cbp5. These 20 structures were selected from 100 structures that were calculated using CYANA-2.1 with minimum target function values. The structures were fitted with an r.m.s.d. value of 0.26 ± 0.04 Å with respect to the backbone atoms in the range from Met-80 to Lys-171 of Csk-SH2 (green) and from Tyr-296 to Lys-319 of Cbp5 (khaki). B, ribbon diagram of the representative structure shows (coral color) an α-helix from Glu-307 to Met-313 and a turn from Arg-299 to Asp-302.
TABLE 1.
Statistics of the final 20 best structures of Csk-SH2 and Cbp (289–321)
| NOE upper distance limits | |
| Total | 2935 |
| Intraresidue (|i − j| = 0) | 604 |
| Sequential (|i − j| = 1) | 779 |
| Medium range (1 〈|i − j | >5) | 552 |
| Long range (|i − j| > = 5) | 1000 |
| Intermolecular | 202 |
| Hydrogen bonds | 33 |
| Disulfide bond | 1 |
| Dihedral angle restraints | 230 |
| Distance restraint violations | |
| Number >0.2 Å | 0 |
| Mean r.m.s. violations from distance restraints | 0.0043 ± 0.0005 Å |
| Dihedral angle restraint violations | |
| Number >5° | 0 |
| Mean r.m.s. violations from dihedral angle restraints | 0.3307 ± 0.0655° |
| Ramachandran statistics | |
| Residues in preferred regions | 93.4% |
| Residues in allowed regions | 4.8% |
| Residues in disallowed regions | 1.8% |
| r.m.s deviation to the mean coordinatesa | |
| Backbone atoms | 0.26 ± 0.04 Å |
| Heavy atoms | 0.68 ± 0.05 Å |
a Pairwise r.m.s.d. was calculated for residues 80–172 and 296–318 among 20 refined structures.
Conventional Interaction Mode
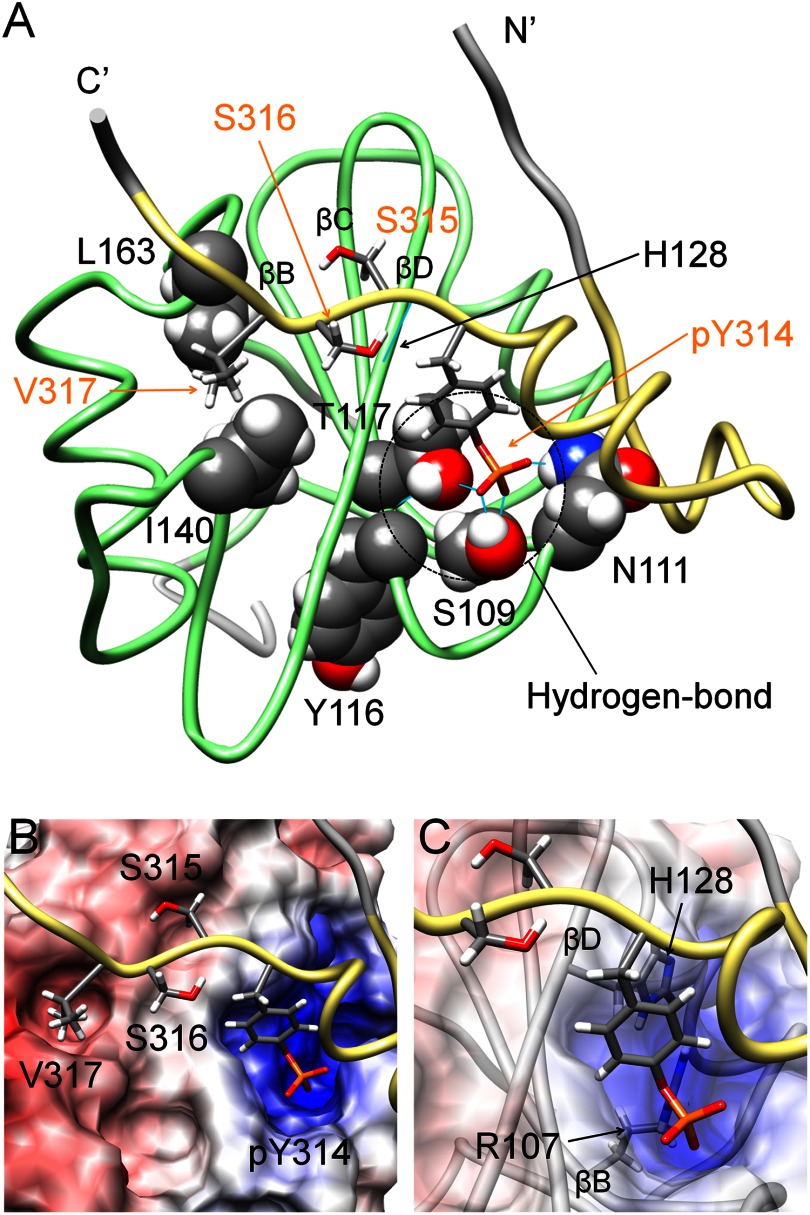
As shown in Fig. 4A, the region of Cbp5 that was expected to be specifically recognized by Csk-SH2, pYSSV, adopted an extended conformation in the complex. In the conformation, the aromatic and phosphate groups of Tyr(P)-314 were accommodated in a positively charged pocket of Csk-SH2. The electrostatic potential of this pocket was provided by Arg-107 in βB and His-128 in βD (Fig. 4, B and C). Both residues are conserved in most SH2 domains and are known to be involved in interactions with phosphotyrosines of associated ligands (45). In addition, the phosphate group of Tyr(P)-314 formed hydrogen bonds with the hydroxyl group of Ser-109 in βB, the amino (–NH2) group of Asn-111 in βC, and the amide group of Tyr-116 through the hydroxyl group of Thr-117 in βC (Fig. 4A). Another intermolecular hydrogen bond was found between the amide group of the next (Tyr(P)+1) residue Ser-315, and the carbonyl group of His-128. The (Tyr(P)+3) residue Val-317 was accommodated in a pocket containing Ile-140 and Leu-163 through hydrophobic interactions (Fig. 4, A and B). This binding mode was the same as that commonly observed for other SH2 domains and their associated phosphopeptides (1, 46–49).
FIGURE 4.
Conventional interaction mode is observed in the structure of the complex of Csk-SH2 with Cbp5. A, tube representation of the complex of Csk-SH2 with Cbp5. Amino acid residues involved in the interaction are represented using a stick model for Cbp5 and a sphere model for Csk-SH2. Hydrogen bonds are shown as cyan lines and those around Tyr(P)-314 are highlighted with dotted circles. Amino acid residues of Cbp5 and Csk-SH2 are labeled in orange and black, respectively. B, binding pockets on the surface of Csk-SH2 with residues around Tyr(P)-314 and Val-317 of Cbp5. The electrostatic potential was calculated using Delphi and mapped on the surface of the structure of Csk-SH2. Positive and negative charges are indicated in blue and red, respectively. C, Arg-107 and His-128 of Csk-SH2, which generates the positive charge around Tyr(P)-314, interact with the phosphate group of Tyr(P)-314 in the conventional manner.
Another Novel Interaction Region between Csk-SH2 and Cbp5
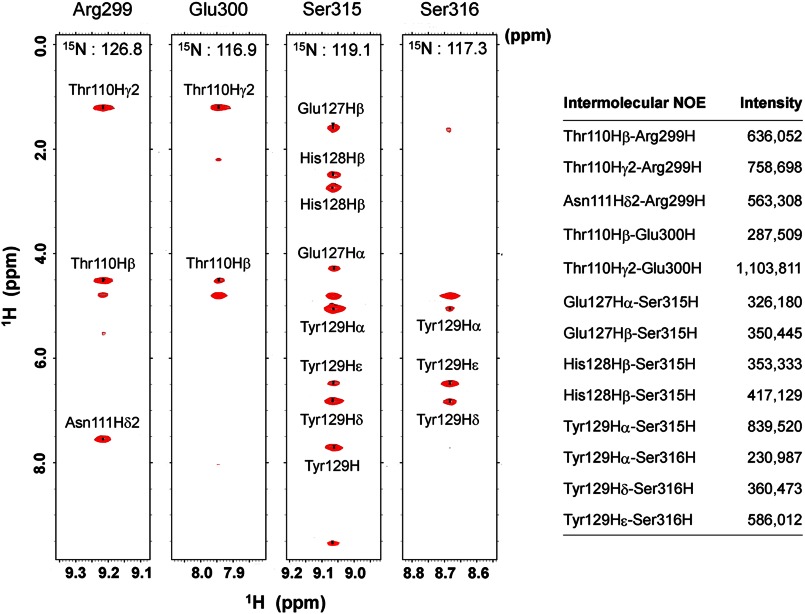
Consistent with the results of our gel filtration experiments, structure determination of the complex revealed an additional interaction region in the N-terminal direction from Tyr(P)-314 of Cbp5 (Fig. 5). This region contained a unique α-helix between Glu-307 and Met-313 and a hydrophobic core comprising aliphatic Lys-297 and Arg-299 and side chains of Leu-305, Ile-310, and Met-313. The hydrophilic side chains of Lys-297, Arg-299, Glu-301, Asp-302, Glu-307, Glu-308, and Glu-309 were exposed to the solvent (Fig. 5B). In addition, the residues ranging from Arg-299 to Asp-302 took a turn conformation that exposed the hydrophilic side chains of Glu-300, Glu-301, and Asp-302 to the solvent. In the structure of Cbp5, at least in its complex with Csk-SH2, Tyr-296 and Tyr(P)-314 were positioned such that both side chains were inserted together into the pocket of Csk-SH2 with their aromatic rings arranged side by side (Fig. 5C). Most regions (99 ± 1%) of Tyr-296 were buried in the interior of the complex structure, and the accessible surface area of its main chain and side chain was 1 ± 1 (%). This is in contrast to the accessible surface area of Tyr-296 in the free state, which is 30 ± 3 (%). Fig. 5D shows the residues of Csk-SH2 that were involved in the interaction with Tyr-296. The position of the Tyr-296 hydroxyl group allowed electrostatic interactions with Csk-SH2 via the phosphate group of Tyr(P)-314, the main-chain amide group of Thr-110, and the main-chain amide and side-chain amino groups of Asn-111. It also formed a hydrogen bond with the guanidinium group of Arg-107. These data explain the decreased affinity observed in gel filtration experiments when Tyr-296 was replaced with Phe lacking this hydroxyl group. Phosphorylation of Tyr-296 caused decreased affinity between Cbp and Csk-SH2, probably because of steric hindrance and electrostatic repulsion of bulky negatively charged phosphate groups on Tyr-296 and Tyr-314. In addition, the complex structure revealed van der Waals contacts between Arg-299/Glu-300 and Thr-110/Asn-111, and the filtered NOESY experiments exhibited strong intermolecular NOE peaks between these residues. These data indicate that Arg-299 and Glu-300 are also involved in the interaction (Figs. 5E and 6).
FIGURE 5.
Another interaction region found in the structure of the complex of Csk-SH2 with Cbp5. A, interaction region between Csk-SH2 and Cbp5 is circled in black in the ribbon diagram of the complex of Cbp5 with Csk-SH2. Amino acid residues involved in the interaction are represented using a stick model. B, Cbp5 exhibits an amphipathic character, with hydrophilic residues exposed to the solvent and hydrophobic residues and aliphatic chains oriented inward. Positively and negatively charged residues are labeled in cyan and red, respectively, and hydrophobic residues are labeled in black. C, Tyr-296 and Tyr(P)-314 of Cbp5 (in khaki) are accommodated in the binding pocket of Csk-SH2. The two residues are represented using a stick model. An α-helix and a turn of Cbp formed on Csk-SH2 bring the two Tyr residues close to each other. D, residues of Csk-SH2 interacting electrostatically with the hydroxyl group of Tyr-296. Hydrogen bonds are shown by cyan lines. Amino acid residues of Cbp5 and Csk-SH2 are labeled in orange and black, respectively. E, residues additionally found as contributors to the second interaction site. Amino acid residues of Cbp5 and Csk-SH2 are labeled in orange and black, respectively. Hydrogen bonds are shown as cyan lines. Csk-SH2 is shown in green and Cbp5 in khaki.
FIGURE 6.
Intermolecular NOEs observed for Arg-299 or Glu-300 and those for Ser-315 or Ser-316. The latter two residues are involved in the well known canonical binding to Csk-SH2. The corresponding regions are shown from a 13C,15N-filtered, 15N-edited NOESY spectrum of 13C,15N-labeled Cbp5 in complex with unlabeled Csk-SH2. The peak intensities represent the peak heights of the corresponding residues, as estimated by Sparky.
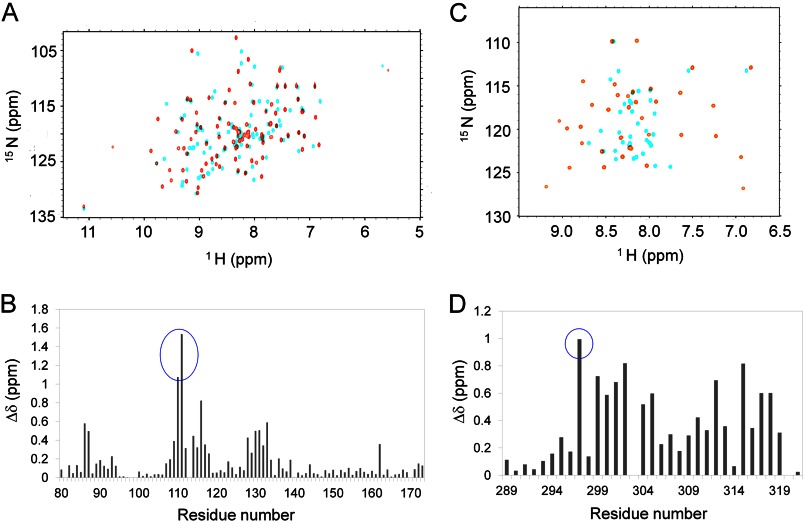
We compared the two-dimensional 1H–15N HSQC spectra of Csk-SH2 before and after complex formation (Fig. 7A) and plotted chemical shift differences against the amino acid sequence (Fig. 7B). No significant difference was observed in the overall peak distribution in the spectra. However, Thr-110 and Asn-111 exhibited the weighted average chemical shift deviations over 1.0 ppm (Fig. 7B). These data correspond to interactions of Thr-110 with Tyr-296 and Arg-299 as well as Glu-300 of Cbp and Asn-111 with Tyr-296, Arg-299, and Tyr(P)-314 of Cbp (Figs. 4 and 5). Because we labeled Cbp5 with isotopes, we compared the two-dimensional 1H-15N HSQC spectra of Cbp5 before and after complex formation (Fig. 7C) and plotted chemical shift differences against the amino acid sequence (Fig. 7D). The complex formation led to a wider distribution of Cbp5 amide 1H-15N cross-peaks (Fig. 7C). In particular, Lys-297 exhibited a chemical shift change as large as 1.0 ppm (Fig. 7D). The structure of the complex indicated a hydrogen bond between the carbonyl group in the main chain of Lys-297 and the amino group of the side chain of Asn-111, indicating that Lys-297 also contributes to the interaction of Cbp5 with Csk-SH2 (Fig. 5E). Among known ligands of SH2 domains, no residues that correspond to Tyr-296, Lys-297, Arg-299, and Glu-300 of Cbp and Thr-110 or Asn-111 of Csk-SH2 are definitely conserved (Fig. 2C). Likewise, in other SH2 domains, the residues that correspond to Thr-110 and Asn-111 are various and not conserved as expected (45).
FIGURE 7.
Chemical shift perturbation experiments of [15N]Csk-SH2 or [15N]Cbp5 in the complex. Δδ indicates the weighted chemical shift deviation that was calculated according to the following equation: Δδ = ((ΔδH)2/2 + (ΔδN/5)2/2)1/2, where ΔδH and ΔδN indicate the chemical shift changes of the amide proton and nitrogen, respectively. A, overlay of the two-dimensional 1H-15N HSQC spectra of [15N]Csk-SH2 with (orange) and without (cyan) unlabeled Cbp5. B, chemical shift differences of the two spectra are plotted against the residue number. The peaks that showed >1.0 ppm changes are highlighted with a blue circle. C, overlay of the two-dimensional 1H-15N HSQC spectra of [15N]Cbp5 with (orange) and without (cyan) unlabeled Csk-SH2. D, chemical shift differences of the two spectra are plotted against the residue number. The peak that showed ∼1.0 ppm change is highlighted with a blue circle.
Involvement of Tyr-296 in the Cbp-Csk Interaction in Cells
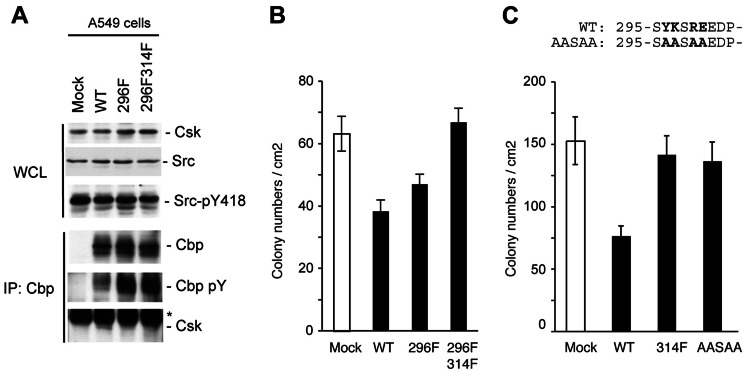
To evaluate the physiological role of Tyr-296 in stabilizing the Cbp-Csk interaction, we examined the function of a Cbp mutant (Y296F) with a substitution of Tyr-296 by Phe in human lung cancer A549 cells with marked up-regulation of c-Src. We have recently shown that the expression of Cbp is extensively down-regulated in these cells and that the re-expression of Cbp inactivates c-Src and suppresses tumor growth by facilitating the recruitment of Csk to the membrane (44). In Cbp-expressing cells, the Cbp-Csk interaction was readily detected by immunoprecipitation (Fig. 8A). However, in mutant cells, Y296F bound very weakly to Csk. Furthermore, a double mutant, in which Tyr-296 and Tyr-314 were replaced with Phe (Y296/314F), completely failed to interact with Csk. Inversely, the Tyr phosphorylation levels of Cbp, which reflect the activation status of c-Src, were significantly augmented when Cbp mutants were expressed. These observations demonstrate that Tyr-296 contributes to stabilizing the Cbp-Csk interaction even in cells. We next assessed the role of Cbp in cellular function by examining the effects of the expression of Cbp on the colony-forming ability of tumor cells. As described previously, the expression of Cbp significantly suppressed colony formation in A549 cells (Fig. 8B). However, this growth suppression was significantly attenuated when Y296F was expressed. The expression of Y296/314F did not induce significant suppression of the colony-forming ability of these cells, revealing that Tyr-296 supports the tumor-suppressive function of Cbp by stabilizing the Cbp-Csk interaction.
FIGURE 8.
Tyr-296 is required for a stable Cbp-Csk interaction and tumor-suppressive function of Cbp. A, whole cell lysates (WCL) from A549 cells and Cbp-expressing wild-type cells, Y296F mutant, and Y296F/Y314F double mutants were immunoblotted with the indicated antibodies. Cbp was then immunoprecipitated (IP) from whole cell lysates with anti-Cbp and then immunoblotted with the indicated antibody. Asterisks indicate the location of the immunoglobulin. B, effects of the expression of Cbp and its YF mutants on the tumor growth of A549 cells were examined using colony formation assays in soft agar. Colonies were stained with 3-(4,5-dimethylthiazol-2–2-yl)-2,5-diphenyltetrazolium bromide 10 days after plating. The colony numbers per cm2 are presented as mean ± S.E. from three independent experiments. C, effects of the expression of Cbp and another mutant, AASAA, on the tumor growth of A549 cells were examined as described in B. The mutated amino acids are shown in bold letters in the sequence above the graph.
Interestingly, another mutant, in which Tyr-296, Lys-297, Arg-299, and Glu-300 were replaced with Ala (AASAA), attenuated growth suppression more than Y296F did. This effect was comparable with that of mock and Y314F despite retention of Tyr-314, which is critical for the Cbp-Csk interaction (Fig. 8C). As described above, these residues in the wild-type specifically interact with Thr-110 or Asn-111 of Csk-SH2 and form the core of the second interaction region. Hence, it is clear that simultaneous replacement of all of these residues in AASAA led to decreased Csk-binding affinity and that the second recognition region is at least required for tumor suppression via the Cbp-Csk interaction.
Cbp Binding to the SH2 Domain Changes Its Structure to the More Active Form
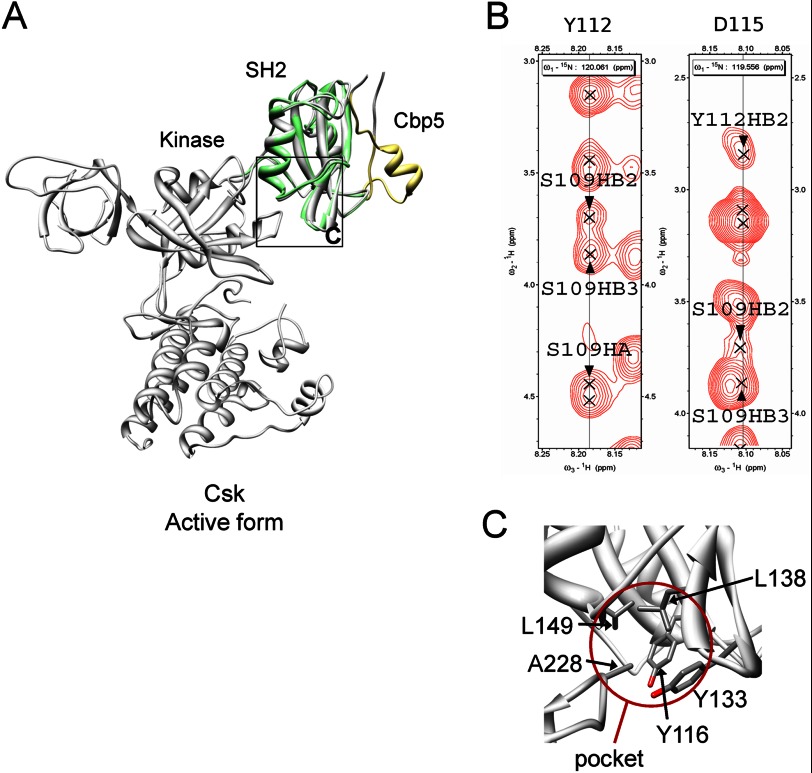
Previous x-ray analyses of intact Csk demonstrated crystal structures of both active and inactive forms, which probably establish an equilibrium in solution (14). Thus, in order for Csk to be completely active, its conformation must shift to the active form and be maintained at least during the kinase reaction. Interestingly, past experiments showed that Csk activity increased upon interaction with a Cbp phosphopeptide consisting of only six residues (AMpYSSV) or 10 residues (ISAMpYSSVNK) (13, 16–19). However, overlaying the solution structure of the complex on intact crystal structures of the free form showed that the Cbp5 peptide was distant from the catalytic domain (Fig. 9A). Therefore, it is conceivable that the catalytic domain is activated through a possible conformational change in Csk-SH2 triggered by Cbp binding to it.
FIGURE 9.
Connection between Cbp5 and the kinase domain through Csk-SH2. A, relative configuration between Cbp5 and the kinase domain revealed by overlaying of the solution structure of Csk-SH2 in complex with Cbp5 and the crystal structure of intact Csk (PDB code 1K9A). Csk-SH2 and Cbp5 in the complex are colored in green and khaki, respectively, and intact Csk is colored in gray. The region corresponding to C is shown with a black square. B, intramolecular NOEs observed among Ser-109, Tyr-112, and Asp-115 supporting the presence of the hydrogen bonds between them. The corresponding regions are shown from a 15N-edited NOESY spectrum of 15N-labeled Csk-SH2 in complex with unlabeled Cbp5. C, insertion of Ala-228 into the hydrophobic pocket formed by Tyr-116, Tyr-133, Leu-138, and Leu-149, as observed in the crystal structure of Csk. These residues are shown with a stick model. The pocket is highlighted with a brown circle.
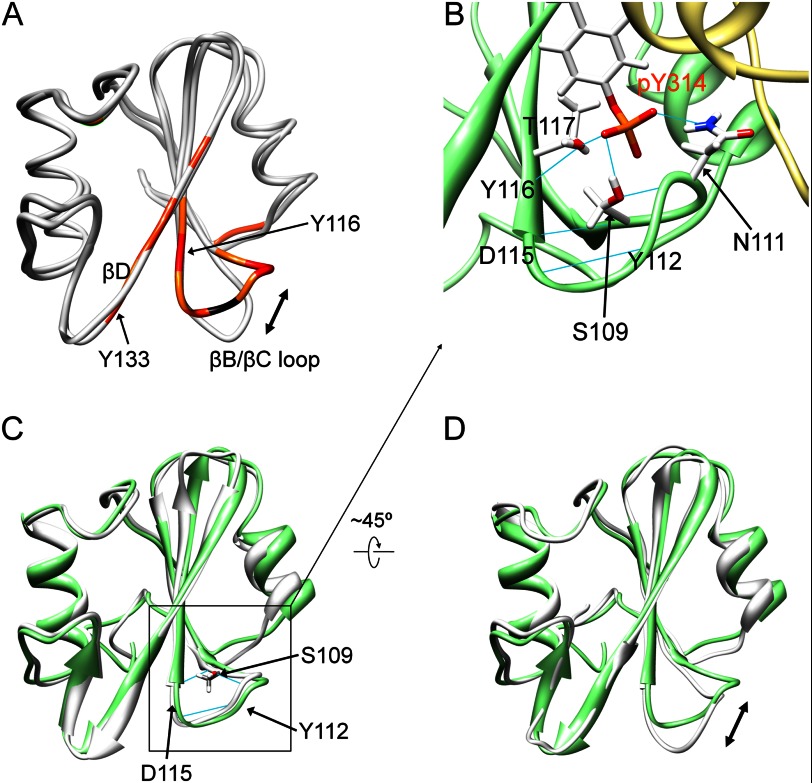
To investigate how the effect of Cbp binding is structurally transmitted to the SH2 domain, we examined the chemical shift perturbation observed for Csk-SH2 upon binding with Cbp5. Thus, mapping of the perturbed residues on the structure of Csk-SH2 suggested that its βB/βC loop and βD strand underwent conformational or dynamic changes following Cbp5 binding (Fig. 10A). We next compared the structure of Csk-SH2 of the complex with the crystal structure of the free form, which had previously been determined by x-ray analysis (PDB code 3EAC). Consistent with the above-mentioned chemical shift changes, these structures in the βB/βC loops were distinctly different from each other (Fig. 10A). Furthermore, in our complex structure, hydrogen bonds were found in the βB/βC loop around Tyr(P)-314, including those between the amide group of Ser-109 and the carbonyl group of Asp-115, the hydroxyl group of Ser-109 and the amide group of Tyr-112, and the carbonyl group of Tyr-112 and the amide group of Asp-115 (Fig. 10B). The presence of these hydrogen bonds was supported by intramolecular NOEs between these residues (Fig. 9B). Interestingly, these were also observed in the crystal structure of the active form (Fig. 10C). The complex structure also showed that the phosphate group of Tyr(P)-314 forms hydrogen bonds with Ser-109 and Asn-111 as well as with Tyr-116 through the hydroxyl group of Thr-117 (Fig. 10B). It is noteworthy that the novel binding sites, Tyr-296, Lys-297, Arg-299, and Glu-300, of Cbp bound with Thr-110 and Asn-111 in the same βB/βC loop of Csk-SH2 (Fig. 5, D and E). These novel binding sites apparently contribute to maintenance of a network of hydrogen bonds around the βB/βC loop, with the phosphate group of Tyr(P)-314 as the center.
FIGURE 10.
Characterization of the structure of Csk-SH2 in complex with Cbp5. A, mapping of the chemical shift deviations in Csk-SH2 upon binding with Cbp5 on the structure of Csk-SH2 and comparison of the solution structure of Csk-SH2 of the complex with the crystal structure of the free form (gray; PDB code 3EAC). The residues with weighted chemical shift deviations larger than 0.8 ppm are colored in red; those between 0.8 and 0.4 ppm are reddish orange, and those between 0.4 and 0.3 ppm are orange. Pro residues are colored in black. Both Tyr-116 and Tyr-133, which are involved in the hydrophobic pocket in contact with Ala-228 in the kinase domain of active Csk (Fig. 9), underwent significant chemical shift changes that were not due to direct contact with Cbp5. B, hydrogen bond network generated between Tyr(P)-314 of Cbp5 and its nearby residues in the βB/βC loop of Csk-SH2. The side chains of these residues are represented by a stick model and hydrogen bonds are shown as blue lines. Amino acid residues of Cbp5 and Csk-SH2 are labeled in orange and black, respectively. C, comparison between the solution structures of the complex of Csk-SH2 with Cbp5 (green) and the crystal structure of intact Csk-SH2 in the active form (gray; PDB code 1K9A). Hydrogen bonds are shown as cyan lines. The r.m.s.d. value of the heavy atoms in the main chains was 0.81 Å. βB/βC loops, which have almost the same conformation in both structures, are highlighted with black squares. D, comparison between the solution structure of the complex and the crystal structure of intact Csk-SH2 in the inactive form (PDB code 1K9A). The r.m.s.d. value of the heavy atoms in the main chains was 0.92 Å. βB/βC loops, indicated with a double-headed arrow, take different conformations in the two structures.
In contrast, these hydrogen bonds were not observed in the crystal structure of the inactive form. The residue pairs in the βB/βC loop in the inactive form are too distant to form such hydrogen bonds. Thus, the structure of Csk-SH2 in complex with Cbp5 was more similar to the crystal structure of Csk-SH2 in the active form than to the one in the inactive form (Fig. 10, C and D). Taken together, these observations show that Cbp binding to the SH2 domain generated a hydrogen bond network around the βB/βC loop, shifting its conformation to the more active form.
NMR Relaxation Analysis
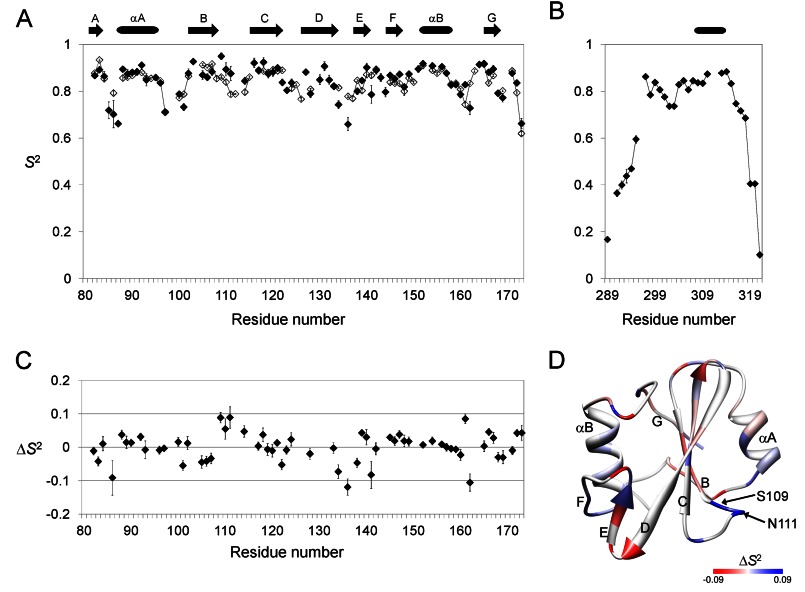
To evaluate changes in the structural dynamics of Csk-SH2 induced by Cbp binding, we measured 15N R1 and R2 relaxation rates, and {1H}-15N steady-state NOE values of uniformly 15N-labeled Csk-SH2 with and without uniformly 15N-labeled Cbp5 peptide. Resonances showing severe overlap or low intensity were excluded from the study. The average rotational correlation time τm was estimated as 5.8 and 8.4 ns for Csk-SH2 in the free and complex forms, respectively. Using these relaxation data, we analyzed residue-specific internal motions of Csk-SH2 in the two forms in terms of the order parameters squared (S2) by the model-free approach (Fig. 11, A and B). Fig. 11C shows the difference between the S2 values of Csk-SH2 with and without Cbp5. The two forms of Csk-SH2 exhibited no significant difference in overall dynamics. The average S2 values for Trp-82–Val-172 of Csk-SH2 were 0.85 ± 0.01 (one S.D.) in the free form and 0.83 ± 0.01 in the complex form. However, the S2 values in loop regions exhibited significant differences between the two forms. In particular, Leu-86 in the N-terminal region, Ser-136 in the βD/βE loop (turn), Asp-141 in the βE/βF loop, and Gly-162 in the αB/βG loop had S2 values that were ∼0.1 less in the bound form. In contrast, Ser-109 and Asn-111 in the βB/βC loop and Asp-161 in the αB/βG loop had greater S2 values (about 0.1) in the bound form. These differences are mapped on the structure of Csk-SH2 (Fig. 11D). The S2 values of Cbp5 in the complex exceeded 0.65 (mean; 0.80 ± 0.01) over the range of 22 amino acids from Tyr-297 to Asn-318 (Fig. 11D). These data indicate that a wide range of the region, including a secondary interaction site, stably bound with Csk-SH2.
FIGURE 11.
Relaxation analyses of 15N-labeled Csk-SH2 of the free and complex form and 15N-labeled Cbp5 of the complex form. The data were obtained at a 15N resonance frequency of 50.68 MHz, which corresponded to a 1H resonance frequency of 500.13 MHz. These relaxation data indicate that Cbp5 in the complex takes a stable conformation across a wide range that includes the N-terminal region from Tyr(P) (residues between Tyr-296 and Asn-318). A–C, plots of the generalized order parameter squared (S2) obtained from the model-free analysis of the 15N-spin relaxation rates and {1H}-15N steady-state NOE data at 11.7 tesla for Csk-SH2 (A) and Cbp5 (B) and of the changes in S2 induced by Cbp-binding (ΔS2 = S2complex − S2free) (C) as a function of residue number. Open diamonds correspond to the free form of Csk-SH2 and filled diamonds the complex form of Csk-SH2 and Cbp5. D, mapping of the changes in the generalized order parameter squared induced by Cbp-binding (ΔS2) on the structure of Csk-SH2. Positive to negative changes are gradually colored in blue to red. Residues showing ΔS2 above +0.09 or below −0.09 are colored in blue or red, respectively.
zz-Exchange Spectroscopy
To evaluate the affinity of Csk-SH2 for the secondary binding site of Cbp, we performed zz-exchange spectroscopy of 15N-labeled Csk-SH2 mixed with each of Cbp5, Cbp6, and Cbp7. Kinetic data were analyzed from well resolved peaks of two residues: Gly-162, which binds directly to Cbp, and Val-172. Although Val-172 is located at the opposite side of the binding region, it exhibited an NMR peak pattern similar to that of Gly-162 because of a possible small conformational change synchronized with Cbp binding. As a result, Cbp5, the longest peptide that includes the secondary binding site, exhibited distinctly lower exchange rates (koff = 0.80 s−1 for Gly-162), whereas both Cbp6 and Cbp7 exhibited ∼8–25 times higher koff values for Gly-162 (koff = 6.69 s−1 for Cbp6 and koff = 23.1 s−1 for Cbp7) (Table 2). These results evidently indicated that longer peptides interacted more strongly and that Cbp5 had the highest affinity because of the additional interaction, which Cbp6 and Cbp7 lack.
TABLE 2.
Chemical exchange parameters of Csk-SH2 in the presence of Cbp5, Cbp6, or Cbp7
| Peptide | SH2 | pB/pA | koff | kex | R1 |
|---|---|---|---|---|---|
| s−1 | s−1 | s−1 | |||
| Cbp5 | Gly-162 | 1.04 ± 0.02 | 0.80 ± 0.05 | 1.65 ± 0.10 | 1.66 ± 0.09 |
| Val-172 | 0.86 ± 0.08 | 1.78 ± 0.15 | 1.69 ± 0.13 | ||
| Cbp6 | Gly-162 | 1.22 ± 0.02 | 6.69 ± 0.37 | 15.0 ± 0.8 | 1.13 ± 0.04 |
| Val-172 | 6.72 ± 0.69 | 15.1 ± 1.5 | 1.23 ± 0.06 | ||
| Cbp7 | Gly-162 | 0.90 ± 0.05 | 23.1 ± 3.2 | 44.0 ± 5.6 | 1.56 ± 0.07 |
| Val-172 | 16.4 ± 6.0 | 31.2 ± 11.4 | 1.24 ± 0.13 |
DISCUSSION
It is generally accepted that SH2 domains bind to ligands by specifically recognizing a Tyr(P) along with the +1 to +3 amino acid residues that follow in the C-terminal direction. Our complex structure of Csk-SH2 with Cbp5 showed that Tyr(P)-314 of Cbp and the three residues that follow toward the C terminus interacted with Csk-SH2 in the same manner as observed in many other phosphopeptides that interact with normal SH2 domains. This observation was supported by our gel filtration experiments and indicates that Csk-SH2 is a normal SH2 domain that mediates typical interaction patterns. However, we have identified a novel binding region of Cbp with Csk-SH2. Analyses of intermolecular NOE and 15N-spin relaxation rates indicated that the region of Cbp that interacts with Csk-SH2 spans the 23 amino acid residues between Tyr-296 and Asn-318.
Even when nonlabeled Csk-SH2 was added to a solution of nonphosphorylated [15N]Cbp5, the two-dimensional 1H–15N HSQC spectrum of Cbp5 exhibited neither chemical shift changes nor signal broadening in its amide resonance peaks. None of the amide peaks that were assigned to a region close to Tyr-296 in the sequence underwent any chemical shift perturbation either. This indicates that the novel N-terminal interaction region of Cbp alone is not sufficient to enable Cbp5 to bind to Csk-SH2. In fact, addition of a phosphate group to Tyr-314 induced amide chemical shift changes of not only Tyr-314 but also Asp-302 and Lys-289, which are distant from Tyr-314 in the context of the amino acid sequence. In addition, our preliminary molecular dynamic simulation suggested a bent conformation of Cbp5, with Tyr(P)-314 close to Tyr-296 even in its free state, rather than a randomly fluctuating extended conformation as often seen in short peptides (data not shown). Thus, there may be interference between Tyr-314 and Tyr-296, and when Tyr-314 is not phosphorylated, Tyr-296 may also fail to interact with Csk-SH2. In either case, the result shows that phosphorylation of Tyr-314 dominantly controls association and dissociation with Csk-SH2.
Although the second binding region containing Tyr-296 certainly contributes to the interaction, it appears to enhance the selectivity of Csk-SH2 for this ligand. Intact Cbp possesses six Tyr residues that can undergo phosphorylation (13). Among them, the three residues following Tyr-224 (pYASV) are almost the same as those following Tyr-314 (pYSSV) except for Ala and Ser at the +1 position in the respective sequences. In fact, another ligand for Csk-SH2, SIT, possesses this pYASV sequence as the binding site (50). In addition, the short peptide GDGpYXXXSPLLL, in which pYXXX corresponds to any of the sequences pY(T/A/S)(K/R/Q/N)(M/I/V/R), is reportedly recognized by Csk-SH2 (51). Furthermore, sequential alignment of Cbp5 with other ligands containing Tyr(P) residues showed that the N-terminal interaction region around Tyr-296 of Cbp5 is unique and that the corresponding regions are rich in variety in the sequences of other ligands (Fig. 2B). These results indicate that the sequence that includes a few residues following Tyr(P)-314 is too short to ensure strict selectivity by Csk-SH2. Through much discussion of the specificity of many SH2 domains and corresponding ligands, it has been concluded that SH2 domains generally have no distinctive selectivity for short ligands (52). However, this type of discussion has been based on peptide ligands that include about 10 amino acids as the longest region (2, 51, 53), probably because such short regions of peptides, exclusively analyzed by x-ray crystallography or NMR, take particularly rigid conformations in complex with corresponding SH2 domains. In reality, however, a region other than that containing Tyr(P) may contribute to the specificity of binding and may play an essential role in the biological and physiological function of each SH2 domain, as demonstrated by the complex structure of Csk-SH2 and Cbp5. Indeed, it is reported that the SH2 domain of phospholipase Cγ recognizes a region of the ligand wider than the conventional recognition sequence (54). Such wide range recognition is expected as a common characteristic of SH2 domains once longer peptide ligands or intact proteins are examined in structural analyses of these complexes.
According to a sequence alignment of various SH2 domains, including Csk-SH2, a part of the additional interaction region found in Csk-SH2 (Thr-110 and Asn-111 in the βB/βC loop) is unique to Csk-SH2 and is not conserved in other SH2 domains (45). Interestingly, it is reported that the N-terminal SH2 domain of phospholipase Cγ interacts with the activated Tyr kinase domain of FGFR1. This occurs through Thr-590–Val-592 residues in the βB/βC loop of the SH2 domain as a secondary binding site with Gln-606, Asp-755, Val-758, and Ala-759 of FGER1 (54). This region in the βB/βC loop matched the one that we identified as the secondary interaction site in our experiments. In particular, Thr-590 of phospholipase Cγ-SH2 structurally corresponded to Asn-111 of Csk-SH2. Furthermore, in various ligands of SH2 domains, the residues that corresponded to Tyr-296, Lys-297, Arg-299, and Glu-300 of Cbp that interacted with Thr-110 or Asn-111 of Csk-SH2 are definitely not conserved (Fig. 2C). Taken together, residues in the βB/βC loops in many other SH2 domains may also be involved in interactions with their ligands, and the versatility of the residues in these loops may enhance the ligand specificity of each SH2 domain.
A two-dimensional 1H-15N HSQC spectrum of Cbp5 in the free form exhibited most of the main-chain amide signals crowding within a 1H resonance range between 8.0 and 8.5 ppm, suggesting that no distinct conformation exists dominantly in the free state of Cbp (Fig. 5C). Although, as described above, particular conformations may be taken, these are unstable. However, Cbp5 in the complex state had a hydrophobic core (Leu-305, Ile-310, and Met-313 and the aliphatic parts of Lys-297 and Arg-299) and hydrophilic residues on the surface (Lys-297, Arg-299, Glu-301, Asp-302, Glu-307, Glu-308, and Glu-309) and was stabilized by adopting a characteristic conformation (Fig. 5B). These characteristics eluded studies of shorter peptide ligands in complex with SH2 domains. Importantly, this structural compaction allowed the Cbp5 peptide to interact with a limited region in the βB/βC loop of Csk-SH2, in which a characteristic hydrogen bond network was observed.
Our NMR study revealed that Cbp binding to Csk-SH2 formed a hydrogen bond network around the βB/βC loop, with the phosphate group of Tyr(P)-314 as the center. According to the crystal structure of intact Csk in the active form, Tyr-116 in this hydrogen bond network forms a hydrophobic pocket with Tyr-133, Leu-138, and Leu-149 that interacts with the catalytic domain through the methyl group of Ala-228 (Fig. 9C). However, these residues are geometrically distant from each other in the inactive form. Interestingly, a recent study reported that any mutation in Tyr-116, Leu-138, or Leu-149 in the hydrophobic pocket or in Ala-228 in the kinase domain to Ala or Gly decreases Csk activity to 10–20% of the wild type in the absence and presence of a Cbp peptide (18). These experiments demonstrate that binding of Ala-228 to this hydrophobic pocket is essential for Csk activation. Cumulatively, these observations suggest that a hydrogen bond network between Tyr(P)-314, probably involving the novel binding sites, of Cbp and the above-mentioned residues in Csk-SH2 maintains the active form, stabilizes packing between the hydrophobic pocket, including Tyr-116 of the SH2 domain and Ala-228 in the catalytic domain, and consequently leads to allosteric activation of Csk (Figs. 10B and 9C).
In conclusion, we determined the solution structure of the complex of Csk-SH2 with Cbp. This structure revealed the presence of a novel binding region in Cbp that is separate from the canonical binding region. Furthermore, mutational analysis in cells showed that both canonical and novel binding sites are required for tumor suppression via the Cbp-Csk interaction. These findings indicate that conventionally known interaction sites in target ligands do not explain the biological functions of the associated SH2 domains alone.
Acknowledgments
We thank Dr. Alexander Tarakhovsky for critical reading of the manuscript. We also thank Tomoyo Takai, Eriko Chikaishi, Keiko Uchida, and Youko Monobe for helping us prepare the samples; Hironobu Yoshimura for helping us conduct 15N-relaxation experiments, and Dr. Kris Frost for modifying the title and capsule.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2rsy) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SH2
- Src homology 2
- Csk
- C-terminal Src kinase
- Cbp
- Csk-binding protein
- PAG
- phosphoprotein associated with glycosphingolipid
- HSQC
- heteronuclear single quantum correlation
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- SFK
- Src family kinase.
REFERENCES
- 1. Waksman G., Shoelson S. E., Pant N., Cowburn D., Kuriyan J. (1993) Binding of a high-affinity phosphotyrosyl peptide to the Src Sh2 domain–Crystal structures of the complexed and peptide-free forms. Cell 72, 779–790 [DOI] [PubMed] [Google Scholar]
- 2. Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J., et al. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 [DOI] [PubMed] [Google Scholar]
- 3. Schlessinger J., Lemmon M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12. [DOI] [PubMed] [Google Scholar]
- 4. Pawson T., Nash P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 5. Piccione E., Case R. D., Domchek S. M., Hu P., Chaudhuri M., Backer J. M., Schlessinger J., Shoelson S. E. (1993) Phosphatidylinositol 3-kinase p85 SH2 domain specificity defined by direct phosphopeptide/SH2 domain binding. Biochemistry 32, 3197–3202 [DOI] [PubMed] [Google Scholar]
- 6. Ladbury J. E., Arold S. (2000) Searching for specificity in SH domains. Chem. Biol. 7, R3–R8 [DOI] [PubMed] [Google Scholar]
- 7. Bradshaw J. M., Mitaxov V., Waksman G. (2000) Mutational investigation of the specificity determining region of the Src SH2 domain. J. Mol. Biol. 299, 521–535 [DOI] [PubMed] [Google Scholar]
- 8. Nada S., Yagi T., Takeda H., Tokunaga T., Nakagawa H., Ikawa Y., Okada M., Aizawa S. (1993) Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 73, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 9. Nada S., Okada M., MacAuley A., Cooper J. A., Nakagawa H. (1991) Cloning of a complementary DNA for a protein-tyrosine kinase that specifically phosphorylates a negative regulatory site of p60c-src. Nature 351, 69–72 [DOI] [PubMed] [Google Scholar]
- 10. Chow L. M., Fournel M., Davidson D., Veillette A. (1993) Negative regulation of T-cell receptor signalling by tyrosine protein kinase p50csk. Nature 365, 156–160 [DOI] [PubMed] [Google Scholar]
- 11. Kawabuchi M., Satomi Y., Takao T., Shimonishi Y., Nada S., Nagai K., Tarakhovsky A., Okada M. (2000) Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature 404, 999–1003 [DOI] [PubMed] [Google Scholar]
- 12. Brdicka T., Pavlistová D., Leo A., Bruyns E., Korínek V., Angelisová P., Scherer J., Shevchenko A., Hilgert I., Cerný J., Drbal K., Kuramitsu Y., Kornacker B., Horejsí V., Schraven B. (2000) Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein-tyrosine kinase csk and is involved in regulation of T cell activation. J. Exp. Med. 191, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeuchi S., Takayama Y., Ogawa A., Tamura K., Okada M. (2000) Transmembrane phosphoprotein Cbp positively regulates the activity of the carboxyl-terminal Src kinase, Csk. J. Biol. Chem. 275, 29183–29186 [DOI] [PubMed] [Google Scholar]
- 14. Ogawa A., Takayama Y., Sakai H., Chong K. T., Takeuchi S., Nakagawa A., Nada S., Okada M., Tsukihara T. (2002) Structure of the carboxyl-terminal Src kinase, Csk. J. Biol. Chem. 277, 14351–14354 [DOI] [PubMed] [Google Scholar]
- 15. Ia K. K., Mills R. D., Hossain M. I., Chan K. C., Jarasrassamee B., Jorissen R. N., Cheng H. C. (2010) Structural elements and allosteric mechanisms governing regulation and catalysis of CSK-family kinases and their inhibition of Src-family kinases. Growth Factors 28, 329–350 [DOI] [PubMed] [Google Scholar]
- 16. Wong L., Lieser S. A., Miyashita O., Miller M., Tasken K., Onuchic J. N., Adams J. A., Woods V. L., Jr., Jennings P. A. (2005) Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J. Mol. Biol. 351, 131–143 [DOI] [PubMed] [Google Scholar]
- 17. Lin X., Ayrapetov M. K., Lee S., Parang K., Sun G. (2005) Probing the communication between the regulatory and catalytic domains of a protein-tyrosine kinase, Csk. Biochemistry 44, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 18. Mikkola E. T., Gahmberg C. G. (2010) Hydrophobic interaction between the SH2 domain and the kinase domain is required for the activation of Csk. J. Mol. Biol. 399, 618–627 [DOI] [PubMed] [Google Scholar]
- 19. Lin X., Wang Y., Ahmadibeni Y., Parang K., Sun G. (2006) Structural basis for domain-domain communication in a protein-tyrosine kinase, the C-terminal Src kinase. J. Mol. Biol. 357, 1263–1273 [DOI] [PubMed] [Google Scholar]
- 20. Lin Z., Xu Y., Yang S., Yang D. (2006) Sequence-specific assignment of aromatic resonances of uniformly 13C,15N-labeled proteins by using 13C- and 15N-edited NOESY spectra. Angew. Chem. Int. Ed. Engl. 45, 1960–1963 [DOI] [PubMed] [Google Scholar]
- 21. Bax A. (1994) Multidimensional nuclear-magnetic-resonance methods for protein studies. Curr. Opin. Struct. Biol. 4, 738–744 [Google Scholar]
- 22. Piantini U., Sorensen O. W., Ernst R. R. (1982) Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104, 6800–6801 [Google Scholar]
- 23. Shaka A. J., Freeman R. (1983) Simplification of NMR-spectra by filtration through multiple-quantum coherence. J. Magn. Reson. 51, 169–173 [Google Scholar]
- 24. Derome A. E., Williamson M. P. (1990) Rapid-pulsing artifacts in double-quantum-filtered cosy. J. Magn. Reson. 88, 177–185 [Google Scholar]
- 25. Yamazaki T., Formankay J. D., Kay L. E. (1993) 2-Dimensional Nmr experiments for correlating C-13-β and H-1-δ/ϵ chemical-shifts of aromatic residues in C-13-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115, 11054–11055 [Google Scholar]
- 26. Zwahlen C., Legault P., Vincent S. J., Greenblatt J., Konrat R., Kay L. E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage λN-peptide/boxB RNA complex. J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 27. Umitsu M., Morishita H., Murata Y., Udaka K., Akutsu H., Yagi T., Ikegami T. (2005) 1H, 13C, and 15N resonance assignments of the first cadherin domain of cadherin-related neuronal receptor (CNR)/protocadherin α. J. Biomol. NMR 31, 365–366 [DOI] [PubMed] [Google Scholar]
- 28. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma D., Rajarathnam K. (2000) C-13 NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR 18, 165–171 [DOI] [PubMed] [Google Scholar]
- 30. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 31. Koradi R., Billeter M., Wuthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 32. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 33. Gilson M. K., Sharp K. A., Honig B. H. (1988) Calculating the electrostatic potential of molecules in solution–Method and error assessment. J. Comput. Chem. 9, 327–335 [Google Scholar]
- 34. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 35. Goddard T. D., Kneller D. G. (1999) SPARKY 3. University of California, San Francisco, CA [Google Scholar]
- 36. Lipari G., Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 37. Dosset P., Hus J. C., Blackledge M., Marion D. (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 38. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 39. Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 [DOI] [PubMed] [Google Scholar]
- 40. Wider G., Neri D., Wüthrich K. (1991) Studies of slow conformational equilibria in macromolecules by exchange of heteronuclear longitudinal 2-spin-order in a 2D difference correlation experiment. J. Biomol. NMR 1, 93–98 [Google Scholar]
- 41. Wang H., He Y., Kroenke C. D., Kodukula S., Storch J., Palmer A. G., Stark R. E. (2002) Titration and exchange studies of liver fatty acid-binding protein with 13C-labeled long-chain fatty acids. Biochemistry 41, 5453–5461 [DOI] [PubMed] [Google Scholar]
- 42. Akagi T., Sasai K., Hanafusa H. (2003) Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. U.S.A. 100, 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oneyama C., Hikita T., Enya K., Dobenecker M. W., Saito K., Nada S., Tarakhovsky A., Okada M. (2008) The lipid raft-anchored adaptor protein Cbp controls the oncogenic potential of c-Src. Mol. Cell 30, 426–436 [DOI] [PubMed] [Google Scholar]
- 44. Kanou T., Oneyama C., Kawahara K., Okimura A., Ohta M., Ikeda N., Shintani Y., Okumura M., Okada M. (2011) The transmembrane adaptor Cbp/PAG1 controls the malignant potential of human non-small cell lung cancers that have c-Src upregulation. Mol. Cancer Res. 9, 103–114 [DOI] [PubMed] [Google Scholar]
- 45. Liu B. A., Jablonowski K., Raina M., Arcé M., Pawson T., Nash P. D. (2006) The human and mouse complement of SH2 domain proteins–establishing the boundaries of phosphotyrosine signaling. Mol. Cell 22, 851–868 [DOI] [PubMed] [Google Scholar]
- 46. Ogura K., Tsuchiya S., Terasawa H., Yuzawa S., Hatanaka H., Mandiyan V., Schlessinger J., Inagaki F. (1999) Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J. Mol. Biol. 289, 439–445 [DOI] [PubMed] [Google Scholar]
- 47. Narula S. S., Yuan R. W., Adams S. E., Green O. M., Green J., Philips T. B., Zydowsky L. D., Botfield M. C., Hatada M., Laird E. R., et al. (1995) Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide. Structure 3, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 48. Lee C. H., Kominos D., Jacques S., Margolis B., Schlessinger J., Shoelson S. E., Kuriyan J. (1994) Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure 2, 423–438 [DOI] [PubMed] [Google Scholar]
- 49. Eck M. J., Shoelson S. E., Harrison S. C. (1993) Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362, 87–91 [DOI] [PubMed] [Google Scholar]
- 50. Pfrepper K. I., Marie-Cardine A., Simeoni L., Kuramitsu Y., Leo A., Spicka J., Hilgert I., Scherer J., Schraven B. (2001) Structural and functional dissection of the cytoplasmic domain of the transmembrane adaptor protein SIT (SHP2-interacting transmembrane adaptor protein). Eur. J. Immunol. 31, 1825–1836 [DOI] [PubMed] [Google Scholar]
- 51. Songyang Z., Shoelson S. E., McGlade J., Olivier P., Pawson T., Bustelo X. R., Barbacid M., Sabe H., Hanafusa H., Yi T., et al. (1994) Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14, 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuriyan J., Cowburn D. (1997) Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 26, 259–288 [DOI] [PubMed] [Google Scholar]
- 53. Songyang Z., Cantley L. C. (1995) Recognition and specificity in protein-tyrosine kinase-mediated signalling. Trends Biochem. Sci. 20, 470–475 [DOI] [PubMed] [Google Scholar]
- 54. Bae J. H., Lew E. D., Yuzawa S., Tomé F., Lax I., Schlessinger J. (2009) The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 138, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]