Summary
This paper focuses on the problem of choosing a prior for an unknown random effects distribution within a Bayesian hierarchical model. The goal is to obtain a sparse representation by allowing a combination of global and local borrowing of information. A local partition process prior is proposed, which induces dependent local clustering. Subjects can be clustered together for a subset of their parameters, and one learns about similarities between subjects increasingly as parameters are added. Some basic properties are described, including simple two-parameter expressions for marginal and conditional clustering probabilities. A slice sampler is developed which bypasses the need to approximate the countably infinite random measure in performing posterior computation. The methods are illustrated using simulation examples, and an application to hormone trajectory data.
Keywords: Dirichlet process, Functional data, Local shrinkage, Meta-analysis, Multi-task learning, Partition model, Slice sampling, Stick-breaking
1. Introduction
Random effects models are commonly used in the analysis of longitudinal and functional data. Suppose that data on subject i (i = 1, …, n) consist of ni measurements, yi = (yi1, …, yini)T, collected at times ti = (ti1,…, tini)T, and let
| (1) |
where ηi is a function for subject i, are basis functions, σ2 is the measurement error variance, θi = (θi1,…, θip)T are basis coefficients for subject i and heterogeneity in the curves is characterized through the random effects distribution P. A potential concern is sensitivity to the choice of random effects distribution. As described by Heard et al. (2006), one strategy is to use a latent class model with , where δΘ denotes a degenerate distribution with all its mass on Θ, πh is the probability that θi = Θh and k is the number of functional clusters. Dirichlet process priors (Ferguson, 1973, 1974) allow k = ∞ and increasing numbers of clusters with sample size, and have been widely used for random effects distributions (Bush & MacEachern, 1996; Müller & Rosner, 1997).
Although much of the recent literature on mixture models has focused on clustering as a primary goal, such models are useful even if there is no interest in clustering. For example, in the analysis of longitudinal data, one may be interested in inferences on the average trajectory over time, in estimating variability in the trajectories across subjects, or in inferences on fixed effects coefficients, with the random effects considered a nuisance. In each of these cases, it is desirable to allow the random effects distribution, P, to be unknown. In many applications, p is moderate to large, so that it is necessary to obtain a sparse approach for characterizing the unknown P due to the curse of dimensionality.
Sparsity can be formalized through favouring a small number of components. This can be accomplished through a Dirichlet process prior, P ~ DP(α P0), where α is a concentration parameter and P0 is a base measure. The number of unique values of increases at a rate proportional to α log n, so that the number of occupied components tends to be small relative to n. However, a drawback of the Dirichlet process is the assumption of global partitioning. In particular, θij = Θhj implies that θij′ = Θhj′ for all j′ ǂ j. Hence, two subjects in the same cluster for any of their random effects are forced to be in the same cluster for all of their random effects. In functional data analysis, it is common for two subjects to have similar trajectories across certain regions while having local deviations. Under the Dirichlet process, either such subjects will be inappropriately clustered together, obscuring local differences, or they will be allocated to separate clusters.
This paper proposes a class of local partition process priors for parsimonious modelling of unknown random effects distributions. The general structure can be characterized as follows:
| (2) |
where ψij ∈ {1, …, ∞} denotes the cluster index for the jth random effect from subject i, θi = Θψi = (Θψi11, …, Θψipp)T, the elements of are independent and identically distributed, and Q is a probability distribution over {1, 2, …, ∞}p.
In specifying Q, it is convenient to let Θ = Θ0 ∪ Θ1, with global coefficient vectors and local coefficient vectors. As a convention, let Θ0 denote the odd-numbered elements of Θ, with Θ1 the even-numbered elements. To induce ψi ~ Q, let
| (3) |
where zij = 1 denotes allocation of θij to a global component, zij = 0 denotes allocation to a local component, ϕi0 ∈ {1, 2, …, ∞} is a global cluster index for subject i and ϕij ∈ {1, 2, …, ∞} is a local cluster index for subject i and element j. From (2)-(3), the ith subject’s random effects vector is equivalent to Θ0ϕi0 for those elements having zij = 1, while the remaining elements are selected from a variety of local coefficient vectors. This specification allows a combination of global and local borrowing of information. Proofs are included in an Appendix.
2. Dependent local partition process
2·1. Formulation 1
To choose G and H in (3), let
| (4) |
where νj is the probability of allocation to the global component for the jth random effect, β is a hyperparameter controlling the overall weight on the local components and the elements of ϕi are assigned independent stick-breaking priors, with the hyperparameter α controlling how rapidly the weights πjh decrease towards zero as the index h increases. The stick-breaking priors are chosen following the Sethuraman (1994) representation of the Dirichlet process for simplicity, though extensions to more general stick-breaking priors (Ishwaran & James, 2001) are straightforward. Let P ~ LPP1(α, β, P0) denote the prior on the unknown random effects distribution induced through (2)-(4).
The prior P ~ LPP1(α, β, P0) induces borrowing of information across subjects through both global and local clustering. In particular, if subjects i and i′ are assigned to the same global cluster, then ϕi0 = ϕi′0, and it becomes more likely that these subjects will have identical values for multiple elements of their random effects vectors, particularly if β is not large. For small β and high-dimensional random effects vectors, subjects in the same global clusters will have identical values for most of their basis coefficients, while having occasional local deviations. This structure addresses the limitations of Dirichlet process priors mentioned in § 1. In a recent application using Dirichlet process priors for the distribution of a high-dimensional latent factor vector underlying gene expression measurements, we noticed a tendency of the Dirichlet process to induce a large number of clusters, as even individuals that were quite similar overall had important local differences. In contrast, the local partition process induced a small number of global and local clusters, leading to substantial gains in efficiency.
One of the appealing characteristics of the Dirichlet process prior is the availability of simple expressions for the prior probability of clustering marginalizing out P. In particular, θi ~ P with P ~ DP(α P0) implies that pr(θi = θi′) = 1/(1 + α). This illustrates the role of the precision parameter α in controlling the tendency of individuals to be clustered together. As described in Proposition 1, a similarly simple expression is obtained for the marginal prior probability of pr(θij = θi′ j) under P ~ LPP1(α, β, P0).
Proposition 1
Assuming θi ~ P with P ~ LPP1(α, β, P0),
Hence, α and β are key hyperparameters controlling the clustering probability ρ with ρ monotone decreasing in α and β. In the limit as α, β → 0, ρ = 1 and all individuals are automatically grouped into a single cluster with θi = θ for all i. To allow the data to provide information about appropriate values for α, β, let α ~ Ga(aα, bα) and β ~ Ga(aβ, bβ).
If subjects i and i′ are in the same cluster for the j′th random effect, so that θij′ = θi′ j′, one has information that these subjects are similar. Ideally, such information would be propagated to other random effects, increasing the probability of θij = θi′ j for j ǂ j′. Proposition 2 demonstrates that the prior P ~ LPP1(α, β, P0) automatically induces the desired dependence in local clustering. In addition, the dependence structure has a simple form, which provides insight into the flexibility of the approach and role of the hyperparameters.
Proposition 2
Assuming θi ~ P with P ~ LPP1(α, β, P0),
The joint probability in Proposition 2 is strictly larger than the product of the marginal probabilities from Proposition 1, which implies that pr(θij = θi′ j) increases given knowledge that θij′ = θi′ j′. The degree of positive dependence in local clustering is monotonely increasing with α and decreasing with β, being controlled by the size of 4α/(1 + β)2.
As P ~ LPP1(α, β, P0) provides a prior for the unknown random effects distribution P, it is important to assess basic properties, such as the mean and variance. Following common convention, the notation P is used to denote both a random probability measure and the corresponding distribution. It is assumed that P0 is a probability measure over a measurable Polish space (Ω, B), with Ω the sample space and B the corresponding Borel σ-algebra. Let θij ~ Pj, with Pj the jth marginal from the joint prior, P ~ LPP1(α, β, P0). Then, Pj is a probability measure over (Ωj, Bj), where Ωj is the sample space for the jth component of θi, and Bj is the corresponding Borel σ-algebra. For any B ∈ Bj, Pj(B) is a random variable, with , where W ~ Be(1, β) and Xl ~ Be[α P0j(B), α{1 − P0j(B)}], for l = 1, 2, are independent random variables. Although the density of Pj(B) does not have a simple form, the expectation and variance can be derived as
so that the prior is centred on the base probability measure P0j and the variance is P0j(B){1 − P0j(B)}ρ, with ρ the probability of clustering shown in Proposition 1.
Nonparametric Bayes procedures allow uncertainty in parametric specifications through priors with large support. The proposed prior, P ~ LPP1(α, β, P0), has weak support on the space of probability measures over (Ω, B). In addition, as formalized in Theorem 1, the proposed process induces a highly flexible prior on the joint distribution of the local cluster indices, ψi = (ψi1, …, ψip)T ~ Q, with Q a probability measure on {1, 2, …, ∞}p.
Theorem 1
Let θi ~ P with P ~ LPP1(α, β, P0), with hyperpriors chosen for α and β having support on ℜ+. Then, the following properties are satisfied:
Property 1. pr{Q(ψ) > ε} > δ for all ψ ∈ {1, 2, …, ∞}p and some ε, δ > 0;
Property 2. pr{Q(ψij = ψi′ j) ∈ A} > ε for all Borel subsets A ⊂ [0, 1], ψl ~ Q, l = i, i′, and some ε > 0;
-
Property 3. , for all j, j′ ∈ {1, …, p}, j′ ǂ j; and
Property 4. , for all Borel subsets A ⊂ [Q(ψij = ψi′ j), 1], j, j′ ∈ {1, …, p}, j′ ǂ j and for some ε > 0.
2·2. Formulation 2
In applications involving large p, it is appealing for computational reasons to consider sparse alternatives to P ~ LPP1(α, β, P0) that avoid the incorporation of a separate stick-breaking process for each component. As a simplification of (4), replace the second line with
This modified case is denoted as P ~ LPP2(α, β, P0). This specification assumes a common prior distribution for each of the elements of ϕi, resulting in a more parsimonious model. Under P ~ LPP2(α, β, P0), the local clustering probability pr(θij = θi′ j) is identical to the expression shown in Proposition 1, so the hyperparameters α and β have a similar role under both specifications. The main question is whether one maintains sufficient flexibility in characterizing dependence in local partitioning, which can be addressed through modifying Proposition 2 as follows.
Proposition 3
Assuming θi ~ P with P ~ LPP2(α, β, P0), then
As in Proposition 2, the joint probability is strictly larger than the product of the marginal clustering probabilities, implying positive dependence in local clustering. To investigate the flexibility in the degree of positive dependence, one can vary the values of α and β widely. Ideally, as α and β are varied, any degree of positive dependence could be obtained, ranging between independence of θij = θi′ j and θij′ = θi′ j′ to perfect dependence, with θij′ = θi′ j′ implying θij = θi′ j. Under Property 4 in Theorem 1, the prior P ~ LPP1(α, β, P0) allows any degree of positive dependence. In contrast, it follows from Proposition 3 that the prior P ~ LPP2(α, β, P0) induces a value for pr(θij = θi′ j ∣ θij′ = θi′ j′) that is constrained to fall in a subset of the interval [pr(θij = θi′ j), 1], so that Property 4 is not satisfied. Because this subset comprises almost the entire interval, with only values very close to the left boundary excluded, this constraint is not important from a practical perspective.
2·3. Special cases and connections
A variety of priors that have been proposed previously arise as special cases of the local partition process formulation in (2) and (3). Considering the case in which G = δ1, so that zij = 1 for all i, j, one obtains ψij = ψi, with ψi ~ H*, where H* is induced from H. By choosing H* to have a stick-breaking form, one can obtain any prior in the class proposed by Ishwaran & James (2001), which includes the Dirichlet process and two-parameter Poisson Dirichlet process (Pitman & Yor, 1997) as special cases. For the priors P ~ LPP1(α, β, P0) and P ~ LPP2(α, β, P0), G = δ1 corresponds to the limiting case β → 0 in which we obtain P ~ DP(α P0).
Another important special case corresponds to G = δ0, so that zij = 0 for all i, j, which occurs in the limit as β → ∞ for the priors P ~ LPP1(α, β, P0) and P ~ LPP2(α, β, P0). In the P ~ LPP1(α, ∞, P0) case, one obtains θij ~ Pj, with Pj ~ DP(α P0j) independently for j = 1, …, p, with P0j the jth marginal of P0. Potentially, dependence can be incorporated through the base measure P0, as in Cifarelli & Regazzini (1978), though this is a restrictive type of dependence. In the P ~ LPP2(α, ∞, P0) case, instead of independent Dirichlet process priors for Pj (j = 1, …, p), one obtains the following dependent Dirichlet process (MacEachern, 1999),
| (5) |
with Pj ~ DP(α P0j) marginally. Expression (5) corresponds to the fixed-π formulation of the dependent Dirichlet process (De Iorio et al., 2004).
Previous nonparametric Bayes methods for discrete random probability measures consider a single cluster index ψi, while local partition processes provide a methodology for multivariate ψi = (ψi1, …, ψip)T. A competing formulation relies on a matrix stick-breaking process (Dunson et al., 2008), which induces dependent local clustering through including stick-breaking random variables for every subject and predictor. This leads to substantial computational difficulties for large n or p, while requiring pr(θij = θi′ j ∣ θij′ = θi′ j′) > 0·5.
3. Posterior computation
Assume that yi ~ g(θi, τ), where yi is an ni × 1 vector of measurements for subject i, θi ~ P is a vector of parameters specific to subject i, and τ is a vector of population parameters. Initially letting P ~ LPP1(α, β, P0), we propose to use an adaptation of the slice sampling approach of Walker (2007) to implement posterior computation. This slice sampler avoids the need for a finite approximation, and is simpler to implement than the retrospective sampler of Papaspiliopoulos & Roberts (2008).
In addition to the latent variables defined in (4), we introduce latent variables (i = 1, …, n), where the complete data joint likelihood of y, u and z is
| (6) |
where the uijs are constrained to fall in (0, 1). The sampler proceeds as follows.
Step 1. For the latent uij, the conditional is Unif(0, πjϕij) (j = 0, 1, …, p).
- Step 2. For the latent zij, the conditional distribution is Ber(pij), with
where θi(zij=l) denotes the current value of θi with the jth element set equal to Θ0ϕi0j for l = 1 and Θ1ϕijj for l = 0. -
Step 3. For the stick-breaking variable , the conditional density is proportional toLetting , the conditional distribution of corresponds to the Be(1, α) prior for , while for the posterior is a Be(1, α) distribution truncated to fall in the (ajh, bjh) interval with
For all h such that , we have ajh = 0. In addition, bjh = 1 for .
- Step 4. The conditional probability of ϕij = h is proportional to 1(h ∈ Aij)g(yi; θi(ϕij=h), τ), where Aij = {h : πjh > uij} is a finite subset of {1, 2, …, ∞} obtained by first sampling , for h = 1, …, ϕ̃j, with ϕ̃j the smallest value satisfying
where . - Step 5. The conditional distribution of Θlh is proportional to
assuming the probability measure P0 has density g0 with respect to Lesbesgue measure. Step 6. For the probability νj, the conditional density is Be{1 + Σi zij, β + Σi (1 − zij)}.
Step 7. For the parameters τ, the conditional distribution is proportional to .
- Step 8. For the hyperparameter β, the conditional distribution is
- Step 9. For the hyperparameter α, the conditional distribution is
Each of these steps is simple to implement, and good rates of mixing and convergence have been observed in several applications. Steps 5 and 7 simplify when conjugate priors are chosen. For example, when g(yi; θi, τ) is the likelihood for a Gaussian linear model and P0 is chosen to obey a Gaussian law, the conditional distribution in Step 5 is Gaussian.
To modify the algorithm for formulation 2, drop the first subscript on each of the πs in (6) and in Steps 1 and 2. In Step 3, the conditional density for is proportional to
Letting ϕ* = max{ϕij, i = 1, …, n, j = 0, 1, …, p}, the conditional distribution of is Be(1, α) for h > ϕ*, while for h ≤ ϕ* the posterior is Be(1, α) truncated to (ah, bh) with
Step 4 is modified to let Aij = {h : πh > uij}, where is sampled for h = 1, …, ϕ̃, with ϕ̃ the smallest value satisfying . Steps 5–8 are unchanged, and in Step 9 the conditional distribution of α is
The major computational advantage relative to formulation 1 occurs in Step 3.
4. Simulation example
To illustrate the approach, a simulation example was considered following (1)-(4), with tis ∈ [0, 1] and basis functions b1(s) = 1, bj+1(s) = exp(−ψ∥s − ξj∥2) (j = 1, …, p − 1), with ξ1, …, ξp−1 equally spaced kernel locations, ψ = 25 and p = 20. To generate n = 16 curves, let θij = zijΘ0j + (1 − zij)Θ1j for i = 1, …, 16 and j = 1, …, 20, with zij ~ Ber(0·5) and the elements of Θ0 and Θ1 sampled independently from a mixture of a point mass at zero, having probability 0·5, and a standard normal density. The point mass allowed exclusion of basis functions. There were 40 equally spaced observations along each curve with σ2 = 0·2.
The data were analyzed using P ~ LPP1(α, β, P0), P ~ LPP2(α, β, P0) and P ~ DP(αP0), with Ga(1, 1) hyperpriors for α and β, a Ga(0·1, 0·1) hyperprior for σ−2, and P0 chosen to correspond to the multivariate Gaussian distribution with zero-mean and identity covariance. The slice sampler was run for 25 000 iterations, with the first 5 000 samples discarded as a burn-in and every twentieth sample collected to thin the chain. In each case, the sampler appeared to converge rapidly and to mix efficiently based on examination of trace plots of α, β, σ2 and function values at a variety of locations and for a variety of individuals. Due to label switching, it is not reliable to monitor the elements of Θh in assessing convergence and mixing.
Figure 1 shows the data, true curves and estimates under P ~ LPP1(α, β, P0). The estimates are close to the truth, with average absolute bias = 0·192, mean square error = 0·060, maximum absolute bias = 0·506 and an average 99% pointwise credible interval width of 1·01. The estimated posterior mean of σ2 was 0·21, with a 95% credible interval of [0·18, 0·26]. The estimate for α was 0·17, with 95% credible interval [0·10, 0·29]. The small value of α suggests the individuals are assigned to few local and global clusters. For component j (j = 0, 1, …, p), all subjects are allocated to the first clusters. The posterior means of are less than 2 for j = 0, 1, …, p, with and , for j = 1, …, p. The probability of allocation to the local component for a random element of a subject’s random effects vector is 1/(1 + β). Hence, β provides a measure of the relative importance of the local and global components, with both components receiving equal weight for β = 1 and the global component dominant for β < 1. For the simulation, we obtained β̂ = 1·23, with a 95% credible interval of [0·51, 2·42], so that the two components receive close to equal weight.
Fig. 1.
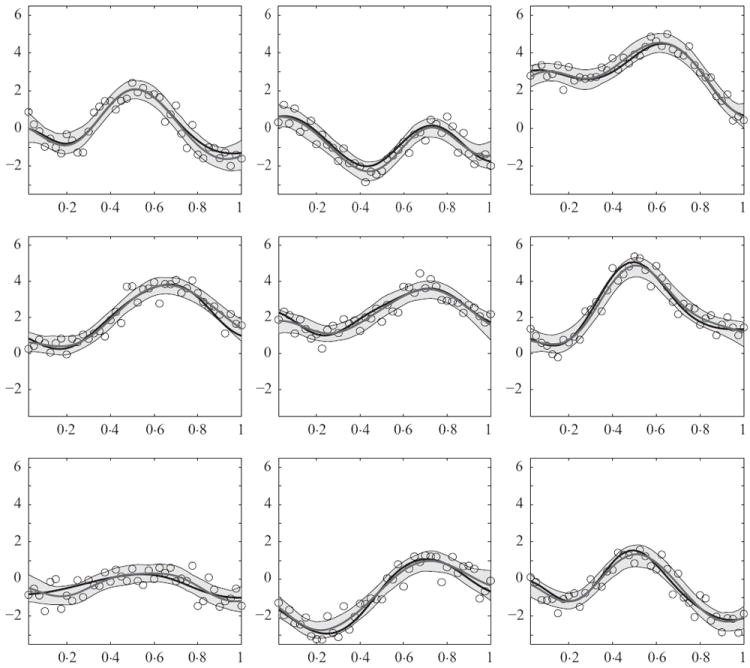
Data and results for 9 of the 16 subjects in the simulation example. Each panel corresponds to one subject in the study, the true functions are represented with black lines, the posterior means with grey lines and 99% pointwise credible intervals are shaded.
For the analysis under P ~ LPP2(α, β, P0), there was a substantial reduction in computational time and the results were improved, with average absolute bias = 0·168, mean square error = 0·046, maximum absolute bias = 0·520 and an average 99% credible interval width of 0·793. The estimated posterior mean of β was 1·56, with a 95% credible interval of [0·64, 3·25]· In addition, σ̂2 = 0·20, with a 95% credible interval of [0·18, 0·23]. It does not appear that the more concentrated posterior distributions reflected a lack of proper account for uncertainty in that the true functions were all entirely enclosed in the 99% pointwise intervals. Hence, the narrower intervals more likely reflect an improvement in efficiency due to the incorporation of fewer parameters in the second formulation.
For the Dirichlet process, there was minimal borrowing of information across the different functions, with posterior probability of allocating the 16 functions to 13 clusters greater than 0·99. The resulting function estimates were still reasonable, but the performance was not nearly as good as for the local partition process. The average absolute bias was increased by 26%, the mean square error by 60% and the maximum absolute bias by 57% relative to the first formulation. The average credible interval widths were similar, but the true functions fell well outside the 99% intervals in two out of 16 of the cases. In addition, the estimated residual variance was σ̂2 = 0·27, with 99% credible interval [0·23, 0·31] not including the true value of 0·20. This overestimation of the residual variance suggests a poor job at recovering the signal. This was the first simulation scenario considered, but there was general improvement for the local partition process over the Dirichlet process in a broad variety of scenarios, with the gain very substantial in many cases. Gains are most notable when data for each function are sparse.
5. Hormone curve application
5·1. Background and motivation
The approach is applied to post-ovulatory progesterone data collected in early pregnancy for n = 165 women (Wilcox et al., 1988). The progesterone metabolite, PdG, was measured in urine. Letting yis denote the sth measurement of log(PdG) for woman i occurring tis days after the estimated day of ovulation, we assume yis ~ tκ {ηi (tis), σ2}, where ηi is a smooth trajectory in log(PdG) for woman i and tκ (μ, σ2) denotes the t-density, with mean μ, degrees of freedom κ and scale σ2. Expression (1) is generalized to allow t-distributed measurement errors. The data contain between ni = 4 and ni = 40 observations per woman, with an average of n̄ = 23·1. In general, the data are collected daily in the morning, but there are gaps, with most of these gaps corresponding to censoring in which the woman stopped collecting urine prior to day 40. Censoring occurred if menses resumed following an early pregnancy loss or the six-month collection period ended. Given the plausible missingness mechanisms, it is reasonable to assume that missingess is conditionally independent of the missing PdG values given the observed PdG values, implying data are missing at random.
The goal is to obtain a flexible, yet parsimonious representation of the progesterone trajectories. Bigelow & Dunson (2009) analyzed these data using a Dirichlet process prior for the distribution of the woman-specific basis coefficients in a spline model assuming normally distributed measurement errors. They estimated 31 PdG trajectory clusters, with 18 of these being singletons containing just one woman. Given that many of their reported trajectory clusters have similar shapes with only local deviations, it is our expectation that replacing the Dirichlet process prior on the basis coefficients with a local partition process prior can produce a more parsimonious representation.
In analyzing the data, we used the approach applied in § 4 for the simulated data after standardizing time to the [0, 1] interval and adapting the approach to accommodate t-distributed measurement errors. This adaptation was straightforward by expressing the t-distribution as a scale mixture of normals by letting , with ξi ~ Ga(κ/2, κ/2) and with the degrees of freedom κ assigned a Ga(1, 1) hyperprior to favour very heavy tails to correspond with our prior knowledge. Under this structure, the full conditional posterior distribution for ξi is gamma, while κ is updated using Metropolis–Hastings steps.
5·2. Analysis and results
We focus initially on the first formulation of the local partition process. Using multiple chains with widely distributed starting points, the slice sampler was observed to exhibit good rates of convergence and mixing. Figure 2 shows the results under P ~ DP(αP0) and P ~ LPP1(α, β, P0) for five women. It is clear from this plot, and from examination of plots for the other women in the study, that the local partition process method produces a good fit. The estimated value of the hyperparameter α was α̂ = 0·41, with a 95% credible interval of [0·25, 0·63]. This value suggests that few global and local clusters are needed, so that a sparse representation of the data is obtained. Indeed, the posterior probability of was 0·97, suggesting that subjects were allocated to three global clusters. The number of local clusters also tended to be small, having an estimated average value of 2·91.
Fig. 2.
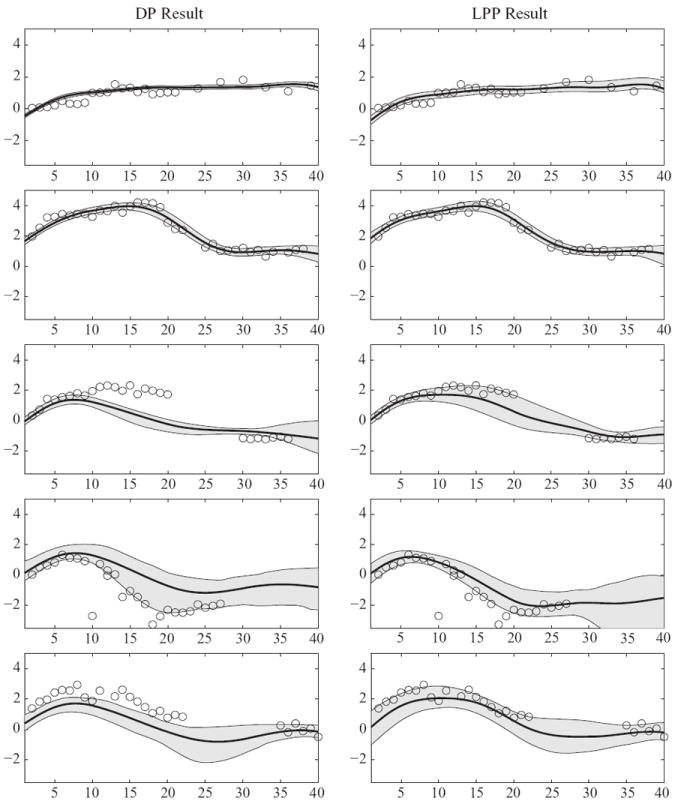
Log PdG function estimates for selected women in the Early Pregnancy Study. The posterior means are solid lines and 95% pointwise credible intervals are shown in grey. The x-axis scale is time in days starting at the estimated day of ovulation.
The estimated value of the hyperparameter β was 1·05, with a 95% credible interval of [0·63, 1·59]. Recall that values of β close to zero provide support for a joint Dirichlet process prior on the distribution of the basis coefficients, while large values provide support for independent Dirichlet process priors for the different coefficients. A value close to one instead provides evidence for a local partition process balanced between the two extremes, with a 50–50 chance of a randomly selected basis coefficient being drawn from a global versus local component. Hence, there is clear evidence in the data favouring our proposed approach over the Dirichlet process.
The estimated degrees of freedom in the t-distribution was 2·05, with a 95% credible interval of [2·01, 2·11], suggesting very heavy tails. This is as expected, since most of the values are tightly distributed about a smooth trajectory, but there are extreme outlying values in the dataset. Commenting further on the estimated hormone curves, many of the trajectories increase rapidly after ovulation and then flatten out within a week or two, while other trajectories increase initially and then decrease. It is likely that the increasing trajectories correspond to healthy pregnancies, while the trajectories that peak a week or more after ovulation and then decline correspond to early pregnancy losses.
Repeating the analysis for P ~ DP(αP0), the posterior mean of α was 3·60, with a 95% credible interval of [1·84, 6·32]. The α parameter was slow-mixing in the Markov chain Monte Carlo implementation, with high autocorrelation. However, because traceplots of the subject-specific basis coefficients and function estimates were well behaved, the mixing problems with α did not appear to impact our inferences. The posterior mean number of clusters was 21·4, with a 95% interval of [15,37], reflecting large posterior uncertainty in clustering and a substantially larger number of clusters than in the local partition process analysis. In most cases, the estimates are similar to those obtained for P ~ LPP1(α, β, P0). However, as illustrated in Fig. 2, there are multiple exceptions in which the Dirichlet process approach produced an estimate inconsistent with the data. We noted particularly poor performance for women having relatively few observations. This likely reflects a tendency of the Dirichlet process to overly favour clustering together of subjects unless there are abundant data available to suggest that this clustering is not supported. Hence, in sparse data situations, one anticipates dramatic gains for the local partition process.
Finally, we implemented the sparse variant of the local partition process. The estimate of α was 0·38 with a 95% interval of [0·10, 0·95], and the estimate of β was 1·87 with a 95% interval of [1·10, 2·92]. The larger estimate of β suggests that P ~ LPP2(α, β, P0) places more weight on the local component than P ~ LPP1(α, β, P0). This is likely due to the fact that there is less of a penalty to be paid for introducing additional local clusters under P ~ LPP2(α, β, P0) due to the incorporation of common stick-breaking weights. The estimates for the overall number of clusters and the log(PdG) curves were very similar to those for the LPP1(α, β, P0). However, there was a substantial decrease in the time required per iteration of the slice sampler. The first formulation took 112 seconds per 100 iterations in Matlab on a MacBook Pro laptop, while the Dirichlet process implementation took 30 seconds and the sparse local partition process took 69 seconds. We repeated each of the analyses for a variety of hyperparameter values, with the variance multiplied by 2 and divided by 2 for α, β, P0 and κ. There were no noticeable differences in the results.
6. Discussion
This paper proposes a generalization of the Dirichlet process, which allows dependent local clustering and borrowing of information. The emphasis is on functional data analysis applications in which each function is characterized as a linear combination of basis functions, and the goal is to flexibly borrow information to more efficiently estimate the individual functions. In order to favour more borrowing of information across the individual functions and obtain a more parsimonious representation of the data, one can allow the basis coefficients for an individual to be locally selected from a small number of vectors of unique basis coefficients. The vectors of unique basis coefficients can be viewed as representing underlying commonalities across the different functions, resulting in a discrete nonparametric analogue of functional principal components. This type of idea seems very promising as a tool for generating sparse characterizations of complex multivariate and functional data in a variety of settings. The proposed local partition process provides a simple, yet flexible approach for characterizing the local selection process. The slice sampling implementation is quite simple and efficient to implement, while allowing posterior computation for the infinite-dimensional nonparametric process instead of a finite approximation.
Appendix
Proof of Proposition 1
In order to derive the marginal probability of local clustering, pr(θij = θi′ j), let
with Proposition 1 following directly, where f(πj) denotes the Be(1, β) prior distribution for πj and B(a, b) denotes the beta(a, b) function.
Proof of Proposition 2
The joint probability of local clustering follows along similar lines:
Proof of Proposition 3
Express the joint probability of local clustering as
Proof of Theorem 1
To demonstrate property 1, first let
where ψ ∈ {1, 2, …, ∞}p, Io = {1, 3, 5, …} and Ie = {2, 4, 6, …}. Let Q̄(ψ) denote the median of the distribution of Q(ψ) for fixed ψ. Note that Q̄(ψ) is expressed as a product of finitely many random variables in (0, 1), so that pr{Q(ψ) > 0} = 1. It follows that Q̄(ψ) > 0 and hence pr{Q(ψ) > εψ} = 0·5, with εψ = Q̄(ψ) > 0. Letting ε = min[εψ, ψ ∈ {1, 2, …, ∞}p], we have pr{Q(ψ) > ε} ⩾ 0·5.
To demonstrate Property 2, first let
where ρ(α, β) is defined in Proposition 1, with the dependence on α and β now explicit, and f(α, β) is the prior density for α, β on (0, ∞) × (0, ∞). For any point a ∈ A, there exists a corresponding region d(a) ⊂ (0, ∞) × (0, ∞) such that ρ(α, β) = a for all (α, β) ∈ d(a). Letting d(A) = ∪a∈A d(a), pr{Q(θij = θi′ j) ∈ A} = ∫d(A) f(α, β)dα dβ. For all Borel subsets A ⊂ (0, 1), the area of d(A) is greater than 0, so pr{Q(θij = θi′ j) = A} > 0 as long as f(α, β) > 0 for all (α, β) ∈ (0, ∞) × (0, ∞), which holds for independent gamma priors on α and β.
Property 3 follows from Propositions 1 and 2. To demonstrate Property 4, first let
where ρ2(α, β) = pr(θij = θi′ j ∣ θi j′ = θi′ j′, α, β). Hence, given that f(α, β) > 0 for all (α, β) ∈ (0, ∞) × (0, ∞), it suffices to show that there exists a region d2(a) ⊂ (0, ∞) × (0, ∞) of (α, β) values that result in ρ2(α, β) = a, with d2(a) ǂ 0̸ for all a ∈ (0, 1). This condition follows directly if there exists an (α, β) solution to the equations ρ(α, β) = a, ρ2(α, β) = b, for every point (a, b) in 0 < a < b < 1, which is easily verified.
References
- Bigelow JL, Dunson DB. Bayesian semiparametric joint models for functional predictors. J Am Statist Assoc. 2009;104:26–36. doi: 10.1198/jasa.2009.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush CA, MacEachern SN. A semiparametric Bayesian model for randomised block designs. Biometrika. 1996;83:275–85. [Google Scholar]
- Cifarelli DM, Regazzini E. Nonparametric statistical problems under partial exchangeability: The use of associative means (in italian) Annali del’Instituto di Matematica Finianziara dell’Universitá di Torino, Serie III. 1978;12:1–36. [Google Scholar]
- De Iorio M, Müller P, Rosner G, MacEachern SN. An ANOVA model for dependent random measures. J Am Statist Assoc. 2004;99:205–15. [Google Scholar]
- Dunson DB, Xue Y, Carin L. The matrix stick-breaking process: Flexible Bayes meta analysis. J Am Statist Assoc. 2008;103:317–27. [Google Scholar]
- Ferguson TS. A Bayesian analysis of some nonparametric problems. Ann Statist. 1973;1:209–30. [Google Scholar]
- Ferguson TS. Prior distributions on spaces of probability measures. Ann Statist. 1974;2:615–29. [Google Scholar]
- Heard NA, Holmes CC, Stephens DA. A quantitative study of gene regulation involved in the immune response of anopheline mosquitoes: An application of Bayesian hierarchical clustering of curves. J Am Statist Assoc. 2006;101:18–29. [Google Scholar]
- Ishwaran H, James LF. Gibbs sampling methods for stick-breaking priors. J Am Statist Assoc. 2001;96:161–73. [Google Scholar]
- MacEachern SN. Proc Sect Bayesian Statist Sci. Alexandria, VA: American Statistical Association; 1999. Dependent nonparametric processes; pp. 50–55. [Google Scholar]
- Müller P, Rosner GL. A Bayesian population model with hierarchical mixture priors applied to blood count data. J Am Statist Assoc. 1997;92:1279–92. [Google Scholar]
- Papaspiliopoulos O, Roberts GO. Retrospective MCMC for Dirichlet process hierarchical models. Biometrika. 2008;95:169–86. [Google Scholar]
- Pitman J, Yor M. The two parameter Poisson–Dirichlet distribution derived from a stable subordinator. Ann Prob. 1997;25:855–900. [Google Scholar]
- Sethuraman J. A constructive definition of Dirichlet priors. Statist Sinica. 1994;4:639–50. [Google Scholar]
- Walker SG. Sampling the Dirichlet mixture model with slices. Comm Statist B. 2007;36:45–54. [Google Scholar]
- Wilcox A, Weinberg CR, O’Connor J, Baird D, Schlatterer J, Canfield R, Armstrong E, Nisula B. Incidence of early loss of pregnancy. New Engl J Med. 1988;319:189–94. doi: 10.1056/NEJM198807283190401. [DOI] [PubMed] [Google Scholar]


