Abstract
The current paradigm in the amyloid hypothesis brands small β-amyloid (Aβ) oligomers as the toxic species in Alzheimer’s disease (AD). These oligomers are fibril-like; contain β-sheet structure, and present exposed hydrophobic surface. Oligomers with this motif are capable of penetrating the cell membrane, gathering to form toxic ion channels. Current agents suppressing precursor Aβ cleavage have only met partial success; and to date, those targeting the peptides and their assemblies in the aqueous environment of the extracellular space largely fail in clinical trials. One possible reason is failure to reach membrane-embedded targets of disease-‘infected’ cells. Here we provide an overview, point to the need to account for the lipid environment when aiming to prevent the formation of toxic channels, and propose a combination therapy to target the species spectrum.
1. Introduction
Alzheimer’s disease (AD) is the most common type of dementia that causes memory loss. Currently there is no cure for the disease; the symptoms persist over time and eventually lead to death. In the early 90’s, it was suggested that depositions of β-amyloid (Aβ) peptides in the brain were the fundamental cause of AD.1,2 The Aβ peptide, 39 – 43 amino acids in length, is a fragment of the amyloid precursor protein (APP). APP cleavage is driven by two enzymes;3,4 β-secretase (BACE) outside the cell and γ-secretase within the cell membrane. Early studies indicated that the extracellular plaques were fibrillar deposits of Aβ peptides and associated with the disease. However, removal of the amyloid plaques in AD did not prevent progressive neurodegeneration,5 suggesting that fibrils were not toxic to cells or tissues in general.
Over the past few decades, there have been many therapeutic efforts to intervene in AD.6,7 Research attention has focused on the reduction of Aβ production by blocking the proteases in the process of APP cleavages. To inhibit the first cleavage process, Ghosh et al.8 introduced β-secretase inhibitors. They demonstrated that these drugs were selective and able to rescue age-related cognitive decline in transgenic AD mice. Alternatively, at the final stage of cleavage, γ-secretase inhibitors, e.g. semagacestat,9,10 were shown to block the subsequent formation of Aβ. However, semagacestat has failed to slow disease progression and has led patients to a high risk of skin cancer. Additionally, several therapeutic agents that reduce Aβ production have failed to suppress the disease in Phase III clinical trials.11
Another strategy to remedy AD has been immunotherapy. For patients with mild-to-moderate AD, a clinical study showed that immunotherapy with CAD106 exhibited a favorable safety profile and did not elicit Aβ-specific T-cell response.12 The Aβ vaccine CAD106 has been designed to stimulate the host immune system to attack a small Aβ peptide fragment (Aβ1–6) acting as a B-cell epitope. CAD106 has been found to be more effective than AN1792 which targets T-cell epitopes-carrying full-length Aβ1–42. Although CAD106 is currently in clinical trials, previous immunotherapy with AN1792 has been suspended due to meningoencephalitis.13 Lastly, several anti-aggregation agents have been introduced to prevent Aβ aggregation. However, these drugs mainly aim to inhibit the formation of Aβ fibrils.14,15
Aβ toxicity is linked to the disruption of the cell’s calcium ion homeostasis which triggers Aβ-induced neuronal apoptosis.16–18 The amyloid hypothesis in AD forcefully points to the oligomeric species as the cytotoxic intermediate in Aβ aggregation in brain cells.19,20 Oligomer toxicity is supported by several observations: (i) Amyloid monomers and fibrils show little cytotoxicity in contrast to intermediate aggregates,21 (ii) transgenic mouse models show disease-like phenotypes far earlier than the appearance of fibrils in such protein deposition diseases including Alzheimer’s, Huntington’s, and Parkinson’s disease,22,23 and (iii) non-fibrillar soluble oligomers promote neuronal dysfunction and neuron death.20,24,25 However, it still remains unclear how these oligomers are kinetically assembled, which types of secondary oligomer conformations are involved in the disease, and through which mechanism of cytotoxicity these oligomers work on/in the cell membranes. These questions may be helpful to consider with the reality that so far traditional drugs that remove or disassemble amyloids seem to fail in clinical trials.
The amyloid cascade hypothesis has gradually shifted the focus of amyloid toxicity toward smaller oligomers.11 This prompts us to reexamine amyloid toxicity and drug discovery in AD. Here we provide a brief overview of Aβ oligomer toxicity focusing on details of molecular-level conformations of toxic Aβ oligomers and channels in the lipid bilayers and the implications for therapeutics. The ion channel hypothesis26 and its further validation27 have long argued that toxicity is mediated by channel formations. Due to the unfavorable chemistry and high energetic cost of complete preformed channels sliding into the membrane, we have previously suggested28 that Aβ oligomers can irreversibly insert into a membrane and spontaneously form an ion channel, leading to cell death (Fig. 1). While the channels can be blocked by drugs,29 their broad conformational heterogeneity leads us to suggest that Aβ-directed therapeutics should consider a combination therapy that targets the toxic Aβ oligomers on the membrane before they are inserted, and, in parallel, targets the oligomers that have already penetrated into the membrane, where these agents could prevent toxic channel formations. While this suggestion currently presents a daunting challenge, the development of therapeutic agents to target already formed toxic channels is further hampered by the channels’ highly polymorphic nature, encompassing different sizes, shapes, and chemistries, suggesting that a “one size fits all” blocker may, or may not work. A combination therapy has important implications for the development and the clinical trials of new therapeutic drugs in the pathogenesis of AD. The free energy landscapes of Aβ monomers, oligomers, and membrane-embedded oligomers and channels are all highly polymorphic,30,31 emphasizing the hurdles facing drug development, and pointing to the need to rethink drug strategies. Such Aβ-direct ‘attacks’ are in addition to a focus on the complex cellular network controlling and mediating Aβ expression and processing. We believe that it is unlikely that a single, universal, approach will suppress all Aβ-related pathologies.
Fig. 1.
Schematic diagram of cell toxicity by Aβ oligomers. Small fibril-like oligomers with the parallel β-sheet structures and an exposed hydrophobic surface are believed to be toxic through a channel formation in the cell membranes.
2. Aβ oligomer toxicity
2.1 Ion channel hypothesis
Arispe et al.26,32–35 first reported the ground-breaking discovery, where Aβ induced unregulated ionic flux across model membranes in planar lipid bilayer (PLB) experiments. They concluded that the ionic flux conducted through non-gated ion channels. This initiated the Aβ ion channel hypothesis, subsequently extended for other amyloids. The Aβ channels exhibited cation selectivity, Tris (tromethamine) and zinc inhibitions, and multiple and large single channel conductances. The heterogeneity of single channel conductances suggested that the channels are formed by multiple molecular species in the membrane. Subsequently, single channel conductances for islet amyloid polypeptide (IAPP),36 prion protein fragment,37 polyglutamine,38 β2-microglobulin,39 transthyretin (TTR),40 and serum amyloid A (SAA),41 were reported by several electrophysiology groups. These amyloid channels were normally cation selective and blocked by zinc.35,39,42 Zinc can block early permeabilization by Aβ channel formations, but not the leakage by Aβ fiber-induced membrane fragmentation.43 It was noted that metal-induced Aβ aggregation and cytotoxicity can be modulated through a metal chelation.44–49
Atomic force microscopy (AFM) has successfully demonstrated a remarkable ability to capture the images of channels formed by Aβ peptides. In 2001, Lin et al.42 presented AFM images that showed channel-like structures of Aβ1–42 peptides when reconstituted in a planar lipid bilayer. They further showed that the channels exhibited multiple single channel conductances, calcium uptake, neuritic degeneration, and blocked by zinc. Besides AFM, using electron microscopy (EM), Lashuel et al.50 reported pore-like structures of Aβ Arctic mutant (E22G) and A53T and A30P mutants of α-synuclein associated with Parkinson’s disease. Based on these observations, it became increasingly clear that channel formation is a general feature for amyloids. Quist et al.51 presented a series of AFM images of ion channels for a series of disease-related amyloid species, including Aβ1–40, α-synuclein, ABri, ADan, SAA, and amylin. AFM resolution could determine that the amyloid channels have outer diameter of 8 – 12 nm and inner diameter ~2 nm. More interestingly, the AFM images revealed that the amyloid channels were assembled by several subunits, yielding various channel shapes from rectangular with four subunits to octahedral with eight subunits.
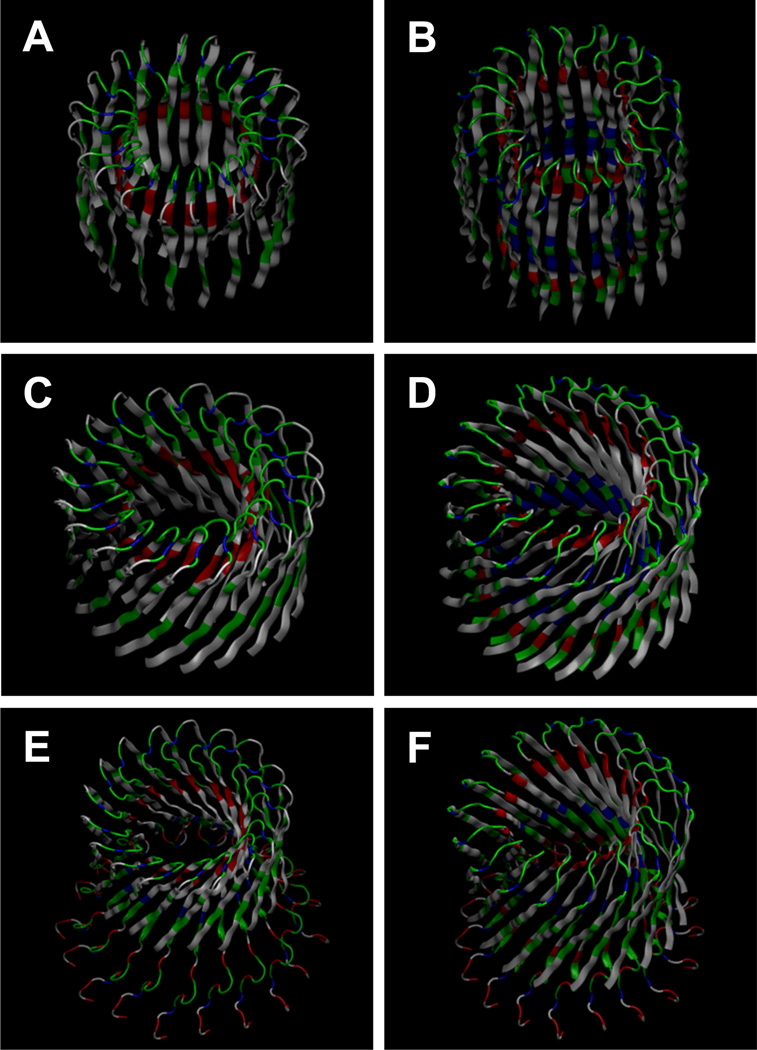
An increasing body of evidence has implicated amyloid channel formations. Recently, a series of molecular dynamics (MD) simulations presented Aβ channels structures at atomic-level resolution for two truncated Aβ peptides, Aβ17–42 (p3) and Aβ9–42 (N9),52–57 and a full-length Aβ1–42 peptide58–61 in the lipid bilayers composed of various types of lipids. The conceptual designs of Aβ channels showed a perfect annular shape with either the conventional or β-barrel-like β-strands arrangement (Fig. 2). The former has the β-stands parallel to the membrane normal, while the latter has an inclination angle between the β-strands and the membrane normal. The annular channels gradually evolved in the lipid bilayer during the simulations and the relaxed channel structures exhibited heterogeneous shapes. The simulations revealed that Aβ channels consisted of β-sheet-rich subunits with the morphologies and dimensions in good agreement with the imaged AFM channels.42,51 The computational studies reported that the misfolded amyloid channels consist of loosely and dynamically associated subunits in the fluidic lipid bilayer in contrast to the stable, function-optimized and evolution-preserved conventional gated ion channels, which fold into their native state. In the amyloid channel hypothesis, Aβ peptides directly form toxic ion channels in cell membranes leading to death of neurons in AD.
Fig. 2.
Computational models of Aβ channels using truncated (A–D) and full-length Aβ peptides (E,F). In the truncated Aβ channels, the β-strands are arranged in a conventional β-sheet without tilt for the (A) Aβ17–42 (p3) and (B) Aβ9–42 (N9) channels, and in a β-barrel topology with tilt for the (C) p3 and (D) N9 barrels. Full-length Aβ1–42 peptides are arranged in a β-barrel topology with tilt for the (E) conformer 1 and (F) conformer 2 Aβ1–42 barrels. The Aβ channels consist of the U-shaped peptides with the β-strand-turn-β-strand motif. The Aβ17–42 and conformer 1 Aβ1–42 peptides contain the turn at Ser26-Ile31, and the Aβ9–42 and conformer 2 Aβ1–42 peptides contain the turn at Asp23-Gly29.
2.2 Receptor binding hypothesis
The toxic Aβ oligomers referred to as amyloid-derived diffusible ligands (ADDLs) attached to synapses and interfered with the mechanism of synaptic plasticity, leading to disruption of neuronal communication.62 These large oligomers were capable of inducing neuronal oxidative stress63 and neuronal tau hyperphosphorylation.64 Such indirect Aβ binding to cell membrane receptors, which subsequently open existing ion channels or transporters, is stereospecific. Ciccotosto et al.65 reported that Aβ toxicity through receptor interaction with the phosphatidylserine lipid flipped to the extracellular side of the lipid bilayer, suggesting stereospecific interactions of L-Aβ peptide. However, cellular stereospecificity can be only expected from receptor-ligand interactions with the peptides made of L-amino acids, since D-enantiomers do not bind to cell receptors. Recent comprehensive studies demonstrated that in the absence of stereospecific interactions, the Aβ D-enantiomers could directly form ion channels in the lipid bilayers.58,60 This suggested that Aβ oligomer toxicity could take place through a receptor independent, nonstereoselective mechanism.
2.3 Membrane thinning hypothesis
The membrane thinning hypothesis was supported by several electrophysiology studies which observed that membrane destabilization by intermediate-to-large amyloid oligomers induced nonselective ion leakage through the low dielectric barrier with consequent thinning of bilayer.66,67 The measured ionic currents gradually increased across the membranes. In contrast, stepwise or spike-like fluctuations of membrane currents represented the typical ion channels characteristics. The two different behaviors of membrane ionic flux have been a debated issue.68 However, the controversy seems to have been resolved, since the gradual increase in conductance, which was later also observed in the absence of Aβ, was attributed to the solvent hexafluoroisopropanol (HFIP),69 which is commonly used for the Aβ sample preparation. It was noted that Aβ behavior strongly depended on experimental conditions, with the different preparations of the peptide exhibiting altered aggregation kinetics.70
2.4 Fiber-dependent membrane disruption
Besides the Aβ oligomer hypothesis, a mechanism of Aβ toxicity involving cell membrane damage has recently proposed fiber-dependent membrane disruption.43,71 Non-specific leakages across the membrane were observed due to the membrane fragmentations from the amyloid fiber growth on the membrane surface. Similar observations of membrane damage by amyloid fibrils were also reported for other amyloids such as β2-microglobulin72 and human islet amyloid polypeptide (hIAPP).73–75
3. Aβ architectures
3.1 Monomer conformations
In an aqueous environment, Aβ monomer generally folded into an α-helical structure. Crescenzi et al.76 reported three-dimensional nuclear magnetic resonance (NMR) structure of Aβ1–42 monomer in non-polar microenvironment (pdb id: 1IYT). The peptide showed two α-helices (composing of residues 8–25 and 28–38) connected by a β-turn (Fig. 3A). They noted that the kinked α-helical structure, as well as the sequence of the C-terminal moiety, was similar to the fusion domain of influenza hemagglutinin, which led them to suggest a direct mechanism of neurotoxicity. The reversible conformational transitions of Aβ1–42 between α-helix and β-sheet structures were observed by using circular dichroism (CD) and NMR spectroscopy in media of varying polarities (pdb id: 1Z0Q).77 The peptide retained its α-helices in solution with the appropriate amount of HFIP (Fig. 3B), while β-sheet developed in solutions with very high water content compared to HFIP. By monitoring the transitions, they found out that while the long N-terminal helix was retained, the C-terminal helix was lost. In a recent NMR study, Vivekanandan et al.78 showed that Aβ1–40 adopted a collapsed and partially folded helical structure in an aqueous environment (pdb id: 2LFM). The peptide contained 310 helix in the central hydrophobic region (residues 13–23) and collapsed in the N- and C-termini (Fig. 3C). They noted that helical intermediates in early fibrillogenesis events could convert into β structures upon binding to the membrane. It has been further demonstrated that the presence of lipid membranes can catalyze β-sheet formation.79
Fig. 3.
NMR-based monomer conformations of Aβ1–42 peptides (A) in an apolar microenvironment (pdb id: 1IYT) and (B) in solution with the appropriate amount of HFIP (pdb id: 1Z0Q), and (C) Aβ1–40 peptide in an aqueous environment (pdb id: 2LFM).
3.2 U-shaped peptides with the β-strand-turn-β-strand motif
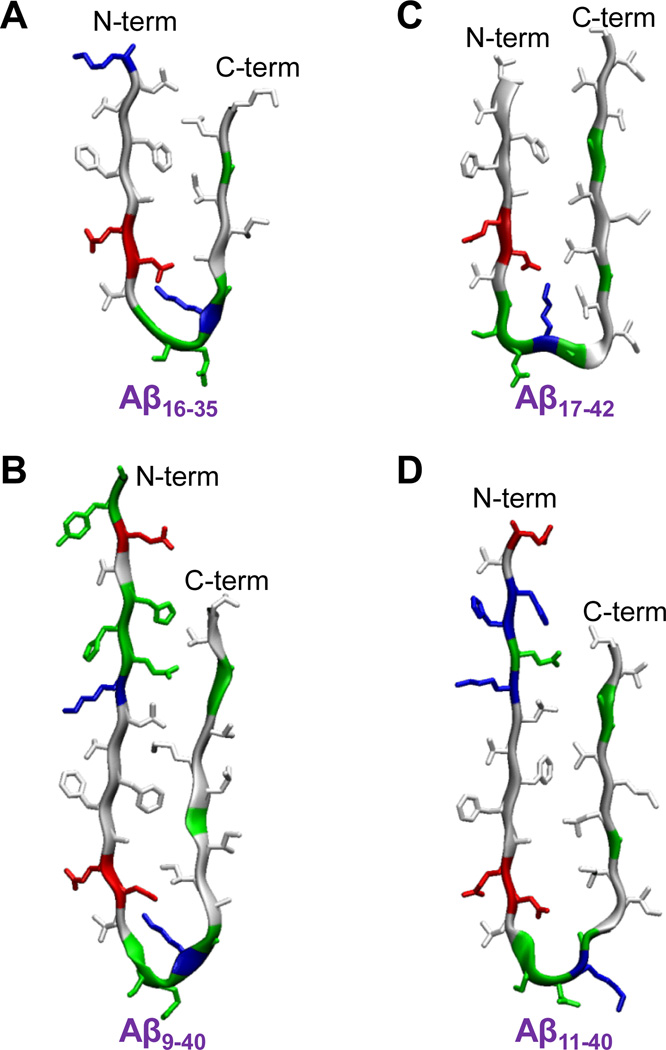
Ma and Nussinov80 first introduced a computational model of U-shaped Aβ peptide using MD simulations. The solvated oligomers of Aβ16–35 peptides with the β-strand-turn-β-strand motif presented a protofibril in the parallel β-sheet organization. The Aβ16–35 peptide contained an intramolecular salt bridge between residues Asp23 and Lys28 near a turn at Val24-Asn27 (Fig. 4A). Subsequently, an updated solid state NMR (ssNMR) model for small Aβ1–40 protofibrils (pdb ids: 2LMN and 2LMO) verified the side chain orientations in the C-terminal strand as predicted by the computational model.81 The U-shaped Aβ1–40 peptide in the protofibrils had a turn at Asp23-Gly29 and the same salt bridge as the Aβ16–35 peptide (Fig. 4B). The N-terminal coordinates (residues 1–8) were missing due to disorder. Lührs et al.82 reported a similar U-shaped peptide but with a slightly different turn motif, located at Ser26-Ile31 including the salt bridge of Asp23/Lys28. They obtained the Aβ1−42 fibril structure from a combination of hydrogen/deuterium-exchange NMR data, side-chain packing constraints from pairwise mutagenesis, ssNMR and EM (pdb id: 2BEG). The Aβ1−42 peptide provided the coordinates for residues 17–42, while the N-terminal coordinates (residues 1–16) were missing due to disorder (Fig. 4C). Recently, Bertini et al.83 reported a structural model of Aβ1−40 fibrils using comprehensive ssNMR techniques. The Aβ1−40 peptide with the the β-strand-turn-β-strand motif exhibited a turn at Val24-Ala30, which is similar to the previous Aβ1–40 model,81 but has a shifted inter β-strand contacts within the U-shaped motif (Fig. 4D). Unlike the previous NMR models, the peptide did not contain the salt bridge of Asp23/Lys28, and the N-terminal coordinates (residues 1–10) were missing due to disorder (coordinates, I. Bertini, pers. Comm.). It has been noted that the U-shaped motif is a general feature of amyloid organization, since other amyloids, such as β2-microglobulin fragment (K3 peptide)84,85 and CA150 WW domain,86 also exhibit the U-shaped with the β-strand-turn-β-strand motif.
Fig. 4.
The U-shaped Aβ peptides with the β-strand-turn-β-strand motif for (A) the computational Aβ16–35 peptide, (B) the ssNMR Aβ9–40 peptide from the Aβ1–40 protofibrils (pdb ids: 2LMN and 2LMO, coordinates for the residues 1–8 were missing due to disorder), (C) the NMR-derived Aβ17–42 peptide from the Aβ1–42 pentamer (pdb id: 2BEG, coordinates for the residues 1–16 were missing due to disorder), and (D) the ssNMR Aβ11–40 peptide from the Aβ1–40 fibrils (coordinates, I. Bertini, pers. Comm., coordinates for the residues 1–10 were missing due to disorder).
3.3 Truncated Aβ channels
It was known that the APP cleavages involving γ- and α-secretases generate a nonamyloidogenic Aβ17–42 (p3) peptide. Cleavage by γ and BACE between residues 10 and 11 creates another nonamyloidogenic Aβ11–42 peptide.87 Adding two more residues to the N-terminal of Aβ11–42 peptide, one can obtain Aβ9–42 (N9) peptide. These truncated Aβ peptides were found in amyloid plaques of AD, and significantly, the p3 peptide was known to be the main constituent of cerebellar preamyloid lesions in Down syndrome.88,89 Since these short Aβ peptides were thought to be nontoxic to neurons, drugs to inhibit β-secretase were used to enhance the production of the N-terminal truncated Aβ peptides. However, recent studies using complementary techniques of MD, AFM, PLB, cell calcium imaging, neuritic degeneration, and cell death assays demonstrated that p3 and N9 peptides formed toxic ion channels in the lipid bilayers.52–57 Unlike the classical ion channel, these channels consisted of loosely attached mobile β-sheet subunits (Fig. 5). A dose- and time-dependent degeneration of neurites in human cortical neurons upon incubation with p3 further suggested that the p3 peptide is a toxic species.55
Fig. 5.
Truncated Aβ channel conformations in the lipid bilayer. (A) Simulated barrel structure with an embedded pore structure, (B) barrel structure with highlighted subunits (Jang et al.57; reprinted with permission), and (C,D) AFM images (Jang et al.55; reprinted with permission) for the Aβ17–42 (p3) channels. (E–H) The same for the Aβ9–42 (N9) channels.
3.4 Full-length Aβ channels
As we noted above, AFM images of full-length Aβ peptides (Aβ1–40/42) showed heterogeneous ion channel structures with the outer diameter between 8 and 10 nm and inner pore diameter about 2.0 nm for the Aβ channels.42,51 In the images, the channel shapes varied from rectangular to hexagonal, corresponding to 4–6 subunits. Subsequently, recent AFM and MD studies also provided the detailed 3D structures of Aβ1–42 ion channels consisting of all L- or all D-amino acid residues.60 In parallel, the PLB electrophysiological recordings showed that both L- and D-Aβ isomers could conduct cations.58 Taken together, D-Aβ1–42 formed ion channels with a behavior indistinguishable from the naturally occurring L-Aβ1−42, suggested Aβ neurotoxicity by a direct pathway through an ion channel.
To model the Aβ1–42 channels, two U-shaped monomer conformations based on ssNMR models81,82 were used to construct the channel in a β-barrel topology. However both NMR models did not provide the N-terminal coordinates due to conformational disorder; they only presented the coordinates for the residues 17–42 or 9–40. The missing N-terminal coordinates were complemented by the solution NMR structure of Aβ1−16, following removal of Zn2+ (pdb id: 1ZE7).90 For each combination of the N-terminal structure with the U-shaped motifs two Aβ1–42 conformers were generated: Conformer 1 has a turn at Ser26-Ile31 and conformer 2 at Asp23-Gly29. In the MD simulations, the model Aβ1–42 barrels exhibited 3–5 subunits in agreement with AFM images (Fig. 6). The outer and inner pore sizes measured for the 18-mer Aβ barrels were also found to be in the AFM ranges. However, the MD study noted that while presumably AFM provided channel images covering all ranges of channel sizes, the simulated Aβ barrels were limited to sizes defined by peptide count.
Fig. 6.
Full-length Aβ1–42 channel conformations in the lipid bilayer. Simulated barrel structure with an embedded pore structure and highlighted subunits for (A) the conformer 1 and (B) the conformer 2 Aβ1–42 barrels (Connelly et al.60; reprinted with permission), (C) AFM images of Aβ1–42 reconstituted in the lipid bilayer. Individual Aβ1–42 channels are enclosed by circles. (D,E) High-resolution images of individual channels as indicated by the circles. The number of subunits is resolved and indicated for each channel. Image sizes are 18.6 nm (D) and 17.1 nm (E).
4. Tentative therapeutic targets/agents
4.1 Fibril-like Aβ oligomers
Wu et al.91 reported that small soluble Aβ oligomers, known as fibrillar oligomers (FOs), contained 3–10 Aβ monomers and exhibited fibril-like morphology rich in β-sheet structure. They noted that FOs could increase toxicity, since the oligomers were able to replicate and expose the hydrophobic sheet surfaces. It has been suggested that a more toxic Aβ oligomer has a solvent exposed hydrophobic face, while a less toxic Aβ oligomer is deficient in β-sheet conformation.92 Using ssNMR and EM, it was observed that the small fibril-like Aβ oligomer intermediates have predominantly parallel β-sheet structures.93 A recent MD study demonstrated that parallel β-sheet Aβ oligomers with the U-shaped peptide motif were capable of penetrating the membrane.28 Taken together, it is not surprising that fibril-like oligomers with exposed hydrophobic surface are cytotoxic, since they spontaneously insert into the membranes and several of these gather to form toxic ion channels. Thus, drugs aimed at inhibiting the production of these oligomers, or inhibitors preventing the oligomers attachment to the membranes would attenuate the Aβ toxicity, indicating that the fibril-like β-sheet oligomers would be tentative therapeutic targets in AD.
4.2 Ion channel blockers
The amyloid hypothesis of AD implicated the toxic ion channel formation in membranes on the basis of large body of experimental and computational observations.26,32–35,42,50–61 Ion channel blockers were thought to be intuitive therapeutic agents that lead to blockage of conducting pores. Diaz et al.29 introduced two small molecular blockers of Aβ channel; MRS2481 and an enatiomeric species, MRS2485. While both blockers could potentially protect neurons from Aβ toxicity, MRS2485 was found to be virtually irreversible in the channel inhibition. Developing pharmacologic inhibitors with high specificity aimed to block the pore would be a major challenge,94 since structural modeling of Aβ channels has suggested that channels are mobile and flexible in the lipid bilayers, with varying sizes and shapes.54,56,57 Amyloid channels are highly polymorphic, suggesting that “one size fits all” blockers may not be effective in comprehensive treatment of amyloid channels.
Indirect strategies to inhibit Aβ channel activity prompted development of drugs that can control existing channels. Liu et al.95 introduced a drug, known as diazoxide, a potassium ATP channel activator. They demonstrated that diazoxide improved memory and reduced Aβ and tau pathology in a transgenic AD mouse model. Since Aβ channels tend to the lower membrane potential due to their relative lack of selectivity, membrane hyperpolarization by the potassium opener can suppress the depolarizing effect of the Aβ channel. A similar approach also demonstrated that voltage-gated calcium channel blockers, such as verapamil, diltiazem, isradipine and nimodipine, exerted protective effects on cultured neurons from Aβ toxicity.96 Thus, drugs that hyperpolarize membranes would be possible therapeutic agents in AD.
4.3 Point mutations
Naturally occurring point mutations of Aβ clustered at residues 22 and 23 were related to familial forms of AD.97 These Aβ mutations were found in fibrillar aggregates on a membrane98 and related to cerebral amyloid angiopathy (CAA) and presenile dementia.99,100 Unlike the disease-related Aβ mutants, proline mutation in the central region of Aβ recently demonstrated that substitution of Phe19 with Pro (F19P) in both truncated Aβ17−42 (p3) and full-length Aβ1−42 channels could prevent bilayer channel activity and cellular toxicity.55,59,61 Proline, known as β-sheet breaker, destabilized pore-lining β-strands based on the computational models, producing kinks at the locations of Pro19. As a result, Aβ channels formed with collapsed pores consequently inhibited ions crossing through the pore (Fig. 7). It was reported that the F19P mutant does not aggregate.101,102 This indicates that the mutant exhibits no toxic oligomer, and hence no ion channel in the membrane.
Fig. 7.
Collapsed pores induced by F19P point mutation. (A) Simulated channel structure with an embedded pore structure and highlighted subunits for the p3-F19P (from the truncated Aβ17–42) mutant channel (Jang et al.55; reprinted with permission). Simulated barrel structure with an embedded pore structure for (B) the conformer 1 and (C) the conformer 2 F19P (from the full-length Aβ1–42) mutant barrels (Connelly et al.61; reprinted with permission). (D) AFM image of F19P Aβ1–42 reconstituted in the lipid bilayer for Individual Aβ1–42 channels, here enclosed by circles. (E) High-resolution images of heterogeneous F19P channels characteristic of the wild type. Image sizes are 17.46 nm, 13.15nm, and 14.54 nm (from top to bottom) respectively.
5. Conclusions
Current therapeutic drugs targeting Aβ peptides were designed to inhibit proteases in the process of APP cleavages. Such β- and γ-secretase inhibitors were aimed at reducing Aβ production.6,7 Yet the development of safe secretase inhibitor drugs is not promoted due to side effects; the enzymes have additional physiological substrates in the cell, most prominent among these is Notch which is cleaved by γ-secretase.103 In the amyloid hypothesis, small oligomers gained interest as a toxic species.104 Fibril-like oligomers rich in β structure were identified as active cytotoxic molecules, spontaneously inserting into the membrane and assembling to form toxic ion channels.28,91–93 The mechanisms of their membrane insertion/disruption are similar to those observed for antimicrobial peptides (AMPs).28,94 Therapeutic development efforts for amyloid-removing or disassembling agents have not been rewarded, to date largely failing in clinical trials. This failure of the current agents appears to at least partly stem from poor permeability of cell membranes. Toxic Aβ oligomers are irreversibly inserted into the cells and form channels in the cell membrane or (possibly) in the mitochondrial membrane in the cytoplasm.79 Under such circumstances, therapeutic agents might be unable to penetrate the cell membrane to block or disassemble the channels inside the membrane and the cell. To compound the challenge, amyloid channels are as highly polymorphic as amyloid fibrils. To date, an effective blocker is yet to be discovered. Here, we argue for a combination of drugs, targeting oligomers on, and in, the membrane. The properties of the drugs will need to differ, given the varied chemical environment, a solvated extracellular milieu and a lipidic bilayer. Combined, these drugs would prevent Aβ insertion into the membrane and act to retain a healthy membrane, preventing channel formation by already ‘infected’ membranes. The heterogeneity may call for drugs which recognize the prevailing conformational species. While here we focused on Aβ, similar strategies may be used in other amyloid species.
Acknowledgments
We thank Dr. Bertini for providing us with the coordinates of the Aβ11–40 oligomer. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. This research was supported by the National Institutes of Health (National Institute on Aging AG028709 to RL). All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Notes and references
- 1.Hardy J, Allsop D. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 2.Polvikoski T, Sulkava R, Haltia M, Kainulainen K, Vuorio A, Verkkoniemi A, Niinisto L, Halonen P, Kontula K. N Engl J Med. 1995;333:1242–1247. doi: 10.1056/NEJM199511093331902. [DOI] [PubMed] [Google Scholar]
- 3.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Neet KE, Thinakaran G. J Biol Chem. 2008;283:29613–29614. doi: 10.1074/jbc.R800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes C, Boche D, Wilkinson D, Yadegarfar G, Hopkins V, Bayer A, Jones RW, Bullock R, Love S, Neal JW, Zotova E, Nicoll JA. Lancet. 2008;372:216–223. doi: 10.1016/S0140-6736(08)61075-2. [DOI] [PubMed] [Google Scholar]
- 6.Citron M. Nat Rev Neurosci. 2004;5:677–685. doi: 10.1038/nrn1495. [DOI] [PubMed] [Google Scholar]
- 7.Teich AF, Arancio O. Biochem J. 2012;446:165–177. doi: 10.1042/BJ20120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh AK, Brindisi M, Tang J. J Neurochem. 2012;120(Suppl 1):71–83. doi: 10.1111/j.1471-4159.2011.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman RJ, Siemers ER, Mawuenyega KG, Wen G, Browning KR, Sigurdson WC, Yarasheski KE, Friedrich SW, Demattos RB, May PC, Paul SM, Holtzman DM. Ann Neurol. 2009;66:48–54. doi: 10.1002/ana.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi P, Hadden C, Kulanthaivel P, Calvert N, Annes W, Brown T, Barbuch RJ, Chaudhary A, Ayan-Oshodi MA, Ring BJ. Drug Metab Dispos. 2010;38:554–565. doi: 10.1124/dmd.109.030841. [DOI] [PubMed] [Google Scholar]
- 11.Karran E, Mercken M, De Strooper B. Nat Rev Drug Discov. 2011;10:698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 12.Winblad B, Andreasen N, Minthon L, Floesser A, Imbert G, Dumortier T, Maguire RP, Blennow K, Lundmark J, Staufenbiel M, Orgogozo JM, Graf A. Lancet Neurol. 2012;11:597–604. doi: 10.1016/S1474-4422(12)70140-0. [DOI] [PubMed] [Google Scholar]
- 13.Orgogozo JM, Gilman S, Dartigues JF, Laurent B, Puel M, Kirby LC, Jouanny P, Dubois B, Eisner L, Flitman S, Michel BF, Boada M, Frank A, Hock C. Neurology. 2003;61:46–54. doi: 10.1212/01.wnl.0000073623.84147.a8. [DOI] [PubMed] [Google Scholar]
- 14.Lashuel HA, Hartley DM, Balakhaneh D, Aggarwal A, Teichberg S, Callaway DJ. J Biol Chem. 2002;277:42881–42890. doi: 10.1074/jbc.M206593200. [DOI] [PubMed] [Google Scholar]
- 15.Parker MH, Chen R, Conway KA, Lee DH, Luo C, Boyd RE, Nortey SO, Ross TM, Scott MK, Reitz AB. Bioorg Med Chem. 2002;10:3565–3569. doi: 10.1016/s0968-0896(02)00251-1. [DOI] [PubMed] [Google Scholar]
- 16.Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loo DT, Copani A, Pike CJ, Whittemore ER, Walencewicz AJ, Cotman CW. Proc Natl Acad Sci U S A. 1993;90:7951–7955. doi: 10.1073/pnas.90.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smale G, Nichols NR, Brady DR, Finch CE, Horton, Jr WE. Exp Neurol. 1995;133:225–230. doi: 10.1006/exnr.1995.1025. [DOI] [PubMed] [Google Scholar]
- 19.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 20.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 21.Pike CJ, Burdick D, Walencewicz AJ, Glabe CG, Cotman CW. J Neurosci. 1993;13:1676–1687. doi: 10.1523/JNEUROSCI.13-04-01676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg MS, Lansbury PT., Jr Nat Cell Biol. 2000;2:E115–E119. doi: 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- 24.Walsh DM, Selkoe DJ. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 25.Ebenezer PJ, Weidner AM, LeVine H, 3rd, Markesbery WR, Murphy MP, Zhang L, Dasuri K, Fernandez-Kim SO, Bruce-Keller AJ, Gavilan E, Keller JN. J Alzheimers Dis. 2010;22:839–848. doi: 10.3233/JAD-2010-101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arispe N, Rojas E, Pollard HB. Proc Natl Acad Sci U S A. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirzabekov T, Lin MC, Yuan WL, Marshall PJ, Carman M, Tomaselli K, Lieberburg I, Kagan BL. Biochem Biophys Res Commun. 1994;202:1142–1148. doi: 10.1006/bbrc.1994.2047. [DOI] [PubMed] [Google Scholar]
- 28.Jang H, Connelly L, Teran Arce F, Ramachandran S, Kagan BL, Lal R, Nussinov R. J Chem Theory Comput. 2012 doi: 10.1021/ct300916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz JC, Simakova O, Jacobson KA, Arispe N, Pollard HB. Proc Natl Acad Sci U S A. 2009;106:3348–3353. doi: 10.1073/pnas.0813355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller Y, Ma B, Nussinov R. Chem Rev. 2010;110:4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma B, Nussinov R. J Mol Biol. 2012;421:172–184. doi: 10.1016/j.jmb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arispe N, Pollard HB, Rojas E. Proc Natl Acad Sci USA. 1993;90:10573–10577. doi: 10.1073/pnas.90.22.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard HB, Rojas E, Arispe N. Ann. N. Y. Acad. Sci. 1993;695:165–168. doi: 10.1111/j.1749-6632.1993.tb23046.x. [DOI] [PubMed] [Google Scholar]
- 34.Arispe N, Pollard HB, Rojas E. Mol Cell Biochem. 1994;140:119–125. doi: 10.1007/BF00926750. [DOI] [PubMed] [Google Scholar]
- 35.Arispe N, Pollard HB, Rojas E. Proc Natl Acad Sci U S A. 1996;93:1710–1715. doi: 10.1073/pnas.93.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirzabekov TA, Lin MC, Kagan BL. J Biol Chem. 1996;271:1988–1992. doi: 10.1074/jbc.271.4.1988. [DOI] [PubMed] [Google Scholar]
- 37.Lin MC, Mirzabekov T, Kagan BL. J Biol Chem. 1997;272:44–47. doi: 10.1074/jbc.272.1.44. [DOI] [PubMed] [Google Scholar]
- 38.Hirakura Y, Azimov R, Azimova R, Kagan BL. J Neurosci Res. 2000;60:490–494. doi: 10.1002/(sici)1097-4547(20000515)60:4<490::aid-jnr7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Hirakura Y, Kagan BL. Amyloid. 2001;8:94–100. doi: 10.3109/13506120109007350. [DOI] [PubMed] [Google Scholar]
- 40.Hirakura Y, Azimova R, Azimov R, Kagan B. Biophys J. 2001;80:129a–129a. [Google Scholar]
- 41.Hirakura Y, Carreras I, Sipe JD, Kagan BL. Amyloid. 2002;9:13–23. doi: 10.3109/13506120209072440. [DOI] [PubMed] [Google Scholar]
- 42.Lin H, Bhatia R, Lal R. Faseb J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 43.Sciacca MF, Kotler SA, Brender JR, Chen J, Lee DK, Ramamoorthy A. Biophys J. 2012;103:702–710. doi: 10.1016/j.bpj.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hindo SS, Mancino AM, Braymer JJ, Liu Y, Vivekanandan S, Ramamoorthy A, Lim MH. J Am Chem Soc. 2009;131:16663–16665. doi: 10.1021/ja907045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi JS, Braymer JJ, Nanga RP, Ramamoorthy A, Lim MH. Proc Natl Acad Sci U S A. 2010;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeToma AS, Salamekh S, Ramamoorthy A, Lim MH. Chem Soc Rev. 2012;41:608–621. doi: 10.1039/c1cs15112f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parthasarathy S, Long F, Miller Y, Xiao Y, McElheny D, Thurber K, Ma B, Nussinov R, Ishii Y. J Am Chem Soc. 2011;133:3390–3400. doi: 10.1021/ja1072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller Y, Ma B, Nussinov R. Proc Natl Acad Sci U S A. 2010;107:9490–9495. doi: 10.1073/pnas.0913114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller Y, Ma BY, Nussinov R. Coordin Chem Rev. 2012;256:2245–2252. [Google Scholar]
- 50.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 51.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Proc Natl Acad Sci U S A. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jang H, Zheng J, Nussinov R. Biophys J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jang H, Zheng J, Lal R, Nussinov R. Trends Biochem Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Jang H, Arce FT, Capone R, Ramachandran S, Lal R, Nussinov R. Biophys J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Proc Natl Acad Sci USA. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. J Phys Chem B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. J Mol Biol. 2010;404:917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capone R, Jang H, Kotler SA, Connelly L, Teran Arce F, Ramachandran S, Kagan BL, Nussinov R, Lal R. J Chem Theory Comput. 2012;8:1143–1152. doi: 10.1021/ct200885r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capone R, Jang H, Kotler SA, Kagan BL, Nussinov R, Lal R. Biochemistry. 2012;51:776–785. doi: 10.1021/bi2017427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Connelly L, Jang H, Arce FT, Capone R, Kotler SA, Ramachandran S, Kagan BL, Nussinov R, Lal R. J Phys Chem B. 2012;116:1728–1735. doi: 10.1021/jp2108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connelly L, Jang H, Arce FT, Ramachandran S, Kagan BL, Nussinov R, Lal R. Biochemistry. 2012;51:3031–3038. doi: 10.1021/bi300257e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, Viola KL, Klein WL. J Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Felice FG, Velasco PT, Lambert MP, Viola K, Fernandez SJ, Ferreira ST, Klein WL. J Biol Chem. 2007;282:11590–11601. doi: 10.1074/jbc.M607483200. [DOI] [PubMed] [Google Scholar]
- 64.De Felice FG, Wu D, Lambert MP, Fernandez SJ, Velasco PT, Lacor PN, Bigio EH, Jerecic J, Acton PJ, Shughrue PJ, Chen-Dodson E, Kinney GG, Klein WL. Neurobiol Aging. 2008;29:1334–1347. doi: 10.1016/j.neurobiolaging.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciccotosto GD, Tew DJ, Drew SC, Smith DG, Johanssen T, Lal V, Lau TL, Perez K, Curtain CC, Wade JD, Separovic F, Masters CL, Smith JP, Barnham KJ, Cappai R. Neurobiol Aging. 2011;32:235–248. doi: 10.1016/j.neurobiolaging.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 66.Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. J Biol Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 67.Sokolov Y, Kozak JA, Kayed R, Chanturiya A, Glabe C, Hall JE. J Gen Physiol. 2006;128:637–647. doi: 10.1085/jgp.200609533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eliezer D. J Gen Physiol. 2006;128:631–633. doi: 10.1085/jgp.200609689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Capone R, Quiroz FG, Prangkio P, Saluja I, Sauer AM, Bautista MR, Turner RS, Yang J, Mayer M. Neurotox Res. 2009;16:1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soto C, Castano EM, Kumar RA, Beavis RC, Frangione B. Neurosci Lett. 1995;200:105–108. doi: 10.1016/0304-3940(95)12089-m. [DOI] [PubMed] [Google Scholar]
- 71.Schauerte JA, Wong PT, Wisser KC, Ding H, Steel DG, Gafni A. Biochemistry. 2010;49:3031–3039. doi: 10.1021/bi901444w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milanesi L, Sheynis T, Xue WF, Orlova EV, Hellewell AL, Jelinek R, Hewitt EW, Radford SE, Saibil HR. Proc Natl Acad Sci U S A. 2012;109:20455–20460. doi: 10.1073/pnas.1206325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sparr E, Engel MF, Sakharov DV, Sprong M, Jacobs J, de Kruijff B, Hoppener JW, Killian JA. FEBS Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 74.Engel MF, Khemtemourian L, Kleijer CC, Meeldijk HJ, Jacobs J, Verkleij AJ, de Kruijff B, Killian JA, Hoppener JW. Proc Natl Acad Sci U S A. 2008;105:6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sciacca MF, Brender JR, Lee DK, Ramamoorthy A. Biochemistry. 2012;51:7676–7684. doi: 10.1021/bi3009888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D'Ursi AM, Temussi PA, Picone D. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 77.Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. Chembiochem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 78.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochem Biophys Res Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kagan BL, Thundimadathil J. Adv Exp Med Biol. 2010;677:150–167. doi: 10.1007/978-1-4419-6327-7_13. [DOI] [PubMed] [Google Scholar]
- 80.Ma B, Nussinov R. Proc Natl Acad Sci U S A. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 84.Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H, Goto Y. Proc Natl Acad Sci U S A. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mustata M, Capone R, Jang H, Arce FT, Ramachandran S, Lal R, Nussinov R. J Am Chem Soc. 2009;131:14938–14945. doi: 10.1021/ja9049299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe TD, Krause G, Flinders J, Petrovich M, Berriman J, Oschkinat H, Fersht AR. Proc Natl Acad Sci U S A. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thinakaran G, Koo EH. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gowing E, Roher AE, Woods AS, Cotter RJ, Chaney M, Little SP, Ball MJ. Journal of Biological Chemistry. 1994;269:10987–10990. [PubMed] [Google Scholar]
- 89.Lalowski M, Golabek A, Lemere CA, Selkoe DJ, Wisniewski HM, Beavis RC, Frangione B, Wisniewski T. J Biol Chem. 1996;271:33623–33631. doi: 10.1074/jbc.271.52.33623. [DOI] [PubMed] [Google Scholar]
- 90.Zirah S, Kozin SA, Mazur AK, Blond A, Cheminant M, Segalas-Milazzo I, Debey P, Rebuffat S. J. Biol. Chem. 2006;281:2151–2161. doi: 10.1074/jbc.M504454200. [DOI] [PubMed] [Google Scholar]
- 91.Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C. J Biol Chem. 2010;285:6071–6079. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ladiwala AR, Litt J, Kane RS, Aucoin DS, Smith SO, Ranjan S, Davis J, Vannostrand WE, Tessier PM. J Biol Chem. 2012;287:24765–24773. doi: 10.1074/jbc.M111.329763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 94.Kagan BL, Jang H, Capone R, Teran Arce F, Ramachandran S, Lal R, Nussinov R. Mol Pharm. 2012;9:708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu D, Pitta M, Lee JH, Ray B, Lahiri DK, Furukawa K, Mughal M, Jiang H, Villarreal J, Cutler RG, Greig NH, Mattson MP. J Alzheimers Dis. 2010;22:443–457. doi: 10.3233/JAD-2010-101017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Anekonda TS, Quinn JF, Harris C, Frahler K, Wadsworth TL, Woltjer RL. Neurobiol Dis. 2011;41:62–70. doi: 10.1016/j.nbd.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Murakami K, Irie K, Morimoto A, Ohigashi H, Shindo M, Nagao M, Shimizu T, Shirasawa T. Biochem Biophys Res Commun. 2002;294:5–10. doi: 10.1016/S0006-291X(02)00430-8. [DOI] [PubMed] [Google Scholar]
- 98.Pifer PM, Yates EA, Legleiter J. Plos One. 2011;6 doi: 10.1371/journal.pone.0016248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murakami K, Irie K, Morimoto A, Ohigashi H, Shindo M, Nagao M, Shimizu T, Shirasawa T. J Biol Chem. 2003;278:46179–46187. doi: 10.1074/jbc.M301874200. [DOI] [PubMed] [Google Scholar]
- 100.de Groot NS, Aviles FX, Vendrell J, Ventura S. FEBS J. 2006;273:658–668. doi: 10.1111/j.1742-4658.2005.05102.x. [DOI] [PubMed] [Google Scholar]
- 101.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, Wetzel R. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 102.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nunan J, Small DH. FEBS Lett. 2000;483:6–10. doi: 10.1016/s0014-5793(00)02076-7. [DOI] [PubMed] [Google Scholar]
- 104.Prangkio P, Yusko EC, Sept D, Yang J, Mayer M. Plos One. 2012;7:e47261. doi: 10.1371/journal.pone.0047261. [DOI] [PMC free article] [PubMed] [Google Scholar]