Abstract
Background
Hyperpigmentation of the visceral peritoneum (HVP) has recently garnered much attention in the poultry industry because of the possible risk to the health of affected animals and the damage it causes to the appearance of commercial chicken carcasses. However, the heritable characters of HVP remain unclear. The objective of this study was to investigate the genetic parameters of HVP by genome-wide association study (GWAS) in chickens.
Results
HVP was found to be influenced by genetic factors, with a heritability score of 0.33. HVP had positive genetic correlations with growth and carcass traits, such as leg muscle weight (rg = 0.34), but had negative genetic correlations with immune traits, such as the antibody response to Newcastle disease virus (rg = −0.42). The GWAS for HVP using 39,833 single nucleotide polymorphisms indicated the genetic factors associated with HVP displayed an additive effect rather than a dominance effect. In addition, we determined that three genomic regions, involving the 50.5–54.0 Mb region of chicken (Gallus gallus) chromosome 1 (GGA1), the 58.5–60.5 Mb region of GGA1, and the 10.5–12.0 Mb region of GGA20, were strongly associated (P < 6.28 × 10-7) with HVP in chickens. Variants in these regions explained >50% of additive genetic variance for HVP. This study also confirmed that expression of BMP7, which codes for a bone morphogenetic protein and is located in one of the candidate regions, was significantly higher in the visceral peritoneum of Huiyang Beard chickens with HVP than in that of chickens without pigmentation (P < 0.05).
Conclusions
HVP is a quantitative trait with moderate heritability. Genomic variants resulting in HVP were identified on GGA1 and GGA20, and expression of the BMP7 gene appears to be upregulated in HVP-affected chickens. Findings from this study should be used as a basis for further functional validation of candidate genes involved in HVP.
Background
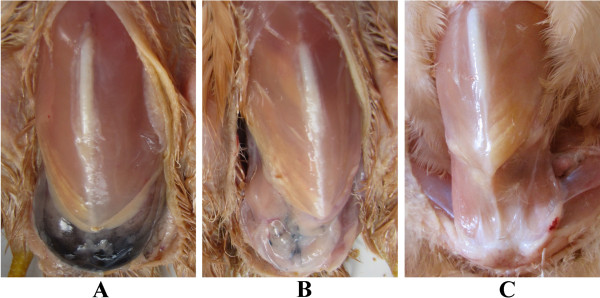
Pigmentation is widespread amongst both plants and animals, and plays important roles in photosynthesis (in plants), camouflage, sex selection, and protection from sunburn. However, abnormal pigmentation in humans and other animals, including hyperpigmentation (e.g. chloasma [1] and melanoma [2,3]) and the absence of pigmentation (e.g. albinism [4] and vitiligo [5,6]), can pose serious health risks. Most pigmentation phenotype variants are affected by genetic factors in both humans [7-9] and animals [10-17]. In chickens, a mutation of the melanocortin 1 receptor (MC1R) gene causes extended dark feathers [15], and a complex genomic rearrangement on chicken (Gallus gallus) chromosome 20 (GGA20) determines dermal hyperpigmentation [14]. However, these studies have mainly focused on pigmentation of the retina, skin, hair, and feathers. Pigmentation of other tissues, including muscular and visceral membranes, is also very important. Recently, more attention has been paid to the hyperpigmentation of the visceral peritoneum (HVP) in chickens, especially the colored chicken breeds, because it affects the carcass appearance of commercial chickens, resulting in economic losses, and may be associated with certain diseases, including melanomas [18]. HVP is similar to fibromelanosis, but the pigmentation is limited to the chicken peritoneum, so it may be peritoneal fibrosis. It is characterized by intense pigmentation of connective tissue in the visceral peritoneum, which results in a dark blue appearance through the skin of the chicken abdomen, and a black connective tissue layer when the skin is removed (Figure 1). A preliminary study showed that HVP was caused by an abnormal distribution of melanin, and that the number of chickens with HVP can increase in cold and humid environments (unpublished data). HVP is distinct from fibromelanosis, which results from a complex genomic rearrangement involving the endothelin 3 (EDN3) locus [14]. In fact, the genetic causes of HVP remain unknown.
Figure 1.
Classification of the hyperpigmentation of visceral peritoneum. A, B, and C represent severe, mild, and absent hyperpigmentation of the visceral peritoneum, respectively.
The development of molecular tools and strategies has allowed the investigation of the genetic basis of HVP. Genome-wide association studies (GWAS) have become an important strategy for investigating the genetic basis of many human diseases, including diabetes, breast cancer, pancreatic cancer, and hypertension, amongst others (http://www.genome.gov/GWAStudies). Livestock breeders have begun to implement GWAS to map economically important quantitative trait loci (QTLs) [19-22]. Significant loci linked to chicken growth traits have been mapped to GGA1 and GGA4 by GWAS [23,24]. Therefore, if HVP is influenced by a major genetic factor, GWAS may be able to dissect its genetic basis.
In this study, we estimated the genetic parameters of HVP to illustrate the inheritance of HVP, and carried out a GWAS analysis of HVP using the chicken 60K single nucleotide polymorphism (SNP) panel in a commercial chicken population with a rich diversity of HVP.
Results
Genetic parameters
HVP was found to have a moderate heritability (h2 = 0.33) through estimating genetic parameters, suggesting that HVP was significantly affected by genetic factors and was not a simple Mendelian trait. As shown in Table 1, HVP had significantly positive genetic correlations with growth and carcass traits in chickens (P < 0.05), such as body weight at day 91 (rg = 0.27), carcass weight (rg = 0.24), net weight (rg = 0.27), breast muscle weight (rg = 0.17), and leg muscle weight (rg = 0.34). Moderately negative genetic correlations were identified between HVP and immune traits, especially the antibody response to Newcastle disease virus (rg = −0.42). These results indicated that the immune capacity of chickens with HVP could be inferior to that of normal non-pigmented chickens, but the growth capacity of chickens with HVP might be greater.
Table 1.
Genetic correlations (rg) between the hyperpigmentation of visceral peritoneum and growth, carcass, and immune traits in chickens
| Traitsa | No.b | Meansc | rg | P-value |
|---|---|---|---|---|
|
BW91, g |
511 |
2,100 ± 16.5 |
0.27 ± 0.14 |
2.43 × 10-2 |
|
CW, g |
511 |
1,847 ± 14.5 |
0.24 ± 0.14 |
4.83 × 10-2 |
|
NW, g |
511 |
1,401 ± 11.7 |
0.27 ± 0.13 |
1.86 × 10-2 |
|
DW, g |
506 |
1,674 ± 13.7 |
0.21 ± 0.16 |
9.47 × 10-2 |
|
BMW, g |
511 |
122 ± 1.09 |
0.17 ± 0.10 |
3.84 × 10-2 |
|
LMW, g |
511 |
168 ± 1.95 |
0.34 ± 0.10 |
4.50 × 10-4 |
|
AFW, g |
510 |
82.6 ± 1.58 |
0.07 ± 0.12 |
2.72 × 10-1 |
|
SIL, cm |
510 |
129 ± 0.59 |
−0.06 ± 0.10 |
2.66 × 10-1 |
|
Ab-NDV |
511 |
3.63 ± 0.07 |
−0.42 ± 0.14 |
1.35 × 10-3 |
|
Ab-AIV |
511 |
1.31 ± 0.05 |
−0.26 ± 0.16 |
5.21 × 10-2 |
| HC | 508 | 13.4 ± 0.38 | −0.25 ± 0.18 | 8.24 × 10-2 |
a BW91 = body weight at day 91, CW = carcass weight, NW = net weight, DW = dress weight, BMW = breast muscle weight, LMW = leg muscle weight, AFW = abdomen fat weight, SIL = small intestine length, Ab-NDV = antibody response to Newcastle disease virus, Ab-AIV = antibody response to avian influenza virus, HC = heterophil count, representing heterophil to lymphocyte ratio.
b Number of samples for estimating genetic correlations.
c Mean ± standard error.
GWAS detection of SNPs associated with HVP
A GWAS was used to dissect the genetic factors associated with HVP. SNP additive effect analysis identified several regions that were significantly (P < 6.28 × 10-7) associated with HVP on both GGA1 and GGA20 (Figure 2A). As shown in Table 2, 20 SNPs with additive effects with genome-wide significance were detected for HVP (P < 6.28 × 10-7). Thirteen SNPs with additive effects were located in the 50.5–54.0 Mb region of GGA1. In addition to this GGA1 region, seven SNPs with functional effects reached genome-wide significance, including two SNPs in the 58.5–60.5 Mb region of GGA1, and five SNPs in the 10.5–12.0 Mb region of GGA20. The extent of linkage disequilibrium (LD) in both GGA1 and GGA20 was about 2 Mb (Additional file 1: Figure S1), which indicated that the three significant regions represented three independent QTLs for HVP. The risk alleles of the QTLs in the 50.5–54.0 Mb region of GGA1 and in the 10.5–12.0 Mb region of GGA20 were from the parent broiler sire line and Huiyang Beard chicken line, respectively. The additive effect in the 50.5–54.0 Mb region of GGA1 was the greatest out of all the regions identified for HVP. The most significantly associated SNP (rs14822943) explained 13% of phenotypic variance for HVP. Because the heritability of HVP reached 0.33, the additive effect of this SNP covered 39% of additive genetic variance for HVP. Together with the most significant SNP in each strongly significant chromosomal region, the three most significant SNPs accounted for >50% of the additive genetic variance for HVP. Unlike the additive effect, no SNP dominance effect reached genome-wide significance (P < 6.28 × 10-7) for HVP (Figure 2B). This result indicated that the genetic factors affecting HVP had much stronger additive effects than dominance effects, and that at least three major genes could influence HVP.
Figure 2.
Manhattan plot of the genome-wide association study for the hyperpigmentation of visceral peritoneum (HVP) in chickens. The green line indicates the threshold P value of the 5% Bonferroni genome-wide significance (P = 6.28 × 10-7). A. Additive effects of GWAS for HVP. B. Dominance effects of GWAS for HVP.
Table 2.
SNPs with statistical significance in the genome-wide association study for hyperpigmentation of visceral peritoneum (HVP)
| SNP | GGAa | Position (bp) | Nearest geneb | Alleles | RAc | FAW | P-value | AE | r2 |
|---|---|---|---|---|---|---|---|---|---|
|
rs14822943 |
1 |
52,576,468 |
0.5 Kb U MAP3K7IP1 |
C/T |
T |
0.21 |
2.65 × 10-14 |
0.05 |
0.13 |
|
GGaluGA017356 |
1 |
52,358,970 |
CACNA1I |
T/C |
T |
0.24 |
2.95 × 10-12 |
0.06 |
0.10 |
|
GGaluGA017598 |
1 |
52,940,122 |
PLA2G6 |
T/C |
C |
0.43 |
9.03 × 10-12 |
0.08 |
0.09 |
|
GGaluGA018011 |
1 |
53,833,467 |
70.8 Kb U MYH9 |
G/A |
G |
0.24 |
1.03 × 10-11 |
0.05 |
0.10 |
|
rs13865536 |
1 |
52,554,283 |
13.3 Kb D MAP3K7IP1 |
C/T |
C |
0.24 |
5.07 × 10-11 |
0.05 |
0.10 |
|
GGaluGA017030 |
1 |
51,619,588 |
0.8 Kb U RANGAP1 |
G/A |
A |
0.49 |
5.80 × 10-11 |
0.08 |
0.08 |
|
rs13652125 |
1 |
50,923,554 |
TTC26 |
A/G |
A |
0.15 |
1.97 × 10-10 |
0.04 |
0.06 |
|
GGaluGA016761 |
1 |
51,087,457 |
SERHL2 |
T/C |
C |
0.48 |
4.62 × 10-9 |
0.07 |
0.06 |
|
GGaluGA016965 |
1 |
51,488,752 |
2.5 Kb D ACO2 |
T/C |
C |
0.25 |
5.75 × 10-9 |
0.05 |
0.07 |
|
GGaluGA017484 |
1 |
52,642,092 |
PDGFB |
G/A |
G |
0.26 |
1.52 × 10-8 |
0.05 |
0.09 |
|
GGaluGA017730 |
1 |
53,094,788 |
5.6 Kb U TRIOBP |
T/C |
T |
0.64 |
4.74 × 10-8 |
0.11 |
0.09 |
|
GGaluGA019336 |
1 |
58,058,352 |
55.3 Kb U HIPK2 |
G/A |
G |
0.70 |
1.55 × 10-7 |
0.10 |
0.04 |
|
rs13873173 |
1 |
60,462,801 |
SLC13A4 |
G/T |
T |
0.75 |
2.37 × 10-7 |
0.10 |
0.05 |
|
rs13865344 |
1 |
52,399,455 |
CACNA1I |
T/C |
T |
0.27 |
4.85 × 10-7 |
0.04 |
0.03 |
|
rs13865892 |
1 |
53,177,756 |
LGALS2 |
G/A |
G |
0.61 |
5.79 × 10-7 |
0.08 |
0.06 |
|
rs16174305 |
20 |
11,707,312 |
30.0 Kb D BMP7 |
C/T |
C |
0.42 |
2.44 × 10-8 |
0.06 |
0.05 |
|
GGaluGA180727 |
20 |
11,075,280 |
36.9 Kb U STX16 |
G/A |
A |
0.36 |
9.25 × 10-8 |
0.06 |
0.05 |
|
rs14278900 |
20 |
10,911,015 |
GNAS |
G/A |
A |
0.37 |
1.24 × 10-7 |
0.05 |
0.05 |
|
GGaluGA180980 |
20 |
11,591,655 |
3.5 Kb U RBM38 |
G/A |
A |
0.44 |
2.59 × 10-7 |
0.06 |
0.03 |
| GGaluGA181122 | 20 | 11,874,967 | 27.0 Kb D TFAP2C | T/C | C | 0.48 | 3.98 × 10-7 | 0.06 | 0.05 |
a GGA = chicken (Gallus gallus) chromosome;
b Gene information from NCBI, 06 July 2011;
c RA = risk allele related to HVP; FAW = frequency of the allele related to HVP in F2 population; AE = additive effect of SNP markers; r2 = contribution of each SNP to HVP.
Positional candidate genes for HVP
As shown in the Table 2, there were 18 genes in close proximity to the 20 significant genome-wide SNP markers. The most significant effect was observed in the promoter region (about 0.5 Kb upstream) of TGF-β activated kinase 1/MAP3K7 binding protein 1 (MAP3K7IP1), located in the 50.5–54.0 Mb region of GGA1. Another SNP, located 13.3 Kb downstream of MAP3K7IP1, also had a genome-wide significant additive effect for HVP. Because HVP was associated with immune traits (Table 1), and because MAP3K7IP1 is involved in some pathways associated with energy metabolism and immunity, such as the MAPK signaling pathway and the Toll-like receptor signaling pathway, MAP3K7IP1 was identified as a potential positional candidate gene for HVP. The most significant effect of the other QTLs linked to HVP on GGA1 was at 55.3 kb upstream of the homeodomain interacting protein kinase 2 (HIPK2) locus. HIPK2 participates in cell development, growth, and apoptosis, such as in the Wnt pathway, and was associated with HVP and chicken growth (Table 1), so was therefore also identified as a candidate gene for HVP. The most significant SNP on GGA20 was located nearest to bone morphogenetic protein 7 (BMP7), which can effect melanocyte growth and melanoma cell metastasis, and therefore BMP7 was also chosen as one of the most important positional candidate genes for HVP. In addition, according to information from the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Map Annotator and Pathway Profiler (GenMAPP), and Biocarta databases, MAP3K7IP1, HIPK2, and BMP7 can all regulate mitogen-activated protein kinase kinase kinase 7 (MAP3K7) expression (Additional file 2: Figure S2), suggesting that each of these genes might affect HVP by mediating MAP3K7 expression.
Expression of chicken MAP3K7IP1, HIPK2, BMP7, and MAP3K7
As shown in Figure 3, MAP3K7IP1, HIPK2, BMP7, and MAP3K7 were all expressed in the visceral peritoneum tissue of normal Huiyang Beard chickens as well as in chicken with HVP. BMP7 mRNA expression was the lowest, only slightly greater than zero in the normal Huiyang Beard chickens, while MAP3K7 mRNA expression was the highest. The mRNA expression of both BMP7 and MAP3K7 in the visceral peritoneum tissue of Huiyang Beard chickens with HVP was significantly higher than that in normal Huiyang Beard chickens (P < 0.05). However, the mRNA expression of MAP3K7IP1 and HIPK2 was not significantly different between the visceral peritoneum tissue of normal Huiyang Beard chickens and those with HVP (P > 0.05).
Figure 3.
Expression of chicken MAP3K7IP1, HIPK2, BMP7, and MAP3K7. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively, in gene expression between the visceral peritoneum of the normal Huiyang Beard chickens and Huiyang Beard chickens with hyperpigmentation of visceral peritoneum.
Discussion
Interestingly, chickens with HVP showed no reduction in body weight, and were actually associated with improved production efficiency. This finding deviated from our hypothesis that HVP was likely associated with disease phenotypes, which could decrease chicken growth. However, HVP-affected birds still had greater health risks because of the observed negative genetic correlations with immune traits (Table 1). The birds with HVP had decreased antibody responses, indicating that they may be more likely to suffer from pathogen infections. In addition, the birds with HVP had larger heterophil counts, resulting in higher heterophil to lymphocyte ratios (H/L). High H/L values in chickens are associated with decreased tolerance of environmental stress [25]. Taken together, our results indicated that birds with HVP should grow faster in a favorable environment, but stressful environmental conditions would more adversely affect the development and growth of birds with HVP compared with those of normal birds. We therefore inferred that the increased emergence of birds with HVP indirectly results from the selection of birds with higher production efficiency in the modern broiler industry.
During our investigation of HVP in chickens, we did not observe this HVP phenomenon in fast-growing birds with white feathers, such as Ross 308 broilers; HVP appeared to be limited to colored chicken breeds, such as Huiyang Beard chickens (unpublished data). The lack of the HVP phenotype in Ross 308 broilers may result from interference of the white dominance or recessive locus with the pigmentation in the abdominal septa of areolar connective tissues. A white recessive locus with a retroviral insertion in the tyrosinase (TYR) gene changes the expression of TYR to interrupt melanin biosynthesis [11,26,27], while a white dominance locus with a mutation in the premelanosome protein gene alters melanosome shape to influence pigmentation [10,28]. In addition, birds with runting and stunting syndrome (RSS) usually also have the HVP phenotype in Chinese farms (personal communication). One of reasons behind RSS may be the fact that birds with HVP have less resistance to pathogens and environmental stress, such as cold temperatures. Therefore, HVP may be one of the traits that mirror non-balance allocation of energy between production and immunity during chicken growth. However, this hypothesis required further experimental validation.
A GWAS was implemented for HVP in this study to attempt to validate the above hypotheses. GWAS are useful for exploring the genetic basis of some special appearance traits, such as pigmentation [29,30]. This was the first study aimed at uncovering the genetic basis of pigmentation of connective tissues in chickens based on a high-density SNP chip panel. We hoped that the findings would increase the genetic knowledge of HVP, and allow us to validate potential HVP candidate genes.
The GWAS identified 20 SNP markers that were significantly (P < 6.28 × 10-7) associated with HVP (Table 2). Based on the extent of LD on GGA1 (Additional file 1: Figure S1), these SNP markers were determined to belong to three different QTLs. The detection of more than one QTL indicates that the causal genes or mutations in these QTLs can affect the same pathway or gene to generate the same phenotype. The MAP3K7IP1, HIPK2, and BMP7 genes were the closest loci to the most significant SNP marker in each of the three QTL regions. MAP3K7IP1, HIPK2, and BMP7 genes are not traditional pigmentation genes, which are generally considered to include MC1R, TYR, tyrosinase-related protein 1, microphthalmia-associated transcription factor (MITF), agouti signaling protein, SRY (sex determining region Y)-box 10 (SOX10), myosin VA (heavy chain 12, myoxin), solute carrier family 45, and member 2 [8,31,32]. Therefore, we hypothesized that HVP does not directly result from mutations in the traditional pigmentation genes, but originates from the upstream genes that can indirectly change pigmentation pathways. GWAS of human pigmentation traits have produced some similar results [33]. Previous studies also suggested that MAP3K7IP1, HIPK2, and BMP7 could influence some pigmentation pathways. For example, Liang et al. found that down-regulation of HIPK2 expression suppressed the expression of MITF, resulting in melanocyte differentiation suppression by increasing C-terminal binding protein 2 levels [34]. BMP7 could inhibit normal melanocyte growth and tumor growth of human uveal melanomas [35,36], and could inhibit metastasis by inducing mesenchymal-to-epithelial transition in melanoma cells [37]. However, evidence indicates that BMP7 is upregulated in the development of melanoma [38,39]. This study also found that upregulation of BMP7 was associated with HVP in Huiyang Beard chickens (Figure 3). In addition, BMP7 could affect pheomelanin generation by interacting with proopiomelanocortin in brown adipocyte differentiation and thermogenesis [32,40].
More interestingly, the MAP3K7 gene was found to be a node linking the MAP3K7IP1, HIPK2, and BMP7 genes according to the pathway maps involving these genes in the KEGG, GenMAPP, and BioCarta databases (Additional file 2: Figure S2). MAP3K7 can also interact with many genes affecting melanocyte development [31,32], such as MITF [41], KIT ligand, B-cell leukemia/lymphoma 2 [42,43], lymphoid enhancer binding factor 1 [44], and epidermal growth factor receptor [45,46]. MAP3K7IP1 is one of the MAP3K7 binding proteins. The MAP3K7IP1 protein interacts with and thus activates MAP3K7 kinase, and may also function as a mediator between TGF-β receptors and MAP3K7 [47-49], suggesting that MAP3K7IP1 can influence the function of the downstream genes of the pathways involving in MAP3K7. Besides the interaction of MAP3K7IP1 and MAP3K7, BMP7 also contacts MAP3K7 in some pathways. Yamaguchi et al. reported that BMP7 activated MAPK signaling through MAP3K7 [50]. Blank et al. verified that BMP7 activated the JNK signaling pathway, and MAP3K7 was required for BMP7-mediated JNK activation [51]. In addition, BMP7 could activate MAP3K7 and enhance Wnt-dependent transcription [52]. Expression of BMP7 was indeed consistent with that of MAP3K7 in this study (Figure 3). It is possible that a specific mutation upregulates expression of BMP7 to result in MAP3K7 upregulation in Huiyang Beard chickens with HVP. Additionally, the MAP3K7-HIPK2 pathway can inhibit c-Myb activity upon Wnt-1 stimulation, affecting the immune response, because c-Myb plays an essential role in the proliferation of immature hematopoietic cells and early T-cell development [53-55]. This is consistent with the fact that this study identified a strong genetic relationship between HVP and the antibody response to Newcastle disease virus (rg = −0.42, Table 1). Importantly, MAP3K7 participates in several pathways related to the immune response, such as the B cell receptor signaling pathway, the toll-like receptor signaling pathway and the IL-6 signaling pathway (http://www.wikipathways.org/index.php/WikiPathways). These findings indicate that MAP3K7IP1, HIPK2, and BMP7 could be candidate genes for HVP, and might affect the development of HVP by regulating the expression of the MAP3K7 gene. Further studies are needed to validate this hypothesis.
Conclusions
HVP was found to be a quantitative trait with moderate heritability. Three independent QTLs for HVP were detected by GWAS on GGA1 and GGA20, and the BMP7 gene was identified as a likely candidate gene for HVP.
Methods
Ethics statement
This study was approved by the Animal Care Committee of the Institute of Animal Science, Guangdong Academy of Agricultural Sciences (Guangzhou, People’s Republic of China), with approval number GAAS-IAS-2009-73. Animals involved in this study were humanely sacrificed as necessary to ameliorate their suffering.
Animals and data collection
A total of 585 commercial chickens were used in this study, consisting of three generations (23 P, 51 F1, and 511 F2 individuals) with an accurate pedigree. All birds were immunized with a commercial avian influenza-inactivated H9 strain vaccine at day 40, and a commercial Newcastle disease virus live LaSota strain vaccine at day 50. At day 91, 511 F2 individuals from six hatches were slaughtered. At this time point, vein blood was collected and a portion transferred into centrifuge tubes containing ethylenediaminetetraacetic acid disodium salt solution, and then stored at −80°C. The remainder was used to prepare serum for measuring antibody responses (S/P values) to Newcastle disease virus and avian influenza virus by enzyme linked immunosorbent assay. At day 91, body weight, carcass weight, net weight, dress weight, breast muscle weight, leg muscle weight, and abdomen fat weight were measured, as was small intestine length. Heterophil count, representing H/L, was measured following the method of Vleck et al. [56]. A higher heterophil count is consistent with a higher H/L value. Because HVP was thought to be a quantitative trait, HVP was classified into three levels, absent, mild, and severe hyperpigmentation, represented by 0, 1, and 2 (Figure 1), respectively, to control for false positives. The absent, mild, and severe hyperpigmentation groups had 352, 132, and 27 individuals, respectively.
SNP genotyping and selection
Genomic DNA extraction from venous blood was performed using the phenol/chloroform method. The quality and concentration of genomic DNA from 511 F2 individuals fulfilled the requirements for the Illumina Infinium SNP genotyping platform. Genotyping using the Illumina 60K Chicken SNP Beadchip [57] was carried out at the Illumina-certified service provider, DNA LandMarks, Saint-Jean-sur-Richelieu, Canada. Quality control was assessed in GenomeStudio v2008.1 [58]. Six samples were excluded as more than 5% of their SNPs had missing genotypes. The final SNP set included 39,833 SNPs for this GWAS under the following SNP selection criteria: low call frequency (>95%), low heterozygosity cluster intensity and separation value (>0.4), and low minor allele frequency (>0.1). Information on the SNP markers on each chicken chromosome is summarized in Additional file 3: Table S1.
Gene expression
The visceral peritoneum tissue from eight normal and eight HVP-positive, 21-day-old Huiyang Beard chickens (a Chinese native chicken breed from Guangdong province) was collected and transferred into RNAlater solution (Life Technologies, Carlsbad, CA, USA) and stored at −80°C. Total RNA was isolated by grinding the tissues to powder under liquid nitrogen and extracting with RNA TRIzol reagent (Life Technologies, Rockville, MD, USA). The RNA was reverse transcribed into cDNA with an M-MLV RTase cDNA Synthesis Kit (Takara Biotechnology Co., Dalian, China). Quantitative real-time polymerase chain reaction (qRT–PCR) analysis was performed to test the expression of MAP3K7IP1, HIPK2, BMP7, and MAP3K7 in the visceral peritoneum tissue of the birds. The primers used for qRT-PCR were designed using Primer Express 2.0 software (Applied Biosystems, Foster City, USA) and their sequences are shown in Table 3. qRT-PCR reactions used SYBR Green Real Time PCR Master Mix (Toyobo Co., Osaka, Japan) according to the manufacturer’s instructions, and contained a passive reference dye, Rox, to correct for well-to-well variation. Reactions were run on a LightCycler 480 Real-Time PCR System (Roche Applied Science, Indianapolis, IN, USA) with the following parameters: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 34 s at 72°C. The relative mRNA expression of the target genes was measured as the number of cycles of PCR required for exceeding threshold fluorescence, and was normalized against that of β-actin, according to the quantitation procedures recommended by Roche Applied Science.
Table 3.
Sequences of primers used for qRT-PCR
| Genes | Forward primers (5′–3′) | Reverse primers (5′–3′) |
|---|---|---|
|
BMP7 |
GAGAACAGCAGCAGCGACC |
CAAAATAGAGCACTGAGATGGC |
|
MAP3K7IP1 |
CCCCACCCTCACTAACCAA |
TCCCTCCTCAGTCTTTTCTCAC |
|
HIPK2 |
CATCCTCGGTTTACCATTTTG |
CGGTGAGTCTGTATCCCTGTT |
|
MAP3K7 |
CCAGGAAACGGACAGCAG |
CTTTGGAGTTCGGGCATG |
| β-actin | CCCCAAAGCCAACAGAGAGA | GGTGGTGAAGCTGTAGCCTCTC |
Statistical analysis
Variance and covariance components were estimated using the average information restricted maximum likelihood algorithm [59] implemented by the DMU package [60]. The variance component estimated model was:
where y was the vector of observations of HVP, e.g. body weight at day 91(a total of nine phenotypes, Table 1); b was the vector of fixed effects, including sex (two levels) and hatch (six levels); a was the vector of animal additive genetic effects; e was the vector of random residuals; and X and Z were corresponding incidence matrices.
Statistical tests of SNP-phenotype association were implemented using the generalized least square version of the epiSNP computer package, which considered sib correlation within each family [61,62]. The statistic model was
where Y was the phenotypic value of HVP, μ was the common mean of HVP, S was the fixed gender effect, H was the fixed hatch effect, f was the random family effect, SNP was the single-locus SNP genotypic effect, and e was the random residual. Additive and dominance effects were tested using linear contrasts of the single-locus SNP genotypic effect [62]. The threshold P value of the 5% Bonferroni genome-wide significance was 6.28 × 10-7 (0.05/39833/2), based on the total number of SNP markers and two SNP genotypic effects (additive and dominance effects) in GWAS. Manhattan plots were produced using SNPEVG version 2.1 [63] to demonstrate the overview of SNP effects.
To evaluate the extent of LD and identify potential regions of causal mutation for HVP, pairwise LD, measured by r2 values for the F2 population, was calculated for GGA1 and GGA20 using Haploview [64]. Pathway analysis was performed using KEGG (http://www.genome.jp/kegg/), GenMAPP (http://genmapp.org/), and BioCarta (http://genmapp.org/) databases.
Differential expression of MAP3K7IP1, HIPK2, BMP7, and MAP3K7 in the visceral peritoneum tissue, between the normal birds and the birds with HVP, was determined using a t-test with SAS 8.0 software (SAS Institute, Cary, NC, USA).
Competing interests
The authors have no competing interests to declare.
Authors’ contributions
CL, HQ, XH, NL, and DS conceived and designed the experiments. CL, HQ, JW, YW, JM, CYL, and DS performed the experiments. CL and HQ analyzed the data. CL, JM, CY, and DS contributed the materials. CL, HQ, and DS wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Pattern of linkage disequilibrium (LD) on chicken (Gallus gallus) chromosomes. A. LD on Chromosome 1. B. LD on Chromosome 20.
Interaction of genes. Blue, green, and red figures indicate the number of pathways that the genes are involved in, based on KEGG, GenMAPP, and BioCarta, respectively. BMP7 interacts with MAP3K7 via the ALK pathway in cardiac myocytes according to the BioCarta database. HIPK2 interacts with MAP3K7 via an enzyme linked receptor protein signaling pathway and Wnt netPath 8 according to the GenMAPP database. MAP3K7IP1 interacts with MAP3K7 via the MAPK and Toll-like receptor signaling pathways according to the KEGG database, by the NF-κB signaling pathway, signal transduction through IL1R, the TGF-β signaling pathway, and the WNT signaling pathway according to the Biocarta database, and by receptor signaling protein activity, the MAPK signaling pathway, and TGF-β-receptor netPath 7 according to the GenMAPP database.
Basic information on the 39,833 SNP markers on the chicken physical map.
Contributor Information
Chenglong Luo, Email: Chenglongluo1981@163.com.
Hao Qu, Email: qhw03@163.com.
Jie Wang, Email: wangjie030@126.com.
Yan Wang, Email: wynew2004@yahoo.com.cn.
Jie Ma, Email: zzitmj@126.com.
Chunyu Li, Email: lichy523@163.com.
Chunfen Yang, Email: yangchunfen06@126.com.
Xiaoxiang Hu, Email: huxx@cau.edu.cn.
Ning Li, Email: ninglcau@cau.edu.cn.
Dingming Shu, Email: shudm@263.net.
Acknowledgments
The authors would like to thank Dr Zheya Sheng (College of Biological Science, China Agricultural University, Beijing, China) for her technical assistance in handling the SNP genotyping data and Dr Yang Da (Department of Animal Science, University of Minnesota, Saint Paul, Minnesota, USA) for his technical support with GWAS. This study was supported by grants from the Guangdong Key Scientific and Technological Projects (Grant No. 2011A020201007), the National Natural Science Foundation of China (Grant No. U0831003), and the Earmarked Fund for Modern Agro-industry Technology Research System (nycytx-42).
References
- Bolanca I, Bolanca Z, Kuna K, Vukovic A, Tuckar N, Herman R, Grubisic G. Chloasma–the mask of pregnancy. Coll Antropol. 2008;32(Suppl 2):139–141. [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Kolata G. Researchers seek melanoma gene. Science. 1986;232(4751):708–709. doi: 10.1126/science.3961500. [DOI] [PubMed] [Google Scholar]
- Stinchcombe J, Bossi G, Griffiths GM. Linking albinism and immunity: the secrets of secretory lysosomes. Science. 2004;305(5680):55–59. doi: 10.1126/science.1095291. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li K, Shi YL, Hamzavi I, Gao TW, Henderson M, Huggins RH, Agbai O, Mahmoud B, Mi X. Systemic analyses of immunophenotypes of peripheral T cells in non-segmental vitiligo: Implication of defective natural killer T cells. Pigment Cell Melanoma Res. 2012;25:602. doi: 10.1111/j.1755-148X.2012.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EE, Timpson NJ, Sikora M, Yee MC, Moreno-Estrada A, Eng C, Huntsman S, Burchard EG, Stoneking M, Bustamante CD. Melanesian blond hair is caused by an amino acid change in TYRP1. Science. 2012;336(6081):554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Shriver MD. Unpacking human evolution to find the genetic determinants of human skin pigmentation. J Invest Dermatol. 2011;131(E1):E5–E7. doi: 10.1038/skinbio.2011.3. [DOI] [PubMed] [Google Scholar]
- Rees JL. The genetics of human pigmentary disorders. J Invest Dermatol. 2011;131(E1):E12–E13. doi: 10.1038/skinbio.2011.5. [DOI] [PubMed] [Google Scholar]
- Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, Fredriksson R, Schutz K, Jensen P, von Heijne G, Okimoto R. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168(3):1507–1518. doi: 10.1534/genetics.104.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CM, Coville JL, Coquerelle G, Gourichon D, Oulmouden A, Tixier-Boichard M. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genomics. 2006;7:19. doi: 10.1186/1471-2164-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom AR, Sundstrom E, Gunnarsson U, Bed'Hom B, Tixier-Boichard M, Honaker CF, Sahlqvist AS, Jensen P, Kampe O, Siegel PB. Sex-linked barring in chickens is controlled by the CDKN2A/B tumour suppressor locus. Pigment Cell Melanoma Res. 2010;23(4):521–530. doi: 10.1111/j.1755-148X.2010.00700.x. [DOI] [PubMed] [Google Scholar]
- Gunnarsson U, Kerje S, Bed'Hom B, Sahlqvist AS, Ekwall O, Tixier-Boichard M, Kampe O, Andersson L. The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment Cell Melanoma Res. 2011;24(2):268–274. doi: 10.1111/j.1755-148X.2011.00825.x. [DOI] [PubMed] [Google Scholar]
- Dorshorst B, Molin AM, Rubin CJ, Johansson AM, Stromstedt L, Pham MH, Chen CF, Hallbook F, Ashwell C, Andersson L. A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011;7(12):e1002412. doi: 10.1371/journal.pgen.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S, Lind J, Schutz K, Jensen P, Andersson L. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim Genet. 2003;34(4):241–248. doi: 10.1046/j.1365-2052.2003.00991.x. [DOI] [PubMed] [Google Scholar]
- Philipp U, Lupp B, Momke S, Stein V, Tipold A, Eule JC, Rehage J, Distl O. A MITF mutation associated with a dominant white phenotype and bilateral deafness in German Fleckvieh cattle. PLoS One. 2011;6(12):e28857. doi: 10.1371/journal.pone.0028857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon HN, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas ER. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39(11):1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Williams SM, Zavala G, Hafner S, Collett SR, Cheng S. Metastatic melanomas in young broiler chickens (Gallus gallus domesticus) Vet Pathol. 2012;49(2):288–291. doi: 10.1177/0300985811415706. [DOI] [PubMed] [Google Scholar]
- Gregersen VR, Conley LN, Sorensen KK, Guldbrandtsen B, Velander IH, Bendixen C. Genome-wide association scan and phased haplotype construction for quantitative trait loci affecting boar taint in three pig breeds. BMC Genomics. 2012;13(1):22. doi: 10.1186/1471-2164-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Onteru SK, Du ZQ, Garrick DJ, Stalder KJ, Rothschild MF. Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs. PLoS One. 2011;6(2):e14726. doi: 10.1371/journal.pone.0014726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn LA, Keele JW, Bennett GL, McDaneld TG, Smith TP, Snelling WM, Sonstegard TS, Thallman RM. Predicting breed composition using breed frequencies of 50,000 markers from the US Meat Animal Research Center 2,000 Bull Project. J Anim Sci. 2011;89(6):1742–1750. doi: 10.2527/jas.2010-3530. [DOI] [PubMed] [Google Scholar]
- Cole JB, Wiggans GR, Ma L, Sonstegard TS, Lawlor TJ, Crooker BA, Van Tassell CP, Yang J, Wang S, Matukumalli LK. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics. 2011. pp. 12–408. [DOI] [PMC free article] [PubMed]
- Gu X, Feng C, Ma L, Song C, Wang Y, Da Y, Li H, Chen K, Ye S, Ge C. Genome-wide association study of body weight in chicken F2 resource population. PLoS One. 2011;6(7):e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Luo C, Zhang C, Zhang R, Tang J, Nie Q, Ma L, Hu X, Li N, Da Y. Genome-Wide Association Study Identified a Narrow Chromosome 1 Region Associated with Chicken Growth Traits. PLoS One. 2012;7(2):e30910. doi: 10.1371/journal.pone.0030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross WB, Siegel HS. Evaluation of the heterophil/lymphocyte ratio as a measure of stress in chickens. Avian Dis. 1983;27(4):972–979. doi: 10.2307/1590198. [DOI] [PubMed] [Google Scholar]
- Chang CM, Furet JP, Coville JL, Coquerelle G, Gourichon D, Tixier-Boichard M. Quantitative effects of an intronic retroviral insertion on the transcription of the tyrosinase gene in recessive white chickens. Anim Genet. 2007;38(2):162–167. doi: 10.1111/j.1365-2052.2007.01581.x. [DOI] [PubMed] [Google Scholar]
- Hearing VJ. Determination of melanin synthetic pathways. J Invest Dermatol. 2011;131(E1):E8–E11. doi: 10.1038/skinbio.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom AR, Watt B, Fard SS, Tenza D, Mannstrom P, Narfstrom K, Ekesten B, Ito S, Wakamatsu K, Larsson J. Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet. 2011;7(9):e1002285. doi: 10.1371/journal.pgen.1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenblith MR, Shi J, Landi MT. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 2010;23(5):587–606. doi: 10.1111/j.1755-148X.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326(5949):150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JL, Harding RM. Understanding the evolution of human pigmentation: recent contributions from population genetics. J Invest Dermatol. 2012;132(3 Pt 2):846–853. doi: 10.1038/jid.2011.358. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Loftus SK, Pavan WJ. Networks and pathways in pigmentation, health, and disease. Wiley Interdiscip Rev Syst Biol Med. 2009;1(3):359–371. doi: 10.1002/wsbm.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candille SI, Absher DM, Beleza S, Bauchet M, McEvoy B, Garrison NA, Li JZ, Myers RM, Barsh GS, Tang H. Genome-wide association studies of quantitatively measured skin, hair, and eye pigmentation in four European populations. PLoS One. 2012;7(10):e48294. doi: 10.1371/journal.pone.0048294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Fekete DM, Andrisani OM. CtBP2 downregulation during neural crest specification induces expression of Mitf and REST, resulting in melanocyte differentiation and sympathoadrenal lineage suppression. Mol Cell Biol. 2011;31(5):955–970. doi: 10.1128/MCB.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Rovinsky SA, Lai CY, Qasem S, Liu X, How J, Engelhardt JF, Murphy GF. Aggressive melanoma cells escape from BMP7-mediated autocrine growth inhibition through coordinated Noggin upregulation. Lab Invest. 2008;88(8):842–855. doi: 10.1038/labinvest.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notting I, Buijs J, Mintardjo R, van der Horst G, Vukicevic S, Lowik C, Schalij-Delfos N, Keunen J, van der Pluijm G. Bone morphogenetic protein 7 inhibits tumor growth of human uveal melanoma in vivo. Invest Ophthalmol Vis Sci. 2007;48(11):4882–4889. doi: 10.1167/iovs.07-0505. [DOI] [PubMed] [Google Scholar]
- Na YR, Seok SH, Kim DJ, Han JH, Kim TH, Jung H, Lee BH, Park JH. Bone morphogenetic protein 7 induces mesenchymal-to-epithelial transition in melanoma cells, leading to inhibition of metastasis. Cancer Sci. 2009;100(11):2218–2225. doi: 10.1111/j.1349-7006.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Rovinsky S, Penmatcha S, Herlyn M, Muirhead D. Bone morphogenetic proteins in melanoma: angel or devil? Cancer Metastasis Rev. 2005;24(2):251–263. doi: 10.1007/s10555-005-1575-y. [DOI] [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 2005;65(2):448–456. [PubMed] [Google Scholar]
- Richard D, Picard F. Brown fat biology and thermogenesis. Front Biosci. 2011;16:1233–1260. doi: 10.2741/3786. [DOI] [PubMed] [Google Scholar]
- Li B, Smith CC, Laing JM, Gober MD, Liu L, Aurelian L. Overload of the heat-shock protein H11/HspB8 triggers melanoma cell apoptosis through activation of transforming growth factor-β-activated kinase 1. Oncogene. 2007;26(24):3521–3531. doi: 10.1038/sj.onc.1210145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Walsh SB, Verney ZM, Kopelovich L, Elmets CA, Athar M. Procarcinogenic effects of cyclosporine A are mediated through the activation of TAK1/TAB1 signaling pathway. Biochem Biophys Res Commun. 2011;408(3):363–368. doi: 10.1016/j.bbrc.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottero V, Kerur N, Sadagopan S, Patel K, Sharma-Walia N, Chandran B. Phosphorylation and polyubiquitination of transforming growth factor beta-activated kinase 1 are necessary for activation of NF-κB by the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. J Virol. 2011;85(5):1980–1993. doi: 10.1128/JVI.01911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang H, Huang T, Wang J, Ding Y, Li Z, Zhang J, Li L. TAB2 scaffolds TAK1 and NLK in repressing canonical Wnt signaling. J Biol Chem. 2010;285(18):13397–13404. doi: 10.1074/jbc.M109.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Shin MS, Singhirunnusorn P, Suzuki S, Kawanishi M, Koizumi K, Saiki I, Sakurai H. TAK1-mediated serine/threonine phosphorylation of epidermal growth factor receptor via p38/extracellular signal-regulated kinase: NF-κB-independent survival pathways in tumor necrosis factor alpha signaling. Mol Cell Biol. 2009;29(20):5529–5539. doi: 10.1128/MCB.00375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhirunnusorn P, Ueno Y, Matsuo M, Suzuki S, Saiki I, Sakurai H. Transient suppression of ligand-mediated activation of epidermal growth factor receptor by tumor necrosis factor-α through the TAK1-p38 signaling pathway. J Biol Chem. 2007;282(17):12698–12706. doi: 10.1074/jbc.M608723200. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Nishi A, Sato N, Mizukami J, Miyoshi H, Sugita T. TAK1-TAB1 fusion protein: a novel constitutively active mitogen-activated protein kinase kinase kinase that stimulates AP-1 and NF-κB signaling pathways. Biochem Biophys Res Commun. 2002;297(5):1277–1281. doi: 10.1016/S0006-291X(02)02379-3. [DOI] [PubMed] [Google Scholar]
- Neil JR, Schiemann WP. Altered TAB1: IκB kinase interaction promotes transforming growth factor β-mediated nuclear factor-κB activation during breast cancer progression. Cancer Res. 2008;68(5):1462–1470. doi: 10.1158/0008-5472.CAN-07-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science. 1996;272(5265):1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270(5244):2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- Blank U, Brown A, Adams DC, Karolak MJ, Oxburgh L. BMP7 promotes proliferation of nephron progenitor cells via a JNK-dependent mechanism. Development. 2009;136(21):3557–3566. doi: 10.1242/dev.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, Haber DA, Settleman J. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148(4):639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18(7):816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci U S A. 2010;107(40):17304–17308. doi: 10.1073/pnas.1004640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RR, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13(9):1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleck CM, Vertalino N, Vleck D, Bucher TL. Stress, corticosterone, and heterophil to lymphocyte ratios in free-living Adelie penguins. The Condor. 2000;102:392–400. [Google Scholar]
- Groenen MA, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RP, Vereijken A, Okimoto R, Muir W, Cheng HH. The development and characterization of a 60K SNP chip for chicken. BMC Genomics. 2011;12(1):274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina: Infinium® genotyping data analysis. 2010. Available: http://www.illumina.com/Documents/products/technotes/technote_infinium_genotyping_data_analysis.pdf. Accessed 30 November 2010.
- Gilmour AR, Thompson R, Cullis BR. Average Information REML: An Efficient Algorithm for Variance Parameter Estimation in Linear Mixed Models. Biometrics. 1995;51(4):1440–1450. doi: 10.2307/2533274. [DOI] [Google Scholar]
- Madsen P, Sørensen P, Su G, Damgaard LH, Thomsen H, Labouriau R. DMU - a package for analyzing multivariate mixed models. Proc. 8th World Congr. Genet. Appl. Livest. Prod. Belo Horizonte, Minas Gerais, Brazil. 2006. pp. 11–27.
- Ma L, Runesha HB, Dvorkin D, Garbe JR, Da Y. Parallel and serial computing tools for testing single-locus and epistatic SNP effects of quantitative traits in genome-wide association studies. BMC Bioinformatics. 2008;9:315. doi: 10.1186/1471-2105-9-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, London NR, Ma L, Dvorkin D, Da Y. Detection of SNP epistasis effects of quantitative traits using an extended Kempthorne model. Physiol Genomics. 2006;28(1):46–52. doi: 10.1152/physiolgenomics.00096.2006. [DOI] [PubMed] [Google Scholar]
- Wang S, Dvorkin D, Da Y. SNPEVG A graphical tool for SNP effect viewing and graphing. 2011. Available: http://animalgene.umn.edu. Accessed 30 June 2011. [DOI] [PMC free article] [PubMed]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pattern of linkage disequilibrium (LD) on chicken (Gallus gallus) chromosomes. A. LD on Chromosome 1. B. LD on Chromosome 20.
Interaction of genes. Blue, green, and red figures indicate the number of pathways that the genes are involved in, based on KEGG, GenMAPP, and BioCarta, respectively. BMP7 interacts with MAP3K7 via the ALK pathway in cardiac myocytes according to the BioCarta database. HIPK2 interacts with MAP3K7 via an enzyme linked receptor protein signaling pathway and Wnt netPath 8 according to the GenMAPP database. MAP3K7IP1 interacts with MAP3K7 via the MAPK and Toll-like receptor signaling pathways according to the KEGG database, by the NF-κB signaling pathway, signal transduction through IL1R, the TGF-β signaling pathway, and the WNT signaling pathway according to the Biocarta database, and by receptor signaling protein activity, the MAPK signaling pathway, and TGF-β-receptor netPath 7 according to the GenMAPP database.
Basic information on the 39,833 SNP markers on the chicken physical map.