Abstract
Endochondral bone formation is a multistep process during which a cartilage primordium is replaced by mineralized bone. Several genes involved in cartilage and bone development have been identified as target genes for the Snail family of zinc finger transcriptional repressors, and a gain-of-function study has demonstrated that upregulation of Snai1 activity in mouse long bones caused a reduction in bone length. However, no in vivo loss-of-function studies have been performed to establish whether Snail family genes have an essential, physiological role during normal bone development. We demonstrate here that the Snai1 and Snai2 genes function redundantly during embryonic long bone development in mice. Deletion of the Snai2 gene, or limb bud-specific conditional deletion of the Snai1 gene, did not result in obvious defects in the skeleton. However, limb bud-specific Snai1 deletion on a Snai2 null genetic background resulted in substantial defects in the long bones of the limbs. Long bones of the Snai1/Snai2 double mutants exhibited defects in chondrocyte morphology and organization, inhibited trabecular bone formation and delayed ossification. Chondrocyte proliferation was markedly reduced, and transcript levels of genes encoding cell cycle regulators, such as p21Waf1/Cip1, were strikingly upregulated in the Snai1/Snai2 double mutants, suggesting that during chondrogenesis Snail family proteins act to control cell proliferation by mediating expression of cell cycle regulators. Snai2 transcript levels were increased in Snai1 mutant femurs, while Snai1 transcript levels were increased in Snai2 mutant femurs. In addition, in the mutant femurs the Snai1 and Snai2 genes compensated for each other's loss not only quantitatively, but also by expanding their expression into the other genes' normal expression domains. These results demonstrate that the Snai1 and Snai2 genes transcriptionally compensate temporally, spatially, and quantitatively for each other's loss, and demonstrate an essential role for Snail family genes during chondrogenesis in mice.
Keywords: SNAIL, SLUG, PRRX1-CRE, FUNCTIONAL REDUNDANCY, ENDOCHONDRAL OSSIFICATION
Introduction
During embryonic long bone development, bone is formed through endochondral ossification, a multistep process in which a cartilage template is replaced by mineralized bone (1,2). To initiate long bone development in the vertebrate limb, lateral plate mesoderm-derived mesenchymal progenitor cells migrate into the limb bud. These cells form mesenchymal condensations that prefigure the skeletal elements of the limb. Cells in the center of the condensation differentiate into chondrocytes, which produce an extracellular matrix largely composed of type II collagen and the chondroitin sulfate proteoglycan aggrecan. The process of chondrocyte maturation, which includes chondrocyte proliferation, production of extracellular matrix, arrangement into longitudinal columns, and finally chondrocyte hypertrophy and apoptosis, is responsible for longitudinal bone growth during development.
The Snail gene family encodes zinc finger proteins that function primarily as transcriptional repressors. Three members of the Snail family have been described in mammals: Snai1 (formerly Snail), Snai2 (formerly Slug), and Snai3. The SNAI1 and SNAI2 proteins are key regulators of the epithelial-mesenchymal transition, directly repressing transcription of genes encoding components of cell-cell adhesive complexes in epithelia. The SNAI1 and SNAI2 proteins also have demonstrated roles in other important developmental and cellular processes, such as the protection of cells from programmed cell death, the establishment of left-right asymmetry and the regulation of cell motility (3–5).
A previous gain-of-function study using a tamoxifen-inducible Snai1 transgenic line demonstrated that upregulation of Snai1 activity in mouse long bones caused a reduction in bone length (6), and several genes involved in cartilage and bone development have previously been demonstrated or implicated as Snail target genes (7–10). However, no in vivo loss-of-function studies have been performed to date, leaving it unclear whether Snail family genes have a physiological role during normal bone development. Here we demonstrate that the Snai1 and Snai2 genes function redundantly during embryonic long bone development, and transcriptionally compensate (temporally, spatially and quantitatively) for each other's loss. These studies demonstrate an essential role for Snail family genes during chondrogenesis in mice.
Materials and Methods
Mice
The Snai2lacZ (formerly called SluglacZ; henceforth designated Snai2−) null allele (11) and Snai1flox conditional allele (12) have been described previously. Prrx1-Cre mice (13) were obtained from the Jackson Laboratory. Animal maintenance and experimental procedures were in accordance with NIH Guidelines for Animal Care and Use, and were approved by the Institutional Animal Care and Use Committee of Maine Medical Center.
Skeletal staining, histology and staining for β-galactosidase
The morning of detection of the vaginal plug was considered as embryonic day (E)0.5. Litters at E13.5 and E16.5 were harvested, and the embryos were weighed. DNA was prepared from the tails for genotyping by PCR. Embryos from different genotype groups were processed for whole-mount Alcian blue-Alizarin red staining of cartilage and mineralized bone, respectively, as described (14). For histology, Bouin's-fixed and paraffin-embedded tissues (humerus, femur, tibia/fibula, radius/ulna) were cut into 7 μm sections, deparaffinized in xylene, rehydrated in PBS, and stained with hematoxylin and eosin. Lengths of the whole bone, growth plate, hypertrophic zone, and proliferating zone were measured. For detection of β-galactosidase (lacZ) expression, femurs were fixed in 4% paraformaldehyde (PFA), suspended in 20% sucrose overnight at 4°C, mounted in OCT medium, and cryosectioned at 12.5 μm. Sections were stained for β-galactosidase activity as described (11), and were counterstained with eosin.
Micromass culture of limb bud mesenchymal cells
Limb buds from E12.5 embryos were dissected, and mesenchymal cells were dissociated by digestion in 300 μl of UltraSaline A buffer (Lonza) containing 10mg/ml dispase (Invitrogen) and 10% fetal bovine serum (FBS) in a 37°C, 5% CO2 incubator for 1.5 hours. Digested limb bud solutions were triturated in 1:1 DMEM/F-12 growth media (Hyclone), passed through a 40 μm cell strainer (Falcon) to obtain a single cell suspension, and briefly centrifuged. Cells were resuspended in growth media at a concentration of 2×107 cell/ml and were plated in 10 μl droplets in a four well culture dish (Delta Nunclon). Cells were incubated 1.5 hours at 37°C-5% CO2 incubator to adhere and then fed with 500 μl of 1:1 DMEM/F-12 growth media containing 10% FBS, 1% Glutamax (Gibco), and 1% Penicillin/Streptomycin (Gibco). Media was changed after 24 hours, and after another 24 hours, media was changed with media containing 50μg/ml ascorbic acid (Sigma). Cell cultures were incubated up to 6 days with media changes every second day. Alcian-blue staining of micromass cultures was performed as described (15).
BrdU incorporation and TUNEL assays
To examine chondrocyte proliferation in embryo limbs, timed pregnant females were injected intraperitoneally with 100 mg/kg body weight of BrdU (5-Bromo-2'-Deoxyuridine) labeling reagent (Sigma) two hours prior to euthanasia. Femurs were dissected and fixed in 4% PFA overnight at 4 °C, embedded in paraffin, and were cut into 7 μm sections. BrdU incorporation was detected as described (16). Briefly, sections were dewaxed, rehydrated and peroxidase activity was quenched in 3% H2O2 in PBS. DNA was denatured using 2N HCl. A purified mouse anti-BrdU antibody (Pharmingen) was used at 1:100 dilution in 10% Donkey serum/TBS-Tween20. It was detected with a peroxidase-conjugated donkey anti-mouse IgG secondary antibody at a 1:100 dilution in TBS-Tween20 (Jackson ImmunoResearch). Filtered diaminobenzidine (DAB, Sigma) substrate with 0.2% NiCl added was used for detection. Sections were counterstained with eosin, and were counted in equivalent areas of at least four separate sections from six embryos for each genotype. Cell proliferation was quantified by the ratio of the number of BrdU-positive chondrocytes to total chondrocytes. Cells were counted in equivalent areas of at least four separate sections from six embryos for each genotype. For TUNEL assay, paraffin sections from femurs were dewaxed, rehydrated, and the TUNEL assay was performed using a Roche In situ Cell Death Detection kit, Fluorescein (Roche, Indianapolis, IN) according to the manufacturer's instructions. Total apoptotic cells were counted in 10 sections per embryo (six embryos from each genotype group).
In situ hybridization and immunofluorescence
For in situ hybridization on glass slides, mouse tissues were fixed in 4% PFA at 4°C overnight, mounted in OCT, and cryosectioned. For whole mount in situ hybridization, embryos were harvested at E12.5, and were fixed in 4% PFA at 4°C overnight. In situ hybridization was performed using digoxigenin-labeled mouse antisense RNA probes, as described (17). RNA probes for mouse Snai1 and Snai2 mRNAs were used as described previously (11,14). Probes for mouse Sox9, Mmp9 and Mmp13 were generated as described (18). Probes for Col1a1, Col2a1, Col10a1, Ihh, Pth1r, bone sialoprotein, and osteocalcin were kindly provided by Drs. Richard Behringer and Gerard Karsenty.
For immunofluorescence, slides containing 7 μm paraffin sections were boiled in 10 mM sodium citrate (pH 6.0) for 10 minutes for antigen retrieval, then incubated with primary and secondary antibodies. Primary antibodies used in this study include: anti-Snai1 (Santa Cruz, E18, 1:50), anti-Snai2 (Santa Cruz, G18, 1:50); anti-p21 (Santa Cruz, M19, 1:50); anti-CDK2 (Santa Cruz, M2, 1:50), anti-Ccnb1 (Millipore, MAB3684, 1:100); anti-MMP13 (Santa Cruz, H90, 1:50), anti-Sox9 (Santa Cruz, H-114; 1:50); Alexa Fluor fluorescent secondary antibodies (Molecular Probes), which included donkey anti-goat Alexa Fluor 546, donkey anti-rabbit Alexa Fluor 555, were applied for 2 hours at room temperature. For detection of PECAM (CD31), frozen sections were fixed in acetone at −20°C for 10 minutes, and stained with rat anti-mouse CD31 (BD Pharmingen, 553370, 1:100), followed by counterstaining with donkey anti-rat Alexa Fluor 488. All slides were mounted using Vectashield mounting medium containing DAPI (Vector Laboratories).
Plate PCR arrays
Gene expression profiling of cell cycle regulatory genes was performed using the RT Profiler PCR Array System (SABiosciences). For PCR array experiments, mouse Cell Cycle RT2 Profiler™ PCR Array (PAMM-020Z) was used to simultaneously examine the mRNA levels of 84 genes in 96-well PCR array plates, according to the manufacturer's instructions. PCR was performed on a Bio-RAD iCycler Real-Time PCR Systems (Life Science Research). Analysis of results was performed using GEArray Expression Analysis Suite software. According to this analysis, transcriptional levels of genes showing fold change values of >1.50 or <0.66 were considered significantly modified. Expression of a subset of these genes was validated by quantitative RT-PCR.
Quantitative RT-PCR
Femurs from E16.5 embryos were dissected and immersed in RNAlater (Ambion). Genotypes were identified by allele-specific PCR. Total RNA was isolated using the Qiagen Mini mRNA Extraction kit. RNA (2.0μg of each sample) was reverse-transcribed with random hexamer primers (Ambion). Six nanograms of cDNA were used for real-time PCR amplification for each well, using primer sequences from Primerbank. qRT-PCR was performed using Super SYBR Green PCR Master Mix on a 7500 Real Time PCR system (Applied Biosystems) using SDS software. For each gene tested we performed three experimental replicates and four biological replicates. Gene expression levels were normalized to the β-actin mRNA level. Primer sequences are included in Supplemental Table 1.
Statistical analysis
Data are presented as mean ± SEM. A two tailed t-test was performed to compare means between two groups, and Analysis of Variance (ANOVA) was performed to compare means of multiple groups. P-values ≤0.05 were considered significant.
Results
Mouse embryos with limb bud mesenchyme-specific deletion of the Snai1 gene on a Snai2 null background display impaired long bone growth and delayed ossification
To delete the Snai1 gene in undifferentiated limb bud mesenchyme prior to the formation of mesenchymal condensations, we utilized the Prrx1-Cre driver line (13) and the Snai1flox (12) conditional allele. Examination of Alcian blue-Alizarin red stained skeletal preparations of Prrx1-Cre; Snai1flox/flox embryos and mice did not reveal any obvious defects in long bone development in the limbs. Similarly, examination of stained skeletons of Snai2−/− embryos and mice also did not reveal long bone defects in the limbs. Since our previous studies analyzing neural crest cell-specific deletion of the Snai1 gene had revealed a phenotype only on the Snai2 null background (14), we assessed limb bud mesenchyme-specific deletion of the Snai1 gene on the Snai2 null background.
For this cross, male Prrx1-Cre; Snai1flox/flox; Snai2+/− mice were mated with female Snai1flox/flox; Snai2+/− mice. Embryos were divided into four different groups for further analysis according to their genotypes: 1. Littermate control (LC): Snai1flox/flox; Snai2+/+; 2. Snai1 single mutant (Snai1 MT): Prrx1-Cre; Snai1flox/flox; Snai2+/+; 3. Snai2 single mutant (Snai2 MT): Snai1flox/flox; Snai2−/−; and 4. Snai1/Snai2 double mutant (Snai1/Snai2 DM): Prrx1-Cre; Snai1flox/flox; Snai2−/−. We carried out our analyses of long bone development on embryos isolated at E16.5, since the Prrx1-Cre; Snai1flox/flox; Snai2−/− mice, as well as many of the Snai1flox/flox; Snai2-−/− mice, died postnatally from cleft palate (11,14).
There was no significant difference in body weight among embryos of the four groups (Supplemental Fig. S1). Measurements of femur length of Alcian blue-Alizarin red stained skeletal preparations revealed no significant differences between the LC, Snai1 MT and Snai2 MT groups. However, mean bone length of femurs of the Snai1/Snai2 DM embryos was reduced approximately 20%, whether the measurements were taken of total bone length, or the length of trabecular bone (Fig. 1C). Similar reductions in length were observed in the other long bones of both the fore- and hindlimbs (Fig. 1, and Fig. S1). All long bones were present, and autopods contained the correct numbers of digits. The distal phalange of digit 5 of the hindlimb characteristically exhibited an altered orientation in Snai1/Snai2 DM embryos (Fig. 1G). The Snai1/Snai2 DM embryos also exhibited delayed ossification in digits of both forelimbs and hindlimbs, as well as in the tibia (Fig. 1E, G). The tibias of Snai1/Snai2 DM embryos, in addition to delayed ossification, characteristically displayed an odd curvature (Fig. 1G). Notably, however, there were no obvious defects in limb patterning in the Snai1/Snai2 DM embryos.
Fig. 1.
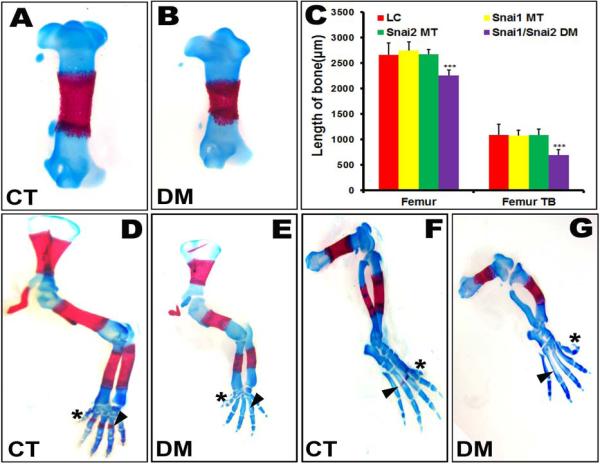
Snai1/Snai2 double mutant embryos exhibit shortened long bones and delayed ossification. (A, B) Alcian blue-Alizarin red staining of E16.5 control (CT) and Snai1/Snai2 double mutant (DM) embryos reveals shortened long bones, such as the femurs shown here, with a reduction of their trabecular bone. *** p < 0.001. (C) Length of the femur and its trabecular bone (TB) at E16.5. The femur and its trabecular bone were significantly shorter in Snai1/Snai2 DM mice. (D–G) Snai1/Snai2 DM embryos exhibited altered orientation in digit 5 of the hindlimbs (asterisks in D, E) and delayed ossification in digits of both forelimbs (arrowheads in D, E) and hindlimbs (arrowheads in F, G), as well as in the tibia (arrows in F, G).
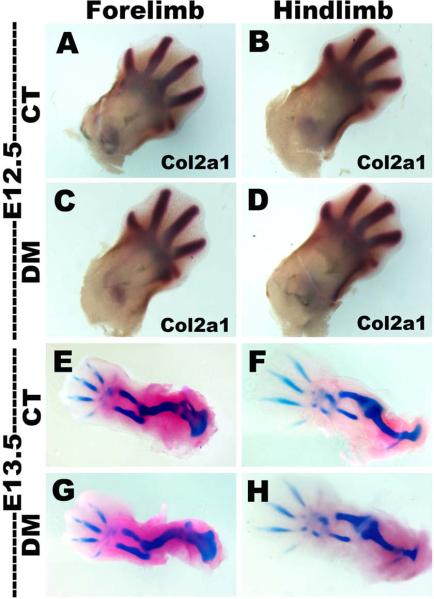
To further analyze early limb bud patterning in Snai1/Snai2 DM embryos, we assessed formation of mesenchymal condensations and generation of cartilaginous precursors of the long bones in embryos isolated at E12.5 and E13.5 (Fig. 2). Both in situ hybridization for Col2a1 expression at E12.5 (Fig. 2A–D) and Alcian blue-Alizarin red staining (Fig. 2E–F) at E13.5 revealed that both the forelimbs and hindlimbs of Snai1/Snai2 DM embryos exhibited normal patterning and were similar in size and stage to limbs of control littermates.
Fig. 2.
Early limb bud patterning is normal in Snai1/Snai2 DM limbs. (A–D) In situ hybridization for Col2a1 expression at E12.5. (E–F) Alcian blue-Alizarin red staining of control and DM embryos at E13.5. Both forelimbs and hindlimbs of Snai1/Snai2 DM embryos exhibit normal patterning and are similar in size and stage to limbs of control littermates.
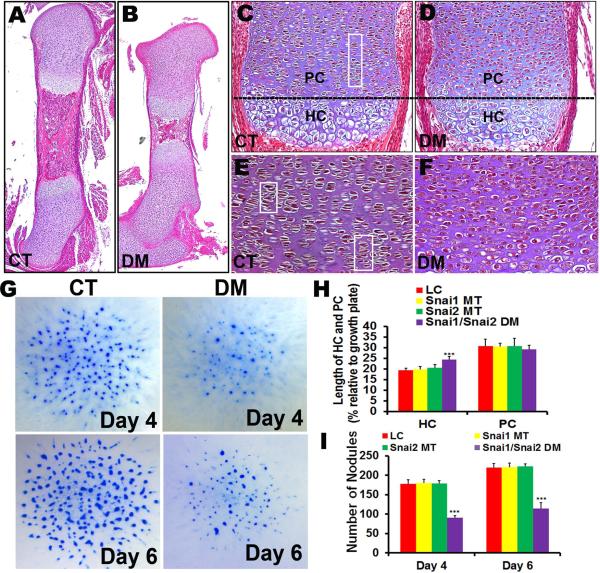
To analyze long bone defects of Snai1/Snai2 DM embryos, we performed histological analyses on E16.5 femurs. In the control group, proliferating chondrocytes of the growth plate were highly organized into vertical columns, and displayed the characteristic flattened lens shape with a normal chondrocyte to lacuna ratio (Fig. 3C, E). In contrast, in Snai1/Snai2 DM growth plates the pattern of well-aligned columnar chondrocytes was disorganized (Fig. 3D, F), and chondrocyte morphology was altered. Proliferating chondrocytes in Snai1/Snai2 DM growth plates were more compact, and exhibited an elliptical shape with a higher chondrocyte to lacuna ratio (Fig. 3F). We measured the lengths of the hypertrophic chondrocyte and proliferating chondrocyte zones in Snai1/Snai2 DM and control littermates (Fig. 3H). We found a small, but statistically significant increase in the length of the hypertrophic chondrocyte zone in Snai1/Snai2 DM growth plates, but did not observe significant differences in the lengths of the proliferating chondrocyte zone (Fig. 3H).
Fig. 3.
Defects in chondrocyte morphology and growth in Snai1/Snai2 DM mutant embryos. (A–F) Hematoxylin-eosin-stained sections of E16.5 control and DM femurs. Snai1/Snai2 DM growth plates lost the columnar arrangement (areas in rectangles) and the flattened lens shape exhibited by control chondrocytes. PC: proliferating chondrocytes; HC: hypertrophic chondrocytes. (G) Snai1/Snai2 DM micromass cultures generated fewer Alcian blue-staining nodules than cultures set up from control limb buds. (H) Quantification of the lengths of the hypertrophic chondrocyte and proliferating chondrocyte zones in growth plates of Snai1/Snai2 DM and control and single mutant littermate femurs at E16.5. The Snai1/Snai2 DM growth plates exhibited an increased length of the hypertrophic chondrocyte zone, but no significant difference in the length of the proliferating chondrocyte zone. *** p < 0.001. (I) Quantification of nodule formation in limb bud micromass cultures revealed that Snai1/Snai2 DM cultures generated approximately half as many Alcian blue-staining nodules as cultures set up from control or single mutant limb buds on both days 4 and 6 of culture. *** p < 0.001.
To analyze growth and differentiation of Snai1/Snai2 DM limb bud mesenchymal progenitor cells, we assessed cartilaginous nodule formation ex vivo using micromass cultures of limb bud mesenchymal cells isolated from Snai1/Snai2 DM and control littermates (Fig. 3G). The LC, Snai1 MT, and Snai2 MT cultures formed nearly identical numbers of Alcian blue-staining nodules after four days of culture, with approximately 30% more nodules after six days of culture. However, micromass cultures from Snai1/Snai2 DM limb buds at either time point yielded approximately half as many nodules as the littermate cultures (Fig. 3I).
Snai1/Snai2 DM growth plates exhibit defects in chondrocyte proliferation
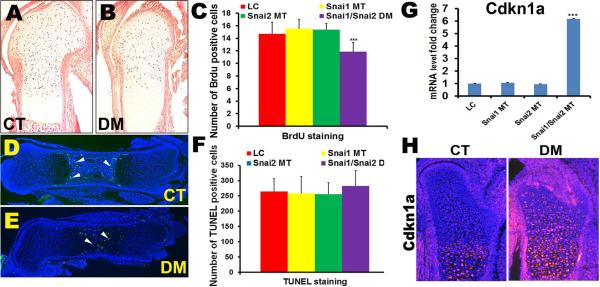
The phenotype observed in the Snai1/Snai2 DM embryos and micromass cultures, with shortened long bones, delayed ossification, and reduced cartilaginous nodule formation, could be the result of reduced chondrocyte proliferation, increased chondrocyte cell death, or altered chondrocyte differentiation in the Snai1/Snai2 DM growth plate. To assess chondrocyte proliferation, we performed in vivo BrdU incorporation assays in femurs of Snai1/Snai2 DM and control embryos at E16.5 (Fig. 4A, B). There were no significant differences in the chondrocyte proliferative index in the LC, Snai1 MT, and Snai2 MT growth plates. However, the proliferative index was significantly decreased in chondrocytes in Snai1/Snai2 DM femurs, indicating that the Snai1 and Snai2 genes are required to maintain the high rate of chondrocyte proliferation in the rapidly growing long bone (Fig. 4C). These observations suggested that reduced chondrocyte proliferation may contribute to the reduction in long bone length observed in Snai1/Snai2 DM femurs.
Fig. 4.
Defects in chondrocyte proliferation in Snai1/Snai2 DM growth plates. (A–C) BrdU incorporation into proliferating chondrocytes was significantly reduced in Snai1/Snai2 DM growth plates, but not in the other genotypes (C). *** p < 0.001. (D–F) Detection of apoptotic cells by TUNEL staining. (D) In the control (CT) group, apoptotic cells were mainly located at regions of terminal differentiation adjacent to the trabecular region. (E) In DM femurs, apoptotic cells were found in throughout the central region of the femur. (F) However, no significant differences in the numbers of TUNEL-positive cells were observed in any genotype group. (G) Quantitative RT-PCR revealed that the Cdkn1a gene (encoding the cyclin-dependent kinase inhibitor p21Waf1/Cip1) was upregulated in DM femurs, but not in control or single mutant femurs. *** p < 0.001. (H) Immunofluorescent staining with an antibody to the p21Waf1/Cip1 protein revealed increased protein expression in DM femurs.
We also assessed by TUNEL assay whether apoptosis was altered in Snai1/Snai2 DM chondrocytes (Fig. 4D, E). No significant difference in the numbers of TUNEL-positive cells was detected between the control and Snai1/Snai2 DM groups (Fig. 4F). In control growth plates, apoptosis was restricted to the regions of terminal differentiation, where hypertrophic chondrocytes are adjacent to mineralizing trabecular bone (Fig. 4D). In Snai1/Snai2 DM femurs, the TUNEL-positive cells were located throughout the central region of the femur (Fig. 4E), although this localization may be the result of the shortening of the trabecular region in the double mutant femurs. No TUNEL-positive cells were detected in the proliferating zone of the growth plate in embryos of any genotype. These data indicate that cell apoptosis is not dramatically affected in Snai1/Snai2 DM femurs.
Since chondrocyte proliferation was reduced in Snai1/Snai2 DM growth plates, we examined expression of major cell cycle regulators by plate-based PCR array. Genes whose expression levels were altered in control versus Snai1/Snai2 DM groups were then validated by quantitative RT-PCR (qRT-PCR). We observed significant alterations in the RNA and protein expression levels of several cell cycle regulators (Supplemental Fig. S2 and S3). Of particular interest was expression of the cyclin dependent kinase inhibitor p21Waf1/Cip1 (encoded by the Cdkn1a gene), a previously described target for repression by the SNAI1 protein (19). Cdkn1a RNA expression was approximately six-fold higher in Snai1/Snai2 DM femurs than in littermate controls (Fig. 4G), and immunofluorescent staining revealed increased p21Waf1/Cip1 protein expression in DM femurs (Fig. 4H).
Snai1/Snai2 DM growth plates exhibit altered chondrocyte differentiation and delayed chondrocyte hypertrophy
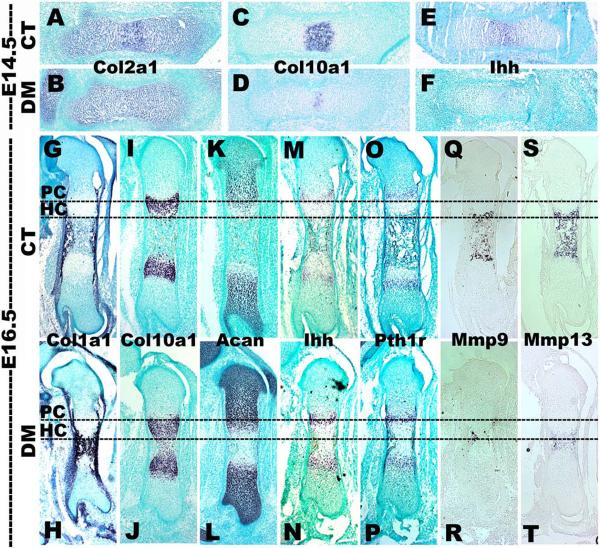
To assess whether Snai1/Snai2 DM embryos exhibited altered chondrocyte differentiation, we examined expression of a panel of stage-specific chondrocyte differentiation markers by qRT-PCR and in situ hybridization in femurs of Snai1/Snai2 DM and control embryos (Fig. 5, 6 and Supplemental Fig. S4). These analyses revealed that alterations in the levels and spatial localization of marker gene expression were observed, but were confined to Snai1/Snai2 DM embryos. The LC, Snai1 MT, and Snai2 MT embryos exhibited no significant differences for any of the markers tested. By qRT-PCR at E16.5, several markers were upregulated in Snai1/Snai2 DM femurs, including Col1a1, Col2a1, Col10a1, Sox9, and Acan (Fig. 5). No quantitative differences in expression of the Ihh and Runx2 genes were observed (Fig. S4). Conversely, genes encoding the matrix metalloproteases Mmp9 and Mmp13 were downregulated (Fig. 5).
Fig. 5.
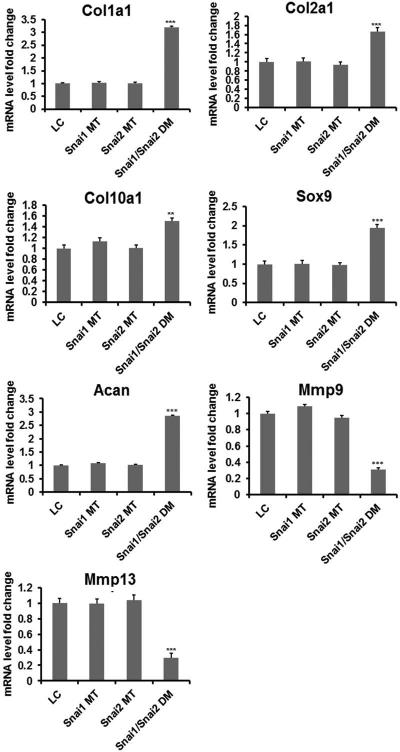
Quantitative RT-PCR of chondrocyte differentiation markers in femurs at E16.5. mRNAs encoding Col1a1, Col2a1, Col10a1, Acan and Sox9 were significantly increased in Snai1/Snai2 DM femurs compared to the other genotypes, whereas Mmp9 and Mmp13 mRNA levels were significantly decreased in DM femurs. ** p<0.05; *** p < 0.001.
Fig. 6.
Altered chondrocyte gene expression in Snai1/Snai2 DM femurs. In situ hybridization of femurs isolated at E14.5 (A–F) and E16.5 (G–T). At E14.5, Snai1/Snai2 DM femurs exhibited reduced expression of Col10a1 (a hypertrophic chondrocyte marker) (C, D) and Indian hedgehog (Ihh) (a prehypertrophic chondrocyte marker) (E, F). Expression of the Col10a1 (I, J) and Ihh (M, N) genes had recovered in DM femurs at E16.5. Expression of the Mmp9 (Q, R) and Mmp13 (S, T) genes (encoding matrix metalloproteases) was strongly decreased in DM femurs at E16.5. PC: proliferating chondrocytes; HC: hypertrophic chondrocytes.
When assessed by in situ hybridization, a number of these genes exhibited altered domains or levels of expression in the Snai1/Snai2 DM growth plates (Fig. 6). Progression into chondrocyte prehypertropy and hypertrophy appeared to be delayed in Snai1/Snai2 DM growth plates. At E14.5, expression of both Col10a1, a hypertrophic chondrocyte marker, and Indian hedgehog (Ihh), a prehypertrophic chondrocyte marker, was markedly delayed in double mutant femurs (Fig. 6D, F). Col2a1, a marker of proliferating chondrocytes, showed no difference in its expression pattern in growth plates of Snai1/Snai2 DM embryos at both E14.5 (Fig. 6A, B) or E16.5 (Fig. S4A). At E16.5 Col1a1, which is normally expressed only in perichondrial cells at this time point (Fig. 6G), was detected in the shortened trabecular bone region of Snai1/Snai2 DM femurs in addition to perichondrial cells (Fig. 6H). Expression of Col10a1 also was expanded into the trabecular bone region of Snai1/Snai2 DM femurs (Fig. 6J). Expression of Acan, which encodes aggrecan, a major component of the chondrocyte extracellular matrix, was only seen in proliferating chondrocytes in control femurs (Fig. 6K), but was also observed in hypertrophic chondrocytes in Snai1/Snai2 DM femurs (Fig. 6L). At E16.5, the expression domains of Ihh and Parathyroid hormone 1 receptor (Pth1r), two prehypertrophic chondrocytes markers (20), were not altered by deletion of both the Snai1 and Snai2 genes (Fig. 6N, P). However, in situ hybridization for the Mmp9 and Mmp13 genes, which encode extracellular matrix metalloproteases, confirmed the dramatic downregulation in expression we observed by qRT-PCR in the Snai1/Snai2 DM femurs (Fig. 6R, Y). These analyses indicate that, in addition to defects in chondrocyte proliferation, growth plates of Snai1/Snai2 DM femurs exhibited altered chondrocyte differentiation and delayed chondrocyte hypertrophy.
Confirming our findings from Alcian blue-Alizarin red staining (Fig. 1), in situ hybridization also demonstrated delayed osteogenesis in the double mutants. Expression of osteoblast marker genes, such as bone sialoprotein and osteocalcin, was delayed and reduced in Snai1/Snai2 DM femurs at E16.5 and E18 (Supplemental Fig. S5). To assess whether defects in angiogenesis might contribute to the phenotypes observed in Snai1/Snai2 DM femurs, we assessed protein expression of the endothelial cell marker Platelet/endothelial cell derived protein 1 (Pecam1; also CD31) in Snai1/Snai2 DM and control littermate femurs at E16.5 and E18. Blood vessel formation and penetration into the trabecular region of the femur was observed in both double mutant and controls (Supplemental Fig. S6), suggesting that defects in angiogenesis did not play a major role in the long bone defects of Snai1/Snai2 double mutants.
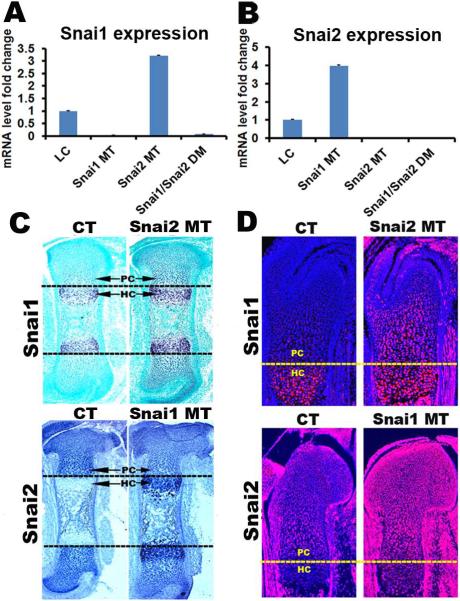
Compensatory regulation of the Snai1 and Snai2 genes during chondrogenesis
Our genetic data indicate that the Snai1 and Snai2 genes function redundantly during long bone chondrogenesis. We therefore examined the expression patterns and levels of the Snai1 and Snai2 genes in all four littermate groups by qRT-PCR, in situ hybridization and immunofluorescence in E16.5 femurs. Quantitative RT-PCR demonstrated that, as expected, no Snai1 transcripts were detected in Snai1 MT femurs, and no Snai2 transcripts were detected in Snai2 MT femurs (Fig. 7A, B). However, Snai2 transcript levels were increased 3.9-fold in Snai1 MT femurs (Fig. 7B), while Snai1 transcript levels were increased 3.3-fold in Snai2 MT femurs (Fig. 7A), indicating that the Snai1 and Snai2 genes can compensate for each other's loss during bone development. Both in situ hybridization (Fig. 7C) and immunofluorescence (Fig. 7D) revealed that in control E16.5 femurs, Snai1 transcripts and protein are highly expressed in hypertrophic chondrocytes, whereas Snai2 transcripts and protein are highly expressed in proliferating chondrocytes. Importantly, the Snai1 and Snai2 genes compensated for each other's loss not only quantitatively, but also by expanding their expression into the other genes' normal expression domain. In Snai2 MT growth plates, Snai1 expression was upregulated in proliferating chondrocytes, while in Snai1 MT growth plates Snai2 expression expanded into the hypertrophic chondrocyte zone (Fig. 7C, D). Expansion of Snai2 expression into the hypertrophic zone in Snai1/Snai2 DM femurs could also be demonstrated by analysis of β-galactosidase expression from the Snai2lacZ null allele (Supplemental Fig. S7). These analyses demonstrate that the Snai1 and Snai2 genes function redundantly during chondrogenesis in the long bones, and transcriptionally compensate temporally, spatially and quantitatively if the other gene is deleted.
Fig. 7.
Compensatory regulation of the Snai1 and Snai2 genes during chondrogenesis. (A) qRT-PCR demonstrates that Snai1 transcript levels were increased 3.3-fold in Snai2 MT femurs. As expected, no Snai1 transcripts were detected in Snai1 MT or Snai1/Snai2 DM femurs. (B) Snai2 transcript levels were increased 3.9-fold in Snai1 MT femurs. No Snai2 transcripts were detected in Snai2 MT or Snai1/Snai2 DM femurs. Both in situ hybridization (C) and immunofluorescence (D) demonstrate that in control (CT) growth plates, Snai1 is expressed in hypertrophic chondrocytes (HC), whereas Snai2 is highly expressed in proliferating chondrocytes (PC). However, in Snai2 MT growth plates, Snai1 expression expanded into proliferating chondrocytes, while in Snai1 MT growth plates Snai2 expression expanded into hypertrophic chondrocytes.
Discussion
Limb chondrogenesis defects are only observed in Snai1/Snai2 double mutant embryos
A previous gain-of-function study using a tamoxifen-inducible Snai1 transgenic line demonstrated that upregulation of Snai1 activity in mouse long bones caused a reduction in bone length (6), and several genes involved in cartilage and bone development had previously been either demonstrated or implicated as Snail target genes (7–10). However, no loss of function analyses examining the roles of the Snai1 and Snai2 genes during endochondral bone formation have been performed, leaving it unclear whether Snail family genes play an essential physiological role during mammalian bone development. The results presented here demonstrate that both Snai1 and Snai2 gene function must be removed in order to cause substantial defects during chondrogenesis in the long bones of the limbs. All long bones of both the fore- and hindlimbs were significantly shorter in Snai1/Snai2 DM embryos, although there were no obvious defects in limb patterning aside from a misorientation of digit 5 of the hindlimb. Chondrocyte morphology in the growth plates was altered, and the organization of the chondrocytes into highly-aligned columns was disrupted.
These findings are reminiscent of our previous study utilizing the Wnt1-Cre driver for neural crest-specific Snai1 gene deletion, where we only detected a craniofacial phenotype if the Wnt1-Cre-mediated deletion of the Snai1 gene occurred on the Snai2 null genetic background (14). We had reported previously the surprising result that neither the Snai1 nor the Snai2 gene, alone or in combination, was required for neural crest cell generation and delamination in mice, at least through 9.5 days of gestation (21). However, these genes are clearly required for neural crest cell differentiation and function. In the Snai1/Snai2 neural crest double mutant embryos, Meckel's cartilage was dramatically shorter than in control littermates, resulting in cleft palate in these embryos (14). Our current analysis of Snai1/Snai2 double mutants generated with the Prrx1-Cre driver suggests that similar mechanisms may be operating in both the Wnt1-Cre and Prrx1-Cre conditional Snai1/Snai2 double mutants.
Snai1/Snai2 double mutants exhibit defects in chondrocyte proliferation, altered chondrocyte differentiation and delayed chondrocyte hypertrophy
Our analyses support the model that impaired chondrocyte proliferation is likely a major contributor to the shortening of the long bones and the reduced cartilaginous nodule formation in micromass cultures of Snai1/Snai2 DM embryos (Supplemental Fig. S8). Since chondrocyte proliferation was reduced in Snai1/Snai2 DM growth plates, we used PCR arrays and qRT-PCR to assess expression of major cell cycle regulators. We observed significant alterations in the transcript levels of numerous cell cycle regulators, including Ccnb1, Ccnb2, Cdk2, Trp53, Ccne1 and Myb. Despite the increase in Trp53 expression, we did not observe increased chondrocyte cell death in these embryos. Particularly noteworthy, however, was the cyclin dependent kinase inhibitor p21Waf1/Cip1 (encoded by the Cdkn1a gene), whose transcript levels were increased six-fold in Snai1/Snai2 DM femurs. Previous work has identified the Cdkn1a gene as a target for repression by the SNAI1 protein (19). These findings suggest that reduced chondrocyte proliferation, along with the disorganization of the chondrocyte columns, are likely key factors contributing to the reduction in length observed in Snai1/Snai2 DM long bones.
Expression and localization of several chondrocyte differentiation markers was altered in Snai1/Snai2 DM femurs. For example, expression of both Col1a1 and Col10a1 was increased quantitatively, and their respective expression domains were expanded into the trabecular region of the femur. Conversely, expression of the Mmp9 and Mmp13 genes was dramatically downregulated in Snai1/Snai2 DM femurs. At E14.5, expression of both Ihh and Col10a1 was markedly delayed in double mutant femurs, indicating delayed chondrocyte prehypertropy and hypertrophy in Snai1/Snai2 DM growth plates. These analyses indicate that, in addition to defects in chondrocyte proliferation, growth plates of Snai1/Snai2 DM femurs exhibited altered chondrocyte differentiation and delayed chondrocyte hypertrophy (Supplemental Fig. S8).
The Snai1 and Snai2 genes compensate for each other's loss during chondrogenesis
In all of the assays utilized in these analyses (morphology, histology, gene and protein expression, and nodule formation in ex vivo micromass cultures) the only genotype group that exhibited an apparent mutant phenotype was the Snai1/Snai2 double mutants. Limb development in the littermate controls, Snai1 conditional single mutants, and Snai2 null single mutants was normal. This finding demonstrates that, of these two genes, the remaining Snail family member can compensate in all ways for deletion of the other family member. Since our genetic data indicated that the Snai1 and Snai2 genes function redundantly during chondrogenesis, we examined expression levels and patterns of the Snai1 and Snai2 genes in femurs of single mutant embryos at E16.5. By qRT-PCR, we found that transcript levels of these genes were increased three to four fold in a mutant for the other gene (i.e., Snai1 transcript levels were increased in the Snai2−/− mutant, and vice versa). Importantly, the Snai1 and Snai2 genes compensated for each other's loss not only quantitatively, but also by expanding their expression into the other genes' normal expression domains. In Snai2 MT growth plates, Snai1 expression was upregulated in proliferating chondrocytes, while in Snai1 MT growth plates Snai2 expression expanded into the hypertrophic chondrocyte zone. These results demonstrate that the Snai1 and Snai2 genes transcriptionally compensate for each other's loss at the temporal, spatial and quantitative levels. Transcriptional compensation by the Snai1 gene in the Snai2 null mutant has previously been observed in endothelial cells during development of the atrioventricular canal and outflow tract of the embryonic mouse heart (22). However, in this case the normal expression domains of the two genes were not affected.
Both the SNAI1 and SNAI2 proteins bind to the E2 class of E box sequences (CAGGTG and CACCTG), and previous work has demonstrated that the SNAI1 protein binds to E-boxes in its own promoter to repress its own expression as part of a negative feedback loop (23). The results from our genetic epistasis experiments utilizing both the Wnt1-Cre (14) and Prrx1-Cre (this report) drivers support a model in which both the SNAI1 and SNAI2 proteins can bind to their own, as well as to the other gene's promoter to repress their transcription during chondrogenesis. Future studies will be directed at determining whether this model is correct.
Supplementary Material
Acknowledgements
The authors thank Richard Behringer and Gerard Karsenty for supplying in situ hybridization probes. This work was supported by NIH grant R01HD034883 to TG. This work also was supported by the Molecular Phenotyping Core Facility supported by NIH grant P20GM103465, and by the Histopathology Core Facility supported by NIH grants P20GM103465 and P30GM103392.
Authors' roles: Study design: YC and TG. Study conduct: YC. Data collection: YC. Data analysis: YC and TG. Data interpretation: YC and TG. Drafting manuscript: YC and TG. Revising manuscript content: YC and TG. Approving final version of manuscript: YC and TG. TG takes responsibility for the integrity of the data analysis.
This work was supported by NIH grant R01HD034883 to TG. This work also was supported by the Molecular Phenotyping Core Facility supported by NIH grant P20GM103465, and by the Histopathology Core Facility supported by NIH grants P20GM103465 and P30GM103392.
Footnotes
Disclosures Both authors state that they have no conflicts of interest.
References
- 1.Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev. 2009;19:437–43. doi: 10.1016/j.gde.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–48. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 3.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 4.Haraguchi M. The role of the transcriptional regulator snail in cell detachment, reattachment and migration. Cell Adh Migr. 2009;3:259–63. doi: 10.4161/cam.3.3.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, Zhou BP. Snail: More than EMT. Cell Adh Migr. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Frutos CA, Vega S, Manzanares M, Flores JM, Huertas H, Martinez-Frias ML, Nieto MA. Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasias. Dev Cell. 2007;13:872–83. doi: 10.1016/j.devcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Seki K, Fujimori T, Savagner P, Hata A, Aikawa T, Ogata N, Nabeshima Y, Kaechoong L. Mouse Snail family transcription repressors regulate chondrocyte, extracellular matrix, type II collagen, and aggrecan. J Biol Chem. 2003;278:41862–70. doi: 10.1074/jbc.M308336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambertini E, Franceschetti T, Torreggiani E, Penolazzi L, Pastore A, Pelucchi S, Gambari R, Piva R. SLUG: a new target of lymphoid enhancer factor-1 in human osteoblasts. BMC Mol Biol. 2010;11:13. doi: 10.1186/1471-2199-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambertini E, Lisignoli G, Torreggiani E, Manferdini C, Gabusi E, Franceschetti T, Penolazzi L, Gambari R, Facchini A, Piva R. Slug gene expression supports human osteoblast maturation. Cell Mol Life Sci. 2009;66:3641–53. doi: 10.1007/s00018-009-0149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Hassan MQ, Li ZY, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J Cell Biochem. 2008;105:487–96. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–85. [PubMed] [Google Scholar]
- 12.Murray SA, Carver EA, Gridley T. Generation of a Snail1 (Snai1) conditional null allele. Genesis. 2006;44:7–11. doi: 10.1002/gene.20178. [DOI] [PubMed] [Google Scholar]
- 13.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 14.Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–97. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 15.Bruce SJ, Butterfield NC, Metzis V, Town L, McGlinn E, Wicking C. Inactivation of Patched1 in the mouse limb has novel inhibitory effects on the chondrogenic program. J Biol Chem. 2010;285:27967–81. doi: 10.1074/jbc.M109.091785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–62. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- 17.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 18.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci U S A. 2009;106:14420–5. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi E, Funato N, Higashihori N, Hata Y, Gridley T, Nakamura M. Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist. Biochem Biophys Res Commun. 2004;325:1136–44. doi: 10.1016/j.bbrc.2004.10.148. [DOI] [PubMed] [Google Scholar]
- 20.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–22. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 21.Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci U S A. 2006;103:10300–4. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol. 2008;182:315–25. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiro S, Escriva M, Puig I, Barbera MJ, Dave N, Herranz N, Larriba MJ, Takkunen M, Franci C, Munoz A, Virtanen I, Baulida J, Garcia de Herreros A. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006;34:2077–84. doi: 10.1093/nar/gkl141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.