Abstract
When bound to the vitamin D receptor (VDR), the active form of vitamin D, 1,25-dihydroxyvitamin D (1,25D ) is a potent regulator of osteoblast transcription. Less clear is the impact of 1,25D on post-transcriptional events in osteoblasts, such as the generation and action of microRNAs (miRNAs). Microarray analysis using replicate (n = 3) primary cultures of human osteoblasts (HOB) identified human miRNAs that were differentially regulated by > 1.5-fold following treatment with 1,25D (10nM, 6 hrs), which included miRNAs 637 and 1228. RT-PCR analyses showed that the host gene for miR-1228, low density lipoprotein receptor-related protein 1 (LRP1), was co-induced with miR-1228 in a dose-dependent fashion following treatment with 1,25D (0.1 – 10nM, 6hrs). By contrast, the endogenous host gene for miR-637, death-associated protein kinase 3 (DAPK3), was transcriptionally repressed by following treatment with 1,25D. Analysis of two potential targets for miR-637 and miR-1228 in HOB, type IV collagen (COL4A1) and bone morphogenic protein 2 kinase (BMP2K) respectively, showed that 1,25D-mediates suppression of these targets via distinct mechanisms. In the case of miR-637, suppression of COL4A1 appears to occur via decreased levels of COL4A1 mRNA. By contrast, suppression of BMP2K by miR-1228 appears to occur by inhibition of protein translation. In mature HOBs, siRNA inactivation of miR-1228 alone was sufficient to abrogate 1,25D-mediated down regulation of BMP2K protein expression. This was associated with suppression of pro-differentiation responses to 1,25D in HOB, as represented by parallel decrease in osteocalcin and alkaline phosphatase expression. These data show for the first time that the effects of 1,25D on human bone cells are not restricted to classical VDR-mediated transcriptional responses but also involve miRNA-directed post-transcriptional mechanisms.
Introduction
Integrated transcriptome and proteome analyses based on new methods for more accurate comparisons indicate that only 40–45% of the transcriptome is actually translated (1–3). This discordance occurs as a consequence of the abundance of noncoding RNA produced, and because mRNA is subject to multi-level, post-transcriptional regulation. These events are largely mediated by regulatory networks established by RNA-binding proteins and RNA species such as small non-coding microRNAs (miRNAs), that orchestrate the concerted production of complex post-transcriptional gene expression networks within specialized cells such as bone-forming osteoblasts (4). Single stranded mature miRNAs are known to regulate the post-transcriptional expression of genes by at least two mechanisms: 1) degradation and gene silencing of target mRNA transcripts, and 2) repression of mRNA translation via (partial) complementary base-pair binding with retainment of the RNA silencing complex (reviewed in (5)). As a single miRNA may target several genes, this is a very efficient mechanism for influencing biological responses. More than 1700 human miRNAs have been identified so far, targeting approximately 40–60% of genes in many cell types (6), but the functions and target genes for most of these miRNAs are still unknown.
Recent data from animal and cellular models suggest that miRNAs play a critical role in skeletal development, osteoclast function, osteogenic lineage progression, adipose tissue-derived stem cell development, and mesenchymal precursor and osteoblastic differentiation under both normal and disease conditions (7). The active form of vitamin D, 1,25-dihydroxyvitamin D (1,25D) also plays a pivotal in bone homeostasis. However, whilst there are extensive data concerning transcriptional regulation by 1,25D and its cognate nuclear vitamin D receptor (VDR) (8), relatively little is known about the interaction between 1,25D and miRNAs in bone cells. Studies using various non-skeletal neoplastic cells have reported induction of miRNAs by 1,25D (9,10), whilst also describing effects of miRNAs on VDR expression (11) and 1,25D signaling (12). In other cells miRNAs have been shown to target enzymes associated with the synthesis (13) and catabolism (11) of 1,25D. With this in mind, the aim of the current study was identify miRNAs in primary human osteoblasts (HOBs) that are induced or suppressed by 1,25D, and to assess the effects of 1,25D-mediated miRNA induction on osteoblastic phenotype and function.
Materials and Methods
Reagents and cell culture
Crystalline 1,25D (Biomol, Plymouth Meeting, PA USA) was reconstituted in absolute ethanol. Cells were maintained at 37°C in a 95% air/5% CO2 atmosphere until 80–90% confluent and then passaged. Primary HOBs and media were purchased from PromoCell GmbH (C-12720; Heidelberg, Germany) and supplemented (10%) with osteoblast growth (C-27001) and mineralization (C-27020) medium when specified. These cells were obtained from a single donor and stored at −80°C at passage 3. Individual aliquots of these cells were thawed and used for separate experiments to provide replicate analyses. HOB passage 8 cells were used in miRNA profiling using technical triplicates. For biological replication and target validation additional normal HOBs were obtained from hipbone biopsies from the UCLA Pediatric Bone Clinic. Human MG-63 osteosarcoma cells (ATCC, Manassas, VA USA) were cultured in a similar manner. For assays, cells were treated with 0–100nM 1,25D incubated with low supplements (1%) in 12-well plates. Three treatment conditions (each n=3) were analyzed per experiment. Ficoll-isolated peripheral blood mononuclear cells (PBMCs) derived from anonymous healthy donors were obtained from the UCLA Center for AIDS Research Virology Core/BSL3 Facility. Monocytes were enriched by adherence by incubating 5×106 PBMCs per well in 12-well plates for 2 hrs in RPMI (Invitrogen, Carlsbad, CA USA) with 10% FBS. Adherent monocytes were then washed and cultured overnight in RPMI with 10% FBS. Cells were then washed and treated with 1,25D in 1% FBS.
miRNA reagents, assays and sequences
Total RNA was purified using the mirVana™ miRNA Isolation Kit (ABI, Carlsbad, CA USA). For miRNA profiling, we utilized the 5th generation (human, mouse, rat) miRCURY LNA™ microRNA Array Kit (Exiqon Inc., Woburn, MA USA). Total RNA recovered from three independent experiments were quality checked using 1% formaldehyde-agarose gel electrophoresis and an Agilent bioanalyzer, and the microarrays were processed (i.e. samples labeled, arrays hybridized, washed and scanned) by the UCLA Clinical Microarray Core facility (http://www.pathology.ucla.edu/cmc). The 5th generation array contains approximately 1891 capture probes, covering all human, mouse and rat microRNAs annotated in miRBase 14.0, as well as all viral microRNAs particular to these species. In addition, this array contains capture probes for 385 new miRPlus™ human microRNA. TaqMan® MicroRNA Assays (U6 snRNA 001973; miR-1228, 002919; ABI) were optimized using 50ng of total RNA with the TaqMan MicroRNA Reverse Transcription Kit (ABI) following the manufacturer’s protocol. For miRNA detection, the PCR conditions were: 95°C (10min), 40 cycles 95°C (15sec; denature) and 60°C (1min; anneal/extend), and measured in quadruplicate. Relative miR-1228 levels were obtained by normalizing to U6 snRNA. Both negative control A (199020-04) and miR-1228 (426683-04) FAM-labeled miRCURY LNA™ microRNA enhanced power inhibitors were purchased from Exiqon and stored as 50μM frozen aliquots. Transfection of inhibitors was performed using the siPORT™ NeoFX™ reagent (ABI) in 12-well (50 pmole) or 6-well (75–150 pmole) tissue culture plates (Corning® Costar®; Sigma Aldrich, St. Louis, MO USA) following the manufacturer’s protocol. Sequences were obtained from the mirbase.org registry: MI0006318 (miR-1228 stem loop), MIMAT0005582 (miR-1228* degraded mature strand), MIMAT0005583 (miR-1228-3p more abundant mature strand, performed target scan), MI0003652 (miR-637 stem loop), MIMAT0003307 (miR-637-3p mature strand, performed target scan).
Cell fractionation, Western blot analysis and fluorescence imaging
Treated cells were harvested and washed with ice-cold PBS. Cell lysates were prepared in RIPA buffer (Sigma Aldrich) with 1X ProteoBlock protease inhibitor cocktail (Fermentas, Glen Burnie, MD USA) and 1mM PMSF. Whole cell lysates (20μg) were separated by SDS-PAGE. Human reactive mouse monoclonal antibody to BMP2K (ab55048; Abcam Inc., Cambridge, MA USA) was used at a 1:1000 dilution. Anti-human β-actin antibody (sc-81178; Santa Cruz Biotechnology, Santa Cruz, CA USA) was used to monitor loading. I-Block™ blocking reagent (ABI) was used during incubation steps. Transfection efficiency of the power LNA inhibitors was monitored using a Nikon Ti Eclipse inverted microscope (Nikon Instruments, Melville, NY USA).
mRNA quantitative real-time PCR (qPCR) analysis
cDNA was synthesized from 200ng total RNA by SuperScript Reverse Transcriptase III (Invitrogen) utilizing random hexamers. qPCR analysis was performed with a Stratagene MX-3005P instrument utilizing TaqMan® system reagents from ABI (Table S1). Target genes were normalized to 18S rRNA expression. All cDNAs were amplified under the following conditions: 50°C for 2 min; 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. All reactions were performed in triplicate. Data presented as comparable arbitrary expression units.
Chromatin immunoprecipitation (ChIP) analyses
Four HOB/ChIP conditions were prepared: 1) IgG/15 min vehicle (ethanol), 2) IgG/15 min 10nM 1,25D3, 3) VDR/15 min vehicle, and 4) VDR/15 min 10nM 1,25D3. Formaldehyde-cross-linked chromatin samples were processed in 1% SDS cell lysis buffer and sonicated to yield fragmented chromatin. Samples were incubated with 10μg of anti-VDR antibody (sc-1008x; Santa Cruz Biotech, CA USA) and IgG (sc-2027). qPCR was performed with primers flanking putative vitamin D response element (VDRE) consensus sequences in the regulatory region of hsa-miR-637 (see Table S2). Primers used were: miR-637-VDRE cgttttccactcctgtcctc (forward) and atcatccctggggttttgag (reverse), and CYP24A1 cgaagcacacccggtgaact (forward) and ccaatgagcacgcagaggag (reverse); and VDRE non-specific hu-calponin agctaagagaagggcggaac (forward) and catctgcggaaagggtatgt (reverse), and hu-TRPV6 tgatgtccaggccctgaacaagt (forward) and gctccggggcagcctccatcagc (reverse). The amplified product was normalized to input DNA content and IgG immunoprecipitated samples, and presented as fold enrichment after vehicle or vitamin D treatments in duplication.
In silico nuclear receptor site prediction
For general nuclear receptor target prediction we used the program NubiScan (www.nubiscan.unibas.ch). Sequences were analyzed using the general weighted matrix for nuclear receptor halfsites (including the VDR canonical halfsites), whereby direct-repeat 3 sites were acquisitioned. For the search parameter, an automatic scan with a raw score threshold of 0.5 was used, where the optimal match would have a raw score of 1.
Multiple miRNA array analysis, target site prediction and functional clustering
Multiple sample analysis to normalize, data adjust, statistically test and cluster was performed with the TM4 microarray software suite (www.tm4.org). For normalization of raw data, we utilized the locally weighted regression method to account for system-related variations in individual array signal intensities to rescale and reveal biological variations using the Midas software. Statistical Analysis of Microarrays (SAM) was performed using TIGR MeV running a two class unpaired analysis with arrays sorted into two groups. For hierarchal cluster analysis, we performed average linkage and Euclidean distance metric methods, and the plots were generated using the MeV software. P-values were computed from the theoretical t-distribution. miRNAs with P-values below a critical 0.05 value were selected for cluster analysis. We used three well-established miRNA target-prediction software: miRanda (14), TargetScan (15), and DIANA-MICROT (16) to predict bone-related targets for the human miRNAs. For miRanda, predictions were ranked based on the predicted efficacy of targeting as calculated using the mirSVR score (i.e. based on site-type, 3′ pairing, local AU, length of UTR and position contributions) of the sites. mirSVR is a regression model that computes a weighted sum of a number of sequence and context features of the predicted miRNA::mRNA duplex, and the scores can be interpreted as an empirical probability of down regulation. The DAVID Gene Functional Classification tool was utilized to determine functional clusters of the miRNA targets (17).
Results
1,25D-dependent miRNA expression in primary human osteoblasts
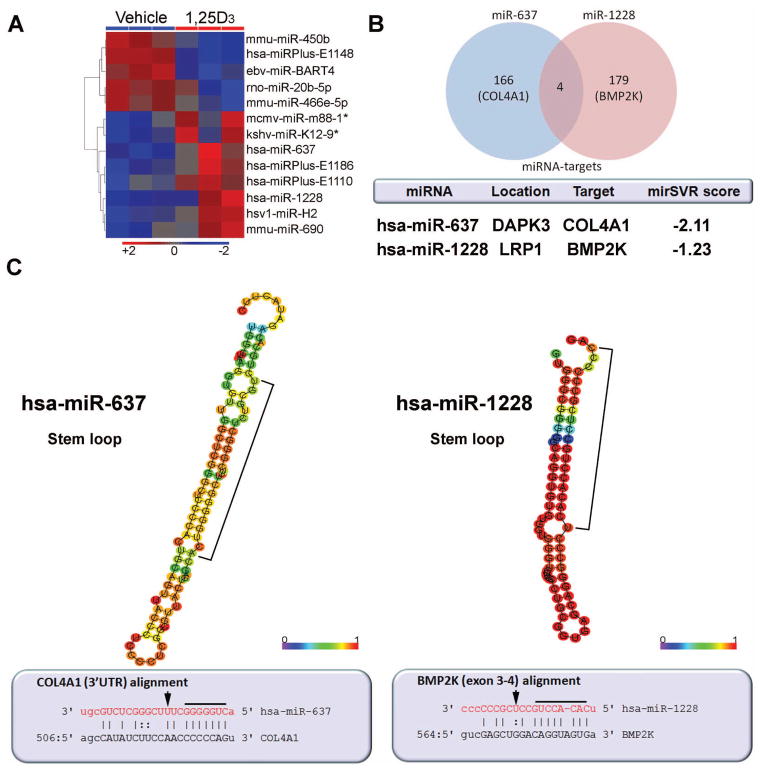
Cultures of primary HOBs at passage (P) 8 were treated with vehicle or 1,25D (10 nM) for 6 hrs, and RNA from these cells was then used for genome-wide miRNA expression profiling. Using n=3 replicate cultures of HOBs we identified 5 human miRNAs (either registered [(www.sanger.ac.uk/Software/Rfam/mirna] or proprietary) that were differentially regulated following treatment with 1,25D (Figure 1A, Table S3, Gene Expression Omnibus repository (www.ncbi.nih.gov/geo) accession number GSE34144). A series of cross-species/viral miRNAs were also identified signifying potential homology to identified/unregistered human miRNAs. These miRNAs may be homologous with either high or low similarity, and may have arisen in an orthologus manner due to speciation events. No further studies were therefore carried out using these miRNAs. Of the five human miRNAs shown to be regulated by 1,25D three were proprietary (has-miRPlus-E1110, -E1148, -E1186), with patented DNA sequences that are not part of the Sanger register. Although these “putative” mature species are detectable in microarray analyses, further studies are necessary to authenticate precursor miRNA forms. The only known miRNAs induced in HOBs following treatment with 1,25D were miR-637 and miR-1228. These miRNAs are documented in the Sanger registry and were therefore chosen for further studies. Analysis of the predicted targets for these miRNAs revealed four targets common to miR-637 and miR-1228 (TSC1, FAM98A, ANKRD52, CCND2), although individually these targets were not high-ranking candidates based on mirSVR scores (Figure 1B).
Figure 1. miRNA profiling of 1,25D-treated human osteoblasts.
A) Regulated miRNAs in primary HOBs treated for 6 hrs with 1,25D (10 nM). Heat plot represents statistically significant miRNA candidates (p<0.05), whereby three replicates were performed per treatment condition. Proprietary miRNAs are referred to as “miRPlus”. B) miRNA targets and salient features of the miRNAs of interest. The mirSVR score represents the predicted level of repression of potential mRNA target (described in Materials and Methods). C) Above, the optimal secondary structures in dot-bracket notation with minimum free energy. The structures are colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. Bracketed regions represent the mature miRNA sequence (RNAfold, rna.tbi.univie.ac.at). Below, miRNA:target pairs derived from miRBase (www.mirbase.org). In red letters are the mature miRNA nucleotides, while the target seed region is underlined. Arrows depict predicted “slicing/cleaving” nucleotides. hsa (Homo sapien), mmu (Mus musculus), ebv (Epstein Barr virus), rno (Rattus norvegicus), mcmv (mouse cytomegalovirus), kshv (kaposi sarcoma-associated herpesvirus), hsv1 (herpes simplex virus 1).
The salient features of miR-637 and 1228, including their host gene locations, potential bone-related biological mRNA targets and their likelihood of target suppression (mirSVR score) are shown in Figure 1B. The range of mirSVR scores for miR-637 and miR-1228 were −2.25-1.0, and −1.95-1.0 respectively. Both miR-637 and 1228 are localized intronically within their respective host genes (death associated protein kinase 3 [DAPK3] and low density lipoprotein receptor-related protein 1 [LRP1] respectively), and both are processed to pre-hairpin structures displaying unique sequence complementarities to the predicted targets once matured (Figure 1C). The 5-prime seed region for miR-637 generates perfect complementarity to the target type 4 collagen alpha 1 (COL4A1) 3′UTR, and the seed region of miR-1228 contains a bulge within its target bone matrix protein 2-inducible protein kinase (BMP2K) sequence spanning exons 3 and 4. Based on multi-organism genome-wide sequence comparisons (BlastN suite, blast.ncbi.nlm.nih.gov, and mirbase.org), human miR-637 belongs to a gene family on chromosome 19 that includes ITGB1BP3, PIAS4, DAPK3, and which is conserved in Pongo pygmaeus and Pan troglodytes, and therefore appears to be a primate-specific miRNA. By contrast, miR-1228 belongs to a gene family (including TMEMK14A, LRP1, STAT6, NAB2) on chromosome 12 which is conserved only in Pongo pygmaeus, but there is a mature miR-1228 ortholog on mouse chromosome 12 identified as mmu-miR-667.
Computational analysis identified COL4A1 and BMP2K as the most high-ranking bone-related targets for miR-637 and miR-1228 in 1,25D-treated HOBs (Figure 1B and Worksheet 1). These targets were identified using the mature miRNA sequences that most likely enter the RNA silencing complex for target degradation (see Materials and Methods). COL4A1 was the second-most likely target of miR-637 (mirSVR score = -2.11), while BMP2K revealed a lower level of predicted suppression with miR-1228 (mirSVR score = −1.23). In silico functional cluster analysis of the diverse miRNA targets revealed overlapping biological themes such as targeting of transmembrane proteins involved in cell adhesion, homeobox binding factors and zinc finger binding proteins in the regulation of transcription as potential repressive pathways regulated by 1,25D treatment (Worksheet 1). Specific to miR-1228 is a cluster of targets known to be serine-threonine protein kinases associated with a range of metabolic processes that includes BMP2K. Other well-conserved target clusters for miR-637/1228 include DNA-binding transcription factors that may play a role in the control of development.
Host gene and miRNA target regulation by 1,25D in primary HOBs
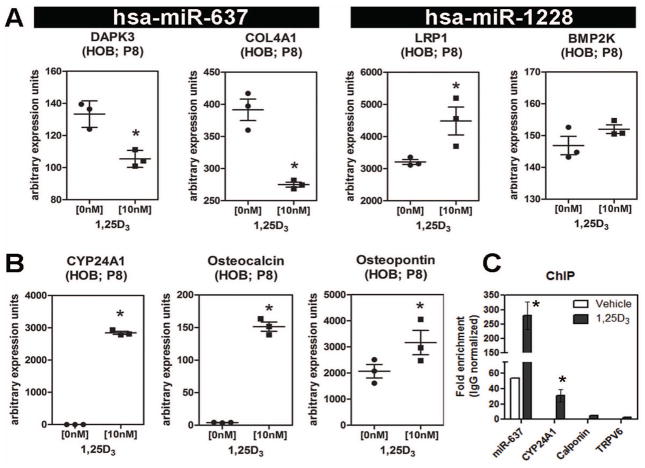
Further studies were carried out to assess 1,25D-mediated regulation of both miRNA host genes and predicted miRNA targets in HOBs. Treatment with 1,25D down regulated mRNA expression for the host gene (DAPK3), and the target for miR-637, (COL4A1) (Figure 2A). By contrast, treatment with 1,25D increased expression of mRNA for the miR-1228 host gene (LRP1), while the predicted target for miR-1228 (BMP2K) showed no change in expression (Figure 2A). The efficacy of 1,25D action in the cultured HOBs was assessed by analysis of markers such as 24-hydroxylase (CYP24A1), osteocalcin and osteopontin, known to be transcriptionally regulated by 1,25D in HOB (Figure 2B). The effects of 1,25D on miRNA host and target gene expression in HOBs were not observed in parallel studies of the human osteosarcoma cell line MG-63 even at relatively high concentrations (100 nM), indicating that the induction of miR-637 and miR-1228 is specific to primary HOBs (Figure S1A–B).
Figure 2. 1,25D-induced regulation of miRNA host genes and miRNA targets.
A) Effect of treatment with 1,25D (10 nM, 6 hrs) on mRNA for the host genes (DAPK3 and LRP1) and target genes (COL4A1 and BMP2K) for miR-637 and miR-1228 respectively in passage 8 (P8) HOBS. B) Effect of treatment with 1,25D (10 nM, 6 hrs) on mRNA for vitamin D metabolism (CYP24A1) and osteoblast (osteocalcin, osteopontin) markers in passage 8 (P8) HOBS. Data and error bars represent mean ± SEM (n=3; * = p≤ 0.05; Student t test) of comparable arbitrary expression units. C) ChIP analysis of the miR-637 promoter after 1,25D treatment (15min, 10nM) showing VDR enrichment. CYP24A1 VDRE positive control and non-specific controls (calponin, TRPV6) lacking VDREs in specific regions are included. Data are mean ± SEM (n=3 HOB cultures; * = p≤ 0.05; Student t test) of comparable arbitrary enrichment units.
Genomic sequence analysis of human DAPK3 revealed two specific “hot spots” for transcription factor regulatory activity (Figure S1C). The promoter region for DAPK3 and the immediate transcription site upstream region of miR-637 were the only two domains to house a cluster of transcription factor binding elements, suggesting that miR-637 is transcribed from its own promoter. To determine whether this was indeed the case, ChIP analysis of VDR binding to the regulatory region of miR-637 was carried out following 1,25D treatment of primary HOBs (Figure 2C). Quantification of data from three separate HOB cultures showed potent enrichment of VDR binding within the miR-637 regulatory region containing the putative VDRE. Interestingly, the VDR may also play a homeostatic gene regulatory role as it was found to be comparatively enriched for miR-637 in the absence of 1,25D. We also surmised that the miR-1228 host gene LRP1 is a direct target of 1,25D, which results in the concomitant up-regulation of the intronic miR-1228. These observations are supported through in silico analysis of the promoter regions of DAPK3 and LRP1 and their respective miRNAs that identified several potential vitamin D response elements (VDREs) of the direct 3 repeat type (Table S2). The promoter regions for the host gene LRP1 and miR-637 contain the most probable and highest-ranked VDR target sites near the transcriptional start site and start of the miRNA sequence, respectively. On the other hand, the upstream regions for DAPK3 and miR-1228 harbored predicted VDREs with low-ranking scores. These in silico data, although predictive in nature, suggest potential mechanisms by which 1,25D is able to regulate expression of miR-637 and miR-1228, and their host genes in primary HOBs.
Analysis of the impact of 1,25D-induced miRNAs on HOB function
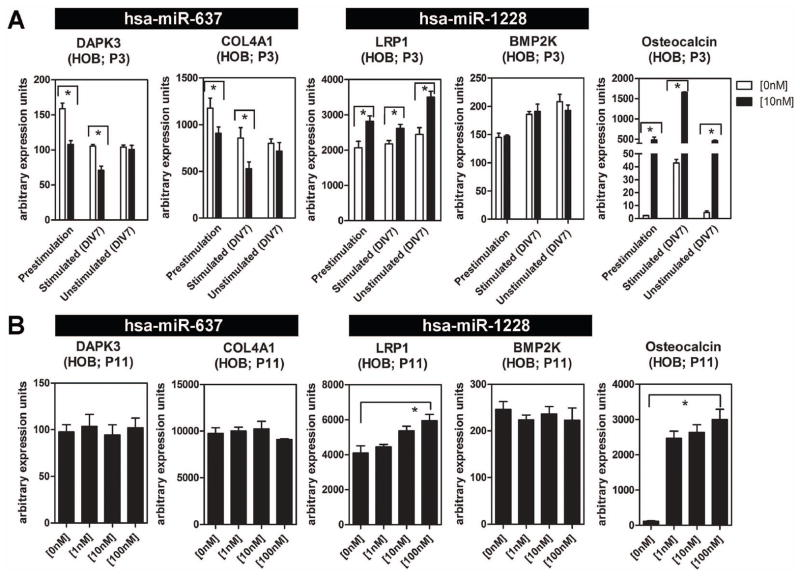
To determine the functional importance of miR-637 and miR-1228 in osteoblasts, studies were carried out to characterize expression and regulation of host and target genes for the miRNAs under different HOB culture conditions. For initial studies, the effects of 1,25D on miRNA host and target were assessed in early (P3) and late (P11) passage HOBs (Figure 3). In each case HOBs were cultured with vehicle or 1,25D (0–10 nM, 6 hrs) under pre-stimulation (proliferating), osteogenic (differentiating) and non-osteogenic media conditions. The non-osteogenic condition represents HOBs cultured for the same period of time as differentiating HOBs but without osteogenic stimulus. The miR-637 host gene DAPK3 and target COL4A1 were down-regulated by 1,25D, but only in early passage P3 cells, and only under pre-stimulation or osteogenic culture conditions. By contrast, the miR-1228 host LRP1 was induced by 1,25D in P3 and P11 HOBs and under all culture conditions. Despite this, expression of mRNA for BMP2K, the miR-1228 target, was unaffected by 1,25D under any HOB culture conditions (Figure 3A and 3B). These differential effects of 1,25D on miRNA host and target gene mRNA occurred despite potently induced expression of the classical 1,25D osteoblastic target gene osteocalcin under all HOB passage and culture conditions (Figure 3A and 3B). Interestingly, primary cultures of monocytes from human peripheral blood mononuclear, which are highly responsive to 1,25D-mediated immune responses, showed no 1,25D-mediated miR-1228 induction (Figure S3B), suggesting cell-type specificity of the miR-1228 effect.
Figure 3. 1,25D effects on miR host and target gene expression under different HOB culture conditions.
A) Effect of treatment with 1,25D (10 nM, 6 hrs) on mRNA for osteocalcin, and the host (DAPK3 and LRP1) and target (COL4A1 and BMP2K) genes for miR-637 and miR-1228 respectively in passage 3 (P3) HOBS. Cells were analyzed prior to osteogenic stimulation (pre-stimulation) and in the presence (stimulated) and absence (unstimulated) of osteogenic treatment. B) Effect of treatment with 1,25D (0 -100 nM, 6 hrs) on mRNA for osteocalcin, and the host genes (DAPK3 and LRP1) and target genes (COL4A1 and BMP2K) for miR-637 and miR-1228 respectively in passage 11 (P11) HOBS. Data and error bars represent mean ± SEM of arbitrary PCR units (n=3; for 3A and 3B) for 3A * = p≤0.05; Student t test, for 3B and 3C * = p≤0.05; 2-factor ANOVA with Bonferroni post hoc test.)
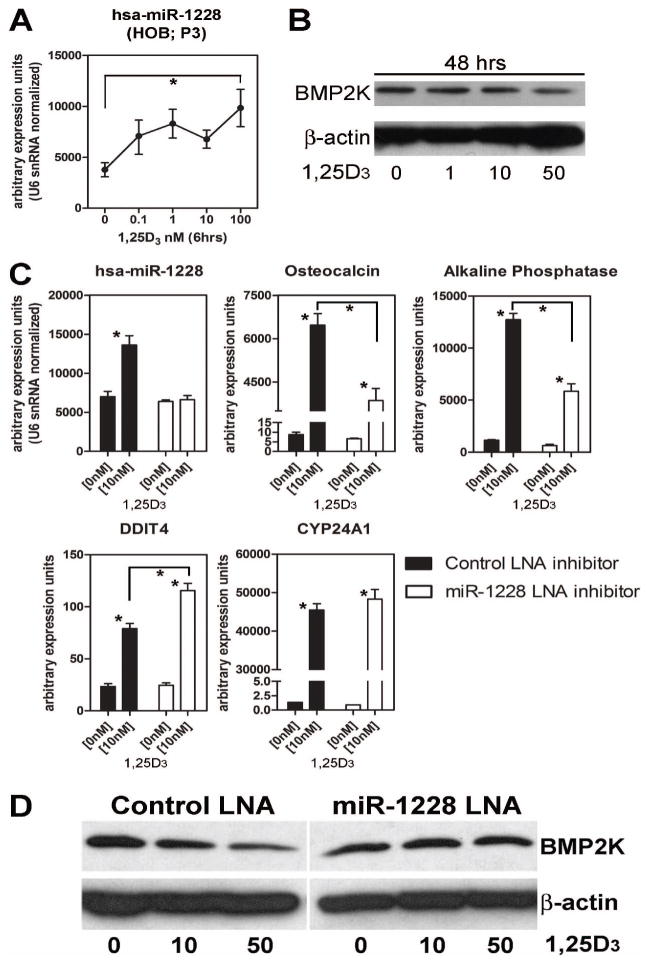
In view of the fact that the miR-1228 host gene LRP1 is regulated by 1,25D under a broad range of HOB culture conditions, further studies were carried out to assess the impact of this miRNA on target gene expression and HOB function. Dose-dependency studies showed that induction of HOB miR-1228 mRNA (Figure 4A), was associated with a concomitant decrease in protein for BMP2K following treatment with 1,25D, indicating that miR-1228 most likely acts via suppression of translation (Figure 4B). Analysis of the broader impact of miR-1228 as a regulator of HOB differentiation and function in the presence of 1,25D was carried out using locked nucleic acid (LNA) antisense oligonucleotide power inhibitors. These inhibitors are ultra-stable due to inclusion of an enzymatic degradation-resistant phosphorothioate backbone for longer lasting antisense activity. After 96 hrs post-transfection, HOBs receiving the non-specific scrambled LNA (control) showed an increase miR-1228 expression following treatment with 1,25D, similar to that observed in non-transfected HOBs. Transfection of the miR-1228 LNA inhibitor had no effect on baseline expression of miR-1228 but prevented its induction by 1,25D (Figure 4C). This effect of LNA inhibition of miR-1228 was paralled by suppression of 1,25D-induced expression of mRNA for osteocalcin and alkaline phopshatase (Figure 4C). By contrast, expression of the 1,25D-induced gene CYP24A1 was unaffected by LNA inhibition of miR-1228, whilst DDIT4 (DNA damage inducible transcript 4), a gene known to be induced by 1,25D in early-stage HOBs (18), showed increased 1,25D-induced expression in the presence of the miR-1228 LNA inhibitor (Figure 4C). Western blot analyses showed that treatment with 1,25D decreased BMP2K protein expression in a dose-dependent fashion in the presence of control scrambled LNA (Figure 4D). By contrast, HOBs treated with 1,25D and the miR-1228 LNA inhibitor showed no change in BMP2K protein. Collectively these data suggest that 1,25D-mediated suppression of BMP2K is dependent on miR-1228 to inhibit BMP2K protein translation. These effects of miR-1228 also appear to be essential for 1,25D-mediated regulation of osteoblast markers such as osteocalcin and alkaline phosphatase, but are independent of some direct transcriptional effects of 1,25D such as induction of CYP24A1.
Figure 4. miR-1228 mediates pro-differentiation effects of 1,25D in HOBs.
A) Effect of treatment with 1,25D (0 -100 nM, 6 hrs) on miR-1228 expression in P3 HOBs. Data represents mean ± SEM (n=3; * = p≤0.05; Student t test) of comparable arbitrary expression units. B) Western blot analysis of BMP2K protein in P3 HOBs treated with 1,25D (50nM) for 48 hrs. C) Effect of LNA inhibition of miR-1228 on 1,25D-induced expression of osteocalcin, alkaline phosphatase, CYP24A1 and DDIT4 mRNAs in P3 HOBs. Data are depicted as arbitrary units of mRNA levels (n = 3, means ± SEM). * = p≤0.05; 2-factor ANOVA with Bonferroni post hoc test. D) Western blot analysis of the effect of LNA inhibition of miR-1228 on BMP2K protein expression. For all experiments, eqi-molar concentrations of control or miR-1228 LNA inhibitors were transfected into P3 HOBs. Samples were collected at 96 hrs post transfection with 72 hrs of 1,25D treatment.
Discussion
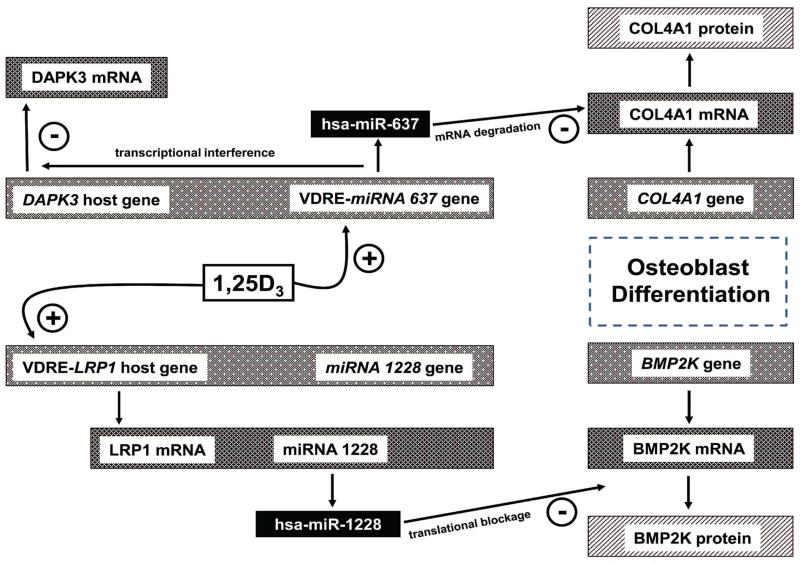
Recent reports have identified a range of regulatory actions for miRNAs in the generation and function of osteoblasts (19–24). However, despite its established effects on osteoblast function (25), little is known about the contribution of vitamin D to miRNA activity in these cells. Data presented in the current study identify two specific miRNAs, miR-637 and miR-1228, that are closely associated with 1,25D responses in HOBs in vitro. As outlined in Figure 5, the generation of these miRNAs from host genes in response to 1,25D is dependent on distinct mechanisms involving either: 1) conventional VDRE-mediated transactivation of a host gene (LRP1), and concomitant induction of its associated miR-1228; and 2) intronic VDRE-mediated induction of miR-637 in the absence of conventional transactivation of its host gene (DAPK3). In a similar fashion, the target gene effects of miR-637 and miR-1228 are mediated via different mechanisms, with miR-637 acting to stimulate degradation of COL4A1 mRNAs, whilst miR-1228 inhibits translation of the BMP2K protein. These observations highlight an entirely new repertoire of 1,25D functions in bone and suggest that miRNAs are key players in fine-tuning the effects of 1,25D on osteoblast differentiation and function.
Figure 5. Regulation and target activities of miR-637 and miR-1228 in 1,25D-treated HOBs.
Schematic representation of miRNA responses to 1,25D in osteoblasts showing: i) induction of miR-637 transcription via candidate intronic VDRE; ii) associated negative regulation of host gene DAPK3 mRNA transcription; iii) associated inhibition of target gene COL4A1 expression at the level of mRNA degradation; iv) 1,25D induced transcription of the miR-1228 host gene LRP1, with parallel induction of the mirtron miR-1228; v) miR-1228 mediated suppression of BMP2K protein expression by inhibition of translation. The effect of 1,25D on regulation of each miRNA activity is influenced by cellular aging as well as osteogenic stimulation. In this way 1,25D promotes HOB differentiation and function via direct effects on gene expression and by indirect regulation of gene expression by specific miRNAs.
The miR-1228 donor LRP1 is a multifunctional endocytic clearance receptor from the low density lipoprotein receptor (LDLR) family that activates signaling pathways through multiple cytosolic adaptor and scaffold proteins (26). Through its extracellular domain, LRP1 mediates endocytosis of more than 40 different ligands, with diverse biological roles (27). Within skeletal tissues LRP1 has been detected in osteoblasts and chondrocytes (28,29), with a potential function in the delivery of lipoproteins and vitamin K to bone (29). To date, no skeletal phenotypes in mice or humans have been attributed to LRP1 knockout; homozygous null mice exhibit embryonic lethality (30). Given the availability of conditional and Cre-specific LRP1 transgenic mouse lines (www.informatics.jax.org; MGI: 96828), it is possible that osteoblast-specific LRP1 knockout will better clarify the role of LRP1 in skeletal function in vivo. Other LDLR family members such as LRP5/6 play a key role in mediating Wnt signaling responses in the regulation of skeletal homeostasis (31), and LRP4 is known to control limb and craniofacial development (32). LRP1 is also known to mediate the canonical Wnt pathway in fibroblasts (33), and so it is possible that, independent of any actions of its miR-1228 product, the induction of HOB LRP1 alone will be sufficient to influence osteogenic potential via the Wnt pathway. Previous studies have described miRNA targeting of LRP1 expression (34), but to the best of our knowledge, our data are the first example of miRNA generation linked to LRP1 expression.
The miR-637 donor gene DAPK3 has been postulated to act as a tumor suppressor serine/threonine kinase (35), but it also plays a role in regulating cell morphology when over-expressed in mammalian cells (36). There have been no reported studies of DAPK3 in bone, but it is possible that suppression of DAPK3 may itself act as a regulatory switch to limit cell death during 1,25D-mediated differentiation of osteoblasts. In contrast to LRP1 and miR-1228, the induction of HOB miR-637 by 1,25D was associated with decreased expression of its host gene DAPK3. A minority of miRNA genes are located in the introns of protein-coding genes, preferentially in the same orientation as the mRNA, and notably both DAPK3 and miR-637 are in the same orientation on the reverse strand. Thus miR-637 is effectively its own VDRE-containing gene which can be transcribed to pre miRNA and then to mature miR-637 independent of its host DAPK3. This will lead to suppression of COL4A1 which is a predicted target of miR-637, but we hypothesize that miR-637 will also act to suppress expression of its host DAPK3 through transcriptional interference. This phenomenon of transcriptional interference has been observed elsewhere as a result of either tandem or convergent transcription of host and nested genes (37).
The miR-637 target, COL4A1, is found primarily in the basal lamina of blood vessels in many distinct skeletal locations (38). Primary HOBs and other osteoblastic cell lines express low levels of COL4A1 in vitro (39), and suppression of COL4A1 has been reported during early stages of osteoblast differentiation (40). In support of our data, miR-29b was found to be up-regulated during osteoblast differentiation and targets many collagens including col4a2 (41), the fibrillation partner of col4a1. It is therefore possible to hypothesize that 1,25D facilitates this process via induction of miR-637 and mRNA targeting of COL4A1. Recently, osteogenin, an extracellular matrix component of bone, was identified as a differentiation factor that initiates endochondral bone formation (42). Importantly, both osteogenin and transforming growth factor beta bind avidly to COL4A1 (43), suggesting a functional role in bone development. It is unclear whether COL4A1 expression during osteoblastic differentiation acts as an inhibitor of matrix mineralization as seen for other bone matrix proteins. Further studies are required to clarify the importance of COL4A1 during vitamin D-induced osteoblastic differentiation.
BMP2K was identified as a gene whose expression was increased during bone morphogenic protein 2 (BMP2)-induced differentiation of a mouse osteoblastic cell line (44). Stable expression of BMP2K (initially referred to as BMP2-inducible kinase) in MC3T3-E1 osteoprogenitor cells, suppressed mature osteoblast function, suggesting that BMP2K plays an important role in attenuating the program of osteoblast differentiation in mineralized tissue. In our study, we show that 1,25D treatment alone can decrease the levels of BMP2K in a BMP-independent manner. This effect was lost following LNA-knockdown of miR-1228, which also impaired 1,25D-mediated induction of alkaline phosphatase and osteocalcin, suggesting a key role for miR-1228 in osteoblast differentiation. In bone, vitamin D and analogs are known to modulate intracellular signaling molecules and the synthesis of ligands and receptors for both TGF-β and BMP pathways (45). Regulation of BMP2K protein by miR-1228 therefore provides another level of complexity by which vitamin D is able to interface with the BMP2 pathway.
Recent reports have highlighted a possible role for miRNAs in bone diseases such as primary osteoporosis (46,47). In this study, we identified two highly evolved miRNAs that are able to mediate novel regulatory responses to vitamin D in osteoblasts. The first, miR-637, is a primate-specific miRNA that was recently identified in the colorectal miRNAome (48). Although it was discovered 6 years ago, its biological role still remains elusive. Endogenous miR-637 was shown to be down-regulated in four hepatocellular carcinoma (HCC) specimens, and over-expression of miR-637 in vitro blocked cell growth and induced apoptosis of HCC (49). This antiproliferative, tumor suppressor function of miR-637 is consistent with the cell cycle arrest and pro-apoptotic effects of 1,25D at supraphysiological concentrations (50). More recent studies have highlighted a musculoskeletal function for miR-637 where its expression was increased during adipocyte differentiation in human mesenchymal stem cells (51). Conversely miR-637 was decreased during osteoblast linage commitment with osterix (osx), an early osteoblast-specific transcription factor, being a direct mRNA target. It is also interesting to note reports demonstrating osx-mediated induction of VDR in osteoblasts (52), suggesting that 1,25D-induced miR-637 may be part of a feedback mechanism controlling osx expression. Overall, these data suggest a potential bi-phasic effect of miR-637 expression, i.e. low during initial osteoblast commitment and higher during vitamin D-induced differentiation to potentially maintain low levels of osx.
The 19 nucleotide mature miR-1228 sequence is homologous to mouse miR-667 (Figure S3C). The 78% similarity between miR-1228 and miR-667 includes perfect similarity within the crucial seed region. Interestingly, mouse miR-667 is located on chromosome 12 within an intergenic region. The mouse LRP1 is located on chromosome 10, and harbors no intronic miRNAs. This suggests that although mice express a miRNA similar to miR-1228, host gene regulation of these miRNAs is likely to be different between the species. In particular it is unclear whether mouse miR-667 is regulated by 1,25D in the same fashion as miR-1228. Importantly, given that the seed region for mouse miR-667 is complementary to human miR-1228, the predicted targets for these miRNAs may be similar. In this way, miR-667 may provide an important future animal model to assess the bone impact of 1,25D-induced miRNA in vivo.
Initial studies identified miR-1228 from a mammalian screen of small RNA libraries from Rhesus macaque and human brain tissues (53), but no evidence of its function has been documented to date. Rather than being a classical miRNA, miR-1228 was predicted to be a mirtron; a short hairpin intron that uses splicing to bypass Drosha cleavage used for the generation of canonical animal miRNAs as an alternative precursor for miRNA biogenesis (54). Spliced mirtrons are exported out of the nucleus and then cleaved by Dicer and incorporated into RNA silencing complexes. Our study shows that miR-1228 affects one of its targets, BMP2K, via repression of protein translation, and this mode of miRNA-translation repression is known to exist in a range of cells (55). In future studies it will be interesting to assess in further detail the miRNA:mRNA interactions that are characteristic of miR-1228 and BMP2K. Notably there was only one miR-1228 binding site within the BMP2K sequence, suggesting that: 1) the singular occupancy of a miR-1228:BMP2K mRNA multiprotein complex is sufficient to ensure aberrant protein translation, or 2) this major duplex in combination with several different miRNAs help facilitate translational repression in an additive manner.
Data presented here provide the first evidence for miRNA involvement in 1,25D-mediated regulatory effects in bone. Further elucidation the role of miRNAs in osteoblast regulation may provide novel strategies for studying the pathogenesis and treatment of vitamin D-associated bone disorders such as rickets and osteoporosis.
Supplementary Material
A) The miR-637 host gene DAPK3 and target COL4A1 message levels were unaffected upon both low (0.1–10nM) and high (100nM) concentrations of 1,25D (6 hrs). Furthermore, the miR-1228 target BMP2K message levels were unaffected upon hormone treatment. Osteopontin, a known vitamin D-responsive target, was induced in the MG-63 cells upon treatment. B) The miR-1228 host gene LRP1 was unaffected by low or high concentrations of 1,25D in MG-63 cells. Furthermore, miR-1228 levels were unaffected upon low or high levels of 1,25D. CYP24A1, osteopontin, and DDIT4 (all known vitamin D target genes) transcripts were up regulated in MG-63 cells upon 1,25D (data not shown). Data are depicted as arbitrary units of mRNA (n=3, means±SEM). *p≤0.001; 2-factor ANOVA with Bonferroni post hoc test. C) Genomic sequence of human DAPK3 contains two “hot spots” for transcription factor activity. A -2KB upstream region from the transcriptional start site of DAPK3 until the 3′UTR was analyzed using MatInspector (www.genomatix.de). NR2F-EREF (nuclear receptor subfamily 2 factors–estrogen response element factor); BRAC (Brachyury gene, mesoderm developmental factor); NF1 (nuclear factor 1–Ebox); SMAD-AP1F (vertebrate SMAD family of transcription factors–AP1 factor); LEFF (lymphoid enhancer-binding factor-1); SREB (sterol regulatory element binding); GATA (GATA-binding factor 1); ETSF-SPF1 (human and murine ETS1 factors/Sweet potato DNA-binding factor with two WRKY-domains)
P3 HOBs were transfected with FAM-labeled LNA inhibitors and analyzed via microscopy post 96 hrs. In conjunction, the cells were treated with either equal volume vehicle (ethanol) or 1,25D for 72 hrs. Upon control LNA inhibitor treatment, there was no striking difference between the level of cytoplasmic labeling per cell between vehicle and 1,25D treatments. On the other hand, the miR-1228 LNA inhibitor revealed an increase in in situ labeling of endogenous miRNA (see enlargements) in the 1,25D-treated cells compared to vehicle. FAM (carboxyfluorescein)
A) Cultured normal primary HOBs (P3) obtained from a bone biopsy independent of the HOBs mainly used in the study show 1,25D regulation of miR-1228. Data are depicted as arbitrary units of miRNA (n=3, means±SEM). *p 0.001; 1-factor ANOVA with Bonferroni post hoc test. B) Primary human PBMCs show no regulation of miR-1228 by 1,25D. C) Predicted optimal secondary structure for mouse (mmu) miR-667 in dot-bracket notation. The structure is colored by base-pairing probabilities (RNAfold). Bracketed region represents the mature miRNA sequences.
Acknowledgments
Grant support: This work is supported by NIH grant 2R01AR037399-21 to JSA
TSL carried out all cell culture, array analyses and in vitro manipulation experiments, and also contributed to data analysis and manuscript writing. RFC carried out HOB cell culture all ChIP analyses and analysis of the resulting data. SR contributed to the analysis of array data and modeling of miRNA structure and target sequences. JSA developed initial experimental strategies and contributed to data analysis and presentation. MH initiated experimental design, analyzed data and co-wrote the manuscript with TSL. We would like to thank Dr. Xinmin Li (UCLA Dept. Pathology), Jamie Zhou (UCLA Dept. Pathology) and Dr. Martin Irmler (Helmholtz Zentrum Munich) for technical assistance with the miRNA microarrays. We would also like to thank Dr. Nancy Liu and Dr. Alejandro Garcia (both UCLA, Department of Orthopaedic Surgery) for assistance with RT-PCR and ChIP analyses. We also thank Dr. Renata Pereira (UCLA, Dept. Pediatric Nephrology) for providing normal HOBs for our analysis. We thank the center for AIDS Research Virology Core for supplying donor PBMCs (supported by National Institutes of Health award AI-28697 and by the UCLA AIDS Institute and the UCLA Council of Bioscience Resources).
References
- 1.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR., 3rd Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100(6):3107–12. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 4.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 5.Shruti K, Shrey K, Vibha R. Micro RNAs: tiny sequences with enormous potential. Biochem Biophys Res Commun. 2011;407(3):445–9. doi: 10.1016/j.bbrc.2011.03.058. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8(4):212–27. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haussler MR, Haussler CA, Bartik L, Whitfield GK, Hsieh JC, Slater S, Jurutka PW. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr Rev. 2008;66(10 Suppl 2):S98–112. doi: 10.1111/j.1753-4887.2008.00093.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1alpha,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol Cancer. 2011;10:58. doi: 10.1186/1476-4598-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8(5):736–41. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125(6):1328–33. doi: 10.1002/ijc.24459. [DOI] [PubMed] [Google Scholar]
- 12.Essa S, Denzer N, Mahlknecht U, Klein R, Collnot EM, Tilgen W, Reichrath J. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J Steroid Biochem Mol Biol. 2010;121(1–2):110–3. doi: 10.1016/j.jsbmb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, Graeber TG, Modlin RL. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. 2012;18(2):267–73. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39(Web Server issue):W145–8. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Lisse TS, Liu T, Irmler M, Beckers J, Chen H, Adams JS, Hewison M. Gene targeting by the vitamin D response element binding protein reveals a role for vitamin D in osteoblast mTOR signaling. FASEB J. 2010 doi: 10.1096/fj.10-172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda A, Amemiya T, Kondoh Y, Tashiro H, Okazaki Y. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368(2):267–72. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, Luo XH. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119(12):3666–77. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24–2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107(46):19879–84. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, Chen J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26(8):1953–63. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 2011;108(15):6139–44. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan G, Li G, Wang H, Lu G, Hu X, Jiang S, Li JN, Lin MC, Zhang YO, Kung H. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8(5) doi: 10.4161/rna.8.5.16043. [DOI] [PubMed] [Google Scholar]
- 25.van Driel M, Pols HA, van Leeuwen JP. Osteoblast differentiation and control by vitamin D and vitamin D metabolites. Curr Pharm Des. 2004;10(21):2535–55. doi: 10.2174/1381612043383818. [DOI] [PubMed] [Google Scholar]
- 26.Franchini M, Montagnana M. Low-density lipoprotein receptor-related protein 1: new functions for an old molecule. Clin Chem Lab Med. 2011;49(6):967–70. doi: 10.1515/CCLM.2011.154. [DOI] [PubMed] [Google Scholar]
- 27.Binder RJ, Han DK, Srivastava PK. CD91: a receptor for heat shock protein gp96. Nat Immunol. 2000;1(2):151–5. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 28.Kawata K, Eguchi T, Kubota S, Kawaki H, Oka M, Minagi S, Takigawa M. Possible role of LRP1, a CCN2 receptor, in chondrocytes. Biochem Biophys Res Commun. 2006;345(2):552–9. doi: 10.1016/j.bbrc.2006.04.109. [DOI] [PubMed] [Google Scholar]
- 29.Niemeier A, Kassem M, Toedter K, Wendt D, Ruether W, Beisiegel U, Heeren J. Expression of LRP1 by human osteoblasts: a mechanism for the delivery of lipoproteins and vitamin K1 to bone. J Bone Miner Res. 2005;20(2):283–93. doi: 10.1359/JBMR.041102. [DOI] [PubMed] [Google Scholar]
- 30.Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71(3):411–21. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 31.Bodine PV. Wnt signaling control of bone cell apoptosis. Cell Res. 2008;18(2):248–53. doi: 10.1038/cr.2008.13. [DOI] [PubMed] [Google Scholar]
- 32.Ohazama A, Porntaveetus T, Ota MS, Herz J, Sharpe PT. Lrp4: A novel modulator of extracellular signaling in craniofacial organogenesis. Am J Med Genet A. 2010;152A(12):2974–83. doi: 10.1002/ajmg.a.33372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terrand J, Bruban V, Zhou L, Gong W, El Asmar Z, May P, Zurhove K, Haffner P, Philippe C, Woldt E, Matz RL, Gracia C, Metzger D, Auwerx J, Herz J, Boucher P. LRP1 controls intracellular cholesterol storage and fatty acid synthesis through modulation of Wnt signaling. J Biol Chem. 2009;284(1):381–8. doi: 10.1074/jbc.M806538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song H, Bu G. MicroRNA-205 inhibits tumor cell migration through down-regulating the expression of the LDL receptor-related protein 1. Biochem Biophys Res Commun. 2009;388(2):400–5. doi: 10.1016/j.bbrc.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brognard J, Zhang YW, Puto LA, Hunter T. Cancer-associated loss-of-function mutations implicate DAPK3 as a tumor-suppressing kinase. Cancer Res. 2011;71(8):3152–61. doi: 10.1158/0008-5472.CAN-10-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998;18(3):1642–51. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21(6):339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker J, Schuppan D, Benzian H, Bals T, Hahn EG, Cantaluppi C, Reichart P. Immunohistochemical distribution of collagens types IV, V, and VI and of pro-collagens types I and III in human alveolar bone and dentine. J Histochem Cytochem. 1986;34(11):1417–29. doi: 10.1177/34.11.3772076. [DOI] [PubMed] [Google Scholar]
- 39.Pautke C, Schieker M, Tischer T, Kolk A, Neth P, Mutschler W, Milz S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 2004;24(6):3743–8. [PubMed] [Google Scholar]
- 40.Hong D, Chen HX, Yu HQ, Liang Y, Wang C, Lian QQ, Deng HT, Ge RS. Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells. Exp Cell Res. 2010;316(14):2291–300. doi: 10.1016/j.yexcr.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(23):15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paralkar VM, Nandedkar AK, Pointer RH, Kleinman HK, Reddi AH. Interaction of osteogenin, a heparin binding bone morphogenetic protein, with type IV collagen. J Biol Chem. 1990;265(28):17281–4. [PubMed] [Google Scholar]
- 43.Paralkar VM, Vukicevic S, Reddi AH. Transforming growth factor beta type 1 binds to collagen IV of basement membrane matrix: implications for development. Dev Biol. 1991;143(2):303–8. doi: 10.1016/0012-1606(91)90081-d. [DOI] [PubMed] [Google Scholar]
- 44.Kearns AE, Donohue MM, Sanyal B, Demay MB. Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J Biol Chem. 2001;276(45):42213–8. doi: 10.1074/jbc.M106163200. [DOI] [PubMed] [Google Scholar]
- 45.Lee HJ, Ji Y, Paul S, Maehr H, Uskokovic M, Suh N. Activation of bone morphogenetic protein signaling by a Gemini vitamin D3 analogue is mediated by Ras/protein kinase C alpha. Cancer Res. 2007;67(24):11840–7. doi: 10.1158/0008-5472.CAN-07-1549. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Li L, Moore BT, Peng XH, Fang X, Lappe JM, Recker RR, Xiao P. MiR-133a in Human Circulating Monocytes: A Potential Biomarker Associated with Postmenopausal Osteoporosis. PLoS One. 2012;7(4):e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26(1):72–8. doi: 10.1002/jbmr.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JF, He ML, Fu WM, Wang H, Chen LZ, Zhu X, Chen Y, Xie D, Lai P, Chen G, Lu G, Lin MC, Kung HF. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54(6):2137–48. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 50.Narvaez CJ, Welsh J. Role of mitochondria and caspases in vitamin D-mediated apoptosis of MCF-7 breast cancer cells. J Biol Chem. 2001;276(12):9101–7. doi: 10.1074/jbc.M006876200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC, Bian XW, Zhou J, Lin MC, Lu G, Poon WS, Kung HF. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011;22(21):3955–61. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Tang W, Li Y, Yang F, Dowd DR, MacDonald PN. Osteoblast-specific transcription factor Osterix increases vitamin D receptor gene expression in osteoblasts. PLoS One. 2011;6(10):e26504. doi: 10.1371/journal.pone.0026504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) The miR-637 host gene DAPK3 and target COL4A1 message levels were unaffected upon both low (0.1–10nM) and high (100nM) concentrations of 1,25D (6 hrs). Furthermore, the miR-1228 target BMP2K message levels were unaffected upon hormone treatment. Osteopontin, a known vitamin D-responsive target, was induced in the MG-63 cells upon treatment. B) The miR-1228 host gene LRP1 was unaffected by low or high concentrations of 1,25D in MG-63 cells. Furthermore, miR-1228 levels were unaffected upon low or high levels of 1,25D. CYP24A1, osteopontin, and DDIT4 (all known vitamin D target genes) transcripts were up regulated in MG-63 cells upon 1,25D (data not shown). Data are depicted as arbitrary units of mRNA (n=3, means±SEM). *p≤0.001; 2-factor ANOVA with Bonferroni post hoc test. C) Genomic sequence of human DAPK3 contains two “hot spots” for transcription factor activity. A -2KB upstream region from the transcriptional start site of DAPK3 until the 3′UTR was analyzed using MatInspector (www.genomatix.de). NR2F-EREF (nuclear receptor subfamily 2 factors–estrogen response element factor); BRAC (Brachyury gene, mesoderm developmental factor); NF1 (nuclear factor 1–Ebox); SMAD-AP1F (vertebrate SMAD family of transcription factors–AP1 factor); LEFF (lymphoid enhancer-binding factor-1); SREB (sterol regulatory element binding); GATA (GATA-binding factor 1); ETSF-SPF1 (human and murine ETS1 factors/Sweet potato DNA-binding factor with two WRKY-domains)
P3 HOBs were transfected with FAM-labeled LNA inhibitors and analyzed via microscopy post 96 hrs. In conjunction, the cells were treated with either equal volume vehicle (ethanol) or 1,25D for 72 hrs. Upon control LNA inhibitor treatment, there was no striking difference between the level of cytoplasmic labeling per cell between vehicle and 1,25D treatments. On the other hand, the miR-1228 LNA inhibitor revealed an increase in in situ labeling of endogenous miRNA (see enlargements) in the 1,25D-treated cells compared to vehicle. FAM (carboxyfluorescein)
A) Cultured normal primary HOBs (P3) obtained from a bone biopsy independent of the HOBs mainly used in the study show 1,25D regulation of miR-1228. Data are depicted as arbitrary units of miRNA (n=3, means±SEM). *p 0.001; 1-factor ANOVA with Bonferroni post hoc test. B) Primary human PBMCs show no regulation of miR-1228 by 1,25D. C) Predicted optimal secondary structure for mouse (mmu) miR-667 in dot-bracket notation. The structure is colored by base-pairing probabilities (RNAfold). Bracketed region represents the mature miRNA sequences.