Abstract
The corticotrophin-releasing hormone (CRH) system integrates the stress response and is associated with stress-related psychopathology. Previous reports have identified interactions between childhood trauma and sequence variation in the CRH receptor 1 gene (CRHR1) that increase risk for affective disorders. However, the underlying mechanisms that connect variation in CRHR1 to psychopathology are unknown. To explore potential mechanisms, we used a validated rhesus macaque model to investigate association between genetic variation in CRHR1, anxious temperament (AT) and brain metabolic activity. In young rhesus monkeys, AT is analogous to the childhood risk phenotype that predicts the development of human anxiety and depressive disorders. Regional brain metabolism was assessed with 18F-labeled fluoro-2-deoxyglucose (FDG) positron emission tomography in 236 young, normally reared macaques that were also characterized for AT. We show that single nucleotide polymorphisms (SNPs) affecting exon 6 of CRHR1 influence both AT and metabolic activity in the anterior hippocampus and amygdala, components of the neural circuit underlying AT. We also find evidence for association between SNPs in CRHR1 and metabolism in the intraparietal sulcus and precuneus. These translational data suggest that genetic variation in CRHR1 affects the risk for affective disorders by influencing the function of the neural circuit underlying AT and that differences in gene expression or the protein sequence involving exon 6 may be important. These results suggest that variation in CRHR1 may influence brain function before any childhood adversity and may be a diathesis for the interaction between CRHR1 genotypes and childhood trauma reported to affect human psychopathology.
Keywords: amygdala, corticotrophin-releasing hormone, genetic association, hippocampus, non-human primate, rhesus macaque
INTRODUCTION
The corticotrophin-releasing hormone (CRH) system mediates the adaptive physiological and behavioral responses to stress and perceived threat.1,2 CRH and related endogenous ligands exert their effects through either of two receptors.2 Several studies indicate that genetic variation within the CRH receptor 1 gene (CRHR1) can affect reactivity to stress and that altered CRHR1 function is associated with stress-related psychopathology, particularly anxiety and depressive disorders.2–4 Importantly, specific CRHR1 haplotypes have been associated with the risk to develop depression following exposure to childhood trauma5–8 and with differential response to antidepressant treatment.9,10 However, despite extensive preclinical and clinical studies, the brain mechanisms by which sequence variation in the CRHR1 gene influences the risk to develop stress-related psychopathology have not been described. Identification of these intermediate mechanisms is critical for understanding the human risk factors associated with the development of anxiety and depressive disorders.
The well-validated rhesus macaque (Macaca mulatta) model11 of childhood anxious temperament (AT) provides an opportunity to characterize the effects of genetic variation in CRHR1 on anxiety-related phenotypes, including the function of the relevant neural circuits. Children with AT are extremely shy, exhibit high levels of behavioral inhibition when exposed to novelty or in the presence of strangers and are at increased risk to develop anxiety, depression and comorbid substance abuse.12,13 In addition to the behavioral phenotype, children with AT commonly display increased pituitary–adrenal and autonomic reactivity.12,13 Young, normally reared rhesus monkeys with high levels of trait-like anxiety show many similarities to children with AT, including behavioral inhibition, or freezing in response to potential threat.11 Furthermore, as in children, the degree of AT expressed by young monkeys is stable, trait-like and significantly heritable.14–16 We recently used fluoro-2-deoxyglucose-positron emission tomography (FDG-PET) to characterize the neural circuit underlying AT, finding that increased metabolic activity in the dorsal amygdala, central nucleus of the amygdala and anterior hippocampus was predictive of AT, whereas activity in the intraparietal sulcus (IPS) and precuneus was negatively correlated with AT.17 The goal of the current study was to use the macaque model to examine the influence of variation in the CRHR1 gene on individual differences in AT and the function of its underlying neural circuit.
MATERIALS AND METHODS
Study subjects and behavioral testing
Two hundred thirty six maternal-reared rhesus macaques (114 males, 122 females) ranging in age from 0.74–4.2 years (mean 2.4 years) were selected from a large, multi-generation single family pedigree and subjected to behavioral testing and FDG-PET brain imaging as previously described.17,18 AT was assessed by exposing each animal to the ‘no eye contact’ (NEC) condition of the human intruder paradigm for 30 min.11,19 In this test, the animal was removed from its home cage and placed alone in a cage in a separate room. An unfamiliar ‘human intruder’ then entered the testing room and stood in profile 2.5 m from the cage, avoiding eye contact with the animal. Previously, we developed and validated a composite measure of AT that incorporates behavioral inhibition (duration of freezing), vocalizations (frequency of ‘coo’ calling) and plasma cortisol levels measured immediately after the 30 min exposure to NEC.11,15
Measurement of brain metabolic activity
18F-labeled FDG (5–7 mCi) was administered intravenously to the subjects immediately before transfer to the test cage for NEC exposure. After the 30-min NEC period, the subject was sedated with ketamine HCl (15 mg/kg intramuscular), blood was drawn for cortisol measurement and the animal was further anesthetized with isoflurane gas for high-resolution microPET imaging (Concorde Microsystems P4 scanner, Knoxville, TN, USA). The use of FDG-PET imaging to quantify local brain metabolic activity optimally matches uptake and kinetic parameters of radiolabeled FDG with the duration of the NEC testing paradigm. Because FDG is trapped in the metabolic cell, it provides a reliable indicator of regional brain metabolic activity occurring during the 30 min exposure to this mildly threatening stressor.17,20,21
Quantification of regional metabolic activity
Based on our previous work,17,20 we focused primarily on regions strongly associated with emotion and reactivity to stress or threat, the hippocampus and amygdala. Secondarily, we also investigated two brain regions exhibiting negative correlations with AT, the IPS and precuneus. For full details of imaging methods, see Oler et al.17 Briefly, each subject’s FDG-PET scan (raw spatial resolution 2 mm3; see Kalin et al.20) was aligned to their T1-weighted MRI (magnetic resonance image) using rigid-body transformation and then aligned to a standard space template based on the T1-weighted MRI image using a nonlinear transformation (FSL; http://www.fmrib.ox.ac.uk/fsl/; Jenkinson et al.22). FDG-PET values were normalized to the whole-brain average and smoothed with a 4 mm full-width half-maximum gaussian kernel. Regional gray-matter probability estimates were based on the T1-weighted MRI and also blurred with a 4mm gaussian kernel.23 Mean metabolism was extracted from the central amygdala, anterior hippocampus, IPS and precuneus, regions that were most predictive of AT, along with 95% spatial confidence intervals.17 These mean metabolic values were then residualized for gray-matter probability and standardized.
Targeted sequencing of exons to identify functional single nucleotide polymorphisms (SNPs)
We used PCR amplification and Sanger methods to sequence all 14 exons and part of the 5′ flanking region of CRHR1 in 236 study subjects (Supplementary Figure 1). The PCR primers and the assay conditions used are available from the authors on request. This sequencing identified 103 SNPs (Supplementary Table 1) with a minimum minor allele frequency of 0.01. Linkage disequilibrium (LD) among SNPs was assessed as r2 using Haploview (Broad Institute, Cambridge, MA, USA).24 After excluding SNPs with ≥25% missing data (n=39), the polymorphisms were clustered into groups consisting of SNPs with pairwise r2≥0.8. Identification of SNP pairs showing high pair-wise LD is necessary because downstream tests of association between individual SNPs and phenotypes can produce inaccurate (false negative) results if ≥2 SNPs in a single analysis are in strong LD. We found 20 of the 103 SNPs were not in high LD (r2≤0.8) with any other SNP, and these singletons were included in subsequent multi-SNP analyses. We also found 10 sets of SNPs constituting high LD clusters. One SNP per cluster was included in the multi-SNP tests.
Annotation of novel SNPs
We used in-house software (Bainbridge, personal communication) to query the Ensembl genome browser and annotate the newly discovered variants according to putative functional classes (intronic, synonymous, nonsynonymous, splice site). Among the 103 SNPs, there were 5 nonsynonymous changes, 3 splice-site variants and 1 SNP that was both a non-synonymous change and a splice-site variant.
Tests of genetic association
Statistical association of individual SNPs with AT and local brain metabolic activity was tested using pedigree-based variance components analysis implemented in SOLAR.25 All phenotypes were adjusted for age before association tests. All tests were performed using age, sex and age × sex as co-variates, but these were not significant parameters in any analysis. These association analyses relating SNP genotypes and individual phenotypes were performed simultaneously using maximum likelihood estimation of the correlation between known pair-wise kinship among animals and pair-wise phenotypic differences among them. In this way, association across all animals between a SNP genotype and a given phenotype was adjusted for background kinship among pedigreed animals and the heritability of the phenotype.
Because we are primarily interested in identifying potentially functional SNPs that drive phenotypic variation, we first tested the effect of the nine annotated putative functional SNPs (amino acid changing non-synonymous variants and splice-site variants). Each of these nine SNPs were evaluated for genetic association with AT, testing one SNP at a time as a co-variate, with simultaneous estimation of the effect of overall kinship on phenotypic differences. Each of the three alternative models was tested for each SNP by coding the genotypes to accommodate a specific genetic model: additive effects of alleles, dominance of the major allele or dominance of the minor allele. Tests on the nine putative functional SNPs include some SNPs in LD with each other. The standard Bonferroni correction assumes that these tests are independent. Considering the LD among these SNPs, the effective number of independent SNPs in the set is seven,26 resulting in a Bonferroni-corrected family-wise 5% significance threshold of 0.007. The three different genetic models tested are also inter-correlated (additive model with either the dominant or recessive model r=0.8; dominance model with recessive model r=0.3) and not independent as required by the Bonferroni procedure. Analyses of these correlations suggests that the effective number of independent tests for each SNP is two,26 and the Bonferroni corrected family-wise 5% significance threshold is 0.025 for genetic models within SNPs. Correcting for both multiple SNPs and multiple genetic models gives a 5% family-wise threshold of 0.0036.
To assess and compare simultaneous effects of multiple SNPs, we next conducted a series of analyses using a variance components multiple regression framework. For each phenotypic trait, we ran measured genotype analyses in SOLAR as maximum-likelihood variance components multiple regressions with multiple SNPs run together as covariates (partial regressions). To avoid dilution of SNP effects (false negatives) resulting from LD among SNPs, we included only one member of each high-LD cluster (that is, one SNP from each set of two or more SNPs that exhibit high pair-wise LD with each other, see Supplementary Figure S3 and Supplementary Table S2). Before assessing the effects of each SNP, we evaluated the overall significance of each of the three multiple regression models. Only multiple regression models that were overall significant (P value ≤0.05) for an initial test of general heritability (h2) were then used to evaluate the effect of each SNP. The multiple regression analyses quantifying multiple SNP effects simultaneously were conducted using either 7 SNPs together (the putative functional non-synonymous and splice-site SNPs, excluding those that exhibit high LD) or 30 SNPs together (one from each of 10 identified LD clusters and 20 singletons, including the SNPs from exon 6 fixed in the model).
Predictions of protein structure
We used the online I-TASSER server27 to predict the three-dimensional structure of rhesus macaque CRHR1-alpha (isoform of the protein that does not include the amino acids encoded by exon 6) and variant DNA sequences putatively encoding altered protein sequences that we identified in this study. The I-TASSER system was the top scoring 3D modeling server in the CASP series of blind protein structure prediction experiments and bench-marking tests.27 We did not use a template for the I-TASSER analyses of the macaque protein sequences, as there were no appropriate full-length crystallized secretin-like G-protein-coupled receptors available. The amino-acid sequences for the seven transmembrane core region plus the C-terminal tail containing the alternative alleles were uploaded to the I-TASSER portal, and the standard I-TASSER protocol was used to generate predicted protein structures. C-scores calculated by I-TASSER were used to assess the quality of the protein structure prediction.
RESULTS
To investigate the influence of variation in the CRHR1 gene on individual differences in AT and related phenotypes in rhesus macaques, we sequenced all 14 exons and part of the 50 flanking region, identifying all SNPs. We next conducted a series of pedigree-based genetic association tests to determine whether that observed variation in the CRHR1 gene influences AT and/or metabolism in the relevant neural circuits during the NEC challenge test.
Gene-wide SNP discovery in rhesus CRHR1
PCR-based Sanger re-sequencing of all exons and a segment of the 50′ putative promoter region of CRHR1 identified 103 SNPs with minor allele frequency ≥0.01. Of these SNPs, nine are potentially functional, being either non-synonymous variants that alter protein amino-acid sequence or splice sites predicted to have possible effects on alternative splicing. The remaining 94 SNPs include 5 synonymous, 50 intronic, 11 3′ UTR SNPs and other flanking and intergenic SNPs. We were primarily interested in the putative functional SNPs because our goal was to identify causal variants with detectable phenotypic effects.
Separate tests of effects of functional SNPs on AT
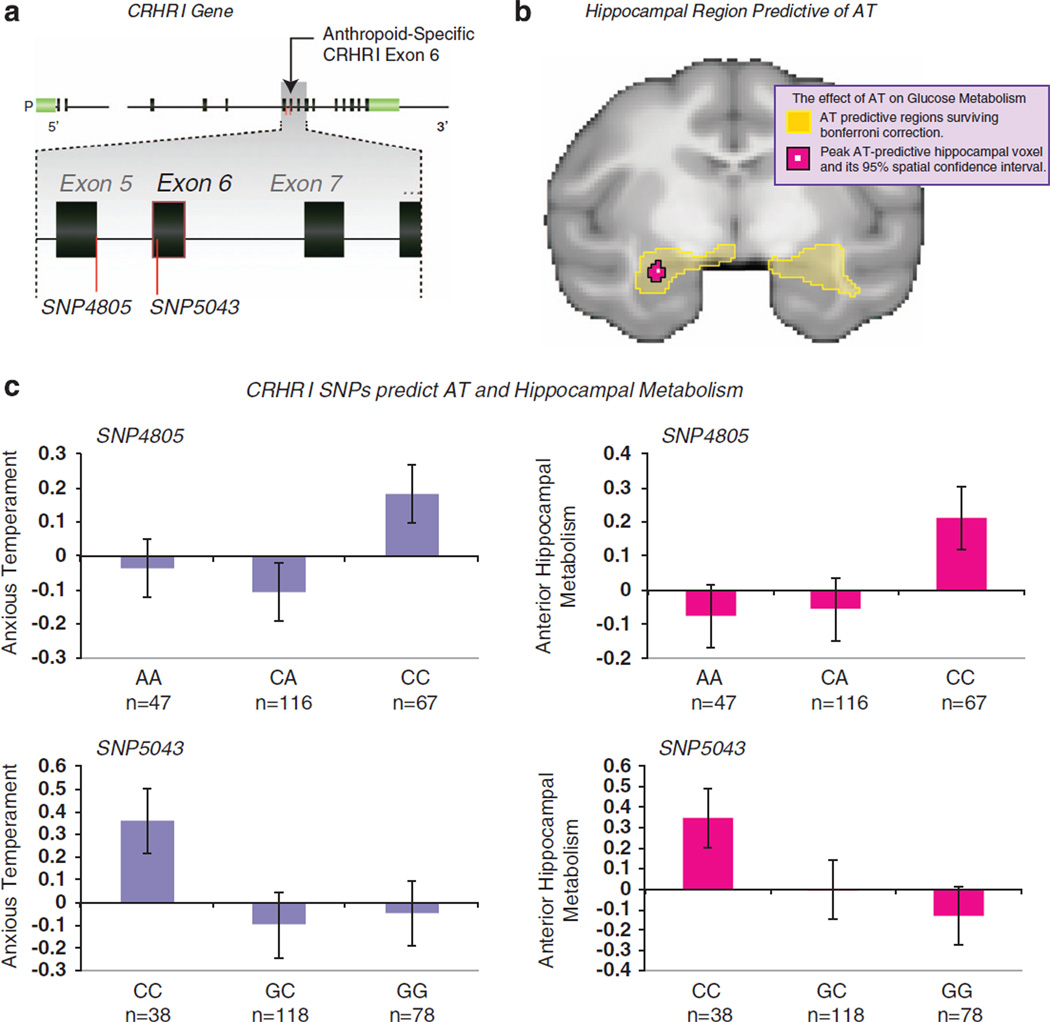
To begin, we tested each of the nine putative functional SNPs separately for genetic association with our composite measure of AT. Testing one SNP at a time, we found that three functional variants (SNP4805, SNP5043 and SNP8689) showed significant association (Table 1). None of these three SNPs exhibit strong pair-wise LD with each other, so these can be considered three independent associations. The most robust association was observed with SNP5043, an amino-acid change encoded by a nucleotide substitution in exon 6 (P=0.0007). This result was obtained when the effect of this SNP was modeled as a dominant system with the major allele (G) dominant to the minor (Figure 1). We note that this result survives conservative Bonferroni correction for multiple testing across both SNPs and genetic models. Probabilities at SNP4805 (P=0.012) and SNP8689 (P=0.017) are also significant when considered against the threshold correcting for number of genetic models. Considering these tests together, and following the approach described by Cheverud,26 we find that these nine SNPs constitute seven statistically independent tests, and the probability of observing three of these independent tests significant at the 0.05 level under the null hypothesis of no significant relationship between CRHR1 and AT is 4.3×10−6. These results demonstrate that variation in CRHR1 influences AT among young rhesus macaques.
Table 1.
Results of association testing for AT, hippocampus and amygdala using functional SNPs in separate tests
| SNP ID |
Phenotype |
|||||
|---|---|---|---|---|---|---|
|
Anxious temperament |
Hippocampus |
Amygdala |
||||
| P= | Genetic model | P= | Genetic model | P= | Genetic model | |
| SNP4805 | 0.012 | D-minor | 0.091 | D-minor | NS | |
| SNP5043 | 0.0007a | D-major | 0.016 | Add. | 0.009 | Add. |
| SNP5094 | 0.085 | Add.andD-minor | NS | 0.008 | Add.andD-minor | |
| SNP5107 | NS | NS | NS | |||
| SNP5886 | NS | NS | NS | |||
| SNP8689 | 0.017 | D-minor | NS | 0.034 | Add. | |
| SNP9006 | NS | NS | NS | |||
| SNP9009 | NS | NS | NS | |||
| SNP9026 | NS | NS | 0.018 | D-minor | ||
Abbreviations: AT, anxious temperament; NS, not significant; SNP, single nucleotide polymorphism.
This table presents individual tests of association for each of nine putatively functional SNPs in CRHR1 for three phenotypes of interest: AT, anterior hippocampal metabolism or metabolism in the central nucleus of the amygdala. The model used to test significance is indicated after the P-value: additive Add.), dominant-major (D-major) or dominant-minor (D-minor). Bolded P-values represent P≤0.05.
The association between SNP5043 and the AT phenotype survived Bonferroni correction for multiple comparisons.
Figure 1.
Association of corticotrophin-releasing hormone receptor 1 gene (CRHR1) single nucleotide polymorphisms (SNPs) with anxious temperament (AT) and hippocampal metabolism. (a) Diagram of the macaque CRHR1 gene showing relative location of SNP4805 and SNP5043. (b) Region within the left anterior hippocampus where glucose metabolism exhibited the strongest correlation with AT (c.f. Oler et al.,17). (c) Genotype plots showing the mean, s.e. and number of animals per genotype for AT and hippocampal metabolic activity across CRHR1 SNP genotypes.
Multiple regression tests of functional SNPs on AT
We next tested association between the CRHR1 gene and AT using multiple regression. This model evaluates multiple SNP variants simultaneously and accounts for multiple testing by using partial regression to estimate the effect of any given SNP within the context of all SNPs in the model. In this approach, SNPs exhibiting high pair-wise LD must be removed to avoid multicolinearity and false-negative results. Two pairs among the nine functional SNPs are in strong LD (r2 ≥ 0.80), so we conducted the multiple regression test using just seven of those functional SNPs. The seven functional SNPs were first tested simultaneously against AT to determine whether the overall multiple regression model was significant. All three genetic models (additive allelic effect, dominant-major effects and dominant-minor effects) produced significant results (Table 2, Panel A). Given those significant results, we then assessed the individual significance of each of the seven functional SNPs controlling for the other six SNPs. In that analysis (Table 2, Panel B), we found that three SNPs (SNP4805, SNP5043 and SNP5094) produce clear evidence of independent genetic association. Recall that SNP5043 is in exon 6 and causes an amino-acid change in the protein. SNP5094 is also in exon 6 and causes a different amino-acid change, while SNP4805 alters a splice site at the 3′ end of exon 5, potentially altering the splicing of exon 6. Interestingly, exon 6 has evolved significantly during primate evolution. Its sequence is quite different in humans, apes and monkeys compared with other more distantly related strepsirrhine primates and other mammals (see Figure 2 and Discussion below).
Table 2.
Results of association testing using the multiple regression approach with seven functional SNPs
|
(A) Multiple regression analysis testing overall genetic model with seven SNPs | |||
|---|---|---|---|
|
Phenotype |
|||
| Anxioustemperament | Hippocampus | Amygdala | |
| P = | P = | P = | |
| Additive | 0.043 | 0.0017 | NS |
| D-major | 0.017 | 0.003 | NS |
| D-minor | 0.032 | 0.0024 | NS |
|
(B) Multiple regression results for seven functional SNPs tested simultaneously | ||||
|---|---|---|---|---|
|
Phenotype |
||||
| AnxiousTemperament | Hippocampus | |||
| SNP4805 | 0.014 | D-minor | 0.017 | D-minor |
| SNP5043 | 0.0003 | D-major | 0.03 | D-major |
| SNP5094 | 0.041 | D-minor | NS | |
| SNP5886 | NS | NS | ||
| SNP8689 | NS | NS | ||
| SNP9009 | NS | NS | ||
| SNP9026 | NS | NS | ||
Abbreviation: SNP, single nucleotide polymorphism.
Using a multiple regression approach after taking linkage disequilibrium into account, seven putatively functional SNPs were assessed for association with anxious temperament (AT), hippocampal metabolism or metabolism in the central amygdala. (A) The significance of the full model for each of three alternative models (additive, dominance-major (D-major) and dominance-minor (D-minor)) was assessed for each phenotype of interest. (B) The partial regressions (SNPs) were assessed for those phenotypes in which the full model was significant (AT and hippocampus). No partial regression results (SNPs) were considered for amygdale metabolism because the overall model was not statistically significant.
Bolded P-values represent P≤0.05.
Figure 2.
Corticotrophin-releasing hormone receptor 1 gene (CRHR1) protein segment encoded by exon 6 is conserved primarily in anthropoid primates. A portion of the aligned protein sequence of CRHR1 containing the segment encoded by exon 6 is illustrated for 12 mammals: five anthropoid primates (human, chimpanzee, gorilla, macaque and marmoset), two more distant primates (bushbaby and mouse lemur), three placental mammals (rat, dog and microbat), one marsupial (opossum) and one monotreme (platypus). The cladogram of the known phylogenetic relationships among these species is presented on the left. The sequence encoded by part of exon 5 and all of exon 6 (bounded by the red square) is shown on the right. Residues identical to the human sequence are shaded for exon 5 (light gray) and exon 6 (blue). Note that while exon 5 is highly conserved across all 12 species, exon 6 is conserved primarily in anthropoid primates.
Multiple regression tests of full SNP data
Out of 103 total SNPs identified, 20 do not exhibit pairwise LD of r2≥0.8 with any other SNP in the data set and were scored in at least 75% of subjects. We also found 10 clusters of SNPs that show significant LD, so the data set for this analysis was reduced by selecting one SNP from each of the 10 clusters of linked markers. To determine whether any additional SNPs exhibit significant effects on AT beyond the documented effects of the three functional exon 6 SNPs discussed above, we tested the data set of 30 SNPs while maintaining the three exon 6 SNPs in the model. We found that three additional polymorphisms were significantly associated with AT, independent of the exon 6 SNPs and each other (Table 3, Panel B). Two of these (SNP2107 and SNP0879) are located in the putative promoter region of this gene.4,28
Table 3.
Results of genetic association testing using 30 gene-wide SNPs in CRHR1
|
(A) Multiple regression analysis of overall genetic models with exon 6 SNPs fixed | ||
|---|---|---|
|
Phenotype |
||
| Anxioustemperament | Hippocampus | |
| P = | P = | |
| Additive | NS | 0.001 |
| D-major | 0.014 | 0.008 |
| D-minor | 0.034 | 0.028 |
|
(B) Multiple regression analysis testing 30 SNPs simultaneously with exon6 SNPs fixed in model | ||||
|---|---|---|---|---|
|
Phenotype |
||||
|
Anxioustemperament |
Hippocampus |
|||
| P= | Geneticmodel | P= | Geneticmodel | |
| SNP5886 | 0.043 | D-minor | NS | |
| SNP9009 | NS | 0.044 | D-minor | |
| SNP2107 | 0.017 | D-major | 0.017 | D-major |
| SNP0879 | 0.012 | D-minor | 0.021 | Add., D-minor |
| SNP4820 | NS | 0.048 | D-minor | |
| SNP5137 | NS | 0.015 | D-minor | |
Abbreviations: CRHR1, corticotrophin-releasing hormone receptor 1; SNP, single nucleotide polymorphism.
After taking linkage disequilibrium into account, 30 SNPs were assessed for association with anxious temperament (AT) and hippocampal metabolism using a multiple regression approach. The SNPs that showed significant association in the previous seven SNP analysis were held fixed in the model (SNP4805, SNP5043 and SNP5094). (A) The significance of the full model for each of the three alternative models (additive (Add.), dominance-major (Dmajor) and dominance-minor (D-minor)) was assessed for AT and hippocampal metabolism. (B) For each multiple regression model that was significant, the partial regressions (SNPs) were assessed. When multiple models were significant for the same SNP, the lowest P-value is presented.
Bolded P-values represent P≤0.05.
Association tests with hippocampal brain metabolism
The statistically significant results for association between AT and specific variants in CRHR1 provide strong justification for investigating possible genetic effects on neural circuits that are correlated with AT.17 We previously showed that metabolic activity in the anterior hippocampus predicts AT in macaques and is heritable.17 Furthermore, we note that altered expression of CRHR1 in the forebrain of genetically engineered mice affects neural function in the hippocampus and amygdala and reduces anxious behavior.29 We tested the nine functional SNPs separately for association with metabolic activity in the anterior hippocampus (Table 1) and found significant association only with SNP5043 (P=0.016, uncorrected). This SNP also showed the strongest individual association with AT. In the next analysis, we included the seven functional SNPs (those that do not show high pairwise LD) in the multiple regression model testing for association with hippocampal activity. The overall regression model was significant (Table 2, Panel A), with two SNPs (SNP4805 and SNP5043) showing significant independent effects (Panel B). Finally, we used the multiple regression framework to assess the simultaneous effects of the full set of 30 SNPs across the gene, while forcing the model to retain the two exon 6 SNPs (SNP4805 and SNP5043) identified to be associated with hippocampal metabolism. This analysis identified five more SNPs with significant association to hippocampal activity (Table 3, panel B). Two of these were also significantly associated with AT in the analogous multiple regression test (SNP2107 and SNP0879) and are in the putative promoter region.4,28
Association tests with amygdala central nucleus metabolism
We next investigated possible association between CRHR1 SNPs and brain metabolism in the central nucleus of the amygdala. Among the nine functional SNPs tested separately (Table 1), four (SNP5043, SNP5094, SNP8689 and SNP9026) show association, with P-values between 0.008 and 0.034. These are not significant under Bonferroni correction across SNPs and genetic models jointly but SNP5043, SNP5094 and SNP9026 are significant when corrected only across models. Considered together, the probability of observing four independent tests significant at the 5% level under the null hypothesis of no association between SNPs and central amygdala metabolism is 2.4 × 10−10. Overall, these results strongly support an association between CRHR1 and amygdala activity, but it is not certain which of the four SNPs could be a false positive. The overall multiple regression testing of CRHR1 SNPs against activity within the central amygdala generated no significant results under any of the three genetic models and thus provides no justification for evaluating individual SNPs within this regression framework. Thus, there is clear evidence for association between CRHR1 SNP genotypes and brain metabolism in the central nucleus of the amygdala, but it remains uncertain which specific SNPs are involved.
Association tests with metabolism in the IPS
We next analyzed genetic association between the identified SNPs and metabolism in the IPS, a cortical region that participates in the integration of sensorimotor tasks with visual inputs and information about the location and orientation of the body in space.30 We previously found IPS metabolism to be significantly negatively correlated with AT.17 When the nine functional SNPs were tested independently against metabolism within the IPS, one polymorphism (SNP5043) generates a P-value of 0.0037, essentially reaching the joint Bonferroni threshold for genetic models and SNPs (threshold=0.0036). As shown in Table 4, three other SNPs (SNP5094, SNP8689 and SNP9006) generate P-values <0.05, but >0.0036. As with the amygdala test above, the probability of observing four SNPs with independent probabilities <0.05 under the null hypothesis of no association between CRHR1 and IPS metabolism is 2.4 × 10−10. When the seven functional SNPs are tested simultaneously, two (SNP5043 and SNP5094) exhibit statistically significant association with IPS metabolism (P=0.0076 and 0.0066, respectively). In the multiple regression test of the 30 SNPs, the multiple regression model is significant, and only one polymorphism (SNP9026) is specifically associated with the phenotype (P=0.036). Overall, these results show that variation in CRHR1 is also associated with local brain activity in the IPS.
Table 4.
Results of association testing for intraparietal sulcus and precuneus using functional SNPs in separate tests
| SNP ID |
Phenotype |
|||
|---|---|---|---|---|
|
Intraparietal sulcus |
Precuneus |
|||
| P = | Genetic model | P = | Genetic model | |
| SNP4805 | NS | n.s | ||
| SNP5043 | 0.0037 | Add. | 0.026 | Add. |
| SNP5094 | 0.012 | Add.and | 0.029 | Add.and |
| D-minor | D-major | |||
| SNP5107 | NS | NS | ||
| SNP5886 | NS | NS | ||
| SNP8689 | 0.029 | Add. | 0.015 | D-minor |
| SNP9006 | 0.0074 | D-major | 0.019 | D-major |
| SNP9009 | NS | NS | ||
| SNP9026 | NS | NS | ||
Abbreviation: SNP, single nucleotide polymorphism.
Individual tests of association for each of the nine putatively functional SNPs in CRHR1 (corticotrophin-releasing hormone receptor 1) for intraparietal sulcus and precuneus. The model used to test significance is indicated after the P-value: additive (Add.), dominant-major (D-major) or dominantminor (D-minor).
Bolded P-values represent P≤0.05.
Association tests with metabolism in the precuneus
Our previous analyses17 also indicate that AT is negatively correlated with brain metabolism in the precuneus. This region of medial parietal cortex is closely integrated with the IPS and is involved in a number of different functions, including integration of information supporting purposeful visually guided body movements in three-dimensional space, episodic memory and visual imagery related to self-representation.31 When the nine CRHR1 functional SNPs were tested independently against precuneus metabolism, none produced P-values <0.01, and thus none survive joint Bonferroni correction for genetic models and SNPs. Two variants (SNP8689 and SNP9006) have P-values <0.02 (Table 4), which survive Bonferroni correction for genetic models alone. Because four variants exhibit p-values <0.05, we once again conclude that the CRHR1 locus influences metabolism in this region, although we cannot identify specific causal SNPs with confidence. Like our analyses of amygdala metabolism, none of the genetic models in the multiple regression analysis of either the sets of seven SNPs or 30 SNPs produce significant overall genetic effects, so there is no justification for evaluating the association between individual SNPs and precuneus metabolism within the multiple regression framework.
Analysis of protein structure
Finally, we used the online I-TASSER software package27 to predict the effects of selected SNPs on the three-dimensional structure of the macaque CRHR1 protein. These results (Supplementary Figure S4) must be considered preliminary because no validated three-dimensional crystal structure is available for any isoform of CRHR1 for any species, and I-TASSER performs best when its predictions are based on templates of known 3D structure. Nevertheless, the results suggest that SNP5043 and SNP5094 individually affect the shape and/or orientation of the first intra-cellular loop of the protein. Definitive conclusions await direct functional assays, but these predictions support the idea that these two mutations do indeed alter 3D protein structure and, therefore, may influence function. We note that normal CRHR1 function may involve both homo-dimerization and hetero-dimerization.32
DISCUSSION
Despite significant progress in the quantitative genetic analysis of the risk to develop anxiety and depressive disorders, knowledge of the specific genes, genetic mechanisms and physiological intermediates underlying this risk remains limited. Several studies have reported associations between CRH-related genes and clinical anxiety or depression,2–4,33 as well as association between CRH genes and related phenotypes in macaques.34 Several analyses report gene versus environment interactions in which early childhood trauma interacts with CRHR1 genotypes to predict stress-related psychopathology later in life.3,5–8,35,36 However, these data do not address the intermediate neurobiological pathways that lead from inherited genetic differences to altered behavioral and physiological reactivity and ultimately to psychopathology. Due to fundamental similarities in the brain structure and behavioral reactivity to stress, rhesus monkey models are well suited for analysis of genetic effects that alter the function of neural circuits implicated in human psychopathology.37–39
In this study, we identified several common SNPs in the rhesus macaque CRHR1 gene that are independently associated with AT. Specifically, we find that SNP4805, a splice-site mutation that may affect the inclusion of exon 6 into the CRHR1 protein, and another variant (SNP5043), which alters the amino-acid sequence within exon 6, are associated with individual differences in AT. We also obtained support for association between another exon 6 variant (SNP5094) and AT. To understand how variation in the CRHR1 gene increases the behavioral and physiological reactivity to threat that is characteristic of individuals with high levels of AT, we examined brain metabolic activity in regions previously shown to be predictive of AT.17 The results indicate that multiple polymorphisms in the CRHR1 locus are significantly associated with individual variation in metabolic activity of the anterior hippocampus and central nucleus of the amygdala occurring during the human intruder challenge. We also find significant association between CRHR1 and metabolic activity in the IPS, with suggestive evidence for association with the precuneus.
We note that three SNPs that show significant effects on AT are predicted to impact exon 6 of the CRHR1 gene (Figure 1). SNP4805 alters a splice site for intron 5, while both SNP5043 and SNP5094 change the amino-acid sequence encoded by exon 6. Figure 2 shows an alignment of partial CRHR1 protein sequences from various species, including several primates and non-primate mammals. The amino-acid sequence of the core of the protein (the segment containing the seven transmembrane domains) is overall highly conserved across all mammals (Figure 2 and Supplementary Figure S2). However, the segment of the protein encoded by exon 6 (part of the first intracellular loop) in anthropoid primates (human, chimpanzee, gorilla, macaque and marmoset (Callithrix jacchus) is much less conserved in the strepsirrhine primates (Microcebus murinus or mouse lemur and Otolemur garnettii or bushbaby) and in non-primate mammals. It appears that human exon 6 evolved its current structure in the ancestor of living anthropoid primates (that is, living New World monkeys, Old World monkeys and apes) and is quite different from non-primate mammals.
Previous functional analysis indicates that the presence or absence of exon 6 has a significant influence on CRHR1 function. Markovic et al.40 demonstrated that CRHR1-alpha, the form of the protein that does not incorporate the amino acids encoded by exon 6 and which is the dominant isoform expressed in humans, responds differently to protein kinase-C induced phosphorylation compared with CRHR1-beta, which is identical to CRHR1-alpha except for the inclusion of those residues. As a result, CRHR1-beta exhibits substantially impaired signaling activity. Teli et al.41 identified specific amino-acid residues within the human exon 6 that account for impaired responsivity of this receptor. The relative levels of expression of CRHR1-alpha and -beta have not been documented in specific structures within the human or nonhuman primate brain (for example, amygdala and hippocampus). Nor have levels of CRHR1-beta been reported in patients with anxiety or mood disorders. The data presented here raise the possibility that genetic variation in the exon 6 splice site (SNP4805) may alter the relative proportion of CRHR1-beta as compared with CRHR1-alpha in individuals with high levels of anxiety and hippocampal or amygdalar metabolism. Additionally, amino-acid changes within exon 6 could alter the function of CRHR1-beta in CRHR1-beta-expressing tissues, leading to increased metabolism, and ultimately AT.
Our results indicate that polymorphisms involving exon 6 are associated with metabolism in the anterior hippocampus, amygdala, IPS and precuneus, as well as with AT. The recent demonstration29 that genetically engineered mice that do not express CRHR1 in the forebrain exhibit impaired propagation of neural signals within the hippocampus and amygdala and also display reduced anxious behavior, indicates that CRHR1 has a critical role in the expression of AT in response to challenge. The relationship between metabolism in the IPS and precuneus and correlated expression of AT is less clear. It is plausible that increased freezing (behavioral inhibition), a component of our composite measure of AT that results in reduced motor activity and locomotion, explains the reduced metabolic activity in these brain regions. However, it is also true that the IPS and precuneus are part of the default mode network,42 and alterations in the function of regions involved in the default mode may be relevant to AT.
When we extended our analysis to include other non-coding SNPs in CRHR1, we found that variation in the 5′ flanking region of CRHR1 is also associated with both anterior hippocampal metabolism and AT. The effects of two putative promoter SNPs (SNP2107 and SNP0879) were statistically significant for both these phenotypes. Neither is in close LD with the exon 6 SNPs. This suggests a possible additional mechanism through which variation in the CRHR1 locus may affect AT, that is, differences in overall levels of gene expression. We note that results from the genetic analysis of complex phenotypes in humans suggest that it is common for a single gene to exhibit several different polymorphisms influencing a single phenotype.43–45
Much of the human CRHR1 genetic data involve gene versus environment interactions, especially interactions with early childhood trauma. Our data were obtained from juvenile monkeys that were normally reared and not exposed to extreme trauma. These findings suggest that children with specific CRHR1 genotypes may exhibit differences in brain activity that precede the expression of clinically significant anxiety and depressive disorders. The interaction between this CRHR1-driven genetic effect on altered early neural circuit activity on the one hand and subsequent child abuse, trauma or other serious adverse environmental events on the other may account for the reported gene-by-environment interaction. More specifically, the early effects of CRHR1 genetic variation that lead to increased activity of the neural circuit, which includes the hippocampus and amygdala, may be the diathesis through which childhood adversity further increases the risk to develop later anxiety and depressive disorders.
Supplementary Material
ACKNOWLEDGEMENTS
This work has been supported by National Institutes of Health Grants MH046729 (to NHK), MH081884 (to NHK and JR), MH084051 (to RJD and NHK), the HealthEmotions Research Institute and the Baylor College of Medicine. We thank the staff at the Wisconsin National Primate Center, the Harlow Center for Biological Psychology, the Waisman Laboratory for Brain Imaging and Behavior, P. Roseboom, H. Van Valkenberg, K. Myer, E. Larson, M. Riedel and J. Storey. We also thank Matthew Bainbridge (BCM) for annotation software and Panagiotis Katsonis (BCM) for assistance with comparative protein alignments. We would like to acknowledge the seminal contributions of Wylie Vale in relation to CRF biology and dedicate this manuscript in fond memory of his friendship, collegiality and support.
Footnotes
CONFLICT OF INTEREST
NHK is the founder and principal owner of a company, Promoter Neuroscience, which is focused on developing tools and finding drugs that affect expression of genes in the CRH family. He also holds the following patents: Promoter sequences for corticotropin-releasing factor CRF2alpha and method of identifying agents that alter the activity of the promoter sequences: US Patent issued on 07-04-06; patent No. 7,071,323 and US Patent issued on 05-12-09; patent No. 7,531,356; Promoter sequences for urocortin II and the use thereof: US Patent issued on 08-08-06; patent No. 7,087,385; and Promoter sequences for corticotropin-releasing factor binding protein and use thereof: US Patent issued on 10-17-06; patent No. 7,122,650. The other authors declare no conflicts of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
REFERENCES
- 1.Arborelius L, Owens MJ, Plotsky PM, Nemeroff CB. The role of corticotrophin-releasing factor in depression and anxiety disorders. J Endocrinol. 1999;160:1–12. doi: 10.1677/joe.0.1600001. [DOI] [PubMed] [Google Scholar]
- 2.Binder EB, Nemeroff CB. The CRF system, stress, depression and anxiety-insights from human genetic studies. Mol Psychiatry. 2010;15:574–588. doi: 10.1038/mp.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, Zhu F, Wang G, Xiao Z, Wang H, Tang J, et al. Association of corticotrophin-releasing hormone receptor1 gene SNP and haplotype with major depression. Neurosci Lett. 2006;404:358–362. doi: 10.1016/j.neulet.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Keck ME, Kern N, Erhardt A, Unschuld PG, Ising M, Salyakina D, et al. Combined effects of exonic polymorphisms in CRHR1 and AVPR1B genes in a case/control study for panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1196–1204. doi: 10.1002/ajmg.b.30750. [DOI] [PubMed] [Google Scholar]
- 5.Bradley RG, Binder EB, Epstein MP, Tang Y, Nair HP, Liu W, et al. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry. 2008;65:190–200. doi: 10.1001/archgenpsychiatry.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heim C, Bradley B, Mletzko TC, Deveau TC, Musselman DL, Nemeroff CB, et al. Effect of childhood trauma on adult depression and neuroendocrine function: sex-specific moderation by CRH receptor 1 gene. Front Behav Neurosci. 2009;3:41. doi: 10.3389/neuro.08.041.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polanczyk G, Caspi A, Williams B, Price TS, Danese A, Sugden K, et al. Protective effect of CRHR1 gene variants on the development of adult depression following childhood maltreatment: replication and extension. Arch Gen Psychiatry. 2009;66:978–985. doi: 10.1001/archgenpsychiatry.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyrka AR, Price LH, Gelernter J, Schepker C, Anderson GM, Carpenter LL. Interaction of childhood maltreatment with the corticotropin-releasing hormone receptor gene: effects on hypothalamic-pituitary-adrenal axis reactivity. Biol Psychiatry. 2009;66:681–685. doi: 10.1016/j.biopsych.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Licinio J, O’Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, et al. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Mol Psychiatry. 2004;9:1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Zhu F, Wang G, Xiao Z, Tang J, Liu W, et al. Association study of corticotropin- releasing hormone receptor1 gene polymorphisms and antidepressant response in major depressive disorders. Neurosci Lett. 2007;414:155–158. doi: 10.1016/j.neulet.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Kalin NH, Shelton SE. Nonhuman primate models to study anxiety, emotion regulation, and psychopathology. Ann NY Acad Sci. 2003;1008:189–200. doi: 10.1196/annals.1301.021. [DOI] [PubMed] [Google Scholar]
- 12.Rapee RM, Schniering CA, Hudson JL. Anxiety disorders during childhood and adolescence: origins and treatment. Annu Rev Clin Psychol. 2009;5:311–341. doi: 10.1146/annurev.clinpsy.032408.153628. [DOI] [PubMed] [Google Scholar]
- 13.Fox NA, Henderson HA, Marshall PJ, Nichols KE, Ghera MM. Behavioral inhibition: linking biology and behavior within a developmental framework. Annu Rev Psychol. 2005;56:235–262. doi: 10.1146/annurev.psych.55.090902.141532. [DOI] [PubMed] [Google Scholar]
- 14.Rogers J, Shelton SE, Shelledy W, Garcia R, Kalin NH. Genetic influences on behavioral inhibition and anxiety in juvenile rhesus macaques. Genes Brain Behav. 2008;7:463–469. doi: 10.1111/j.1601-183X.2007.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox AS, Shelton SE, Oakes TR, Davidson RJ, Kalin NH. Trait-like brain activity during adolescence predicts anxious temperament in primates. PLoS ONE. 2008;3:e2570. doi: 10.1371/journal.pone.0002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalin NH, Shelton SE. Ontogeny and stability of separation and threat-induced defensive behaviors in rhesus monkeys during the first year of life. Am J Primatol. 1998;44:125–135. doi: 10.1002/(SICI)1098-2345(1998)44:2<125::AID-AJP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, et al. Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature. 2010;466:864–868. doi: 10.1038/nature09282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalin NH, Shelton SE, Rickman M, Davidson RJ. Individual differences in freezing and cortisol in infant and mother rhesus monkeys. Behav Neurosci. 1998;112:251–254. doi: 10.1037//0735-7044.112.1.251. [DOI] [PubMed] [Google Scholar]
- 19.Kalin NH, Shelton SE. Defensive behaviors in infant rhesus monkeys: environmental cues and neurochemical regulation. Science. 1989;243:1718–1721. doi: 10.1126/science.2564702. [DOI] [PubMed] [Google Scholar]
- 20.Kalin NH, Shelton SE, Fox AS, Oakes TR, Davidson RJ. Brain regions associated with the expression and contextual regulation of anxiety in primates. Biol Psychiatry. 2005;58:796–804. doi: 10.1016/j.biopsych.2005.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox AS, Oakes TR, Shelton SE, Converse AK, Davidson RJ, Kalin NH. Calling for help is independently modulated by brain systems underlying goal-directed behavior and threat perception. Proc Nat Acad Sci USA. 2005;102:4176–4179. doi: 10.1073/pnas.0409470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuro- Image. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87(Pt 1):52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 27.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parham KL, Zervou S, Karteris E, Catalano RD, Old RW, Hillhouse EW. Promoter analysis of human corticotropin-releasing factor (CRF) type 1 receptor and regulation by CRF and urocortin. Endocrinology. 2004;145:3971–3983. doi: 10.1210/en.2004-0194. [DOI] [PubMed] [Google Scholar]
- 29.Refojo D, Schweizer M, Kuehne C, Ehrenberg S, Thoeringer C, Vogl AM, et al. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science. 2011;333:1903–1907. doi: 10.1126/science.1202107. [DOI] [PubMed] [Google Scholar]
- 30.Grefkes C, Fink GR. The functional organization of the intraparietal sulcus in humans and monkeys. J Anat. 2005;207:3–17. doi: 10.1111/j.1469-7580.2005.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavanna AE, Trimble MR. The precuneus: a review of its functional anatomy and behavioural correlates. Brain. 2006;129(Pt 3):564–583. doi: 10.1093/brain/awl004. [DOI] [PubMed] [Google Scholar]
- 32.Murat B, Devost D, Andres M, Mion J, Boulay V, Corbani M, et al. V1b and CRHR1 receptor heterodimerization mediates synergistic biological actions of vasopressin and CRH. Mol Endocrinol. 2012;26:502–520. doi: 10.1210/me.2011-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smoller JW, Rosenbaum JF, Biederman J, Kennedy J, Dai D, Racette SR, et al. Association of a genetic marker at the corticotropin-releasing hormone locus with behavioral inhibition. Biol Psychiatry. 2003;54:1376–1381. doi: 10.1016/s0006-3223(03)00598-5. [DOI] [PubMed] [Google Scholar]
- 34.Barr CS, Dvoskin RL, Gupte M, Sommer W, Sun H, Schwandt ML, et al. Functional CRH variation increases stress-induced alcohol consumption in primates. Proc Natl Acad Sci USA. 2009;106:14593–14598. doi: 10.1073/pnas.0902863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller MB, Zimmermann S, Sillaber I, Hagemeyer TP, Deussing JM, Timpl P, et al. Limbic corticotropin-releasing hormone receptor 1 mediates anxiety-related behavior and hormonal adaptation to stress. Nat Neurosci. 2003;6:1100–1107. doi: 10.1038/nn1123. [DOI] [PubMed] [Google Scholar]
- 36.Ressler KJ, Bradley B, Mercer KB, Deveau TC, Smith AK, Gillespie CF, et al. Polymorphisms in CRHR1 and the serotonin transporter loci: gene x gene x environment interactions on depressive symptoms. Am J Med Genet B Neuropsychiatr Genet. 153B:812–824. doi: 10.1002/ajmg.b.31052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr CS, Newman TK, Becker ML, Parker CC, Champoux M, Lesch KP, et al. The utility of the non-human primate; model for studying gene by environment interactions in behavioral research. Genes Brain Behav. 2003;2:336–340. doi: 10.1046/j.1601-1848.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 38.Barr CS, Schwandt M, Lindell SG, Chen SA, Goldman D, Suomi SJ, et al. Association of a functional polymorphism in the mu-opioid receptor gene with alcohol response and consumption in male rhesus macaques. Arch Gen Psychiatry. 2007;64:369–376. doi: 10.1001/archpsyc.64.3.369. [DOI] [PubMed] [Google Scholar]
- 39.Vallender EJ, Ruedi-Bettschen D, Miller GM, Platt DM. A pharmacogenetic model of naltrexone-induced attenuation of alcohol consumption in rhesus monkeys. Drug Alcohol Depend. 2010;109:252–256. doi: 10.1016/j.drugalcdep.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markovic D, Papadopoulou N, Teli T, Randeva H, Levine MA, Hillhouse EW, et al. Differential responses of corticotropin-releasing hormone receptor type 1 variants to protein kinase C phosphorylation. J Pharmacol Exp Ther. 2006;319:1032–1042. doi: 10.1124/jpet.106.107441. [DOI] [PubMed] [Google Scholar]
- 41.Teli T, Markovic D, Hewitt ME, Levine MA, Hillhouse EW, Grammatopoulos DK. Structural domains determining signalling characteristics of the CRH-receptor type 1 variant R1beta and response to PKC phosphorylation. Cell Signal. 2008;20:40–49. doi: 10.1016/j.cellsig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, et al. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 43.Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, et al. Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet. 2007;16:1051–1057. doi: 10.1093/hmg/ddm050. [DOI] [PubMed] [Google Scholar]
- 44.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum Mol Genet. 2011;20:R158–R162. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Achkar JP, Duerr R. The expanding universe of inflammatory bowel disease genetics. Curr Opin Gastroenterol. 2008;24:429–434. doi: 10.1097/MOG.0b013e3283009c92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.