Abstract
Background and Aim
Retrospective evaluation of hepatitis C virus (HCV) prevalence in lymphoma tissues has important applications in clarifying the contribution of viral factors to the pathogenesis. Trials for detection of HCV at the cellular level in lymphoma tissues are, so far, minimal with unsatisfactory results. We aimed to study the detection and localization of HCV in the tissues of B-cell non-Hodgkin lymphoma (NHL) patients.
Design
We performed immunohistochemistry to detect the HCV nonstructural 3 protein in paraffin-embedded tissue specimens of B-cell NHL patients, in 39 serum HCV-RNA positive samples and 35 serum HCV-RNA negative samples as controls. The serum analysis was carried out for HCV antibodies using enzyme-linked immunoassay and for HCV-RNA using reverse transcription-polymerase chain reaction. Reverse transcription-polymerase chain reaction was used to detect the HCV-RNA in tissues in immunohistochemically positive cases. We correlated the results with the clinicopathologic characteristics of the patients.
Results
A diffuse cytoplasmic immunohistochemical staining for HCV in the lymphoid cells was detected in 8 of 39 serum positive cases (20.5%), all of which were genotype 4, which is the most prevalent HCV genotype in Egypt. Only 2 out of 35 serum negative control samples showed positive staining and in 1 of them HCV-RNA was detected in tissue. No significant correlation was detected between HCV positive cases and the clinicopathologic features of the patients.
Conclusions
Immunohistochemical detection of HCV proteins in lymphoma tissues supports a potential role of viral replication in lymphomagenesis. The low number of cases showing expression of viral proteins may represent a low viral load in lymphoid tissue and/or restriction of HCV protein expression to certain subtypes of B-cell NHL. Immunohistochemistry can be used as a complementary tool for specific HCV detection in the paraffin-embedded material of lymphoma tissues not suitable for RNA analysis.
Keywords: hepatitis C virus, non-Hodgkin lymphoma, immunohistochemistry
Several viruses have been implicated in the pathogenesis of various types of hematologic malignant neoplasms in humans.1–7 The best examples of these are Epstein-Barr virus in Burkitt lymphoma1 and Hodgkin disease,2 human T-lymphotropic virus I in human T-cell leukemia,3 Kaposi sarcoma-associated herpes virus in primary effusion lymphoma,5 and the lymphomas associated with multicentric Castleman disease in human immunodeficiency virus-infected patients.6 Hepatitis C virus (HCV) is one of the tumor-associated viruses because of its role in hepatocarcinogenesis. HCV-associated lymphomas have been observed, but whether they are caused by HCV remains to be shown definitively.8
The World Health Organization estimates that 170 million people are infected with HCV.9 An estimated 12% to 15% of Egyptians, or 8 to 10 million people, have serologic evidence of HCV infection.10 The HCV genome, a 9.4-kb positive sense single stranded RNA, encodes for a polyprotein precursor that is posttranslationally cleaved into putative structural and nonstructural (NS) proteins. HCV consists of several sequential functional elements: a conserved 5′ untranslated region, followed qby the nucleocapsid (core) protein and 2 envelope glycoproteins (E1 and E2/NS1) that are included in a structural region, and then NS2, NS3, NS4, and NS5 (the viral replicase), which are part of a NS region.11
Epidemiologic studies based on serologic detection of HCV have provided evidence of an association between HCV infection and increased risk of non-Hodgkin lymphoma (NHL).8,12–15 In these studies, HCV infection was found in 9% to 32% of patients with B-cell NHL. In a large multicenter study, a 2-fold increased risk for B-cell NHL and a greater than 3-fold elevated risk for diffuse large B-cell lymphoma (DLBCL) was found in relation to HCV-RNA.14 Evidence from epidemiologic studies is strengthened by clinical data showing a regression of lymphoma after successful treatment of HCV infection.16–18 Chronic HCV infection leads to chronic stimulation of the immune system and has been associated with the development of immunerelated disorders, such as type II mixed cryoglobulinemia (MC).7,19 MC is a nonmalignant lymphoproliferative condition, which may evolve into B-cell NHL in 5% to 8% of cases.19
Few studies were able to detect HCV-RNA sequences in the tissues of patients with B-cell NHL, suggesting a role for HCV infection in lymphomagenesis.20–25 Testing for HCV core protein by immunohistochemistry (IHC) did not yield satisfactory results.24,25 We studied the prevalence and distribution of HCV proteins in lymphoma tissues using a commercially available monoclonal antibody, directed against a synthetic NS3 peptide fragment of HCV.
MATERIALS AND METHODS
We performed a retrospective study on 39 formalin-fixed paraffin-embedded tissue specimens of patients serologically positive for HCV and diagnosed as having B-cell NHL in the National Cancer Institute, Cairo University, Egypt between January 2001 and May 2006. All serologically positive cases with adequate paraffin-embedded tissue material were included in the study. Lymphoma tissue samples of 35 patients serologically negative for HCV were included in the study as controls. Data concerning the patient’s age, sex, and location of the primary tumor presentation were obtained from the pathology reports. Hematoxylin and eosin stained sections prepared from the paraffin blocks were reexamined and classified according to the World Health Organization classification of lymphoid tumors.26
Evidence of HCV infection in the 39 patients was based on previously performed serologic testing of blood specimens withdrawn from the patients before the start of treatment. Testing for anti-HCV antibodies was carried out by Abbott HCV enzyme-linked immunoassay (EIA) 3.0 (Abbott Park, IL) according to the manufacturer’s instructions. Samples were initially tested for HCV-RNA by direct nested reverse transcription-polymerase chain reaction (RT-PCR) as described previously, with modifications to increase the sensitivity of the assay.27 The primers used for RT-PCR were derived from the highly conserved 5′-untranslated region of HCV genome.28 The nested primers amplified a 237 base pair fragment. The PCR products were detected on 3% agarose gels in 0.5 × Tris-borate-ethylenediaminetetraacetic acid buffer. The sensitivity of the assay is 50 IU/mL. The sequences of the primers used are shown in Table 1. A positive result by the direct RT-PCR method was considered truly positive and no further investigation was carried out. A sample that was negative by both direct RT-PCR and EIA was considered negative. However, all samples that tested negative by direct nested RT-PCR and positive by EIA were retested by conventional RT-PCR, which differed from direct RT-PCR in that it included an RNA purification step.29
TABLE 1.
The Sequences of Primers Used for HCV-RNA Amplification
| Name | Orientation | Primer Sequence | Position |
|---|---|---|---|
| P1 (NF5) | Outer s | 5′-GTG AGG AAC TAC TGT CTT CAC GCA G-3′ | 47-71 |
| P2 (NR5) | Outer a-s | 5′-TGC TCA TGG TGC ACG GTC TAC GAG A-3′ | 324-348 |
| P3 (KF2) | Inner s | 5′-TTC ACG CAG AAA GCG TCT AG-3′ | 63-82 |
| P4 (NR4) | Inner a-s | 5′-CTA TCA GGC AGT ACC ACA AGG-3′ | 279-299 |
a-s indicates antisense; s, sense
We used IHC to detect the HCV-NS3 protein in lymphoma tissue samples of the HCV serologically positive patients and the serologically negative controls. All samples were formalin-fixed, paraffin-embedded tissue material obtained from the archives of the Department of Pathology, National Cancer Institute, Cairo University. Paraffin sections were cut by a microtome at 5 μm thickness and picked up onto poly-L-lysine coated microscope slides and dried overnight at room temperature. After dewaxing and rehydration, the sections were pretreated in citrate buffer (pH 6.0) using microwave (Goldstar 1000 W with digital control) at 130°C for 30 minutes, followed by blocking of endogenous peroxidase. Nonspecific protein binding was blocked through incubation with 10% normal horse serum for 20 minutes at room temperature. Sections were then incubated with the monoclonal mouse antihepatitis C virus antibody HCV-NS3, clone MMM33 (Novocastra, Newcastle, UK); 1:50 dilution overnight at 4°C. Avidin-biotin peroxidase complex detection system from Lab Vision Corporation was applied with 3,3′ diaminobenzidine as chromogen (Lab Vision, Fremont, CA). Liver tissue samples positive for HCV-NS3 protein were used as positive controls for immunohistochemical staining. For the negative control slides, substitution of the monoclonal antibody by diluted normal bovine serum was carried out.
RT-PCR was performed as described before in serum to detect the HCV-RNA in the lymphoma tissue samples positive for HCV by IHC. RNA extraction from formalin-fixed paraffin-embedded tissue was carried out as follows: 2 unstained 5-μm sections were deparaffinized in a series of xylene and graded alcohol. Then, tissue was digested overnight in a lysis buffer containing 500 μg/mL proteinase K, 1% sodium dodecyl sulfate, 20 mmol/L Tris-HCl (pH 8.0), and 5 mmol/L ethylenediaminetetraacetic acid (pH 8.0). After DNase treatment (20 U/μL), RNA was extracted using QIAamp Viral RNA Mini kit procedure (Qiagen, Santa Clarita). The adequacy of the RNA extracts from all specimens was assessed by assaying for glyceraldehyde-3-phosphate dehydrogenase messenger RNA by RT-PCR. Nucleic acids extracted from an HCV-positive liver specimen were included as the positive control and nucleic acids extracted from an HCV-negative liver specimen were used as the negative control.
Descriptive statistics were expressed as mean and SD for continuous variables. For discrete data, it was expressed in percentage. Statistical analysis was performed using the χ2 or Fisher exact test when appropriate, and P<0.05 was taken as the level of significance.
RESULTS
There was no age or sex difference between the serum HCV-RNA positive and the serum HCV-RNA negative groups in B-cell NHL patients. Male predominance was present in both groups; 22 out of 39 in the serum positive patients and 21 out of 35 in the serum negative patients. The mean age for the serum HCV-positive patients was 51.46 ± 11.82 years whereas for the serum HCV-negative patients it was 44.31 ± 15.41 years. Extranodal presentation was documented in 7 out of 39 serum positive patients and 6 out of 35 serum negative patients. Diffuse large-cell histology was the dominant subtype followed by the follicular subtype in serum positive patients (72% and 18%) and also in serum negative patients (77% and 11%). All serum HCV-RNA positive patients had genotype 4 of HCV and all except 1 were positive for anti-HCV antibody in serum. Table 2 shows the data for serum anti-HCV, HCV NS3 immunostaining, and clinicopathologic characteristics for serum HCV-RNA positive patients.
TABLE 2.
Clinicopathologic Characteristics, Serum Anti-HCV, and HCV NS3 Immunostaining in Serum HCV-RN Positive B-cell Non-Hodgkin Lymphoma Patients (n = 39)
| Patient Number |
Age (y) | Sex | EN | Diagnosis | Serum Anti-HCV |
HCV-NS3 Protein |
|---|---|---|---|---|---|---|
| 1 | 68 | M | No | SLL | + | − |
| 2 | 39 | M | No | DLBCL | + | − |
| 3 | 50 | M | No | FL | + | − |
| 4 | 57 | F | No | DLBCL | + | − |
| 5 | 71 | M | Yes | DLBCL | + | − |
| 6 | 51 | M | Yes | DLBCL | + | − |
| 7 | 41 | M | No | DLBCL | + | − |
| 8 | 56 | F | No | TCRLBL | + | − |
| 9 | 51 | F | Yes | DLBCL | + | − |
| 10 | 69 | M | No | DLBCL | − | − |
| 11 | 30 | M | No | DLBCL | + | − |
| 12 | 58 | F | No | DLBCL | + | − |
| 13 | 68 | M | No | DLBCL | + | − |
| 14 | 27 | M | No | DLBCL | + | − |
| 15 | 60 | F | No | DLBCL | + | + |
| 16 | 52 | F | No | TCRLBL | + | − |
| 17 | 34 | F | Yes | DLBCL | + | + |
| 18 | 48 | M | No | DLBCL | + | − |
| 19 | 52 | F | No | DLBCL | + | − |
| 20 | 72 | M | No | DLBCL | + | − |
| 21 | 38 | M | Yes | FL | + | − |
| 22 | 40 | M | No | DLBCL | + | − |
| 23 | 56 | F | No | DLBCL | + | − |
| 24 | 62 | M | No | SLL | + | + |
| 25 | 51 | M | Yes | DLBCL | + | + |
| 26 | 53 | F | No | MZL | + | − |
| 27 | 68 | M | No | FL | + | − |
| 28 | 53 | M | No | DLBCL | + | + |
| 29 | 68 | F | No | DLBCL | + | + |
| 30 | 53 | F | No | FL | + | − |
| 31 | 51 | F | No | DLBCL | + | − |
| 32 | 63 | M | No | DLBCL | + | − |
| 33 | 59 | F | Yes | DLBCL | + | − |
| 34 | 74 | M | No | FL | + | − |
| 35 | 48 | F | No | FL | + | + |
| 36 | 55 | M | No | FL | + | − |
| 37 | 61 | F | No | SLL | + | − |
| 38 | 55 | F | No | DLBCL | + | + |
| 39 | 41 | M | No | DLBCL | + | − |
DLBCL indicates diffuse large B-cell lymphoma; EN, extranodal presentation; F, female; FL, follicular lymphoma; HCV, hepatitis C virus; M, male; MZL, marginal zone lymphoma; SLL, small lymphocytic lymphoma; TCRLBL, T-cell rich large B-cell lymphoma.
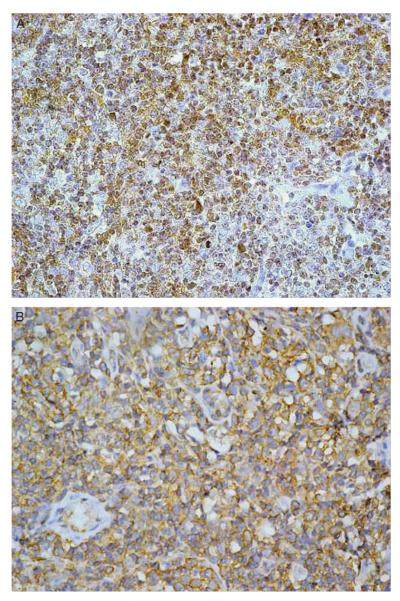
We detected HCV-NS3 protein by IHC in the tissues of 8 cases (20.5%) of serum HCV-RNA positive B-cell NHL patients. These 8 cases comprised DLBCL (6 cases), follicular lymphoma (1 case), and small lymphocytic lymphoma (1 case). All positive cases showed cytoplasmic staining for the HCV-NS3 protein in malignant and reactive benign lymphoid cells (Fig. 1). The percentage of stained cells ranged between 10% and 70% with a mean of 29% stained lymphoid cells. Other nonlymphoid cells were negative except for rare staining in normal endothelial cells. Two DLBCL, out of the 35 serum negative B-cell NHL specimens, were positive for HCV immunostaining (5.7%). No significant correlation was detected between immunostaining results and the age or sex of the investigated B-cell NHL patients. Also no significant correlation was found with the extranodal presentation or the histologic subtype of the B-cell NHL.
FIGURE 1.
Two cases of B-cell non-Hodgkin lymphoma showing immunohistochemical staining for hepatitis C virus nonstructural 3 protein. Avidin-biotin peroxidase complex method. A, Strong cytoplasmic staining in the lymphoid cells. Original magnification 400 ×. B, Diffuse cytoplasmic staining in the lymphoid cells. Original magnification 400 ×.
Only 1 of 10 immunohistochemically positive specimens was positive for HCV-RNA in tissue using RT-PCR (Fig. 2). This tissue RNA positive specimen belonged to the control group in which HCV-RNA was negative in serum. The other 9 specimens did not yield adequate RNA material.
FIGURE 2.

Agarose gel electrophoresis of hepatitis C virus reverse transcription-polymerase chain reaction products from formalin-fixed paraffin-embedded tissue of a seronegative non-Hodgkin lymphoma with positive (PC) and negative (NC) controls.
DISCUSSION
We performed a retrospective analysis of HCV in archival paraffin-embedded blocks of lymphoma tissues. This is one of the few studies correlating the results of IHC with the serologic profile of the patients. The association between HCV and B-cell NHL has been essentially based on serologic testing for HCV.8,12–15 We detected HCV antigens by immunostaining in the tissues of 8 (20.5%) serum HCV-RNA positive and 2 (5.7%) serum HCV-RNA negative B-cell NHL patients. Two previous studies have evaluated the prevalence of HCV in NHL in the serum and tissue simultaneously.24,30 In the study by Paydas et al,24 RT-PCR was used to detect the HCV-RNA in serum and tissue samples of 30 cases of NHL. The anti-HCV antibodies were tested with microparticle enzyme immunoassay. Reactive lymph node samples taken from 30 cases without lymphoma were used as controls. In that study, the HCV-RNA prevalence was higher in tissue samples (37%) than in serum samples (23.3%). HCV core protein was studied in 10 of 11 HCV-RNA positive cases by IHC. Only 2 cases showed detectable HCV core protein, the tissue HCV-RNA was positive in these cases whereas the serum HCV-RNA was negative. In an Egyptian study performed on 29 cases of NHL, HCV-NS3 antigens were found in tissue by IHC in 41% of cases and HCV-RNA in the serum in 27.6% of cases.30 Tissue RT-PCR findings in tissue did not yield satisfactory results. The serum findings of the patients in that study were compared with those of 36 apparently healthy individuals as a control group, whereas 10 nonmetastatic lymph nodes from cancer cases other than NHL were used as a control for IHC. Another earlier study showed HCV-RNA in 26% of lymphoma tissues but none of the cases showed HCV core protein by IHC. Thirty-one cases of B-cell NHLs and lymph nodes from 32 patients with diseases other than B-cell NHL as negative controls were the subjects of that study, but the serologic findings were not investigated.25
The low expression of the viral proteins in lymphoma tissues of otherwise serum HCV-RNA positive cases (20.5%) could represent a low viral load in lymphoid tissue as reported by previous studies.24,25 Such a low viral load might be below the detection threshold of IHC. The expression of HCV gene products may be closely related to certain stages of B-cell differentiation with a possible restriction of HCV infection to certain subtypes of B-cell NHL more than others.20 NHL is now considered as a diverse group of separate entities that differ in their pathologic features, molecular lesions, clinical outcomes, and perhaps etiology.26 We did not find any significant association between serum HCV-RNA prevalence and NHL subtype in our cases when compared with serum HCV-RNA negative controls. Among B-cell NHLs, associations with HCV were seen for diffuse large B-cell, follicular, marginal zone, and chronic lymphocytic leukemia/small lymphocytic lymphoma.13 Other studies did not show an increased risk of follicular lymphoma associated with HCV infection but confirmed the increased risk for DLBCL and marginal zone lymphoma, as well as lymphoplasmacytic lymphoma.15,31 However, in the majority of lymphoma studies, small sample sizes have prevented an analysis of the relationship between HCV and single lymphoma subtypes.32
The detection of HCV antigens in 2 (5.7%) of serum HCV-RNA negative samples in our study may be explained by the long-term persistence of HCV-RNA in lymphatic cells despite HCV-RNA negativity of serum and liver. A role played by HCV lymphotropism in subjects with incomplete viral clearance after antiviral treatment is strongly suggested, even if the exact mechanisms involved are at present unknown.7 It was previously reported that higher HCV-RNA prevalence was found in lymphoma tissue samples compared with serum samples.24 Several reports support the concept of HCV-RNA positive peripheral blood mononuclear cells in the absence of HCV viremia, and as there is acceptance that both hepatitis B virus and HCV can persist in the liver in individuals who have apparently recovered from infection, it is possible that virus persistence is equally possible in other cell types.33 The detection of HCV antigens in lymphoma tissues by IHC was also reported to be more frequent than serum HCV-RNA prevalence.30 However, the use of IHC for the detection of viral proteins by itself leaves open the question of whether positive results might be because of cross reaction of the viral antiserum with a cellular antigen. In this study, we investigated for HCV-RNA in tissues using RT-PCR in 10 lymphoma tissue samples positive for HCV by IHC (8 HCV serum RNA positive and 2 serum negative samples). We found HCV-RNA in the tissues of one HCV-RNA serum negative sample. We could not isolate adequate RNA material from the other 9 tissue samples. This might have been caused by inconsistency in tissue fixation, where some of the samples were fixed in suboptimal dilutions of formaldehyde, with variable pH levels, or subjected to high temperatures during formalin fixation. It has been reported that formalin fixation conditions might have an effect on RT-PCR detectable RNA from tissue blocks.34
The cytoplasmic staining pattern of lymphoid cells detected in this study has also been reported in other studies applying HCV-NS3 antibody for the detection of HCV antigens in lymphoid tissue.20,30,35 In this study, the stained lymphoid cells ranged from 10% to 70%, being randomly distributed among both malignant and reactive small lymphocytes. Comparable results were also reported as regards to the distribution of HCV-related proteins in lymphoid tissue.20 In that study, NS3 and NS4 proteins were detected in lymphoid cells of the inter-follicular areas of reactive lymph nodes and in circulating mononuclear cells of capsular blood vessels. It was suggested that HCV infection precedes neoplastic transformation and consequently HCV may have a role in the development of lymphoma. Another study applying HCV-NS3 antibody for the detection of HCV antigens among NHL cases (B-cell and T-cell lymphomas) reported a range of 11% to 30% positively stained lymphoid cells where most of the stained cells were lymphoma cells.30 In several previous studies, immunoreactivity for HCV antigens in lymphoma tissues was either poor or absent.20,24,25,35,36 The results indicated that the detection rates of HCV in extrahepatic tissues were low and the amount of HCV-positive staining cells in extrahepatic tissues was also obviously lower than that in livers; and there were differences in HCV expression in various cells among different extrahepatic tissues.35
The demonstration of HCV antigens in lymphoma tissue supports a potential role of HCV infection in the etiology of NHL. However, HCV is less likely to cause lymphomagenesis by a direct oncogenic mechanism, as the RNA virus cannot integrate its viral nucleic acid sequences into the lymphocyte genome.32 It has been proposed that the persistence of the virus in lymphocytes can indirectly induce lymphomagenesis by sustained chronic antigenic stimulation of B-cell proliferation in a fashion similar to Helicobacter pylori. The HCV envelope protein E2 is able to bind to CD81, a B-cell receptor. The E2/CD81 complex enables B-cells to respond to lower thresholds of B-cell signaling. The sustained polyclonal expansion of B-cells would be prone to mistakes in immunoglobulin gene rearrangement processes such as the t(14;18) translocation, present in a significant percentage of peripheral blood lymphocytes in HCV infected individuals, particularly with MC. This translocation is responsible for BCL-2 activation, which extends B-cell survival by inhibiting apoptosis. Abnormal B-cell survival would favor the accumulation of genetic mutations, possibly leading to malignant transformation.7 Conversely, a “hit and run” mechanism has been proposed for HCV-induced mutations of immunoglobulin genes and oncogenes in B-cell lines infected by HCV as compared with noninfected cells.37
The extensive epidemiologic studies relying on PCR-based associations along with the laboratory studies using in situ methods for detection of HCV gene products in lymphoma tissues are still not sufficient for a credible case. The specificity of the association of HCV infection to particular subtypes of NHL needs to be addressed especially in populations with high HCV prevalence to strengthen the evidence for a causal role for HCV in the development of NHL. More laboratory research is required to understand how chronic immune stimulation may lead to NHL. This research would help to inform the interpretation of epidemiologic data on subtype-specific associations.32 It will be interesting to investigate to what extent host factors, that control the natural course of HCV infection, may modulate lymphoma risk among HCV infected individuals.31 HCV testing is not routinely recommended or performed for patients with lymphoma, the true prevalence of HCV infection in these patients is unknown. Therefore, the ability to accurately detect HCV viral sequences from archival tissue embedded in paraffin is essential to document HCV infection in such tissues.24 IHC can be used as a complementary tool for specific HCV detection in the paraffin-embedded material of lymphoma tissues not suitable for RNA analysis. Further studies are essential to validate the sensitivity and specificity of various commercially available monoclonal antibodies against HCV antigens in lymphoma tissue. The NS proteins are assumed to be more specific targets as they are acknowledged as markers of ongoing virus replication.33 The results obtained should be related to the data obtained from serum HCV-RNA and to the results of in situ detection of nucleic acids by in situ hybridization or RT-PCR in tissue.
ACKNOWLEDGMENT
The authors thank Lillian Young and William Win for the technical help in the preparation of the immunohistochemical slides.
REFERENCES
- 1.Hausen HZ, Schulte-Holthausen H, Klein G, et al. EBV DNA in biopsies of Burkitt tumors and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 2.Brousset P, Knecht H, Rubin B, et al. Demonstration of Epstein-Barr virus replication in Reed-Sternberg cells of Hodgkin’s disease. Blood. 1993;82:872–876. [PubMed] [Google Scholar]
- 3.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris JDH, Eddleson ALWF, Crook T. Viral infection and cancer. Lancet. 1995;346:54–58. doi: 10.1016/s0140-6736(95)91510-9. [DOI] [PubMed] [Google Scholar]
- 5.Nador RG, Cesarman E, Chadburn A, et al. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma–associated herpes virus. Blood. 1996;88:645–656. [PubMed] [Google Scholar]
- 6.Cesarman E, Chang Y, Moore PS, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Eng J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 7.Zignego AL, Giannini C, Ferri C. Hepatitis C virus-related lymphoproliferative disorders: an overview. World J Gastroenterol. 2007;13:2467–2478. doi: 10.3748/wjg.v13.i17.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowgill K, Loffredo C, Eissa SA, et al. Case-control study of non-Hodgkin’s lymphoma and hepatitis C virus infection in Egypt. Int J Epidemiol. 2004;33:1034–1039. doi: 10.1093/ije/dyh183. [DOI] [PubMed] [Google Scholar]
- 9.Lavanchy D, McMahon B. Worldwide prevalence and prevention of hepatitis C. In: Liang TJ, Hoofnagle JH, editors. Hepatitis C. Academic Press; San Diego, CA: 2000. [Google Scholar]
- 10.Frank C, Mohamed MK, Strickland GT, et al. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet. 2000;355:887–891. doi: 10.1016/s0140-6736(99)06527-7. [DOI] [PubMed] [Google Scholar]
- 11.Houghton M, Weiner A, Kuo G, et al. Molecular biology of hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 12.Duberg AS, Nordstrom M, Torner A, et al. Non-Hodgkin’s lymphoma and other nonhepatic malignancies in Swedish patients with hepatitis C virus infection. Hepatology. 2005;41:652–659. doi: 10.1002/hep.20608. [DOI] [PubMed] [Google Scholar]
- 13.Dal Maso L, Franceschi S. Hepatitis C virus and risk of lymphoma and other lymphoid neoplasms: a meta-analysis of epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2006;15:2078–2085. doi: 10.1158/1055-9965.EPI-06-0308. [DOI] [PubMed] [Google Scholar]
- 14.Nieters A, Kallinowski B, Brennan P, et al. Hepatitis C and risk of lymphoma: results of the European Multicenter Case-Control Study EPILYMPH. Gastroenterology. 2006;131:1879–1886. doi: 10.1053/j.gastro.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 15.Spinelli JJ, Lai AS, Krajden M, et al. Hepatitis C virus and risk of non-Hodgkin lymphoma in British Columbia, Canada. Int J Cancer. 2008;122:630–633. doi: 10.1002/ijc.23105. [DOI] [PubMed] [Google Scholar]
- 16.Hermine O, Lefrere F, Bronowicki JP, et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347:89–94. doi: 10.1056/NEJMoa013376. [DOI] [PubMed] [Google Scholar]
- 17.Gisbert JP, Garcia-Buey L, Pajares JM, et al. Systematic review: regression of lymphoproliferative disorders after treatment for hepatitis C infection. Aliment Pharmacol Ther. 2005;21:653–662. doi: 10.1111/j.1365-2036.2005.02395.x. [DOI] [PubMed] [Google Scholar]
- 18.Svoboda J, Andreadis C, Downs LH, et al. Regression of advanced non-splenic marginal zone lymphoma after treatment of hepatitis C virus infection. Leuk Lymphoma. 2005;46:1365–1368. doi: 10.1080/104281905001028289. [DOI] [PubMed] [Google Scholar]
- 19.Ramos-Casals M, Trejo O, Garcia-Carrasco M, et al. Mixed cryoglobulinemia: new concepts. Lupus. 2000;9:83–91. doi: 10.1191/096120300678828127. [DOI] [PubMed] [Google Scholar]
- 20.Sansonno D, De Vita S, Carnacchiulo V, et al. Detection and distribution of hepatitis C virus-related proteins in lymph nodes of patients with type II mixed cryoglobulinemia and neoplastic or non-neoplastic lymphoproliferation. Blood. 1996;88:4638–4645. [PubMed] [Google Scholar]
- 21.Zignego AL, Ferri C, Innocenti F, et al. Lack of preferential localization of tumoral mass in B-cell non-Hodgkin’s lymphoma associated with hepatitis C virus infection. Blood. 1997;89:3066–3068. [PubMed] [Google Scholar]
- 22.Tursi A, Brandimanta G, Chiarelli F, et al. Detection of HCV RNA in gastric mucosa associated lymphoid tissue by in situ hybridization: evidence of a new extrahepatic localization of HCV with increased risk of gastric malt lymphoma. Am J Gastroenterol. 2002;97:1802–1806. doi: 10.1111/j.1572-0241.2002.05848.x. [DOI] [PubMed] [Google Scholar]
- 23.De Vita S, Dammacco F, Gloghini A, et al. Hepatitis C virus within a malignant lymphoma lesion in the course of type II mixed cryoglobulinemia. Blood. 1995;86:1887–1892. [PubMed] [Google Scholar]
- 24.Paydas S, Ergin M, Tanriverdi K, et al. Detection of Hepatitis C virus RNA in paraffin-embedded tissues from patients with non-Hodgkin lymphoma. Am J Hematol. 2004;76:252–257. doi: 10.1002/ajh.20092. [DOI] [PubMed] [Google Scholar]
- 25.Karavatthayyil SJ, Kalkeri G, Liu HJ, et al. Detection of hepatitis C virus RNA sequences in B-cell non-Hodgkin’s lymphoma. Am J Clin Pathol. 2000;113:391–396. doi: 10.1309/REV9-FDTM-5NGC-HBWY. [DOI] [PubMed] [Google Scholar]
- 26.Jaffee ES, Harris NL, Stein H, et al. WHO classification of tumors of haematopoietic and lymphoid tissues. IARC Press; Lyon: 2001. pp. 10–11. [Google Scholar]
- 27.Abdel-Hamid M, Edelman DC, Highsmith WE, et al. Optimization, assessment, and proposed use of a direct nested reverse transcription-polymerase chain reaction protocol for the detection of hepatitis C virus. J Hum Virol. 1997;1:58–65. [PubMed] [Google Scholar]
- 28.Okamoto S, Okada S, Sugiyama J, et al. Detection of hepatitis C virus RNA by a two-stage polymerase chain reaction with two pairs of primers deduced from the 58-noncoding region. Jpn J Exp Med. 1990;60:215–222. [PubMed] [Google Scholar]
- 29.Habib M, Mohamed MK, Abdel-Aziz F, et al. Hepatitis C virus infection in a community in the Nile delta: risk factors for seropositivity. Hepatology. 2001;33:248–253. doi: 10.1053/jhep.2001.20797. [DOI] [PubMed] [Google Scholar]
- 30.El-Sayed GM, Mohamed WS, Nouh MA, et al. Viral genomes and antigen detection of hepatitis B and C viruses in involved lymph nodes of Egyptian non-Hodgkin’s lymphoma patients. Egypt J Immunol. 2006;13:105–114. [PubMed] [Google Scholar]
- 31.De Sanjose A, Benavente Y, Vajdic CM, et al. Hepatitis C and non-Hodgkin lymphoma among 4784 cases and 6269 controls from the International Lymphoma Epidemiology Consortium. Clin Gastroenterol Hepatol. 2008;6:451–458. doi: 10.1016/j.cgh.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson L, Engels EA. Hepatitis C virus infection and non-Hodgkin lymphoma: interesting association or causal relationship? Int J Cancer. 2008;122:x–xii. doi: 10.1002/ijc.23462. [DOI] [PubMed] [Google Scholar]
- 33.Gowans EJ. Distribution of markers of hepatitis C virus infection throughout the body. Semin Liver Dis. 2000;20:85–102. doi: 10.1055/s-2000-9503. [DOI] [PubMed] [Google Scholar]
- 34.Hamoud M, Villegas P, Williams S. Detection of infectious bursal disease virus from formalin-fixed paraffin-embedded tissue by immunohistochemistry and real-time reverse transcription polymerase chain reaction. J Vet Diagn Invest. 2007;19:35–42. doi: 10.1177/104063870701900106. [DOI] [PubMed] [Google Scholar]
- 35.Yan FM, Chen AS, Hao F, et al. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805. doi: 10.3748/wjg.v6.i6.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vita S, De Re V, Sansonno D, et al. Lack of HCV infection in malignant cells refutes the hypothesis of a direct transforming action of the virus in the pathogenesis of HCV-associated B-cell NHLs. Tumori. 2002;88:400–406. doi: 10.1177/030089160208800510. [DOI] [PubMed] [Google Scholar]
- 37.Machida K, Cheng KT, Sung VM, et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci USA. 2004;101:4262–4267. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]