Abstract
Elevated serum urate concentrations can cause gout, a prevalent and painful inflammatory arthritis. By combining data from >140,000 individuals of European ancestry within the Global Urate Genetics Consortium (GUGC), we identified and replicated 28 genome-wide significant loci in association with serum urate concentrations (18 new regions in or near TRIM46, INHBB, SFMBT1, TMEM171, VEGFA, BAZ1B, PRKAG2, STC1, HNF4G, A1CF, ATXN2, UBE2Q2, IGF1R, NFAT5, MAF, HLF, ACVR1B-ACVRL1 and B3GNT4). Associations for many of the loci were of similar magnitude in individuals of non-European ancestry. We further characterized these loci for associations with gout, transcript expression and the fractional excretion of urate. Network analyses implicate the inhibins-activins signaling pathways and glucose metabolism in systemic urate control. New candidate genes for serum urate concentration highlight the importance of metabolic control of urate production and excretion, which may have implications for the treatment and prevention of gout.
Uric acid is a final breakdown product of purine oxidation in humans and is present in the blood as urate. Elevated concentrations of serum urate—hyperuricemia—can cause gout1. Gout is the most prevalent inflammatory arthritis in developed countries, with an estimated 8.3 million US adults in 2007–2008 having had at least one of the extremely painful attacks2. Prevalence is increasing, owing in part to population aging, dietary and lifestyle factors, and rising levels of obesity and insulin resistance3–5. Chronic gout inflicts a considerable social and economic burden resulting from the associated pain and disability6,7 as well as reduced work-related activity and productivity8.
Blood urate concentrations are determined by a balance between uric acid production, primarily in the liver, and its disposal via the kidney and gut. High circulating levels are mainly due to the net reabsorption of 90% of filtered urate in the renal proximal tubules9. Understanding the control of uric acid homeostasis is critical to improving the management and treatment of patients with hyperuricemia and gout.
The heritability of serum urate concentrations is estimated at 40–70% (refs. 10–12), which justifies the search for its genetic determinants. Previous genome-wide association studies (GWAS) have so far identified 11 genomic loci associated with urate concentrations and gout13–18. Together, SNPs at these loci explained about 5–6% of variance in serum urate concentrations17,18, suggesting that additional loci remain to be identified. We therefore aimed to identify and validate variants associated with serum urate concentrations in >140,000 individuals of European ancestry and with gout in approximately 70,000 individuals in the GUGC. These variants may provide a basis for the identification of new potential therapeutic targets for gout19.
RESULTS
Characteristics of the study samples
An overview of the characteristics of the GUGC study samples is provided in Supplementary Table 1. Our study comprises 110,347 individuals from 48 studies contributing to the discovery GWAS meta-analysis of serum urate concentrations and participants of 14 studies contributing to the meta-analysis of gout (2,115 cases and 67,259 controls). Across studies, mean serum urate concentrations ranged from 3.9 to 6.1 mg/dl (median of 5.2 mg/dl), and the proportion with gout ranged from 0.9% to 6.4% (median of 3.3%). Detailed information about the GUGC studies is provided in Supplementary Table 2. Information about study-specific genotyping, imputation, analysis tools and inflation factors for the individual serum urate concentration GWAS (range of 0.98–1.05) are shown in Supplementary Table 3.
Loci associated with serum urate concentrations
A quantile-quantile plot for the 2,450,547 investigated autosomal SNPs in the discovery GWAS meta-analysis showed many more SNPs with low observed P values than expected, even after excluding SNPs in known urate concentration–associated regions (Supplementary Fig. 1). All 2,201 SNPs associated with serum urate concentrations at P < 5 × 10−8 in the discovery stage are listed in Supplementary Table 4. Overall, 37 different genomic loci were identified that contained SNPs associated with serum urate concentrations at P <1 × 10−6; 26 of these were associated at genome-wide significance (P < 5 × 10−8, 10 known and 16 new loci; Fig. 1). Regional association plots provide a detailed overview of the associated regions (Supplementary Fig. 2). To assess the presence of independently associated SNPs within the regions associated at genome-wide significance and suggestive levels of significance (5 × 10−8 < P < 1 × 10−6), conditional analyses were carried out (Online Methods). This analysis identified only the SLC22A11-NRXN2 region as containing more than one independent signal (Supplementary Table 5). The association at the previously reported LRRC16A locus17 was not independent from that at the SLC17A1 locus in our study. For each of the 37 independent loci, the index SNP with the lowest P value was carried forward for replication.
Figure 1.
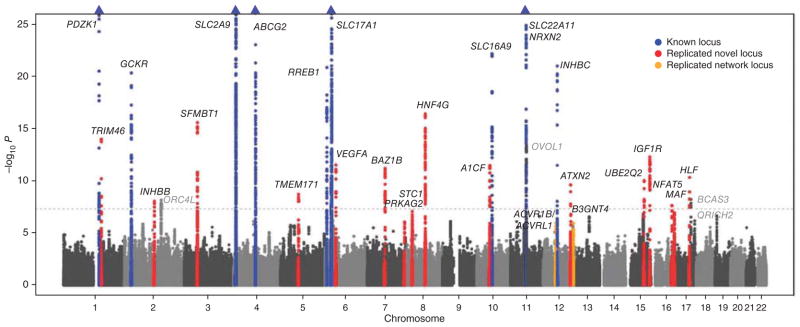
Multiple genomic loci contain SNPs associated with serum urate concentrations. Truncated Manhattan plot showing −log10 (P values) for all SNPs of the urate discovery GWAS ordered by chromosomal position. The gene closest to the SNP with the lowest P value at each locus (index SNP) is listed. Loci in gray met one but not both replication criteria. Blue triangles represent loci containing SNPs with P values below 1 × 10−25.
Replication was attempted in up to 32,813 new study participants. Successful replication was defined as q value < 0.05 in the independent replication sample and genome-wide significance in the combined sample (P < 5 × 10−8; Online Methods). Summary results for the 26 replicated urate concentration–associated loci are shown (Table 1): there were 16 newly identified regions in or near TRIM46, INHBB, SFMBT1, TMEM171, VEGFA, BAZ1B, PRKAG2, STC1, HNF4G, A1CF, ATXN2, UBE2Q2, IGF1R, NFAT5, MAF and HLF in addition to 10 known urate concentration–associated regions in or near PDZK1, GCKR, SLC2A9, ABCG2, RREB1, SLC17A1, SLC16A9, SLC22A11, NRXN2 and INHBC. The proportion of variance in serum urate concentrations explained by these replicated loci in our data was 7.0%, with 5.2% of this estimate explained by the ten previously known loci and 3.4% explained by SLC2A9 and ABCG2 alone. The moderate proportion of trait variance explained is in line with findings from GWAS of other phenotypes20. The proportion of the age- and sex-adjusted variance in serum urate concentrations explained by all common SNPs rather than just the genome-wide significant index SNPs ranged from 0.27 (standard error (s.e.) of 0.04, ARIC Study) to 0.41 (s.e. of 0.07, SHIP Study) (Online Methods).
Table 1.
SNPs associated with serum urate concentrations in individuals of European ancestry
| SNP | Chr. | bp (Build 36) | Closest gene | GRAIL gene | A1 | A2 | Freq. A1 | Serum urate (mg/dl)
|
Gout
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | s.e. | P value | OR | P value | ||||||||
| Discovered loci described previously | ||||||||||||
| rs1471633 | 1 | 144435096 | PDZK1 | PDZK1 | A | C | 0.46 | 0.059 | 0.005 | 1.2 × 10−29 | 1.03 | 2.8 × 10−1 |
| rs1260326 | 2 | 27584444 | GCKR | GCKR | T | C | 0.41 | 0.074 | 0.005 | 1.2 × 10−44 | 1.14 | 8.2 × 10−6 |
| rs12498742 | 4 | 9553150 | SLC2A9 | SLC2A9 | A | G | 0.77 | 0.373 | 0.006 | 0b | 1.56 | 1.9 × 10−31 |
| rs2231142 | 4 | 89271347 | ABCG2 | ABCG2 | T | G | 0.11 | 0.217 | 0.009 | 1.0 × 10−134 | 1.73 | 1.7 × 10−39 |
| rs675209 | 6 | 7047083 | RREB1 | RREB1 | T | C | 0.27 | 0.061 | 0.006 | 1.3 × 10−23 | 1.09 | 1.1 × 10−2 |
| rs1165151 | 6 | 25929595 | SLC17A1 | SLC17A3 | T | G | 0.47 | −0.091 | 0.005 | 7.0 × 10−70 | 0.86 | 5.3 × 10−7 |
| rs1171614 | 10 | 61139544 | SLC16A9 | SLC16A9 | T | C | 0.22 | −0.079 | 0.007 | 2.3 × 10−28 | 0.91 | 1.7 × 10−2 |
| rs2078267 | 11 | 64090690 | SLC22A11 | SLC22A11 | T | C | 0.51 | −0.073 | 0.006 | 9.4 × 10−38 | 0.88 | 2.3 × 10−5 |
| rs478607 | 11 | 64234639 | NRXN2 | SLC22A12 | A | G | 0.84 | −0.047 | 0.007 | 4.4 × 10−11 | 0.97 | 4.1 × 10−1 |
| rs3741414 | 12 | 56130316 | INHBC | INHBE | T | C | 0.24 | −0.072 | 0.007 | 2.2 × 10−25 | 0.87 | 2.7 × 10−4 |
| New loci | ||||||||||||
| rs11264341 | 1 | 153418117 | TRIM46 | PKLR | T | C | 0.43 | −0.050 | 0.006 | 6.2 × 10−19 | 0.92 | 7.4 × 10−3 |
| rs17050272 | 2 | 121022910 | INHBB | INHBB | A | G | 0.43 | 0.035 | 0.006 | 1.6 × 10−10 | 1.03 | 3.9 × 10−1 |
| rs2307394a | 2 | 148432898 | ORC4L | ACVR2A | T | C | 0.68 | −0.029 | 0.005 | 2.2 × 10−8 | 0.94 | 6.3 × 10−2 |
| rs6770152 | 3 | 53075254 | SFMBT1 | MUSTN1 | T | G | 0.58 | −0.044 | 0.005 | 2.6 × 10−16 | 0.90 | 3.0 × 10−4 |
| rs17632159 | 5 | 72467238 | TMEM171 | TMEM171 | C | G | 0.31 | −0.039 | 0.006 | 3.5 × 10−11 | 0.91 | 6.0 × 10−3 |
| rs729761 | 6 | 43912549 | VEGFA | VEGFA | T | G | 0.30 | −0.047 | 0.006 | 8.0 × 10−16 | 0.87 | 4.1 × 10−5 |
| rs1178977 | 7 | 72494985 | BAZ1B | MLXIPL | A | G | 0.81 | 0.047 | 0.007 | 1.2 × 10−12 | 1.14 | 6.7 × 10−4 |
| rs10480300 | 7 | 151036938 | PRKAG2 | PRKAG2 | T | C | 0.28 | 0.035 | 0.006 | 4.1 × 10−9 | 1.09 | 6.5 × 10−3 |
| rs17786744 | 8 | 23832951 | STC1 | STC1 | A | G | 0.58 | −0.029 | 0.005 | 1.4 × 10−8 | 0.93 | 2.1 × 10−2 |
| rs2941484 | 8 | 76641323 | HNF4G | HNF4G | T | C | 0.44 | 0.044 | 0.005 | 4.4 × 10−17 | 1.04 | 1.7 × 10−1 |
| rs10821905 | 10 | 52316099 | A1CF | ASAH2 | A | G | 0.18 | 0.057 | 0.007 | 7.4 × 10−17 | 1.09 | 2.6 × 10−2 |
| rs642803a | 11 | 65317196 | OVOL1 | LTBP3 | T | C | 0.46 | −0.036 | 0.005 | 2.9 × 10−13 | 0.90 | 2.2 × 10−4 |
| rs653178 | 12 | 110492139 | ATXN2 | PTPN11 | T | C | 0.51 | −0.035 | 0.005 | 7.2 × 10−12 | 0.95 | 7.3 × 10−2 |
| rs1394125 | 15 | 73946038 | UBE2Q2 | NRG4 | A | G | 0.34 | 0.043 | 0.006 | 2.5 × 10−13 | 1.03 | 3.6 × 10−1 |
| rs6598541 | 15 | 97088658 | IGF1R | IGF1R | A | G | 0.36 | 0.043 | 0.006 | 4.8 × 10−15 | 1.04 | 2.5 × 10−1 |
| rs7193778 | 16 | 68121391 | NFAT5 | NFAT5 | T | C | 0.86 | −0.046 | 0.008 | 8.2 × 10−10 | 0.92 | 3.4 × 10−2 |
| rs7188445 | 16 | 78292488 | MAF | MAF | A | G | 0.33 | −0.032 | 0.005 | 1.6 × 10−9 | 0.95 | 1.1 × 10−1 |
| rs7224610 | 17 | 50719787 | HLF | HLF | A | C | 0.58 | −0.042 | 0.005 | 5.4 × 10−17 | 0.96 | 1.6 × 10−1 |
| rs2079742a | 17 | 56820479 | BCAS3 | C17orf82 | T | C | 0.85 | 0.043 | 0.008 | 1.2 × 10−8 | 1.04 | 3.5 × 10−1 |
| rs164009a | 17 | 71795264 | QRICH2 | PRPSAP1 | A | G | 0.61 | 0.028 | 0.005 | 1.6 × 10−7 | 1.08 | 8.6 × 10−3 |
P values are corrected for inflation using genomic control. Median imputation quality was calculated across all cohorts for urate concentration–associated SNPs in the overall sample and ranged from 0.83 to 1 (median of 0.98). Allele frequencies are presented for the discovery sample. Gout estimates are provided for the combined discovery and validation samples. Sample sizes available for the individual SNPs are listed in Supplementary Table 6. Chr., chromosome; A1, allele 1, effect allele; freq., frequency; s.e., standard error.
These SNPs met one but not both criteria required for replication.
P value < 1 × 10−700.
Additionally, regions in or near ORC4L, OVOL1 and BCAS3 were associated at genome-wide significance but had replication q values of ≥0.05, whereas the QRICH2 locus had a replication q value of <0.05 but did not reach genome-wide significance in the combined sample. Effect estimates, allele frequencies and heterogeneity measures are presented separately for the discovery, replication and combined analyses for all 37 loci tested for replication (Supplementary Table 6). Although the functions of genes within the associated genomic regions were frequently connected to urate transport for the known loci, newly associated regions contained a series of transcription and growth factors encoding genes potentially connected to the metabolic control of serum urate production and excretion (Supplementary Table 7). Ten of the replicated loci also showed significant association with other complex traits, as listed in the National Human Genome Research Institute (NHGRI) GWAS Catalog21, and VEGFA and ATXN2 showed significant associations with ratios of serum metabolites containing a γ-glutamyl amino acid from a published metabolite-SNP association resource22.
Secondary analyses
Three secondary analyses were conducted to identify additional urate concentration–associated SNPs. First, as rare monogenic syndromes featuring gout can be caused by X-chromosome mutations in PRPS1 (MIM 300661) and HPRT1 (MIM 300322), we queried 54,926 X-chromosomal SNPs for association with serum urate concentrations in a meta-analysis of 72,026 participants from 25 of the GUGC studies (Supplementary Note). No genome-wide significant associations were observed, either in the combined or the sex-specific analyses (Supplementary Fig. 3); all X-chromosomal SNPs associated with serum urate concentrations at P < 1 × 10−4 are listed (Supplementary Table 8).
Second, because of the higher prevalence of gout in men and the known sex-related differences in the effects of urate concentration–associated variants in SLC2A9 and ABCG2 (refs. 14,16), we conducted meta-analyses of GWAS separately for 49,825 men and 60,522 women. In addition to SNPs at loci detected in the combined analysis, SNPs were associated with serum urate concentrations at P < 1 × 10−6 in one region in men and in five regions in women. None of these SNPs was significantly associated with serum urate concentrations in the replication samples (Supplementary Table 6), although the index SNP at HNF1A in women can be considered borderline significant with an association at P = 8.1 × 10−8 in the combined discovery and replication data set. Besides SLC2A9 and ABCG2, no additional regions contained SNPs that differed significantly (P < 5 × 10−8) in their association effect sizes between men and women. Sex-related differences for genome-wide significant and suggestive SNPs of the overall and sex-stratified analyses are shown (Supplementary Table 9).
Third, we conducted a search for associated SNPs in genes that are family members of known urate transporter genes and, to the best of our knowledge, had not yet been connected to urate transport in humans. After correcting for multiple testing (Supplementary Note), SNPs in the SLC22A7 region (SLC22A11- SLC22A12 family member) showed significant association with serum urate concentrations in the discovery (P = 1.9 × 10−5) and replication (q value < 0.05) samples but did not reach the stringent genome-wide significance level in the combined samples (Table 2). It was recently found that the organic anion transporter 2 (OAT2), encoded by SLC22A7, transports urate in HEK293 cells23. Our findings show that variation in SLC22A7 has a measurable effect on serum urate concentrations in humans, supporting the idea that it is a genuine urate concentration–associated GWAS locus of small effect size. Regional association plots for loci implicated through sex-specific analyses or the candidate gene approach are also shown (Supplementary Fig. 2).
Table 2.
Urate concentration–associated SNPs from secondary analyses
| SNP | Chr. | bp (Build 36) | Gene | A1 | A2 | Freq. A1 | Serum urate (mg/dl)
|
Gout
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | s.e. | P value | OR | P value | |||||||
| Urate transporter candidate analysis | |||||||||||
| rs4149178a | 6 | 43380166 | SLC22A7 | A | G | 0.84 | −0.034 | 0.007 | 1.2 × 10−6 | 0.95 | 2.2 × 10−1 |
| Functional association network analysis | |||||||||||
| rs4970988a | 1 | 149216686 | ARNT | A | G | 0.36 | −0.028 | 0.005 | 1.0 × 10−7 | 0.96 | 2.1 × 10−1 |
| rs7976059 | 12 | 50537539 | ACVR1B-ACVRL1 | T | G | 0.35 | 0.032 | 0.005 | 1.9 × 10−9 | 0.95 | 1.8 × 10−1 |
| rs7953704 | 12 | 121191945 | B3GNT4 | A | G | 0.47 | −0.029 | 0.005 | 2.6 × 10−8 | 0.95 | 1.6 × 10−1 |
Allele frequencies are presented for the discovery sample. Chr., chromosome; A1, allele 1, effect allele; freq., frequency; s.e., standard error.
These SNPs met one but not both criteria required for replication.
Gout GWAS
We conducted a genome-wide gout discovery meta-analysis including 2,538,056 autosomal SNPs. Two previously reported loci had associations that reached genome-wide significance in the overall and sex-stratified analyses: ABCG2 (rs1481012, P = 2.0 × 10−32) and SLC2A9 (rs4475146, P = 4.1 × 10−26) (Supplementary Fig. 4 and Supplementary Table 10). The quantile-quantile plot of the P values from the combined analysis showed more observed low P values than expected by chance, even after excluding the previously known urate concentration–associated regions (Supplementary Fig. 5). The index SNPs in SLC2A9 and ABCG2 from the combined analysis of gout were in high linkage disequilibrium (LD; r2 > 0.9) with the index SNPs from the serum urate concentration meta-analysis at these loci.
Serum urate concentration–associated loci and gout
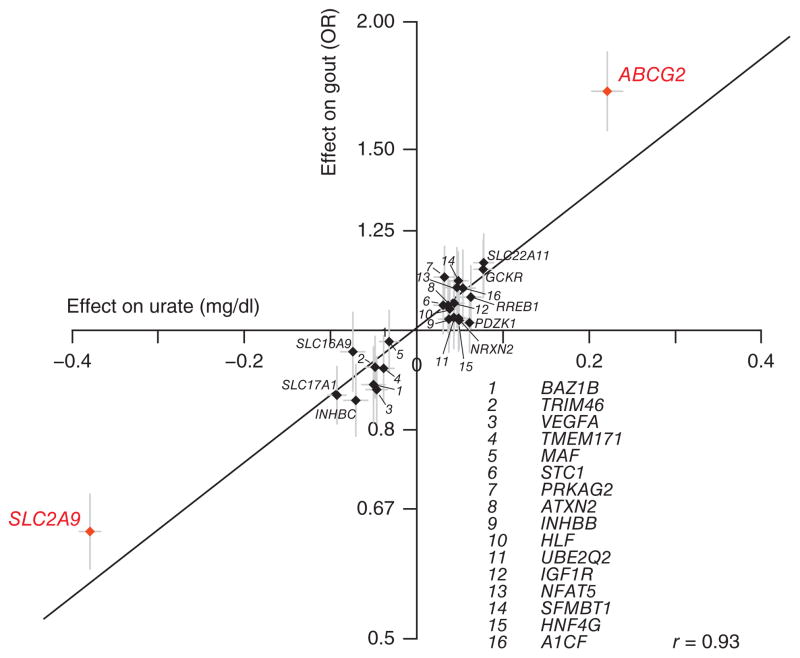
Because elevated serum urate levels are a key risk factor for the development of gout, we investigated the association of the urate concentration–associated SNPs with gout in the data set described. We observed a positive linear correlation between the genetic effect on serum urate concentrations and the log odds of gout for the replicated loci (Pearson’s correlation = 0.93; Fig. 2). In addition, information was available on the associations with incident gout over a period of up to 22 years in two independent studies that were not part of either the gout or serum urate concentration discovery analyses. The 1,036 cases of gout in these studies met the American College of Rheumatology Criteria24,25. In all gout samples combined (3,151 cases and 68,350 controls), 17 out of 26 of the replicated urate concentration–associated SNPs showed nominal association with gout (P < 0.05; Table 1). SNP associations with prevalent and incident gout were of generally comparable direction and magnitude overall, as well as among men and women separately (Supplementary Table 11).
Figure 2.
Minor alleles of all replicated GWAS loci show direction-consistent association with serum urate concentrations and the odds of gout. Loci with significant sex-specific effects are shown in red. Lines correspond to 95% confidence intervals. The Pearson’s correlation coefficient was calculated using the log odds ratio (ln(OR)) for gout.
Genetic urate risk score is associated with gout
A weighted genetic urate score was constructed on the basis of the number of risk alleles across loci from the main analysis as described previously26 and scaled to a risk allele count range as outlined (Supplementary Note). The association between urate scores and gout was evaluated in studies with a large number of cases (prevalent gout: ARIC, SHIP and KORA F4; incident gout: NHS and HPFS). Risk scores ranged from 10 to 45 (mean of 31 ± 5). Risk scores were significantly associated with increased odds of prevalent gout (odds ratio (OR) = 1.11 per risk score unit increase, 95% confidence interval (CI) = 1.09–1.14; P = 2.5 × 10−29; n = 693 cases) and incident gout over a period of up to 22 years (OR = 1.10, 95% CI = 1.08–1.13; P = 3.7 × 10−21; n = 1,036 cases). Gout prevalence increased from <1% to 18% across risk score categories in the population-based studies, and the increased prevalence of gout with higher genetic urate score categories could be replicated in the independent studies recording incident cases of gout (Supplementary Fig. 6).
Characterization of serum urate concentration–associated loci
To gain insight into how the identified variants might contribute to altered serum urate levels, we evaluated their association with the fractional excretion of uric acid (FEUA, n = 6,799; Supplementary Table 12). FEUA is the proportion of urate filtered by the glomeruli that is eventually excreted in the urine. Physiological values are typically well below 10%. SNPs at 10 replicated loci showed nominal association with FEUA (Table 3); SNPs at the SLC2A9, GCKR and IGF1R loci passed a multiple testing–corrected threshold for 26 replicated SNPs (P < 1.9 × 10−3). In all ten instances, the allele associated with higher serum urate concentrations was associated with lower FEUA.
Table 3.
Nominally significant associations between urate concentration–associated SNPs and FEUA
| SNP | Closest gene | A1 | A2 | Serum urate (mg/dl)
|
FEUA (%)
|
|||
|---|---|---|---|---|---|---|---|---|
| Effect | P value | Effect | s.e. | P value | ||||
| rs1260326 | GCKR | T | C | 0.074 | 1.2 × 10−44 | −0.073 | 0.018 | 3.0 × 10−5 |
| rs12498742 | SLC2A9 | A | G | 0.373 | 0a | −0.184 | 0.022 | 1.2 × 10−16 |
| rs2231142 | ABCG2 | T | G | 0.217 | 1.0 × 10−134 | −0.076 | 0.030 | 9.8 × 10−3 |
| rs675209 | RREB1 | T | C | 0.061 | 1.3 × 10−23 | −0.064 | 0.021 | 2.2 × 10−3 |
| rs2078267 | SLC22A11 | T | C | −0.073 | 9.4 × 10−38 | 0.042 | 0.020 | 3.7 × 10−2 |
| rs478607 | NRXN2 | A | G | −0.047 | 4.4 × 10−11 | 0.046 | 0.023 | 4.6 × 10−2 |
| rs1394125 | UBE2Q2 | A | G | 0.043 | 2.5 × 10−13 | −0.059 | 0.023 | 1.1 × 10−2 |
| rs6598541 | IGF1R | A | G | 0.043 | 4.8 × 10−15 | −0.059 | 0.019 | 1.5 × 10−3 |
| rs7193778 | NFAT5 | T | C | −0.046 | 8.2 × 10−10 | 0.086 | 0.032 | 7.2 × 10−3 |
| rs7224610 | HLF | A | C | −0.042 | 5.4 × 10−17 | 0.045 | 0.017 | 9.3 × 10−3 |
Serum urate concentration estimates are provided for the combined discovery and replication sample. Sample size for the SNPs for FEUA ranged from 6,798 to 6,799. A1, allele 1, effect allele; s.e., standard error.
P <1 × 10−700.
Associations in individuals of non-European ancestry
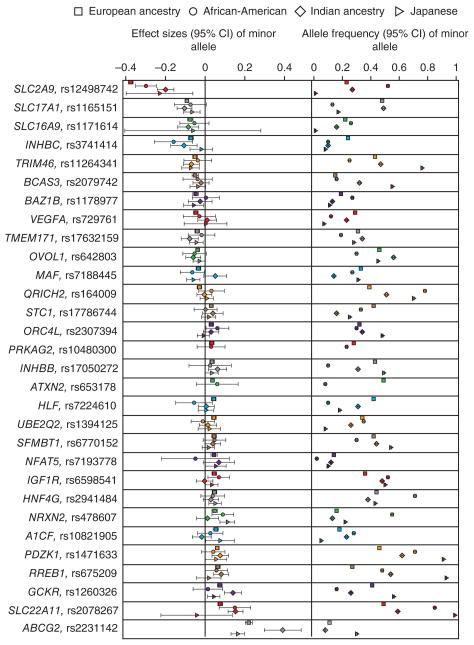
To evaluate whether the observed SNP associations are generalizable to individuals of non-European ancestry, we investigated the replicated and genome-wide significant urate concentration–associated SNPs in 8,340 individuals of Indian ancestry, 5,820 African-Americans and 15,286 Japanese (Supplementary Table 13). Although allele frequencies at the index SNPs varied considerably across the groups (Fig. 3, right), the effects on serum urate concentrations were of comparable magnitude and identical direction for the majority of SNPs (Fig. 3, left), further supporting the idea that these index SNPs represent true urate concentration–associated loci.
Figure 3.
SNP effects on urate concentrations (mg/dl) are similar among individuals of European ancestry, African-Americans, Indians and Japanese, whereas allele frequencies vary. Comparison of SNP effects at the replicated loci and at four additional loci meeting one of the replication criteria. Left, effects on serum urate concentrations per minor allele (defined in individuals of European ancestry) sorted by effect size. Right, comparison of allele frequencies across the four different samples. Symbols are absent if the data could not be provided for reasons related to allele frequency (not polymorphic, low minor allele frequency) or poor imputation quality.
Association with transcript expression in cis
The association of implicated SNPs with transcript expression in cis can potentially provide insights into the gene underlying the association signal. We therefore queried existing expression quantitative trait locus (eQTL) databases containing data from various tissues (Supplementary Note). For ten of the replicated loci, there were significant eSNPs that were associated with the expression of one or more transcript in one or more tissue (Supplementary Table 14). These eSNPs were either the index SNP itself or a variant in high LD with the index SNP (r2 > 0.8); we highlight only instances where the index SNP or a perfect proxy was the best eSNP for a given transcript and tissue. These analyses identify the INHBB gene rather than any of the neighboring genes as being causative at 2q13 because only INHBB transcript levels were significantly associated with the index SNP in the different data sets queried (P value range of 3 × 10–5 to 2 × 10−14).
Association with correlated traits
The 26 identified and replicated urate concentration–associated loci were evaluated for their association with complex traits that are phenotypically correlated with serum urate concentrations (blood pressure, cardiovascular outcomes and glucose homeostasis; Supplementary Table 15). SNPs at the GCKR, BAZ1B and ATXN2 loci showed genome-wide significant associations with one or more of these traits. The direction of association between serum urate and plasma C-reactive protein (CRP) concentrations at GCKR and BAZ1B was consistent with the epidemiologically observed associations, as was the case for the pleiotropic ATXN2 locus and blood pressure.
In aggregate, a weighted risk score composed of the 26 replicated urate concentration–increasing alleles was significantly associated (P < 4.5 × 10−3) only with plasma CRP concentrations after correcting for the number of traits investigated (Supplementary Note). This association was driven by the pleiotropic GCKR locus rather than being a general effect of urate concentration–associated variants on the inflammatory marker CRP; the association between the score and CRP concentrations was abolished in a sensitivity analysis when rs1260326 in GCKR was excluded from the genetic urate score. The absence of significant associations between the genetic urate score and the investigated traits, including blood pressure, might suggest a lack of causal relationships between serum urate concentrations and these traits, given that the present study had adequate statistical power to detect significant associations, as shown previously in a smaller subset18. However, the methodology used has limitations, and alternative explanations cannot be ruled out.
Network analysis identifies additional loci
To identify further urate concentration–associated genomic regions and in order to place these into a biologically interpretable context, we incorporated previous knowledge on molecular interactions in which the gene products of implicated genes operate. First, GRAIL27 was used to infer the most likely gene underlying the association at each of the 37 loci from the discovery analyses of the sex-combined sample (see Supplementary Note for details and Supplementary Table 16 for an overview of GRAIL results). We then constructed a functional association network using the most likely genes as seed genes (Supplementary Note). The complete networks of one-, two- and three-edge neighborhoods are provided as online supplements (see URLs) and highlight genes that are implicated by forming functional associations, mostly protein-protein interactions, with one of the seed genes. To systematically identify urate concentration–associated SNPs in these genes, we selected the SNP with the lowest urate concentration– associated P value in the new candidate genes implicated by the network analyses that passed a multiple testing–corrected threshold (P < 6.8 × 10−5, Bonferroni correction for SNPs in all implicated genes). Replication was attempted for the resulting 17 SNPs in up to 23,078 participants (Supplementary Table 6). Replication criteria, defined as in the discovery analysis, were met for two regions: B3GNT4 and ACVR1B-ACVRL1 (Fig. 1 and Table 2). Moreover, ARNT had a suggestive replication q value of <0.05 but did not reach genome-wide significance. Regional association plots are shown (Supplementary Fig. 7).
A specific subnetwork from the analysis around the inhibins-activins pathway was highlighted as part of the functional association network analysis, in which three seed genes (INHBB, INHBC-INHBE and ACVR2A) were connected with three genes implicated by the functional association network analysis (ACVR1B-ACVRL1, ACVR1C and BMPR2), one of which replicated (Supplementary Fig. 8). The gene products operate in one pathway and are inhibins-activins receptors and their ligands. Detailed information on all replicated genes in the inhibins-activins network is provided in the Supplementary Note. Another pathway linking a large number of GWAS discovery association signals contains genes involved in growth factor signaling through tyrosine kinase receptors implicated in the control of cell growth and differentiation (Supplementary Fig. 9). One of these genes, PKRL, is also highlighted in the Supplementary Note, which provides information about identified associated loci that include candidate genes with a role in glucose metabolism. The functional network associations that formed the basis for Supplementary Figures 8 and 9 are provided in Supplementary Table 17. Pathway analyses using Ingenuity Pathway Analysis software (Supplementary Note) showed functional network associations with gene expression, cellular organization, carbohydrate metabolism, molecular transport and endocrine system disorders (lowest P = 1 × 10−28; Supplementary Table 18).
DISCUSSION
In GUGC, we identified and replicated 28 genome-wide significant urate concentration–associated loci, 18 of which were newly identified, using GWAS (n = 26) and pathway (n = 2) approaches. The loci from our GWAS meta-analysis were characterized in detail, including analyzing their associations with serum urate concentrations in samples of non-European ancestry and with the fractional excretion of urate. The serum urate concentration–increasing allele at all loci was associated with higher risk of gout. The functional association network analysis supports a new role for inhibins-activins pathways in regulating urate homeostasis.
We confirmed the presence of urate concentration–associated SNPs from previous GWAS for all 11 previously reported loci13–18, 6 of which map to genes encoding proteins related to urate transport across membranes. Conversely, none of the genes at the newly associated loci from this study seem to be obvious candidates for factors involved in urate transport. A primary candidate for having an influence on urate generation is PRPSAP1, which encodes a protein involved in the regulation of purine synthesis28. Many of the remaining index SNPs map to genes coding for transcription or growth factors with broad downstream responses. Some of these may ultimately affect the activity of known urate transporters, such as the transcription factor HNF-1α, encoded by a suggestive urate concentration–associated locus that has response elements upstream of ABCG2, SLC17A3, SLC22A11, SLC16A9 and PDZK1.
Many of the newly identified loci can be connected into functional or biochemical pathways. All pathway approaches evaluated in our analyses highlight terms related to carbohydrate metabolism, such as regulation of glycolysis, glucose, insulin and pyruvate (Supplementary Tables 16 and 18). In addition to the previously identified GCKR locus, the gene products of five of the newly discovered replicated loci (Supplementary Note) directly modulate glucose flux. The association of serum urate concentrations with these glucose metabolism–related loci may be explained by increased flow of glucose-6-phosphate through the pentose phosphate pathway and/or high levels of aerobic glycolysis with lactate production as observed in proliferating cells (Warburg effect)29–31. The pentose phosphate pathway generates ribose-5-phosphate, a key precursor of de novo purine synthesis and thereby of uric acid production. The amount of lactate generated can influence urate transport across membranes and can therefore alter urate elimination by the kidney. Detailed gene information and an illustration of how several of these genes are connected in regulatory networks targeting the rate-limiting glycolytic enzymes glucokinase and pyruvate kinase are provided (Supplementary Note). Moreover, insulin and insulin-like growth factor 1, the receptors of which showed suggestive and significant association with urate concentrations, induce glucokinase gene (GCK) expression in the liver and pancreas, respectively32.
In addition to known urate transporters expressed in the renal proximal tubules, three loci linked to glucose metabolism and/or insulin response (GCKR, IGF1R and NFAT5) were nominally associated with FEUA. The urate concentration–increasing alleles at all loci were associated with lower FEUA, which could either be owing to increased urate reabsorption in the kidney by the mechanisms discussed or possibly to the reported effect of insulin on decreasing renal urate clearance and sodium excretion in healthy people33,34.
Despite the connection of these new candidate genes for urate regulation in networks related to glucose metabolism, there was no significant aggregate effect of urate concentration–associated loci on glycemic traits or measures of insulin resistance. A possible explanation might be the absence of a sizeable effect of SNPs in heterogeneous pathways on a single related physiological process or the lack of statistical power of current GWAS efforts to detect small effect sizes.
Another major pathway highlighted by our analyses is the inhibins-activins growth factor system (Supplementary Fig. 8 and Supplementary Note). This pathway is thought to mediate a very wide range of processes, which could affect urate levels through a variety of known functions, such as energy balance, insulin release, apoptosis, inflammation and sex hormone regulation. In addition, our approach demonstrates the value of the functional association network analyses in identifying additional loci from conventional GWAS results.
The mainstay of gout therapy is allopurinol, which inhibits the formation of urate and is an inexpensive and effective preventative agent for chronic gout. Although other therapeutic options exist, the overall number of urate concentration–lowering agents approved for clinical use is limited. Data from a clinical trial indicate that only 21% of patients offered the most common dose of allopurinol (300 mg/d) achieve optimal levels of serum urate35. In addition, there are rare, serious side-effects, and reduced doses of allopurinol are recommended in individuals with impaired renal function. The identification of additional therapeutic options to lower serum urate levels is therefore an active area of research. Our finding that gene loci implicated in glucose homeostasis influence serum urate concentrations fits with the observation that drugs that decrease insulin resistance, such as thiazolidinediones and metformin, tend to decrease serum urate levels36–40. Thus, the identification of new gene loci in the present study may open new avenues for research to improve the treatment and prevention of gout.
The major strengths of our study are the large sample size with >110,000 individuals for the discovery step, the independent replication of associations and the possibility to examine identified loci for their relationship to prevalent and American College of Rheumatology Criteria–confirmed incident gout. The comprehensive evaluation of urate concentration–associated variants provided additional insights by detecting associations with fractional urate excretion and transcript expression.
Limitations of our study include the relatively modest sample size available for the replication step, which compromises power and potentially results in an inability to validate true urate concentration–associated loci such as ORC4L, OVOL1 and BCAS3. The definition of gout varied across several cohorts, which may have led to some misclassification but is unlikely to result in false positive results. As with other GWAS, our analysis focused mostly on common SNPs, and we could therefore not assess the contributions of rare variants, such as a recently described risk variant for gout41.
We identified 28 genome-wide significant loci associated with serum urate concentrations. We found that alleles associated with increased serum urate concentrations were also associated with increased risk of gout. The modulation of urate production and excretion by signaling processes that influence metabolic pathways, such as glycolysis and the pentose phosphate pathway, seem to be central pathways including the genes from the newly identified associated loci. These findings may have implications for further research into urate concentration–lowering drugs to treat and prevent the common inflammatory arthritis gout.
URLs
Complete results from network analyses can be found at http://www.gwas.eu/gugc/. R, http://www.r-project.org/; Quanto, http://hydra.usc.edu/gxe/.
ONLINE METHODS
Participating studies
The overall study comprises data of >70 study samples as detailed in Supplementary Table 1. The meta-analysis of serum urate levels combined genome-wide scans of 48 studies totaling 110,347 individuals, and the meta-analysis of gout comprised 14 studies totaling 2,115 cases and 67,259 controls.
Selected SNPs from the serum urate analysis were followed up in 12 studies with in silico GWAS data totaling 18,821 individuals, as well as 3 studies in which de novo genotyping was conducted (HYPEST, KORA S2 and Ogliastra Genetic Park, totaling 13,992 individuals). All individuals were of European descent.
Additional study samples consisted of two nested case-control studies of incident gout (HPFS and NHS, totaling 1,036 cases and 1,091 controls), as well as 8,340 participants of Indian ancestry (LOLIPOP), 5,820 African-Americans (ARIC, CARDIA and JHS) and 15,288 samples of Japanese ancestry (BioBank Japan).
A list of all studies contributing to the different analyses is provided in the Supplementary Note. Information on study design, study-specific exclusions, urate concentration measurement and gout definition for each study is provided in Supplementary Table 2. The individual studies obtained written informed consent from their study participants, and the studies were approved by their respective local ethics committees.
Power calculations and explained variance
Power calculations were performed a priori, separately for discovery and replication, using Quanto (see URLs). The standard deviation of serum urate concentrations for the calculations was taken from the population-based ARIC Study42, one of the largest contributing studies. For a discovery analysis in 110,000 individuals (urate), there was >80% power to detect a SNP explaining 0.04% or more of the variance in serum urate concentration at P < 5 × 10−8 (corresponding to a ~0.04 mg/dl change in serum urate per allele). Assuming a 2% population prevalence of gout, there was >80% power to detect a risk variant conferring 30% increased odds of gout in 3,000 cases and ~67,000 controls at P < 5 × 10−8 for a SNP with minor allele frequency (MAF) of 10%.
We further used the method by Park et al.43 to estimate that 16 new genome-wide significant urate concentration–associated loci would be detected, assuming a study of 100,000 individuals and effect, frequency and statistical power of previously published loci18 as the reference. The proportion of the explained variance in serum urate concentrations for this study was estimated at 7.7%.
For the replication analyses, 40,546 samples were estimated to be required to independently replicate a SNP with MAF of 10% and effect of 0.06 mg/dl per allele with 80% power (or 15,201 samples for a SNP with MAF of 0.4). For this analysis, we assumed 20 SNPs carried forward to replication, with statistical significance defined as a multiple testing–corrected one-sided P value.
The proportion of variance in serum urate concentrations explained by all 26 independent replicated SNPs from the overall analysis was calculated as the sum of the mean proportion of variance explained by each SNP separately under an additive model adjusted for sex and age, calculated within 31 cohorts. The proportion of the age- and sex-adjusted variance in serum urate concentration explained by all common (MAF > 0.01) genotyped SNPs that met quality control criteria was calculated in three large population-based studies (ARIC, n = 9,049; CoLaus, n = 5,409; SHIP, n = 4,067) using the REML method in GCTA software44.
Statistical analyses in the individual studies
In each study of the discovery GWAS, genotyping was performed on genome-wide chips, and imputation was conducted using HapMap 2 data as the reference. Quality control before imputation was applied in each study separately. Detailed genotyping and imputation information for each study is provided (Supplementary Table 3).
In the individual-study GWAS, outcomes were related to the SNP dosages using linear (urate concentration, mg/dl) or logistic (gout) regression adjusted for age and sex as well as study-specific covariates, if applicable (for example, principal components and study center). Secondary sex-stratified analyses were also conducted. An additive genetic model was used.
Quality control
Each file of genome-wide per-SNP summary statistics underwent extensive quality control before meta-analysis. Examination of file formatting, data plausibility and distributions of test statistics and quality measurements was facilitated by the gwasqc() function of the GWAtoolbox package v1.0.0 in R45. Additionally, the direction and magnitude of effect at the known urate concentration–associated SNP rs16890979 in SLC2A9 was investigated, and the minor allele was consistent in frequency and associated with lower serum urate concentrations in all studies.
Before all meta-analyses, monomorphic SNPs were excluded, and all study-specific results were corrected by the genomic inflation factor of the study if it was >1, calculated by dividing the median of the observed χ-square distribution of the GWAS by the median of the expected χ-square distribution under the null hypothesis of no association. Study-specific inflation factors for urate analyses are shown in Supplementary Table 3. For meta-analyses of gout, only cohorts with >50 gout cases were included, and SNPs with an effect size of |β| > 1,000 were excluded to remove only a minimum number of SNPs with implausibly large effects that could systematically influence the results.
Meta-analyses
All meta-analyses were carried out in duplicate by two independent analysts. Meta-analysis was performed on the results of all genome-wide scans using a fixed-effects model applying inverse variance weighting as implemented in METAL46. Results were confirmed by comparing the results to those from a z score–based meta-analysis. SNPs that were present in <75% of all samples contributing to the respective meta-analysis were excluded, and the remaining SNPs were used as the basis for all subsequent analyses.
Consequently, data for 2,450,547 genotyped or imputed autosomal SNPs were available for the primary analysis of serum urate concentrations, and data for 2,538,056 SNPs were available for the primary gout analysis. After all meta-analyses, a second genomic control correction was applied to primary analyses (genomic inflation factor = 1.12 for urate and 1.03 for gout) as well as secondary sex-stratified analyses (genomic inflation factor for urate = 1.07 (men), 1.08 (women) and gout = 1.04 (men), 1.03 (women)). The top three SNPs from the gout meta-analysis results among women were removed because they were thought to represent false positive results on the basis of low MAF (0.02), very low P values and an I2 heterogeneity measure of >99%.
Genome-wide significance was defined as a P value of <5 × 10−8 after the second correction for genomic control, corresponding to a Bonferroni correction of 1 million independent tests47. All loci containing SNPs of suggestive significance (P < 1 × 10−6) in the discovery-stage meta-analyses were considered for further analysis and assessed for the presence of independent SNPs within each locus.
Assessment of the presence of independent signals at each locus
Two steps were performed to identify independent SNPs in the same region that associated with serum urate concentration in the overall analyses. First, SNPs with P values of <1 × 10−5 were aggregated on the basis of the LD structure from the HapMap release 28 Utah residents of Northern and Western European ancestry (CEU) data set using PLINK48 (settings r2 > 0.01, 1-Mb distance). Second, to verify the potential independent associations from the first step, a multiple-regression model was calculated in 32 of the studies, adjusting for all lead SNPs at once, and meta-analysis was performed, followed by a comparison of the effect estimate for each SNP with those from the single-SNP association model using a t test (Supplementary Table 5). Independent SNPs were defined as those with (i) a t-test P value of >0.05 and (ii) a difference of the effect estimates between the single- and the multiple-SNP models of ≤20% compared to the single-SNP model. rs1178977 in BAZ1B did not fulfill the criteria for independence (20.7% change in effect estimate). Yet, it was treated as an independent SNP, as the adjacent associated genomic region was located >75 Mb away, suggesting that the change in β estimate might be due to other factors than a nearby SNP in LD. Analyses were performed in R.
Subsequently, 37 independent loci of suggestive significance (P < 1 × 10−6) were carried forward for replication.
Secondary analyses
To further identify genetic variants associated with serum urate concentrations and gout, several additional analyses were carried out. These included X-chromosome analyses, sex-stratified analyses, a gene-based test and urate transporter candidate analyses, as well as secondary analyses to improve the characterization of associated SNPs. To this end, we investigated associations with FEUA, associations in individuals of non-European ancestry, associations with transcript expression, associations with serum metabolite concentrations, risk score analyses and associations with other urate-related phenotypes. Secondary analyses are described in detail in the Supplementary Note.
As a result of the secondary analyses, seven additional SNPs were identified for further replication testing: one from the analysis in men only, five from the analysis in women and one from the urate transporter candidate analysis.
A new part of the secondary analyses was the implementation of functional association networks as described in the Supplementary Note. As detailed there, the approach based on functional associations among the urate genes, mostly protein-protein interactions, led to the identification of 17 additional independent SNPs in newly identified genomic regions that were subjected to replication testing. Altogether, we therefore tested 61 SNPs (37 overall, 6 sex specific, 1 urate transporter candidate and 17 network) for replication in additional study samples.
Replication analyses
Up to 18,821 participants contributed information to the in silico replication analyses of the 61 independent variants tested, and additional de novo genotyping was conducted in up to 13,992 individuals. The cohorts are described in Supplementary Table 1. We excluded SNPs with call rate of <0.9 or imputation quality of <0.3 (MACH) or <0.4 (Impute, BEAGLE). The statistical methods used to estimate the associations were identical to the ones used in the discovery cohorts.
Details about de novo genotyping, including platforms, array design and quality control, are provided in the Supplementary Note.
To assess evidence for replication, test statistics of all in silico and de novo replication cohorts were combined using fixed-effects and inverse variance weighting using METAL46. The false discovery rate was estimated in the independent replication samples as a q value on the basis of the P-value distribution of all SNPs tested for replication using the qvalue package in R49. Replication was defined, for each SNP, as q value < 0.05 in the replication samples alone, indicating that <5% false positives would be expected among these variants. To place an additional filter on these results, only SNPs that were associated with genome-wide significance (P < 5 × 10−8) in the combined analysis were considered to have been successfully replicated; combined estimates were obtained by meta-analysis of the double-corrected test statistics of the discovery GWAS with the replication estimates.
Supplementary Material
Acknowledgments
A detailed list of acknowledgments is provided in the Supplementary Note.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Study design: A. Köttgen, C.G., E.A. and M. Caulfield.
Design and/or management of the individual studies: A.A.H., A. Tenesa, A.F.W., A.L., B.M.P., C.G., D.I.C., D.R., E.G.H., E.O., E. Trabetti, G.C., G.P., H. Campbell, H.-E.W., H. Snieder, I.J.D., J.A., J.C., J.F.W., J.V., L.M.R., M. Ciullo, M. Caulfield, M.F., M. Kubo, M.L., M.V., N.H., N.J.S., N. Kamatani, O.M.W., O.P., O.R., P.B.M., P.D., P.G., P.K., P. Mudgal, P.M.R., P.P.P., P.V., R.M.P., R. Sorice, S.H.W., S.M.F., S.U., T.E., T.L., Toshihiro Tanaka, V.S., W.H.L.K., Y.N., Y.O. and Z.K.
Phenotype collection: A.A.H., A.J.G., A.v.E., B.M.P., E.O., G.C., G.G., G.K.G., G.W., H. Snieder, I.J.D., I.K., I. Persico, J.C., J.F.W., J.V., L. Frogheri, M. Ciullo, M. Caulfield, M.G.D., M.G.P., M.J.B., M. Kähönen, M. Kubo, M.L., M. Pirastu, M.V., N.J.S., O.D., O.M.W., O.P., P. Sharma, P.B.M., P.K., P. Mudgal, P.P.P., P.V., S. Schipf, S.H.W., S.M.F., S.T., S.U., T. Zemunik and V.S.
Genotyping: A.A.H., A. Tenesa, A. Teumer, C. Hayward, D.I.C., E.L., F.E., G.C., G.D., G.W.M., I. Persico, J.F.W., M. Ciullo, M. Caulfield, M.E.K., M.G.D., M.G.P., M. Kubo, M.L., M. Putku, M.W., N.J.S., N. Klopp, O.R., P.B.M., P.D., P.M.R., P.P.P., P.v.d.H., R.J.S., S.M.F., T.E., T.L., T. Zeller and T. Zemunik.
Statistical methods and analysis: A. Tenesa, A. Tin, A. Köttgen, A. Teumer, A. Demirkan, C. Hayward, C. Hundertmark, C.G., C. Schurmann, D.C., D.I.C., D.R., E.A., E.G.H., F.M., F.T., G.A.T., G.K.G., G.L., G.M., G.P., I.M.L., I. Prokopenko, J.H., J.K., L.M.L., L.M.R., L.P., M.A.N., M. Steri, M. Bochud, M.E.K., M.F., M. Kähönen, M. Stumvoll, M. Putku, N.P., O.D., P. Mudgal, P.N., P.v.d.H., R.M.P., R.P.S.M., S.C., S.H.W., S. Sanna, T.E., T.H., T.L., V.V., W.H.L.K., X. Li, Y.O. and Z.K.
Interpretation of results: A. Tenesa, A. Tin, A. Köttgen, A.L.G., A. Teumer, B.M.P., C.G., D.R., E.A., G.W.M., H. Campbell, H. Snieder, J.K., M. Ciullo, M.A.N., M. Bochud, M. Caulfield, O.M.W., P.v.d.H., R.M.P., S.H.W., T.H., T.L., Toshihiro Tanaka, V.V., W.H.L.K., Y.O. and Z.K.
Manuscript review: A.A.H., A.B.S., A. Tenesa, A. Dehghan, A.D.J., A. Tin, A. Grotevendt, A. Goel, A.G.U., A.H., A.I., A. Jula, A. Köttgen, A.L., A.L.G., A. Kraja, A.M., A. Döring, A. Tönjes, A.P., A.R.S., A.S., A. Johansson, A. Teumer, A.V.S., B.B., B.H.R.W., B.M.P., B.O.B., B.R.W., B.W.P., C. Hundertmark, C. Hengstenberg, C. Sala, C.L., C.M., C.M.v.D., C.O., C.M.O., C.P.N., C. Schurmann, C.S.F., D.I.C., D.R., D.R.J., D.S.S., D.T., E.B., E.G.H., E. Theodoratou, F.C., F.E., F.R., F.T., G.A.T., G.C., G.G., G.N., G.W.M., H. Campbell, H. Choi, H. Schmidt, H.L.H., H.O., H. Snieder, H.V., H.W., I.B.B., I.K., I.M.L., I.M.N., I. Prokopenko, I.R., J.A., J.B.W., J.C., J.C.C., J.C.M.W., J.E.M., J.F.M., J.F.P., J.F.W., J.H.S., J.H.Z., J.I.R., J.K., J.S., J.S.K., J.V., K.B., K.L., K.S., K.T.K., L.J.L., L. Ferrucci, L.Y., M. Bruinenberg, M.A.N., M. Bochud, M. Caulfield, M. Ciullo, M.F., M.F.F., M.G.D., M.I., M. Burnier, M. Stumvoll, M. Kähönen, M. Kirin, M.M., M.N., M. Perola, M. Struchalin, M. Schallert, M.W., N.B.-N., N.G.M., N.J.W., N. Kamatani, N.M.P.-H., N.S., O.D., O.P., O.R., P. Sharma, P.F., P.K., P. McArdle, P.P.P., P. Salo, P.S.W., P.V., P.v.d.H., Q.Y., Q.Z., R. Schmidt, R.J.F.L., R.J.S., R.N., R.P.S., R. Sorice, S.B., S.H.W., S.J.L.B., S.K., S.L., S.R., S. Sanna, S.-Y.S., T.B.H., T.D.S., T.H., T.L., Toshiko Tanaka, T. Zemunik, U.G., V.G., V.L., V.V., W.H.L.K., W.H.v.G., W.M., W.Z., X. Liu, Y.N., Y.O. and Z.K.
Analysis group: A. Köttgen, A. Teumer, C.G., C. Hundertmark, C.S.F., D.R., E.A., G.P., J.K., Q.Y., T.H., Toshiko Tanaka, V.V. and W.H.L.K.
Writing group: A. Köttgen, A. Teumer, C.G., C.M.O., C.S.F., E.A., J.K., M. Bochud, M. Caulfield, M. Ciullo and V.V.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.So A, Thorens B. Uric acid transport and disease. J Clin Invest. 2010;120:1791–1799. doi: 10.1172/JCI42344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011;63:3136–3141. doi: 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- 3.Arromdee E, Michet CJ, Crowson CS, O’Fallon WM, Gabriel SE. Epidemiology of gout: is the incidence rising? J Rheumatol. 2002;29:2403–2406. [PubMed] [Google Scholar]
- 4.Wallace KL, Riedel AA, Joseph-Ridge N, Wortmann R. Increasing prevalence of gout and hyperuricemia over 10 years among older adults in a managed care population. J Rheumatol. 2004;31:1582–1587. [PubMed] [Google Scholar]
- 5.Yoo HG, et al. Prevalence of insulin resistance and metabolic syndrome in patients with gouty arthritis. Rheumatol Int. 2011;31:485–491. doi: 10.1007/s00296-009-1304-x. [DOI] [PubMed] [Google Scholar]
- 6.Singh JA. Quality of life and quality of care for patients with gout. Curr Rheumatol Rep. 2009;11:154–160. doi: 10.1007/s11926-009-0022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker MA, et al. Quality of life and disability in patients with treatment-failure gout. J Rheumatol. 2009;36:1041–1048. doi: 10.3899/jrheum.071229. [DOI] [PubMed] [Google Scholar]
- 8.Brook RA, et al. The economic burden of gout on an employed population. Curr Med Res Opin. 2006;22:1381–1389. doi: 10.1185/030079906X112606. [DOI] [PubMed] [Google Scholar]
- 9.Hediger MA, Johnson RJ, Miyazaki H, Endou H. Molecular physiology of urate transport. Physiology (Bethesda) 2005;20:125–133. doi: 10.1152/physiol.00039.2004. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield JB, Martin NG. Inheritance and alcohol as factors influencing plasma uric acid levels. Acta Genet Med Gemellol (Roma) 1983;32:117–126. doi: 10.1017/s0001566000006401. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, et al. Genome-wide search for genes affecting serum uric acid levels: the Framingham Heart Study. Metabolism. 2005;54:1435–1441. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Nath SD, et al. Genome scan for determinants of serum uric acid variability. J Am Soc Nephrol. 2007;18:3156–3163. doi: 10.1681/ASN.2007040426. [DOI] [PubMed] [Google Scholar]
- 13.Li S, et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doring A, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 15.Vitart V, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 16.Dehghan A, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolz M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodward OM, et al. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci USA. 2009;106:10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato M, et al. Renal secretion of uric acid by organic anion transporter 2 (OAT2/SLC22A7) in human. Biol Pharm Bull. 2010;33:498–503. doi: 10.1248/bpb.33.498. [DOI] [PubMed] [Google Scholar]
- 24.Wallace SL, et al. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977;20:895–900. doi: 10.1002/art.1780200320. [DOI] [PubMed] [Google Scholar]
- 25.Choi HK, Atkinson K, Karlson EW, Willett W, Curhan G. Purine-rich foods, dairy and protein intake, and the risk of gout in men. N Engl J Med. 2004;350:1093–1103. doi: 10.1056/NEJMoa035700. [DOI] [PubMed] [Google Scholar]
- 26.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizuka T, et al. The human phosphoribosylpyrophosphate synthetase–associated protein 39 gene (PRPSAP1) is located in the chromosome region 17q24-q25. Genomics. 1996;33:332–334. doi: 10.1006/geno.1996.0207. [DOI] [PubMed] [Google Scholar]
- 29.Luo Z, Saha AK, Xiang X, Ruderman NB. AMPK, the metabolic syndrome and cancer. Trends Pharmacol Sci. 2005;26:69–76. doi: 10.1016/j.tips.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Tong X, Zhao F, Mancuso A, Gruber JJ, Thompson CB. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc Natl Acad Sci USA. 2009;106:21660–21665. doi: 10.1073/pnas.0911316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 32.Iynedjian PB. Molecular physiology of mammalian glucokinase. Cell Mol Life Sci. 2009;66:27–42. doi: 10.1007/s00018-008-8322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinones Galvan A, et al. Effect of insulin on uric acid excretion in humans. Am J Physiol. 1995;268:E1–E5. doi: 10.1152/ajpendo.1995.268.1.E1. [DOI] [PubMed] [Google Scholar]
- 34.Ter Maaten JC, et al. Renal handling of urate and sodium during acute physiological hyperinsulinaemia in healthy subjects. Clin Sci (Lond) 1997;92:51–58. doi: 10.1042/cs0920051. [DOI] [PubMed] [Google Scholar]
- 35.Becker MA, et al. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N Engl J Med. 2005;353:2450–2461. doi: 10.1056/NEJMoa050373. [DOI] [PubMed] [Google Scholar]
- 36.Iwatani M, et al. Troglitazone decreases serum uric acid concentrations in type II diabetic patients and non-diabetics. Diabetologia. 2000;43:814–815. doi: 10.1007/s001250051380. [DOI] [PubMed] [Google Scholar]
- 37.Gokcel A, et al. Evaluation of the safety and efficacy of sibutramine, orlistat and metformin in the treatment of obesity. Diabetes Obes Metab. 2002;4:49–55. doi: 10.1046/j.1463-1326.2002.00181.x. [DOI] [PubMed] [Google Scholar]
- 38.Seber S, Ucak S, Basat O, Altuntas Y. The effect of dual PPARα/γ stimulation with combination of rosiglitazone and fenofibrate on metabolic parameters in type 2 diabetic patients. Diabetes Res Clin Pract. 2006;71:52–58. doi: 10.1016/j.diabres.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Tsouli SG, Liberopoulos EN, Mikhailidis DP, Athyros VG, Elisaf MS. Elevated serum uric acid levels in metabolic syndrome: an active component or an innocent bystander? Metabolism. 2006;55:1293–1301. doi: 10.1016/j.metabol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 40.Rizos CV, Liberopoulos EN, Mikhailidis DP, Elisaf MS. Pleiotropic effects of thiazolidinediones. Expert Opin Pharmacother. 2008;9:1087–1108. doi: 10.1517/14656566.9.7.1087. [DOI] [PubMed] [Google Scholar]
- 41.Sulem P, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43:1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 42.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 43.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuchsberger C, Taliun D, Pramstaller PP, Pattaro C. GWAtoolbox: an R package for fast quality control and handling of genome-wide association studies meta-analysis data. Bioinformatics. 2012;28:444–445. doi: 10.1093/bioinformatics/btr679. [DOI] [PubMed] [Google Scholar]
- 46.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.