Abstract
Genetically determined capacity for NER may modulate both cancer risk and prognosis. Thus, we evaluated associations of seven selected variants in the NER core genes with recurrence risk in 658 SCCOP patients treated principally by radiation. The seven polymorphisms in the core NER genes (XPC-rs2228000, XPC-rs2228001, XPD-rs1799793, XPD-rs13181, XPG-rs17655, ERCC1-rs3212986, and XPA-rs1800975) were genotyped using PCR-RFLP method and log-rank test and multivariable Cox models were used to evaluate the associations in both dominant and recessive genetic models. In a dominant model, we found that polymorphisms of XPC-rs2228000, XPD-rs1799793, and XPG-rs17655 were significantly associated with disease-free survival (log-rank, P = 0.014; P = 0.00008; and P = 0.0007, respectively), and these polymorphisms were significantly associated with recurrence risk of SCCOP (HR = 1.6, 95% CI, 1.1–2.3 for XPC-rs2228000; HR = 0.4, 95%, 0.3–0.6 for XPD-rs1799793; and HR = 0.5, 95% CI, 0.4–0.8 for XPG-rs17655) after multivariable adjustment. Moreover, the borderline significant or significant associations were also found for these three polymorphisms in HPV16/18-positive SCCOP patients (HR= 1.6, 95% CI, 1.0–4.1 for XPC-rs2228000; HR = 0.2, 95%, 0.1–0.5 for XPD-rs1799793; and HR = 0.1, 95% CI, 0.0–0.9 for XPG-rs17655). However, similarly significant associations were not found for these polymorphisms in a recessive model. These findings suggest that polymorphisms of XPC-rs2228000, XPD-rs1799793, and XPG-rs17655 in the NER core genes may contribute to recurrence risk of SCCOP, particularly HPV-positive SCCOP, in a dominant but not in a recessive model. However, validation of these results is warranted.
Keywords: genetic variants, nucleotide excision repair, human papillomavirus, oropharyngeal cancer, recurrence, DNA repair
Introduction
Approximately 52,610 new cases of squamous cell carcinoma of the head and neck occurred with 11,500 deaths in 2012 in the United States. 1 The incidence of squamous cell carcinoma of the oropharynx (SCCOP), a subset of squamous cell carcinoma of the head and neck, continues to increase, particularly in young adults.2 The growing incidence of SCCOP may be attributed to viral infection, as human papillomavirus (HPV) has been established as an etiologic risk factor for SCCOP.3–7 SCCOP is characterized by local tumor aggressiveness, a moderately high recurrence rate, a high frequency of second primary malignancies, and a high frequency of medical comorbidities.8 Surgery, radiotherapy, and chemotherapy have been successfully used both individually and in combination to treat SCCOP, however recurrence remains a major problem resulting in disease-specific mortality. As diagnostic and therapeutic approaches for SCCOP continue to improve, accurately predicting recurrence in patients with this disease would facilitate intensive surveillance or targeted intervention for patients with high recurrence risk.
Nucleotide excision repair (NER) proteins function synergistically to repair a wide variety of DNA damage, including damage caused by cancer therapy. Because DNA damage from treatments, such as chemotherapy and radiotherapy, can initiate cellular processes including DNA repair, cell cycle control, and apoptosis, common single nucleotide polymorphisms (SNPs) within the NER core genes may cause interindividual differences in DNA repair capacity, and thus differences in susceptibility to genotoxic effects of cancer therapy. Genetic variations in the NER pathway have been widely studied in association with many types of cancers,9–18 and SNPs in genes regulating NER have previously been studied as potential risk factors involved in genetic predisposition to SCCHN and second primary tumors.19,20 However, few large studies have examined the association between genetic variations in the NER pathway and risk of recurrence of SCCOP. In the current study, we evaluated the impact of seven selected variants in the NER core genes on recurrence risk among 658 patients with SCCOP. Considering genetic models of inheritance of traits associated with alleles of the selected common variants, the results may differ depending on the models used. We explored the associations using alternative genetic models including a dominant and a recessive model.
Materials and methods
Study Subjects
This study included 658 patients with newly diagnosed, previously untreated, and histopathologically confirmed SCCOP who were consecutively recruited between May 1995 and April 2007 as part of an ongoing molecular epidemiological study at The University of Texas MD Anderson Cancer Center. 9,21 Patients were eligible regardless of age, sex, ethnicity, or cancer stage (except those < 18 years of age or with distant metastasis were excluded) and interviewed to collect the relevant information on demographic, epidemiologic, and clinical characteristics as well as blood samples for genetic testing at the time of initial presentation to our institution.
All subjects completed an Institutional Review Board-approved informed consent form before enrollment. Approximately 95% of contacted patients consented to enrollment in the study. Patients were excluded from this study if they 1) had known distant metastases; 2) had any prior cancer history except nonmelanoma skin cancer; 3) had a primary sinonasal tumor, a salivary gland tumor, cervical metastases of unknown origin, or a tumor outside the upper aerodigestive tract; 4) had no blood samples available for genotyping (this was the case for some patients who were recruited early in the parent study); 5) had treatment performed outside of our institution; or 6) underwent only palliative treatment.
Patients were followed up throughout their treatment and posttreatment course with scheduled regular clinical and radiographic examinations. Patients were considered disease free if absence of disease was documented at the date of the last visit with the head and neck surgeon, head and neck radiation oncologist, or head and neck medical oncologist. There were no universal standards for imaging. Typically patients had either routine serial imaging, or follow-up imaging on the basis of symptoms or findings on physical examinations. Recurrent disease was defined as appearance of a new lesion of the same histology verified by biopsy (incisional, excisional, or needle biopsy), reappearance of any lesion that had disappeared, or development of tumor-related symptoms.
Clinical data, including stage at presentation of the index tumor, site and histologic subtype of the index tumor and any recurrence, comorbidity, and treatment, were obtained from review of the medical records. Alcohol use and smoking status data were collected at the initial interview. Patients who had drunk at least one alcoholic beverage per week for at least one year during their lifetime were categorized as “ever drinkers,” and patients who had never had such a pattern of drinking were categorized as “never drinkers.” Patients who had smoked at least 100 cigarettes in their lifetime were categorized as “ever smokers,” and patients who had smoked fewer than 100 cigarettes in their lifetime were categorized as “never smokers.”
Selection of candidate genes and SNPs
Among 1098 SNPs identified to date within eight core genes in the NER pathway, 40 SNPs are non-synonymous, which cause different polypeptide sequences, and five of these 40 SNPs have a minor allele frequency greater than 0.05 in non-Hispanic whites: XPC rs2228000, XPC rs2228001, XPD rs1799793, XPD rs13181, and XPG rs17655.9 In addition to these five non-synonymous SNPs, another two common regulatory SNPs at the 3′-untranslated region of ERCC1 rs3212986, and the 5′-untranslated region of XPA rs1800975, were reported to be correlated with the DNA repair capacity phenotype.22
Genotyping
Genomic DNA was extracted from patient peripheral blood samples drawn at the time of patient registration. These DNA samples were used to genotype for seven potentially functional SNPs of the NER pathway: ERCC1 rs3212986, XPA rs1800975, XPC rs2228000, XPC rs2228001, XPD rs1799793, XPD rs13181, and XPG rs17655. The detailed methods for genotyping these SNPs (e.g., polymerase chain reaction conditions and restriction enzymes used) have previously been described.9 These SNPs were genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The primers used for genotyping were: 1) for XPD rs1799793: 5′-CTGTTGGTGGGTGCCCGTATCTGTTGGTCT-3′ and 5′-TAATATCGGGGCTCACCCTGCAGCACTTCCT-3′ and for XPD rs13181: 5′-TCAAACATCCTGTCCCTACT-3′ and 5′-CTGCGATTAAAGGCTGTGGA-3′; 2) for ERCC1 rs3212986: 5′-TACACAGGCTGCTGCTGCAGCT-3′ and 5′-GCCAGAGACAGTGCCCCAAGAG-3′; 3) for XPA rs1800975: 5′-CTAGGTCCTCGGAGTGGTCC-3′ and 5′-GCCCAAACCTCCAGTAGCC-3′; 4) for XPC rs2228000: 5′-TAAGGACCCAAGCTTGCCCG-3′ and 5′-CCCACTTTTCCTCCTGCTCACAG-3′ and XPC rs2228001: 5′-ACCAGCTCTCAAGCAGAAGC-3′ and 5′-CTGCCTCAGTTTGCCTTCTC-3′; and 5) for XPG rs17655: 5′-GACCTGCCTCTCAGAATCATC-3′ and 5′-CCTCGCACGTCTTAGTTTCC-3′. Positive and negative controls were used in each genotyping assay, and 10% of samples were randomly selected and assayed in duplicates, and the concordance between duplicates was 100%.
Tumor HPV16/18 Detection
Paraffin-embedded tissues were tested for HPV16/18 DNA using polymerase chain reaction (PCR)-based, type-specific assays with modification and quality control for the E6 and E7 regions. 23,24 Assays of the samples were run in triplicate, with positive and negative controls (Siha and TPC-1 cell lines, respectively). β-actin was used as a DNA quality control. Specificity for HPV16/18 E6 and E7 was confirmed by Southern blot analysis of paraffin-embedded tissue samples using a Roche Diagnostics labeling and hybridization system(Roche Applied Science, Indianapolis, IN). HPV16/18 E6 and E7 specificity was confirmed by retesting 10% of the samples using restriction digestion of the PCR products with BanII and MspI to verify the presence of E6- and E7-specific fragments. 24 The results of both methods were 100% concordant.
Statistical Analysis
The mean age and follow-up time for patients with and without recurrence were first compared using Student’s t test. The chi-squared test was used to evaluate differences in ethnicity, sex, smoking status, and alcohol use, index tumor site, tumor stage, comorbidity, treatment, genotype distributions, and allele frequencies between patients with and without recurrence. The primary endpoint of the study was recurrence. Time to recurrence was computed from the date of presentation to the date of clinically detectable recurrence (local, regional, or distant). Participants who remained recurrence free or were lost to follow-up or died of other/unknown cause were considered censored. The associations between individual epidemiological factors, clinical characteristics, and treatment variables, and time to recurrence, were initially assessed using univariate Cox proportional hazards regression models. The data were consistent with the assumptions of the Cox proportional hazards regression model from the examination of Kaplan-Meier survival curves and log-minus-log survival plots.25, 26 The log-rank test was used to determine the associations between various variables and disease-free survival (DFS). The associations between individual epidemiologic risk factors, clinical characteristics (including tumor site, stage, comorbidity, and treatment variables), and time to recurrence were assessed using both univariate and multivariable Cox proportional hazards regression models. Associations were quantified using hazard ratios (HRs) and their 95% confidence intervals (CIs) for recurrence risk. The Cox model included adjustment for potential confounders including age, sex, ethnicity, smoking, alcohol use, tumor stage, comorbidity, and treatment. We evaluated the individual variants in a recessive genetic model, in which we compared the variant homozygous genotype to the combined variant heterozygous and wild-type homozygous genotypes. We also explored the effect of individual variants on recurrence risk in an alternative dominant model, in which we added the variant homozygous genotype and the variant heterozygous genotype and compared to the wild-type homozygous genotype. For all analyses, statistical significance was set at P < 0.05, and all tests were two-sided. SAS software (version 9.2.3; SAS Institute) was used to perform all statistical analyses.
Results
A total of 807 SCCOP patients were recruited from May 1995 to April 2007, and 149 patients were excluded from the final analysis due to lack of information on follow-up and treatment status as well as unavailability of blood samples. We first compared the distribution of the characteristics in Table 1, no significant differences for these selected variables were found between 658 study cases and 149 excluded patients except for tumor HPV16/18 status (most of the patients had no tumor specimens available). Therefore, our final analysis included 658 patients with newly diagnosed, previously untreated SCCOP. These patients were followed from May 1995 to October 2011, and the overall median follow-up time was 55.2 months (range 2 to 171 months), during which period 132 patients had disease recurrence. The overall median follow-up time was 61.3 and 12.6 months for recurrence-free patients and patients with recurrence, respectively. Of the patients with recurrence, 51 (38.6%) had distant recurrence, 39 (29.5%) had local recurrence, 11 (8.4%) had regional recurrence, and 31 (23.5%) had recurrence of more than one category.
Table 1.
Characteristics of patients with SCCOP (n = 658).
| Variable | Overall | Patients with Rec. | 5-year Rec. rate | P1 | |
|---|---|---|---|---|---|
|
|
|
||||
| No. | % | No. | % | ||
| No. of patients | 658 | 100 | 132 | 0.20 | |
| Age | 0.001 | ||||
| ≤ median (57 years) | 420 | 63.8 | 66 | 0.16 | |
| > median (57 years) | 238 | 46.2 | 66 | 0.27 | |
| Sex | 0.616 | ||||
| Male | 563 | 85.6 | 115 | 0.20 | |
| Female | 95 | 14.4 | 17 | 0.22 | |
| Ethnicity | 0.001 | ||||
| Non-Hispanic white | 591 | 89.8 | 105 | 0.17 | |
| Other | 67 | 10.2 | 27 | 0.42 | |
| Smoking | 0.010 | ||||
| Never | 241 | 36.6 | 37 | 0.14 | |
| Ever | 417 | 63.4 | 95 | 0.24 | |
| Alcohol use | 0.010 | ||||
| Never | 171 | 26.0 | 22 | 0.13 | |
| Ever | 487 | 74.0 | 110 | 0.23 | |
| Comorbidity | 0.195 | ||||
| None-Mild | 587 | 89.2 | 115 | 0.19 | |
| Moderate-Severe | 71 | 10.8 | 17 | 0.25 | |
| Index cancer stage | 0.473 | ||||
| 1 or 2 | 49 | 7.5 | 8 | 0.18 | |
| 3 or 4 | 609 | 92.5 | 124 | 0.20 | |
| Treatment | 0.148 | ||||
| X/XC/SX* | 606 | 92.1 | 121 | 0.20 | |
| SXC | 52 | 7.9 | 11 | 0.21 | |
| Tumor HPV16/18 status | 0.002 | ||||
| Positive | 102 | 69.4 | 20 | 0.13 | |
| Negative | 45 | 30.6 | 3 | 0.21 | |
P: Log-rank test for DFS (disease-free survival) between the two groups.
X: radiation, C: chemotherapy, and S: surgery.
Table 1 summarizes the demographics, risk factors, and clinical characteristics for the overall cohort of patients with associated 5-year actuarial recurrence rates. The mean age at diagnosis for the overall cohort was 55.3 years (median, 54 years). The mean age at diagnosis was significantly greater for those patients who developed recurrence than for those patients who did not develop recurrence (57.7 years vs. 54.6 years; P = 0.006). Patients in the overall group were predominantly male (85.6%) and non-Hispanic white (89.8%). Ethnicity was also significantly different between the patients with and without disease recurrence (P = 0.00091). Compared with the recurrence-free group, patients with recurrence were older (P = 0.0002) and more likely to be smokers (P = 0.021) and alcohol drinkers (P = 0.006) and HPV16/18-negative tumors (P = 0.043). However, we did not observe significant differences between patients with and without recurrence with regard to clinical characteristics including comorbidity (P = 0.387), index cancer stage (P = 0.498), or treatment (P = 0.838).
Table 2 shows univariate Kaplan-Meier survival analysis, genotype distributions of the seven SNPs, 5-year actuarial recurrence rates, and multivariable Cox proportional hazards regression analysis of the association between the seven SNPs and recurrence risk among SCCOP patients in both genetic models. As shown in Figure 1 in a dominant model, Kaplan-Meier survival estimates showed significantly worse disease-free survival (DFS) in SCCOP patients with XPC rs2228000 Ala/Val+Val/Val, XPD rs1799793 Asp/Asp, and XPG rs17655 His/His genotypes than in SCCOP patients with XPC rs2228000 Ala/Ala, XPD rs1799793 Asp/Asn+Asn/Asn, and XPG rs17655 His/Asp+Asp/Asp, respectively. However, no significant differences in DFS were observed for SNPs of ERCC1 rs3212986 (log-rank P = 0.666), XPA rs1800975 (log-rank P = 0.670), XPC rs2228001 (P = 0.131), and XPD rs13181 (P = 0.100). Moreover, the similarly significant differences were not found in a recessive model. Estimates of association were adjusted for potential confounders, including age, sex, ethnicity, smoking and alcohol status, comorbidity, stage, and treatment. In a dominant model, a moderately increased risk of cancer recurrence was observed for SCCOP patients with the XPC rs2228000 Ala/Val+Val/Val, XPD rs1799793 Asp/Asp, and XPG rs17655 His/His genotypes compared to patients with the XPC rs2228000 Ala/Ala, XPD rs1799793 Asp/Asn+Asn/Asn, and XPG rs17655 His/Asp+Asp/Asp genotypes. However, no significant associations were observed between recurrence risk and SNPs of ERCC1 rs3212986, XPA rs1800975, XPC rs2228001, and XPD rs13181 among SCCOP patients. Furthermore, we did not find any significant associations of each of the seven SNPs with recurrence risk in the recessive model.
Table 2.
Multivariable analysis of 7 variants of NER genes and SCCOP recurrence in both dominant and recessive genetic models
| Genotype | Patients with SCCOP
|
||||
|---|---|---|---|---|---|
| Rec. No./patient No. | 5-year Rec. rate | Log-rank tests | aHR*, (95% CI) | P | |
| ERCC1 rs3212986 | |||||
| CC | 82/392 | 0.19 | 1.0 | ||
| CA/AA (D) | 50/266 | 0.20 | 0.666 | 0.9 (0.6–1.3) | 0.628 |
| CA/CC | 126/624 | 0.20 | 1.0 | ||
| AA (R) | 6/34 | 0.19 | 0.795 | 0.9 (0.4–2.1) | 0.825 |
| XPA rs1800975 | |||||
| GG | 61/296 | 0.20 | 1.0 | ||
| GA/AA (D) | 71/362 | 0.20 | 0.670 | 0.9 (0.6–1.2) | 0.311 |
| GA/GG | 114/574 | 0.20 | 1.0 | ||
| AA (R) | 18/84 | 0.23 | 0.788 | 0.9 (0.6–1.7) | 0.978 |
| XPC rs2228000 | |||||
| Ala/Ala | 59/359 | 0.16 | 1.0 | ||
| Ala/Val or Val/Val (D) | 73/299 | 0.24 | 0.014 | 1.6 (1.1–2.3) | 0.009 |
| Ala/Ala or Ala/Val | 113/594 | 0.20 | 1.0 | ||
| Val/Val (R) | 19/64 | 0.26 | 0.100 | 1.6 (0.9–2.6) | 0.100 |
| XPC rs2228001 | |||||
| Lys/Lys | 56/242 | 0.23 | 1.0 | ||
| Lys/Gln or Gln/Gln (D) | 76/416 | 0.18 | 0.131 | 0.8 (0.5–1.1) | 0.112 |
| Lys/Gln or Lys/Lys | 120/579 | 0.20 | 1.0 | ||
| Gln/Gln (R) | 12/79 | 0.16 | 0.254 | 0.7 (0.4–1.3) | 0.258 |
| XPD rs1799793 | |||||
| Asp/Asp | 89/301 | 0.29 | 1.0 | ||
| Asp/Asn or Asn/Asn (D) | 43/357 | 0.13 | 0.00008 | 0.4 (0.3–0.6) | 0.0001 |
| Asp/Asn or Asp/Asp | 125/579 | 0.14 | 1.0 | ||
| Asn/Asn ® | 7/79 | 0.20 | 0.126 | 0.4 (0.2–1.1) | 0.222 |
| XPD rs13181 | |||||
| Lys/Lys | 65/276 | 0.23 | 1.0 | ||
| Lys/Gln or Gln/Gln (D) | 67/382 | 0.18 | 0.100 | 0.8 (0.6–1.1) | 0.149 |
| Lys/Gln or Lys/Lys | 118/574 | 0.19 | 1.0 | ||
| Gln/Gln (R) | 14/84 | 0.20 | 0.505 | 0.8 (0.5–1.5) | 0.540 |
| XPG rs17655 | |||||
| His/His | 97/397 | 0.24 | 1.0 | ||
| His/Asp or Asp/Asp (D) | 35/261 | 0.14 | 0.0007 | 0.5 (0.4–0.8) | 0.0008 |
| His/Asp or His/His | 124/613 | 0.20 | 1.0 | ||
| Asp/Asp (R) | 8/45 | 0.24 | 0.713 | 0.8 (0.4–1.6) | 0.462 |
D: Dominant genetic model and R: Recessive genetic model.
Adjusted for age, sex, race, smoking, alcohol use, stage, comorbidity, and treatment.
P: significance for adjusted HRs in both dominant and recessive genetic models.
Figure 1.
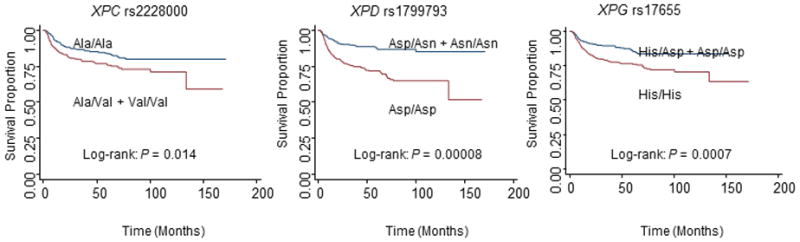
Kaplan-Meier estimates for DFS by genotypes in 658 patients with SCCOP in a dominant model.
Because human papillomavirus (HPV) is the strongest risk factor for SCCOP, we then evaluated the univariate Kaplan-Meier survival analysis and associations between genotypes of 7 SNPs and risk of SCCOP recurrence among those in whom the tumor HPV16/18 status was available in both genetic models (Table 3). In a dominant model, the Kaplan-Meier survival as stratified by the genotypes of 7 SNPs among tumor HPV16/18-positive patients with SCCOP was performed, and a borderline significant or significant difference in DFS was observed among the patients with different genotypes of XPC-rs2228000 (log-rank P = 0.061), XPD-rs1799793 (log-rank P = 0.001), and XPG-rs17655 (log-rank P = 0.0006) polymorphisms as shown in Figure 2. Furthermore, we found that the associations of these three polymorphisms with risk of recurrence were statistically borderline significant or significant for the polymorphisms of XPC-rs2228000 (HR, 1.6, 95% CI, 1.0–4.1), XPD-rs1799793 (HR, 0.2, 95% CI, 0.1–0.5), and XPG-rs17655 (HR, 0.1, 95% CI, 0.0–0.9) in the dominant model among 102 patients with a HPV16/18-positive SCCOP, while no significant associations were observed in a recessive model (Table 3). In addition, we did not find any significant associations of the 7 polymorphisms in the NER genes with recurrence risk among the patients with HPV16/18-negative SCCOP since there was no enough sample size or outcome events of these patients for such analysis (only 45 HPV16/18-negative SCCOP patients were included in this study).
Table 3.
Associations between 7 variants of NER genes and HPV16/18-positive SCCOP recurrence in both dominant and recessive genetic models
| Genotype | Patients with SCCOP
|
||||
|---|---|---|---|---|---|
| Rec. No./patient No. | 5-year Rec. rate | Log-rank tests | aHR*, (95% CI) | P | |
| ERCC1 rs3212986 | |||||
| CC | 16/64 | 0.20 | 1.0 | ||
| CA/AA (D) | 4/38 | 0.14 | 0.113 | 0.4 (0.1–1.3) | 0.138 |
| CA/CC | 20/99 | 0.20 | 1.0 | ||
| AA (R) | 0/3 | n/a | 0.412 | n/a | 0.993 |
| XPA rs1800975 | |||||
| GG | 10/49 | 0.22 | 1.0 | ||
| GA/AA (D) | 10/53 | 0.21 | 0.640 | 1.0 (0.4–2.4) | 0.933 |
| GA/GG | 18/94 | 0.17 | 1.0 | ||
| AA (R) | 2/8 | 0.25 | 0.124 | 2.5 (0.6–9.6) | 0.188 |
| XPC rs2228000 | |||||
| Ala/Ala | 7/54 | 0.12 | 1.0 | ||
| Ala/Val or Val/Val (D) | 13/48 | 0.28 | 0.061 | 1.6 (1.0–4.1) | 0.051 |
| Ala/Ala or Ala/Val | 17/91 | 0.20 | 1.0 | ||
| Val/Val (R) | 3/11 | 0.25 | 0.139 | 2.6 (0.8–8.7) | 0.110 |
| XPC rs2228001 | |||||
| Lys/Lys | 7/30 | 0.25 | 1.0 | ||
| Lys/Gln or Gln/Gln (D) | 13/72 | 0.20 | 0.643 | 0.7 (0.3–2.0) | 0.523 |
| Lys/Gln or Lys/Lys | 18/95 | 0.25 | 1.0 | ||
| Gln/Gln (R) | 2/7 | 0.28 | 0.551 | 1.0 (0.2–4.6) | 0.988 |
| XPD rs1799793 | |||||
| Asp/Asp | 16/44 | 0.31 | 1.0 | ||
| Asp/Asn or Asn/Asn (D) | 4/58 | 0.10 | 0.001 | 0.2 (0.1–0.5) | 0.002 |
| Asp/Asn or Asp/Asp | 18/94 | 0.18 | 1.0 | ||
| Asn/Asn (R) | 2/8 | 0.19 | 0.756 | 0.8 (0.1–6.5) | 0.840 |
| XPD rs13181 | |||||
| Lys/Lys | 11/45 | 0.21 | 1.0 | ||
| Lys/Gln or Gln/Gln (D) | 9/57 | 0.17 | 0.312 | 0.4 (0.2–1.1) | 0.100 |
| Lys/Gln or Lys/Lys | 17/93 | 0.18 | 1.0 | ||
| Gln/Gln (R) | 3/9 | 0.19 | 0.193 | 1.8 (0.5–6.5) | 0.404 |
| XPG rs17655 | |||||
| His/His | 19/70 | 0.26 | 1.0 | ||
| His/Asp or Asp/Asp (D) | 1/32 | 0.10 | 0.0006 | 0.1 (0.0–0.9) | 0.036 |
| His/Asp or His/His | 20/96 | 0.20 | 1.0 | ||
| Asp/Asp (R) | 0/6 | n/a | 0.228 | n/a | 0.993 |
D: Dominant genetic model and R: Recessive genetic model.
Adjusted for age, sex, race, smoking, alcohol use, stage, comorbidity, and treatment.
P: significance for adjusted HRs in both dominant and recessive genetic models.
Figure 2.
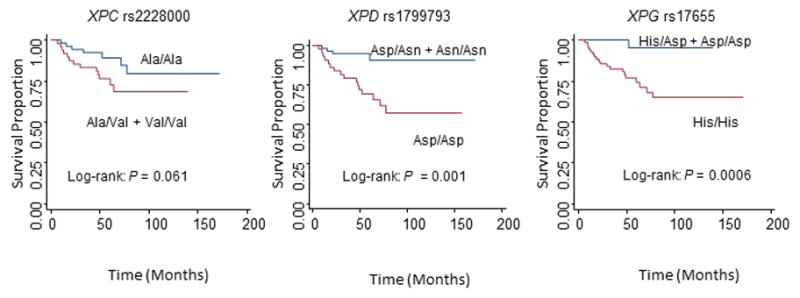
Kaplan-Meier estimates for DFS by genotypes in 102 HPV16/18-positive SCCOP patients in a dominant model.
Discussion
In this study, we comprehensively assess the associations between seven potentially functional SNPs in the NER pathway and recurrence risk among 658 patients with incident SCCOP. We did observe significant associations in the assumption of dominant genetic model, while we did not observe a significant effect in a recessive genetic model. Our results suggest that SCCOP patients with XPC rs2228000 Ala/Val + Val/Val, XPD rs1799793 Asp/Asp, and XPG rs17655 His/His genotypes had a higher risk of cancer recurrence, particularly for patients with HPV16/18-positive SCCOP.
It is well established that DNA repair capacity phenotype is associated with cancer risk and clinical outcome.27,28 Genetic variations in the DNA repair pathway genes are thought to modulate the DNA repair capacity phenotype, and have been suggested to affect risk and prognosis for various cancers.26–31 Therefore, it is plausible that genetic variants in DNA repair pathway genes may significantly influence clinical cancer outcomes, particularly for cancer such as SCCOP with definitive radiotherapy. Ultimately, such knowledge may help identify patients who can benefit from various treatments or by consideration of alternative treatment/intensified therapy. The NER pathway specifically excises bulk base damage induced by environmental carcinogens, such as tobacco compounds, and certain types of cancer treatment.32
Overall, the functional significance of these seven variants in the NER pathway is still largely uncertain. Several studies have reported conflicting findings regarding the association of polymorphisms in the NER pathway with clinical outcomes of cancers, but studies focusing on risk of recurrence of SCCOP only in the NER pathway are limited. For ERCC1 rs3212986 SNP, others reported that the CC genotype was associated with an increased risk of recurrence compared with the CA or AA genotypes among patients with squamous cell carcinoma of the head and neck or esophageal cancer, whereas in contradistinction, the A allele was associated with poorer survival among patients with advanced non-small-cell lung cancer (NSCLC).33,34 In the current study, we did not found a significant association between this ERCC1 polymorphism and recurrence risk among SCCOP. XPA is an essential DNA-binding protein in the NER pathway,35 and individuals with the XPA rs1800975 G allele exhibit more efficient DNA repair than individuals with the homozygous XPA rs1800975 AA genotype. Although lung and ovarian cancer patients with the variant genotypes (XPA rs1800975 AA and GA) have previously been reported to have shorter survival and higher recurrence risk than those with homozygous XPA rs1800975 GG genotype,36,37 we did not find an association between XPA rs1800975 and recurrence risk in patients with SCCOP. Although previous studies have explored the association of XPC polymorphisms with risk of lung cancer, bladder cancer, and head and neck cancers,9,38 few studies have analyzed the association of XPC SNPs with cancer outcome, particularly for SCCOP. One study found that patients with acute myeloid leukemia carrying the XPC rs2228000 variant allele had a greater risk of death or recurrence than patients with the wild-type genotype,39 and this finding is consistent with what we observed in our current study. However, the functional significance of the XPC rs2228000 variant, which causes amino acid change, is still unclear. The XPD rs1799793 at exon 10 and the XPD rs13181 at exon 23, both of which cause amino acid changes, are the two most frequently studied XPD SNPs. Colorectal cancer patients with XPD rs13181 wild-type genotype had a longer survival than patients with the heterozygous and homozygous variants,40 whereas NSCLC patients with the variant allele of XPD rs1799793 had a shorter overall survival.41 However, a similar study in NSCLC patients did not find an association of either SNP with survival.42 In ovarian cancer, carriers of at least one variant allele of both SNPs had significantly reduced overall survival.37 In the current study, we found an association only for XPD rs1799793 but not for XPD rs13181. Conflicting results have also been reported for XPG rs17655 between a study of ovarian cancer patients37 and a study of patients with bladder cancer.43 In contrast, our analysis of this same XPG SNP showed that patients with the homozygous wild-type genotype had a significantly increased risk of recurrence compared with patients with variant genotypes.
Different genetic backgrounds and patient characteristics in the aforementioned studies might explain, to some extent, the somewhat conflicting results with respect to the impact of these NER pathway SNPs in different cancers and different populations. Other factors in these studies could also contribute to the inconsistent results, including small sample size, different cancer types, variations in stage, different treatments, interactions between functional variants of these SNPs and therapeutic agents used, inclusion of different ethnic groups in a single study, and inadequate adjustment for other confounding factors.
We also observed that the modifying effect of genotypes of XPC rs2228000, XPD rs1799793, and XPG rs17655 polymorphisms was statistically borderline significant or significant among the patients with tumor HPV16/18-positive SCCOP. These observations are biologically plausible because virtually all (99.7 %) of these SCCOP patients in our study had definitive radiotherapy or chemoradiotherapy, and such treatments lead to mixed types of DNA damage, including those that need to be repaired by the NER pathway. HPV16/18-positive tumors, especially those occurring in never smokers are much less likely to have a p53 mutation. While the tumor cells harboring such intact p53 might activate DNA repair pathways including the NER pathway, and such patients who received radiotherapy or chemoradiotherapy typically also have accumulated more DNA damage induced by reactive oxygen species (ROS) than other patients, these patients thus are at higher risk for recurrence or progression. Therefore, genetic variants of these genes may lead to interindividual differences in DNA repair capacity phenotype, in turn resulting in different susceptibility to the genotoxic effects of radiation and /or cancer drugs resulting in different clinical outcomes after such treatments.44,45 However, the interaction between tumor HPV16/18 status and combined risk genotypes on risk of recurrence was not statistically significant (Pint. = 0.621 for dominant model and Pint. = 0.983 for the recessive model). This lack of significance could be either because there was no such interaction effect in these subgroups or because the small sample sizes in each substratum limited the statistical power to detect a significant interaction effect. Therefore, the significance and degree of such interaction in each subgroup needs to be further investigated in future studies with larger sample sizes.
There is some uncertainty as to what would be the best genetic model to represent genetic effect for these variants in the NER pathway. We analyzed the data in the current study first assuming a dominant model and did find significant associations. In contrast, when we assumed a recessive genetic model, we did not identify a significant association of these putatively functional SNPs with risk of recurrence. It becomes obvious that the results could vary depending on the model used in nominal statistical significance, particularly for weak associations of individual SNPs such as the results we found in the current study.
This study has certain limitations. For future studies on associations between genetic variants in the NER pathway and patient outcome, information on radiotherapy doses, drugs, and drug doses, and fields or their combinations will be important, because the treatments may cause different types of DNA damage and DNA repair pathways might have cross-functionality between pathways, which could be differentially regulated and activated in different tissues treatments. Unfortunately, in the present study, the treatment our patients received in this study was not homogeneous. These patients were treated with either different amounts of radiation doses or adjuvant therapy with diverse DNA-damaging drugs, or determined by the multidisciplinary team treating the patients at the time of presentation rather than a single uniform clinical trial. Other limitations included the selection of a limited number of polymorphisms, small sample size in some strata, and the lack of complete information on HPV status of the patients’ tumors. In conclusion, genetic variants of XPC rs2228000, XPD rs1799793, and XPG rs17655 in the NER pathway might modify risk of the recurrence of SCCOP, particularly those which are HPV16/18-positive. However, we need confirm such findings in future studies that are warranted to further explore the utility of genetic variants as clinical prognostic biomarkers.
Novelty and impact statements.
Variants of XPC rs2228000, XPD rs1799793, and XPG rs17655 in the NER core genes modify the risk of SCCOP recurrence, and may be a marker of genetic susceptibility to recurrence of SCCOP, particularly in HPV-positive SCCOP patients.
Acknowledgments
The authors wish to thank Ms. Margaret Lung, Ms. Liliana Mugartegui, Ms. Kathryn Tipton and Jessica Fiske for assistance with patient recruitment.
Funding
National Institute of Environmental Health Sciences grant R01 ES-11740 (to Q.W.); and National Institutes of Health grant CA133099 (to G.L.).
Abbreviations
- NER
nucleotide excision repair
- SCCOP
squamous cell carcinoma of the oropharynx
- HR
hazard ratio
- CI
confidence interval
- SNP
single nucleotide polymorphism
- HPV
human papillomavirus
- PCR-RFLP
polymerase chain reaction-restriction fragment length polymorphism
Footnotes
Disclosure
The authors declare no conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA-Cancer J Clin 2012. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Vineis P, Alavanja M, Buffler P, Fontham E, Franceschi S, Gao YT, Gupta PC, Hackshaw A, Matos E, Samet J, Sitas F, Smith J, et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst. 2004;96:99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 3.Dahlstrom KR, Adler-Storthz K, Etzel CJ, Liu Z, Dillon L, El-Naggar AK, Spitz MR, Schiller JT, Wei Q, Sturgis EM. Human papillomavirus type 16 infection and squamous cell carcinoma of the head and neck in never-smokers: a matched pair analysis. Clin Cancer Res. 2003;9:2620–6. [PubMed] [Google Scholar]
- 4.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers. Cancer. 2007;110:1429–35. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 5.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–9. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 7.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–94. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 9.An J, Liu Z, Hu Z, Li G, Wang LE, Sturgis EM, El-Naggar AK, Spitz MR, Wei Q. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007;16:1633–8. doi: 10.1158/1055-9965.EPI-07-0252. [DOI] [PubMed] [Google Scholar]
- 10.Brewster AM, Alberg AJ, Strickland PT, Hoffman SC, Helzlsouer K. XPD polymorphism and risk of subsequent cancer in individuals with nonmelanoma skin cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1271–5. [PubMed] [Google Scholar]
- 11.Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, Lee JE, Ross MI, Mansfield PF, Cormier JN, Prieto VG, Duvic M, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–32. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- 12.Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, He X, Bonner MR, Tian L, Yeager M, Welch R, Chanock S, Zheng T, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116:768–73. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- 13.Rybicki BA, Conti DV, Moreira A, Cicek M, Casey G, Witte JS. DNA repair gene XRCC1 and XPD polymorphisms and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:23–9. doi: 10.1158/1055-9965.epi-03-0053. [DOI] [PubMed] [Google Scholar]
- 14.Kumar R, Höglund L, Zhao C, Försti A, Snellman E, Hemminki K. Single nucleotide polymorphisms in the XPG gene: Determination of role in DNA repair and breast cancer risk. Int J Cancer. 2003;103:671–5. doi: 10.1002/ijc.10870. [DOI] [PubMed] [Google Scholar]
- 15.Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ. Genetic variation in the nucleotide excision repair pathway and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:2263–9. doi: 10.1158/1055-9965.EPI-06-0449. [DOI] [PubMed] [Google Scholar]
- 16.García-Closas M, Malats N, Real FX, Welch R, Kogevinas M, Chatterjee N, Pfeiffer R, Silverman D, Dosemeci M, Tardón A, Serra C, Carrato A, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–42. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 17.Xing D, Qi J, Miao X, Lu W, Tan W, Lin D. Polymorphisms of DNA repair genes XRCC1 and XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population. Int J Cancer. 2002;100:600–5. doi: 10.1002/ijc.10528. [DOI] [PubMed] [Google Scholar]
- 18.Le Morvan V, Longy M, Bonaïti-Pellié C, Bui B, Houédé N, Coindre JM, Robert J, Pourquier P. Genetic polymorphisms of the XPG and XPD nucleotide excision repair genes in sarcoma patients. Int J Cancer. 2006;119:1732–5. doi: 10.1002/ijc.22009. [DOI] [PubMed] [Google Scholar]
- 19.Sturgis EM, Wei Q. Genetic susceptibility—molecular epidemiology of head and neck cancer. Curr Opin Oncol. 2002;14:310–7. doi: 10.1097/00001622-200205000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Zafereo ME, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Nucleotide excision repair core gene polymorphisms and risk of second primary malignancy in patients with index squamous cell carcinoma of the head and neck. Carcinogenesis. 2009;30:997–1002. doi: 10.1093/carcin/bgp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Spitz MR, Lee JJ, Huang M, Lippman SM, Wu X. Nucleotide excision repair pathway genes and oral premalignant lesions. Clin Cancer Res. 2007;13:3753–8. doi: 10.1158/1078-0432.CCR-06-1911. [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, Terry MB. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1614–9. doi: 10.1158/1055-9965.EPI-06-0218. [DOI] [PubMed] [Google Scholar]
- 23.Guan X, Sturgis EM, Lei D, Liu Z, Dahlstrom KR, Wei Q, Li G. Association of TGF-beta1 genetic variants with HPV16-positive oropharyngeal cancer. Clin Cancer Res. 2010;16:1416–22. doi: 10.1158/1078-0432.CCR-09-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji X, Sturgis EM, Zhao C, Etzel CJ, Wei Q, Li G. Association of p73 G4C14-to-A4T14 polymorphism with human papillomavirus type 16 status in squamous cell carcinoma of the head and neck in non-Hispanic whites. Cancer. 2009;115:1660–8. doi: 10.1002/cncr.24184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collett D. Modelling Survival Data in Medical Research. 2. Bristol, United Kingdom: Chapman & Hall; 2003. [Google Scholar]
- 26.Cox DR. Regression models and life tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 27.Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non-small-cell lung cancer. J Natl Cancer Inst. 2002;94:1091–9. doi: 10.1093/jnci/94.14.1091. [DOI] [PubMed] [Google Scholar]
- 28.Wang LE, Yin M, Dong Q, Stewart DJ, Merriman KW, Amos CI, Spitz MR, Wei Q. DNA repair capacity in peripheral lymphocytes predicts survival of patients with non-small-cell lung cancer treated with first-line platinum-based chemotherapy. J Clin Oncol. 2011;29:4121–8. doi: 10.1200/JCO.2010.34.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509:165–74. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility--a mini review. Int J Cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin M, Yan J, Martinez-Balibrea E, Graziano F, Lenz HJ, Kim HJ, Robert J, Im SA, Wang WS, Etienne-Grimaldi MC, Wei Q. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17:1632–40. doi: 10.1158/1078-0432.CCR-10-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, Gurubhagavatula S, Liu G, Park S, Neuberg DS, Wain JC, Lynch TJ, Su L, Christiani DC. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10:4939–43. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 34.Bradbury PA, Kulke MH, Heist RS, Zhou W, Ma C, Xu W, Marshall AL, Zhai R, Hooshmand SM, Asomaning K, Su L, Shepherd FA, et al. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics. 2009;19:613–25. doi: 10.1097/FPC.0b013e32832f3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.States JC, Reed E. Enhanced XPA mRNA levels in cisplatin-resistant human ovarian cancer are not associated with XPA mutations or gene amplification. Cancer Lett. 1996;108:233–7. doi: 10.1016/s0304-3835(96)04428-x. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Zhao H, Wei Q, Amos CI, Zhang K, Guo Z, Qiao Y, Hong WK, Spitz MR. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis. 2003;24:505–9. doi: 10.1093/carcin/24.3.505. [DOI] [PubMed] [Google Scholar]
- 37.Saldivar JS, Lu KH, Liang D, Gu J, Huang M, Vlastos AT, Follen M, Wu X. Moving toward individualized therapy based on NER polymorphisms that predict platinum sensitivity in ovarian cancer patients. Gynecol Oncol. 2007;107(1 Suppl 1):S223–9. doi: 10.1016/j.ygyno.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 38.Qiu L, Wang Z, Shi X, Wang Z. Associations between XPC polymorphisms and risk of cancers. A meta-analysis. Eur J Cancer. 2008;44:2241–53. doi: 10.1016/j.ejca.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 39.Strom SS, Estey E, Outschoorn UM, Garcia-Manero G. Acute myeloid leukemia outcome: role of nucleotide excision repair polymorphisms in intermediate risk patients. Leuk Lymphoma. 2010;51:598–605. doi: 10.3109/10428190903582804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, Yun J, Sones E, Mallik N, Lenz HJ. A xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8. [PubMed] [Google Scholar]
- 41.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced nonsmall-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 42.Ryu JS, Hong YC, Han HS, Lee JE, Kim S, Park YM, Kim YC, Hwang TS. Association between polymorphisms of ERCC1 and XPD and survival in non-small cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–6. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Gu J, Zhao H, Dinney CP, Zhu Y, Leibovici D, Bermejo CE, Grossman HB, Wu X. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408–15. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 44.Booton R, Ward T, Heighway J, Taylor P, Power F, Ashcroft L, Morris J, Thatcher N. Xeroderma pigmentosum group D haplotype predicts for response, survival, and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer. Cancer. 2006;106:2421–7. doi: 10.1002/cncr.21885. [DOI] [PubMed] [Google Scholar]
- 45.Ambrosone CB, Tian C, Ahn J, Kropp S, Helmbold I, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Chang-Claude J. Genetic predictors of acute toxicities related to radiation therapy following lumpectomy for breast cancer: a case-series study. Breast Cancer Res. 2006;8:R40. doi: 10.1186/bcr1526. [DOI] [PMC free article] [PubMed] [Google Scholar]


