Abstract
The catechol-o-methyl transferase (COMT) 158Val/Met variant has been suggested to play a role in COMT function. Epigenetic regulation of COMT may further influence the prevalence of metabolic syndrome in these patient populations. This study examined the correlation between COMT promoter methylation and metabolic syndrome in schizophrenia patients receiving atypical antipsychotic (AAP) therapy. DNA was extracted from peripheral blood samples of schizophrenia subjects screened for metabolic syndrome. Pyrosequencing was used to analyze two methylation sites of the COMT-s promoter region. Associations between AAP use, lifestyle variables, metabolic syndrome, and COMT genotype with peak methylation values were analyzed. Data are reported in 85 subjects. Methylation on CpG site 1 had a mean of 79.08% (± 4.71) and 12.43% (±1.19) on site 2. COMT genotype proved to be an indicator of COMT methylation status on site 1 (F(2,84) = 5.78, p=0.0044) and site 2 (F(2,84), p=0.027). A significant negative correlation between physical activity and COMT promoter region methylation was found in Val/Val homozygous patients (Site 1: p=0.013 and Site 2: p=0.019). Those homozygous for Met/Met showed a positive correlation between promoter site methylation and physical activity (Site 1: p=0.027, Site 2: p=0.005), and between CpG site methylation and metabolic syndrome (Site 1: p=0.002; Site 2: p=0.001). The results of this study suggest COMT promoter region methylation is largely influenced by COMT genotype and that physical activity plays a significant role in epigenetic modulation of COMT.
Keywords: COMT, Metabolic Syndrome, Methylation, Schizophrenia, Activity, Genotype
1. Introduction
Metabolic Syndrome is a complex disease currently affecting approximately 47 million Americans (1). Associated largely with cardiovascular disease, metabolic syndrome is commonly diagnosed by meeting three or more risk factors as defined by NCEP ATP III guidelines including high LDL/VDL lipid levels, increased insulin resistance, obesity, and high blood pressure (2). Previous studies have found that the incidence of metabolic syndrome in psychiatric patients currently on atypical antipsychotics (AAPs) is two to four fold higher than the general population (3). Recent evidence supports an association between folic acid related enzyme genetic variability (i.e. that of methylenetetrahydrofolate reductase (MTHFR)), aberrant homocysteine metabolism related to catecholamine-o-methyl transferase (COMT), and risk of metabolic syndrome in patients with schizophrenia (4–6). However, other lifestyle issues such as diet and exercise also continue to be implicated in the development of metabolic syndrome within schizophrenia.
Schizophrenia is a wide-spread debilitating psychiatric disease affecting over 3 million Americans. Despite the disease’s high prevalence and commonality, very little of its intricate genetic and environmental etiology is truly understood. Along with external influences, such as living conditions, dietary intake, and past drug use, several genetic factors have been suggested to play a part in the disease’s manifestation in an individual. Interest in COMT has increased among researchers in psychiatry and neuroscience due to its role in regulating catecholamine levels (i.e. dopamine) in brain tissue and cognitive function (7). A deficiency in catecholamine metabolism is likely related to disease states like schizophrenia that are characterized by disruptions in catecholamine neurotransmission. The role of a common variant in the COMT promoter region—158Val/Met (rs4680)— has been extensively examined in multiple psychiatric disorders (8,9). Biochemical kinetic studies have shown that those homozygous with the Val allele, classified as having a “G/G” or “Val/Val” genotype, metabolize dopamine at significantly higher rates than those with the Met variant, ultimately resulting in lower synaptic dopamine levels (10). While current evidence suggests that the presence of the COMT Met variant does not indicate a risk factor for schizophrenia etiology (11), meta-analysis regarding the gene’s actual role in the disease remains inconclusive (12). Although genotype may not directly constitute a schizophrenia risk factor, data indicates that COMT functionality does play a role in the disease manifestation and treatment outcomes (13). Work related to COMT’s methylation suggests that in addition to genotype, epigenetics may also regulate COMT activity (14). Given COMT additional role in homocysteine regulation, which has been linked to cardiovascular disease, a greater understanding of COMT’s role in metabolic syndrome within schizophrenia is needed, specifically in relation to genetically regulated methylation differences.
The aim of this investigation was to assess the relationship between COMT promoter methylation status and metabolic syndrome in patients receiving AAP therapy in a cross-sectional analysis. We have postulated that decreased COMT promoter methylation status in peripheral blood-originated DNA samples is associated with metabolic syndrome in a schizophrenia patient population and that physical activity may modulate this relationship.
2. Methods
2.1 Human Subjects and Collection of Samples
As part of this study, subjects were included if they currently had a DSM-IV diagnosis of schizophrenia, were between the ages of 18–90, and were currently stable on an antipsychotic for at least 6 months. Subjects were excluded if they lacked the ability to give informed consent or had any alcohol addiction or illicit drug use within the month, which was confirmed through the subject’s medical records and consultation with their primary prescriber. Subjects meeting these criteria were then seen in the Michigan Clinical Research Unit (MCRU) at the University of Michigan Hospitals and Clinics where they underwent informed consent and the study assessments. This study was approved by the University of Michigan Institutional Review Board and carried out in accordance with the Declaration of Helsinki (ClinicalTrials.gov Identifier: NCT00815854).
2.2 Clinical Data Collection
Physical and physiological function parameters, including a physical exam, dietary questionnaire, cigarette smoking status, and physical activity assessments, were acquired at the University of Michigan Clinical Research Unit (MCRU) upon subject consent and IRB approval. Pre-laboratory assessment measures included a schizophrenia diagnosis via a Structured Clinical Interview for DSM Diagnoses (SCID) and dietetic assessments of height, weight, and hip, and waist circumference. Blood pressure was measured and a BMI was calculated. Fasting blood glucose, serum folate, B12, homocysteine, insulin, hemoglobin A1c, and lipids (total cholesterol (TC), high and low density lipoproteins (HDL and LDL)), levels were collected via blood samples. The blood sample was also used for genetic and methylation analysis. Upon review of patient data, the status of metabolic syndrome of each patient was assessed. Metabolic syndrome was defined via NCEP ATP III guidelines of having three or more of the following (2): blood pressure (≥ 130/85 mmHg); fasting blood glucose (≥ 100 mg/dL); large waist circumference (men ≥ 40″, women ≥ 35″); low HDL cholesterol (men < 40 mg/dL, women < 50 mg/dL), 0triglycerides ≥ 150 mg/dL.
A medication history including over the counter (OTC) and herbal supplement usage was collected via questionnaire and review of subject records. Subjects receiving clozapine, olanzapine, risperidone, paliperidone or quetiapine were considered to be receiving an AAP. All atypical antipsychotic drugs were standardized by converting dosage regimens to chlorpromazine equivalents based on a mg/kg basis, (i.e., dose in chlorpromazine equivalents multiplied by the # of years used)/100) (15). Each subject was asked to quantify the number of cigarettes smoked per day and the amount of time they have smoked to obtain a pack-year history for each subject. Non-smokers (as defined by our research group as having no cigarette use within the past 12 months) were questioned on their past smoking history, including amount and duration of exposure.
A total activity score was computed based on a previously designed questionnaire (16). The subjects were asked to record the total “strenuous activity” (i.e., jogging, aerobics, swimming, and physical labor), “moderate activity” (i.e., housework, light jogging, painting, etc.), and “mild activity,” (i.e., walking), in minutes per week and the amount of times engaged in such activity per week. When completing this assessment, subjects were asked to report on their physical activity for the week prior to their current study visit which was when the DNA sample was obtained. A final score (in metabolic equivalent (MET/minute) was computed by multiplying the time for each activity (in minutes) by a metabolic equivalent score (i.e. 3, 5 or 7 METs), as defined previously (16).
2.3 Assessing Genotype and Methylation Status
Upon purification of the DNA obtained through whole blood-cell samples, the COMT-s allele—including promoter regions–was amplified via PCR. Val/Met variant status was determined via pyrosequencing (sequence templates available upon request), and samples were stored at −20C. A DNA bisulfite conversion process was conducted in vivo to assess the methylation status of the COMT-s promoter region using the peripheral DNA samples. Using EZ DNA Methylation-Gold™ Kit (Zymo Research, Irvine CA), the methylated CpG sites were converted to a CG group while un-methylated CpG islands were converted to uracil and were detected as thymine following polymerase chain reaction (PCR) and pyro-sequencing protocol. Two primers- a standard forward (5′ – GAT GGG TTG TAG GAT GAA TTC G – 3′) and biotinylated reverse (5′-/5Biosg/AAA CAC TAA CGC CCC TCC CC – 3′) were used and optimized for the PCR amplification of the two methylation sites (Site 1 and Site 2) within the COMT-s promoter gene using pyrosequencing technology based on the sequencing primer 5′ – GTA ATA TAG TTG TTA ATA GTA GA 3′ (14). Pyro-sequencing identified the defined CpG islands as a percentage of methylation for each subject. In general DNA regions with higher methylation may be less apt to be expressed (17).
2.4 Statistical Analysis
Baseline differences between those with metabolic syndrome and those without were determined through the use of simple student t-tests for continuous variables (i.e., weight, waist circumference) and chi-squared for dichotomous variables (i.e,, smoking status, AAP use). ANOVA was used to determine the relationship between COMT promoter region methylation at both sites 1 and 2 and the dependent variables of metabolic syndrome, COMT genotype (i.e Val/Val, Val/Met, and Met/Met), and interactions. Linear regression was used to determine the relationship between percent methylation at sites 1 and 2, total activity score, COMT genotype and interactions. These analyses used an allele load model followed by a secondary correlation analysis to examine methylation differences stratified by genotype. All analyses were controlled for any baseline differences between groups. A two tailed p-value < 0.05 was predetermined as significant with the sample size used in this study. Statistical analysis was performed with JMP 9® and values are reported as mean ± S.D.
3. Results
A total of 85 peripheral DNA samples were sequenced and analyzed. The age range of subjects was 22–70, with a mean of 45.36 (±11.74). Male subjects constituted 64.7% of the study population (n=55) and 80.1% (n=68) were classified as AAP users, and 36% (n=31) met the criteria for metabolic syndrome (2). There were no differences noted in age, race, or sex between genotype groups. Mean methylation on site 1 was 79.08% (± 4.71) (range 66.94% – 88.1%), and mean methylation on site 2 was 12.43% (± 1.19) (9.96% –15.43%). CpG percent methylation at site 1 showed considerably greater methylation than that of site 2 a finding that parallels previous COMT CpG methylation studies (14,18). Every sample showed some degree of methylation at both CpG sites.
3.1 COMT Promoter Methylation and Val/Met Genotype
COMT alleles were broken down into 3 genotypes: 18 Met/Met (“A/A”, lower enzyme activity), 39 Val/Met (“A/G”), and 28 Val/Val (“G/G,” higher enzyme activity) and were found to be in Hardy-Weinberg equilibrium (χ2=0.41. p=0.52) COMT genotype proved to be a strong indicator of COMT methylation status on site 1 (F(2,84) = 5.78, p=0.0044) and site 2 (F(2,84) = 3.79, p=0.027). When examining methylation status as a function of genotype, a relationship is found were individuals with the Met variant on one or both allele exhibit increased percentage of methylation relative to those without variant (Figure 1).
Figure 1.
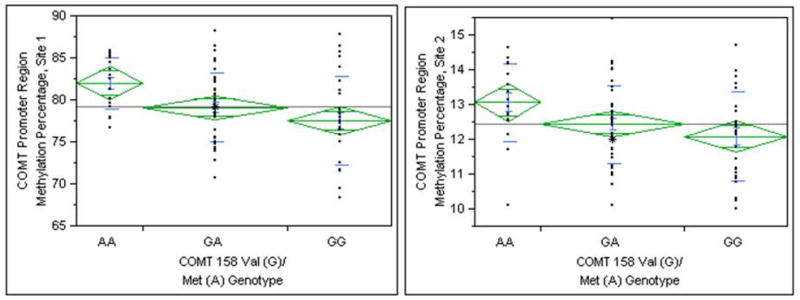
Percent methylation on the COMT promoter region versus COMT 158Val/Met genotype.
3.2 Percent Methylation versus Metabolic Syndrome
When methylation was examined in relation to the presence or absence of metabolic syndrome, percent methylation on either promoter CpG sites revealed a significant relationship in the entire population with the COMT genotype and metabolic syndrome variable interacting to predict methylation at site 1 (F(3,81) = 5.89 p<0.0001) and site 2 (F(3,81) = 5.33 p=0.0002). Specific to both of these models, the interaction between metabolic syndrome and COMT genotype remained significant after controlling for each variable separately, with both the Val/Val and Met/Met genotypes being statistically different (site 1: p= 0.002 and site 2: p=0.001) from each other based on presence or absence of metabolic syndrome. These differences can be seen when the data is broken down by specific genotypes. The Val/Val genotypes with metabolic syndrome had a significantly higher methylation average on site 1 (80.8% ± 4.03) than those without the metabolic syndrome complex (74.97% ±4.8) (p=0.002) (Figure 2). The same trend proved true on site 2, with 13.04 ±0.9 % methylation in those with metabolic syndrome and 11.36 ±1.0 % methylation in those without (p=0.001). Those with the Met/Met genotype did not have significantly higher methylation regardless of metabolic syndrome on site 1 (Yes: 80.943 ±2.99, No: 82.99 ±3.15; p=0.1895) or on site 2 (Yes: 12.87 ±1.5, No: 13.3 ± 0.76; p=0.468.). This finding shows a distinct difference between homozygous carriers of the Val/Met variant with regards to methylation percentages in patients with metabolic syndrome. These data show that each genotype has a unique methylation profile with regards to metabolic syndrome (Figure 2).
Figure 2. Breakdown of methylation versus metabolic syndrome, grouped by COMT 158Val/Met Genotype.
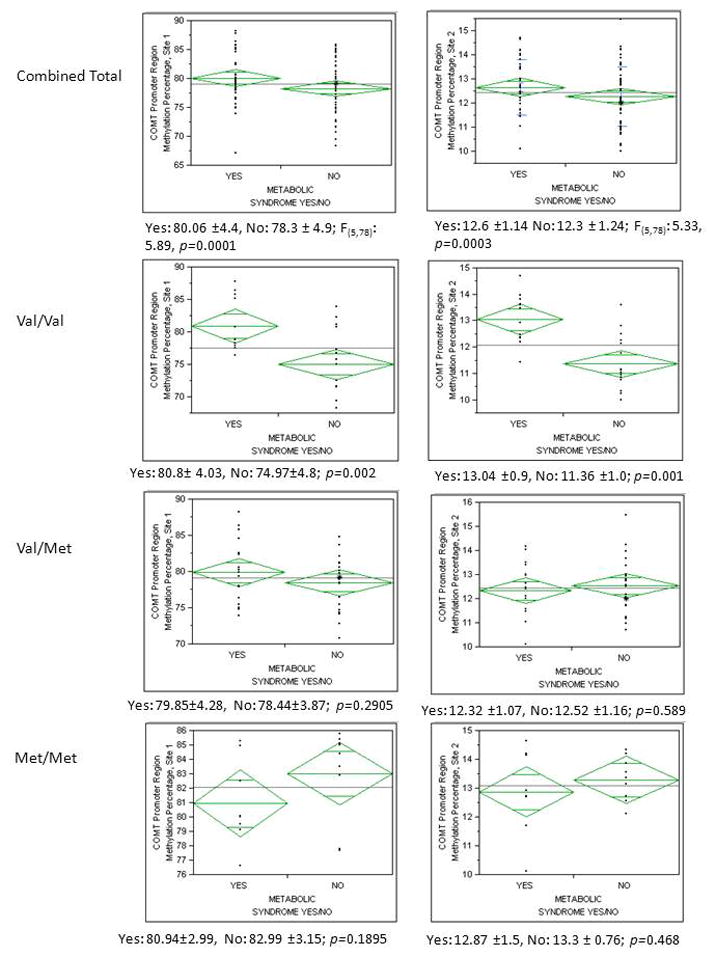
The first row represents the entire group broken out by presence or absence of metabolic syndrome. The next three rows are broken down by the COMT Val/Val (A/A), Val/Met (A/G), and Met/Met (G/G) genotypes. Column 1 represents relationships with COMT methylation at position 1 and column 2 represents COMT methylation at position 2.
3.3 Methylation compared to smoking status, serum folate, fasting glucose, serum homocysteine, and AAP use
No significant relationships were found between methylation status on the two promoter CpG sites analyzed and smoking status, serum folate, fasting glucose, or AAP use for both total populations and when subdivided by genotype (data not shown). When broken down by genotype, individuals with the Val/Val genotype showed a significant relationship between percent methylation on site 1 and homocysteine levels (F(1,26): 5.1214, p=0.032), in which higher homocysteine levels were directly related to higher methylation percentages on CpG site 1. CpG site 2 had a similar, yet non-significant trend (F(1.26): 2.13, p=0.15). The remaining genotypes did not show a significant relationship between homocysteine levels and methylation values.
3.4 Promoter Methylation and Total Activity Score
Total activity score averaged 2645 ± 2473 MET/minute (Range: 0–12420, n=82). Overall a significant relationship was found between COMT promoter methylation, COMT genotype and physical activity using the TAM2 scale (site 1: F(3,81): 7.97, p=0.0001 and site 2: F(3,81): 6.79, p=0.0004) whereas a significant interaction between genotype and physical activity was found (Site 1: p = 0.005, Site 2: p = 0.001). This relationship can be better seen, when broken down by genotype. For those with the Val/Val genotype methylation at both site 1 and site 2 showed a negative correlation with total activity score (p=0.013, r2=0.27 for site 1 and p=0.019, r2=0.39 for site 2) (Figure 3), while for those with the Met/Met genotype a positive correlation was shown between the total activity score and COMT methylation (p=0.027, r2=0.21 for site 1, and p=0.005 r2=0.19 for site 2). Those heterozygous for the variant showed a linear non-significant relationship between activity score and COMT methylation.
Figure 3. Linear correlations between Total Activity Score and COMT promoter methylation, grouped by COMT 158Val/Met Gentoype.
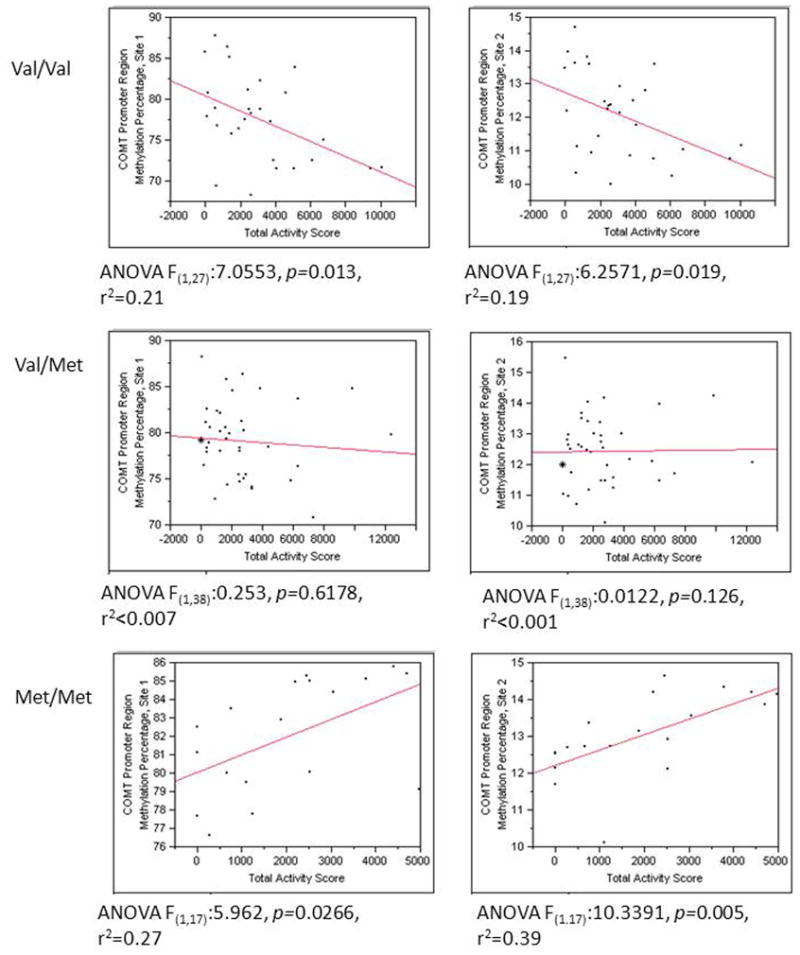
Column 1 represents relationships with COMT methylation at position 1 and column 2 represents COMT methylation at position 2. Overall for those with a Val/Val (A/A) genotype a negative relationship was found between Total Activity score and COMT methylation, while for those with a Met/Met (G/G) genotype a positive relationship was found.
4. Discussion
Epigenetic influences on genomic regulation have become a focal point in schizophrenia research. Studies have suggested that the methylation status of CpG dinucleotide islands in both soluble (S) and membrane bound (MB) COMT promoter regions may play a role in disease manifestation with hypomethylation of promoter region as a major risk factor for schizophrenia (17,19). Such epigenetic alterations of the COMT gene could potentially impair the individual’s ability to transcribe and express adequate levels of the enzyme, which may further impair additional downstream pathways involved in catecholamine and nutrient metabolism.
COMT plays a role in folic acid metabolism, assisting in the conversion of methionine to homocysteine – a key cofactor in the folic acid metabolic pathway. This pathway has been well documented and suggests that high levels of homocysteine may result from decreased levels of folic acid or presence of the COMT Val allele (20). Elevations in homocysteine have been associated with an increased risk of cardiovascular diseases, including congestive heart failure, and thrombotic and atherosclerotic vascular disease (21). The genetic etiology of folate deficiency has been studied extensively and many key variants have been found responsible for differences in folate levels among populations. Specifically, the MTHFR 667C/T (rs1801133) variant has been found to be an important marker in folate metabolism (22). The presence of a MTHFR C667T variant may contribute to a 35% reduction in folate metabolism, while a TT genotype results in as much as 70% reduction (6). Similar variants involved in folate metabolism, such as the COMT Val/Met, as well as the functional status of the COMT promoter gene, could prove to be important biological markers for metabolic syndrome development in schizophrenia (4,6) and may be related to overall Homocysteine levels (20). Decreased functionality and/or transcription of COMT based on promoter CpG dinucleotide methylation may play a role in lowered folate and homocysteine levels in schizophrenia patients. Given COMT’s role in the catabolism of dopamine and the effects of this variant on cognitive function within schizophrenia (13), this key metabolic process may serve as a very important linkage between cardiovascular disease and schizophrenia pathology. Understanding the relationship between environment, lifestyle, and dietary forces that affect metabolic syndrome development in general as well as the epigenetics of the illness itself, will assist clinicians in preventing and/or treating metabolic syndrome in high-risk psychiatric patients.
Overall we found that COMT genotype proved to be a strong indicator of COMT methylation status on both promoter CpG sites; a finding consistent with previous studies (23,24). It is possible that the higher methylation seen on both CpG sites with the Met/Met genotype may be associated with reduced gene expression, which others have hypothesized as well (23).
Additionally this is the first cohort study designed to assess the relationship of COMT promoter regulation methylation status with metabolic syndrome in living schizophrenia spectrum patients receiving atypical antipsychotic pharmacotherapy. Our data show a significant difference in COMT promoter region methylation between those with metabolic syndrome and those without in the Val/Val genotype population. Compared to methylation differences seen based on genotype alone, it appears that presence of the metabolic syndrome within Val/Val genotypes creates COMT methylation levels similar to that seen in Met/Met individuals without metabolic syndrome. This may indicate that physiological presence of metabolic syndrome within individuals with the Val/Val genotype may alter epigenetic responses which may ultimately affect COMT functionality. It is feasible that a differing catecholamine-driven phenomenon occurs between genotype groups relating to metabolic syndrome and COMT function. Previous studies have shown that reduced dopaminergic tone may trigger metabolic syndrome (25), yet our preliminary results appear to contradict these findings in patients with the Val variant. While our data suggests the existence of a relationship between metabolic syndrome, genotype, and methylation, a definite conclusion cannot be drawn.
The significant relationship between total activity level and percent methylation is noteworthy. Prefrontal dopamine is increased during periods of physical activity, and an increase in dopamine may alter the expression of COMT and other dopamine (DA) regulatory processes (26). As COMT expression increases, methylation may decrease in response, which is what the results of this study appear to support in the Val population due to the possible increased dopamine levels from higher exercise levels. Those with the Val/Val genotype, with both higher functioning and expressed COMT product, show a negative relationship between methylation percent and total activity levels on both methylation sites. Conversely, those with the Met/Met genotype, and thus lower functioning COMT, show a positive correlation between COMT methylation and increased activity scores. The lower methylation percentages seen in the Val/Val group with higher activity levels may suggest that as these individuals exercise, their COMT functionality and overall ability to effectively metabolism DA is maintained. What is seen with the Met/Met genotype contradicts the notion that higher DA would trigger COMT transcription, and suggests the functional mutation may respond to dopamine surges in a counter-intuitive fashion. It is important to suggest though that for those with the Val/Val genotype, the lifestyle practice of reduced physical activity may contribute to reduced COMT methylation, which may be related to overall COMT functioning and a reduction in DA metabolism. So for those with the Val/Val genotype, regular exercise may be an important intervention to maintain lower COMT methylation and optimal DA metabolism for more favorable outcomes related to schizophrenia psychopathology as well as metabolic syndrome prevention or reduction. This finding is provocative and warrants further investigation, such as COMT rt-PCR and western blotting in response to activity and the corresponding dopamine surge, along with measurement of a cognitive outcome to assess overall clinical effect and outcome.
A few studies have shown the effects of environment on COMT promoter region methylation, such as physical stress and smoking (23,27), and it is well documented that environment plays a large role in epigenetic DNA modulation (14). We are the first group to examine COMT promoter methylation and physical activity within living schizophrenia subjects. However others have shown a relationship between other physical outcomes (i.e. muscle mass and bone mineral density) and COMT 158Val/Met genotype (28,29). Additionally in a recent study of healthy volunteers, subjects were randomized according to their COMT Val/Met status and placed in exercise and control groups. When exposed to periods of physical activity, Val/Val subjects had significantly greater cognitive performance ability than those in the Met/Met cohort, which the authors attributed to the dopamine regulatory properties of COMT (30). While we did not obtain a neurocognitive assessment on these subjects and cannot delve specifically into these relationships, our data does show a similar relationship and supports our theory that low exercise levels in those with a ValVal genotype may result in higher methylation affecting COMT functioning and altered DA catabolism.
While the results of this study are interesting a few limitations must be discussed. First, the blood samples collected from the patients came from a peripheral artery as brain tissue extraction is currently unfeasible in living human subjects. However, previous studies have shown that blood-originated samples of DNA closely mimic brain-originated samples (18,23), thus our samples should closely represent subject brain tissue methylation. Also akin to cross-sectional studies, a limitation of this study is that we are unable to determine specific cause-effect relationships. It is out hope that future prospective studies will help address this limitation through inclusion of measures accessing COMT gene expression in order to shed light on the potential epigenetic mechanisms related to our data. Our study did not differentiate between S-COMT and MB-COMT isoenzymes, which have been found to have different expression levels based on genotype, methylation, and location (31), however our sequencing primers were derived from the COMT-S sequence. Additionally, we were unable to examine neurocognition or DA metabolic measures (i.e. Homovanillic Acid levels) to determine the phenotype endpoint of these COMT methylation differences between groups. Due to a small sample size and large number of assessments, the possibility of confounding variables may skew results and significance, additionally the use of p-value correction for the number of statistical tests done was not completed. While this may alter some of our minor findings (i.e. homocysteine and methylation relationship), our primary results would withstand this greater statistical scrutiny. Lastly, the primary measure of physical activity (the TAM2) has not been validated within the schizophrenia population and the imputed values are based on subject recall which has its own biases (16). Very similar to this our TAM2 values are based on physical activity over the last 7 days, thus this shorter time frame for estimating exercise may not be the most appropriate and perhaps a longer timeframe (i.e. a month or longer) needs to be utilized in future studies.
5. Conclusion
Our results reveal a significant relationship between COMT promoter region methylation, physical activity, and metabolic syndrome in 158Val/Met patients; findings which may lead to improved understanding of both schizophrenia and metabolic syndrome’s genetic and biochemical etiology. Overall we found that COMT methylation patterns differed based on the 158Val/Met variant and that presence of the metabolic syndrome in Val/Val genotype subjects resulted in a COMT methylation profile for these subjects that resembled that seen in Met/Met genotype subjects without metabolic syndrome. Additionally, our data shows that for the Val/Val genotype group, lower physical activity levels may result in greater COMT methylation which may affect overall COMT activity related to metabolic syndrome risk or DA metabolism. Follow-up studies will use the results of this report in the examination of the true role of physical activity on important phenotypic outcomes of COMT expression as well as neurocognitive measures and outcomes in schizophrenia.
Further study of the role of epigenetics in psychiatric disorders will benefit through analysis of additional DNA methylation sites, global LINE methylation, and the role of other enzymes, such as MTHFR. Additionally, rtPCR analysis of COMT mRNA and western blotting studies will further enhance the results of this study in terms of understanding the ramifications of methylation on enzyme expression and functionality. Continued research elucidating the role of DNA methylation in transcription regulation will aid the advancement of therapeutic methods.
Acknowledgments
The authors would like to acknowledge the subjects who participated in this research, the Washtenaw Community Health Organization (WCHO) and the Detroit-Wayne County Community Mental Health Agency (D-WCCMHA). Lastly we would like to acknowledge Dr. Richard Pilsner who provided guidance with the methylation assay.
Footnotes
Conflicts of Interest
No conflicts of interest are reported by the authors.
Disclosure
The following funding sources were utilized for this publication NIMH (R01 MH082784) and the NIH-NCCR for the Michigan Institute for Clinical and Health Research (MICHR - UL1RR024986) and the Chemistry Core of the Michigan Diabetes Research and Training Center (NIH5P60 DK 20572).
References
- 1.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002 Jan 16;287(3):356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 2.Grundy S, Cleeman J, Daniels S, Donato K, Eckel R, Franklin B, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute scientific statement: Executive Summary. Critical pathways in cardiology. 2005;4(4):198–203. doi: 10.1097/00132577-200512000-00018. [DOI] [PubMed] [Google Scholar]
- 3.Nasrallah HA, Meyer JM, Goff DC, McEvoy JP, Davis SM, Stroup TS, et al. Low rates of treatment for hypertension, dyslipidemia and diabetes in schizophrenia: data from the CATIE schizophrenia trial sample at baseline. Schizophr Res. 2006 Sep;86(1–3):15–22. doi: 10.1016/j.schres.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Ellingrod VL, Taylor SF, Dalack G, Grove T, Bly MJ, Brook R, et al. Risk Factors Associated with Metabolic Syndrome in Bipolar and Schizophrenia Subjects Treated With Antipsychotics: The Role of Folate Pharmacogenetics. Journal of Clinical Psychopharmacology. doi: 10.1097/JCP.0b013e3182485888. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Winkel R, Rutten BP, Peerbooms O, Peuskens J, van Os J, De Hert M. MTHFR and risk of metabolic syndrome in patients with schizophrenia. Schizophr Res. 2010 Jun 12; doi: 10.1016/j.schres.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Ellingrod VL, Miller DD, Taylor SF, Moline J, Holman T, Kerr J. Metabolic syndrome and insulin resistance in schizophrenia patients receiving antipsychotics genotyped for the methylenetetrahydrofolate reductase (MTHFR) 677C/T and 1298A/C variants. Schizophr Res. 2008 Jan;98(1–3):47–54. doi: 10.1016/j.schres.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruder GE, Keilp JG, Xu H, Shikhman M, Schori E, Gorman JM, et al. Catechol-O-methyltransferase (COMT) genotypes and working memory: associations with differing cognitive operations. Biol Psychiatry. 2005 Dec 1;58(11):901–907. doi: 10.1016/j.biopsych.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Hosak L. Role of the COMT gene Val158Met polymorphism in mental disorders: a review. Eur Psychiatry. 2007 Jul;22(5):276–281. doi: 10.1016/j.eurpsy.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Bilder R, Volavka J, Lachman H, Grace A. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29(11):1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- 10.Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995 Apr 4;34(13):4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- 11.Nieratschker V, Frank J, Muhleisen TW, Strohmaier J, Wendland JR, Schumacher J, et al. The catechol-O-methyl transferase (COMT) gene and its potential association with schizophrenia: findings from a large German case-control and family-based sample. Schizophr Res. 2010 Sep;122(1–3):24–30. doi: 10.1016/j.schres.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Okochi T, Ikeda M, Kishi T, Kawashima K, Kinoshita Y, Kitajima T, et al. Meta-analysis of association between genetic variants in COMT and schizophrenia: an update. Schizophr Res. 2009 May;110(1–3):140–148. doi: 10.1016/j.schres.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Goldman D, Weinberger DR, Malhotra AK, Goldberg TE. The role of COMT Val158Met in cognition. Biol Psychiatry. 2009 Jan 1;65(1):e1–2. doi: 10.1016/j.biopsych.2008.07.032. author reply e3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mill J, Dempster E, Caspi A, Williams B, Moffitt T, Craig I. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet. 2006 Jun 5;141B(4):421–425. doi: 10.1002/ajmg.b.30316. [DOI] [PubMed] [Google Scholar]
- 15.Andreasen NC, Pressler M, Nopoulos P, Miller D, Ho BC. Antipsychotic dose equivalents and dose-years: a standardized method for comparing exposure to different drugs. Biol Psychiatry. 2010 Feb 1;67(3):255–262. doi: 10.1016/j.biopsych.2009.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orrell A, Doherty P, Miles J, Lewin R. Development and validation of a very brief questionnaire measure of physical activity in adults with coronary heart disease. Eur J Cardiovasc Prev Rehabil. 2007 Oct;14(5):615–623. doi: 10.1097/HJR.0b013e3280ecfd56. [DOI] [PubMed] [Google Scholar]
- 17.Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006 Nov 1;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy BC, O’Reilly RL, Singh SM. Site-specific cytosine methylation in S-COMT promoter in 31 brain regions with implications for studies involving schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2005 Feb 5;133B(1):37–42. doi: 10.1002/ajmg.b.30134. [DOI] [PubMed] [Google Scholar]
- 19.Singh SM, McDonald P, Murphy B, O’Reilly R. Incidental neurodevelopmental episodes in the etiology of schizophrenia: an expanded model involving epigenetics and development. Clin Genet. 2004 Jun;65(6):435–440. doi: 10.1111/j.1399-0004.2004.00269.x. [DOI] [PubMed] [Google Scholar]
- 20.Tunbridge EM, Harrison PJ, Warden DR, Johnston C, Refsum H, Smith AD. Polymorphisms in the catechol-O-methyltransferase (COMT) gene influence plasma total homocysteine levels. Am J Med Genet B Neuropsychiatr Genet. 2008 Sep 5;147B(6):996–999. doi: 10.1002/ajmg.b.30700. [DOI] [PubMed] [Google Scholar]
- 21.Vizzardi E, Bonadei I, Zanini G, Frattini S, Fiorina C, Raddino R, et al. Homocysteine and heart failure: an overview. Recent Pat Cardiovasc Drug Discov. 2009 Jan;4(1):15–21. doi: 10.2174/157489009787259991. [DOI] [PubMed] [Google Scholar]
- 22.Ueland PM, Hustad S, Schneede J, Refsum H, Vollset SE. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol Sci. 2001 Apr;22(4):195–201. doi: 10.1016/s0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- 23.Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, et al. Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci. 2011 May 4;31(18):6692–6698. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human Molecular Genetics. 2011 Sep 09;20(24):4786–1496. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pijl H. Reduced dopaminergic tone in hypothalamic neural circuits: expression of a “thrifty” genotype underlying the metabolic syndrome? Eur J Pharmacol. 2003 Nov 7;480(1–3):125–131. doi: 10.1016/j.ejphar.2003.08.100. [DOI] [PubMed] [Google Scholar]
- 26.Christine G. Optimal physical performance in athletes: key roles of dopamine in a specific neurotransmitter/hormonal mechanism. Mech Ageing Dev. 1995 Oct 13;84(2):83–102. doi: 10.1016/0047-6374(95)01635-x. [DOI] [PubMed] [Google Scholar]
- 27.Xu Q, Ma JZ, Payne TJ, Li MD. Determination of Methylated CpG Sites in the Promoter Region of Catechol-O-Methyltransferase (COMT) and their Involvement in the Etiology of Tobacco Smoking. Front Psychiatry. 2010 Jun 10;1:16. doi: 10.3389/fpsyt.2010.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawling S, Roodi N, Mernaugh RL, Wang X, Parl FF. Catechol-O-methyltransferase (COMT)-mediated metabolism of catechol estrogens: comparison of wild-type and variant COMT isoforms. Cancer Res. 2001 Sep 15;61(18):6716–6722. [PubMed] [Google Scholar]
- 29.Lorentzon M, Eriksson AL, Nilsson S, Mellstrom D, Ohlsson C. Association between physical activity and BMD in young men is modulated by catechol-O-methyltransferase (COMT) genotype: the GOOD study. J Bone Miner Res. 2007 Aug;22(8):1165–1172. doi: 10.1359/jbmr.070416. [DOI] [PubMed] [Google Scholar]
- 30.Stroth S, Reinhardt RK, Thone J, Hille K, Schneider M, Hartel S, et al. Impact of aerobic exercise training on cognitive functions and affect associated to the COMT polymorphism in young adults. Neurobiol Learn Mem. 2010 Oct;94(3):364–372. doi: 10.1016/j.nlm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Tunbridge EM. The catechol-O-methyltransferase gene: its regulation and polymorphisms. Int Rev Neurobiol. 2010;95:7–27. doi: 10.1016/B978-0-12-381326-8.00002-8. [DOI] [PubMed] [Google Scholar]


