Abstract
A mutasynthetic strategy has been used to generate fluorinated TM-025 and TM-026, two biosynthetically engineered pactamycin analogs produced by Streptomyces pactum ATCC 27456. The fluorinated compounds maintain excellent activity and selectivity toward chloroquine-sensitive and multidrug-resistant strains of malarial parasites as the parent compounds. The results also provide insights into the biosynthesis of 3-aminobenzoic acid in S. pactum.
Pactamycin, a bacterial-derived natural product discovered by the Upjohn Company in the early 1960s, has been shown to have broad-spectrum growth inhibitory activity against bacteria,1 mammalian cells,2 viruses,3 and protozoa.4 This broad-spectrum activity is primarily due to its strong binding to a conserved region within the ribosome of most organisms, leading to non-selective inhibition of protein synthesis.5, 6 Consequently, its wide-ranging cytotoxicity, coupled with stability issues and difficulties to generate analogs of pactamycin through organic synthesis, have hampered its further development. As one of the most complex aminocyclitol natural products, pactamycin had presented great synthetic challenges; although, through seminal work of Hanessian and co-workers, more recently, this densely functionalized aminocyclitol antibiotic has finally surrendered to total synthesis.7, 8 In addition, a number of synthetic methodologies to access the aminocyclopentitol moiety of pactamycin have also been reported.9, 10 However, long synthetic routes and low overall yields remain major limitations of the synthetic approach to generate pactamycin analogs for clinical uses.
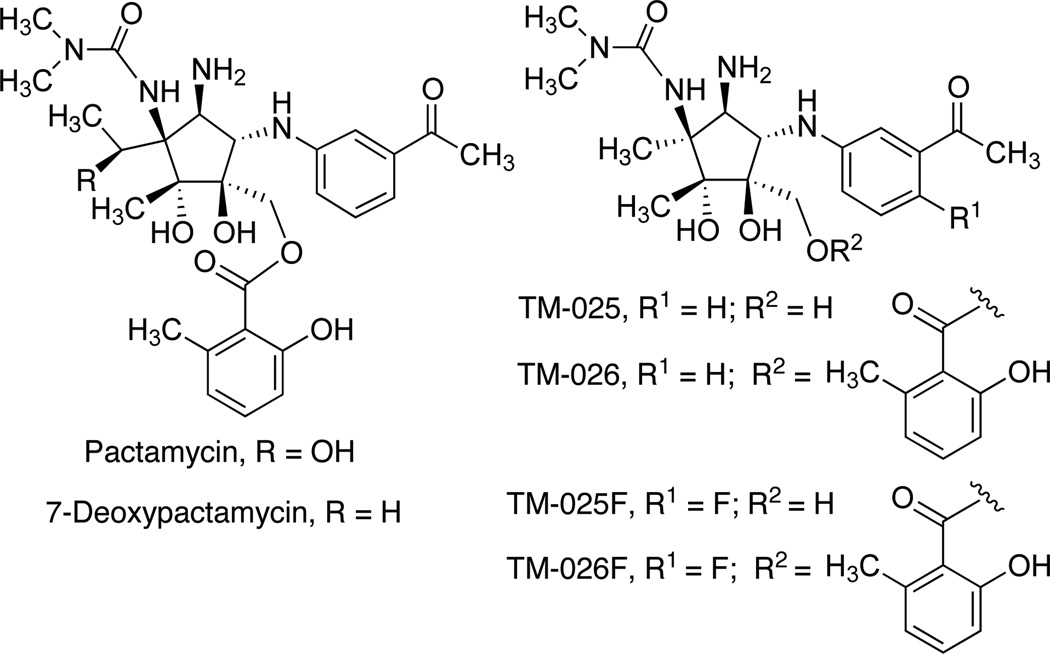
Recently, using biosynthetic engineering technology, we were able to generate a number of mutant strains of Streptomyces pactum ATCC 27456 that produce novel analogs of pactamycin with improved chemical and biological properties.11, 12 One of the mutants, in which ptmH (a gene that encodes a radical SAM-dependent protein) (Figure 1A) was inactivated, produces two novel analogs of pactamycin, TM-025 (de-6MSA-7-demethyl-7-deoxypactamycin) and TM-026 (7-demethyl-7-deoxypactamycin). These new analogs are chemically more stable and are produced in 10- to 20-fold higher yield than pactamycin. More significantly, they showed potent antimalarial activity at low nanomolar concentrations, but in contrast to pactamycin have no significant antibacterial activity and reduced cytotoxicity against human colorectal HCT-116 cells.12
Figure 1.
Identification of genes responsible for the production of pactamycin precursor.
To further improve the pharmacological properties of these analogs, we explored the application of the mutasynthetic approach to generate fluorinated TM-025 and TM-026 by blocking the formation of pactamycin biosynthetic precursor in S. pactum and performing precursor-directed structure modifications. Fluorinated compounds are rare in nature; however, the fluorine atom has been widely used in medicinal chemistry to improve drug properties. An estimated 15–20% of new pharmaceutical leads contains the fluorine atom in their structures.13
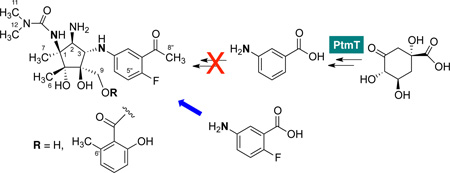
Previously, Rinehart and co-workers demonstrated that feeding of S. pactum var. pactum with 3-amino-5-fluorobenzoic acid resulted in the production of a fluorinated pactamycin analog.14 The product was reported to have a comparable biological activity as pactamycin, but the production yield was inferior, as the fluorinated precursor has to compete with the intrinsic precursor 3-aminobenzoic acid (Figure 1B) in the cells. Therefore, we set out to engineer a mutant strain of S. pactum that lacks the ability to produce 3-aminobenzoic acid.
Based on isotope incorporation studies using 13C-labeled glucose, it has been proposed that 3-aminobenzoic acid is derived from an intermediate of the shikimate pathway (either dehydroquinate or dehydroshikimate) involving a transamination reaction.15 However, no genetic and biochemical data are available to support that notion. Our recent investigations of the pactamycin biosynthetic gene cluster revealed the presence of two genes, ptmA and ptmT, that encode PLP-dependent aminotransferases, in the cluster.11 However, it is unclear if any of these genes is involved in 3-aminobenzoic acid formation.
To investigate the role of the aminotransferase genes in pactamycin biosynthesis, we carried out in-frame deletion of ptmA and ptmT individually from the S. pactum genome and analyzed the metabolic profiles of the resulting mutants. The mutants were constructed by cloning two 1-kb PCR fragments upstream and downstream of the ptmA and ptmT genes, respectively, and separately cloned into the plasmid pTMN002.12 The products pTMW034 (for ptmA) and pTMW037 (for ptmT) were individually introduced into S. pactum by conjugation. Apramycin resistant strains representing single crossover mutants were obtained and subsequently grown on nonselective mannitol-soy flour agar containing MgCl2 to allow the formation of double crossover recombinants. Apramycin sensitive colonies were counter-selected by replica plating onto MS-Mg agar with and without apramycin, and the resulting double-crossover candidate strains were confirmed by PCR amplification (Figure S1).
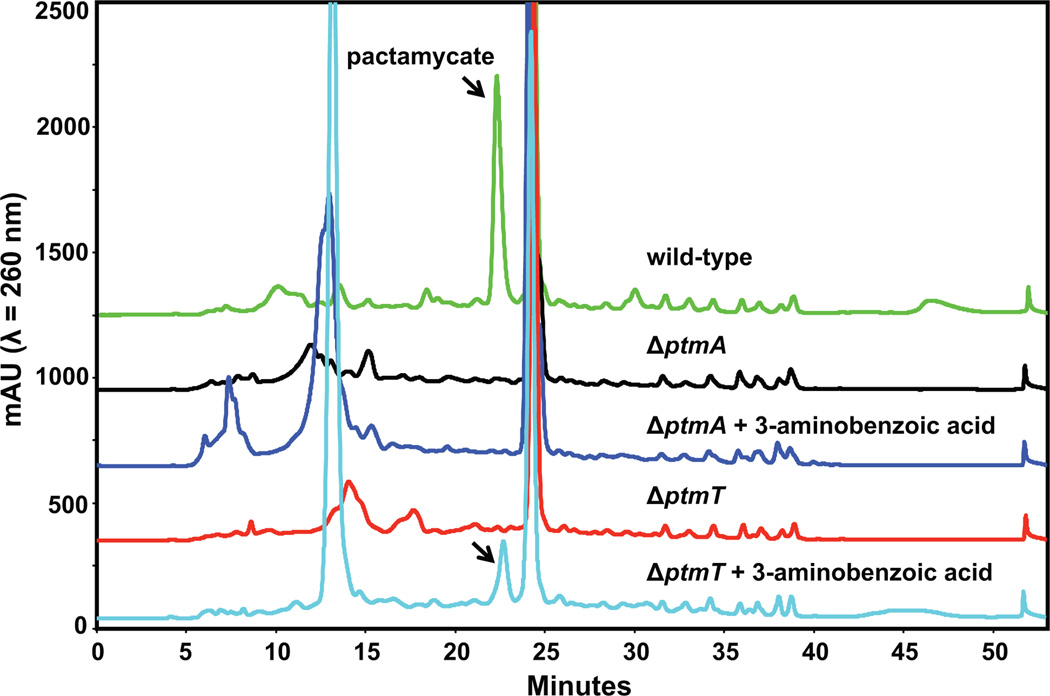
HPLC (Figure 2) and MS analyses of the extracts of those mutants showed that both mutants lack the ability to produce pactamycin or its intermediates. While the results suggest that both ptmA and ptmT are critical for pactamycin biosynthesis it was not clear if ptmA or ptmT are involved in the formation of 3-aminobenzoic acid.
Figure 2.
HPLC analysis of EtOAc extracts of the wild-type and the mutant strains of S. pactum.
To investigate if any of the genes are involved in the formation of 3-aminobenzoic acid, we carried out chemical complementation experiments with 3-aminobenzoic acid. The compound was pulse-fed to the cultures of ΔptmA and ΔptmT mutants and the products were harvested and analyzed by HPLC and MS. The results showed that feeding ΔptmT mutant with 3-aminobenzoic acid did rescue the production of pactamycin (detected as pactamycate),11 whereas that of ΔptmA mutant did not (Figure 2).
The results suggest that ptmT, not ptmA, is the one involved in the formation of 3-aminobenzoic acid. To confirm this incorporation result, we subsequently fed the mutant with [7-13C]-3-aminobenzoic acid, and MS analysis of the EtOAc extract of the culture broths revealed the incorporation of 13C in the final products (Figure 3C), confirming the direct involvement of 3-aminobenzoic acid in pactamycin biosynthesis and PtmT is a dedicated enzyme for 3-aminobenzoic acid biosynthesis.
Figure 3.
Mass spectral data for EtOAc (A – C) and n-BuOH (D – F) extracts of S. pactum mutants with and without chemical complementations.
To produce fluorinated TM-025 and TM-026 as well as other possible analogs by mutasynthesis approach, we generated a ΔptmT/H double mutant strain. This mutant strain was obtained by introducing the ptmT knockout plasmid pTMW037 into ΔptmH mutant (which produces TM-025 and TM-026)12 by conjugation. The product (ΔptmT/H mutant) was cultured in the production medium and confirmed for its lack of TM-025 and TM-026 production (Figure 3D). Further chemical complementation experiment with 3-aminobenzoic acid resulted in the recovery of TM-025 and TM-026 production (Figure 3E). The result confirmed full functionality of the biosynthetic machinery in this mutant except for the genes that have been inactivated.
To explore the possibility of using ΔptmT/H mutant to produce pactamycin analogs with different amino positions of aminoacetophenone, we fed the mutant with 2-, 3-, and 4-aminobenzoic acids, individually. The mutant was pulse-fed with the compounds (50 µmol) at 16, 28, 40, 52, 64 h and the products were isolated using n-BuOH and analyzed by MS. The results showed that only feeding experiments with 3-aminobenzoic acid gave the expected products TM-025 and TM-026 (Figure S2B), whereas feeding with 2- and 4-aminobenzoic acids did not give any corresponding products (Figure S2A and S2C), suggesting substrate rigidity of some of the enzymes in the pathway.
To determine if fluorinated TM-025 and TM-026 analogs can be produced by mutasynthesis, we fed ΔptmT/H mutant with 2-fluoro-5-aminobenzoic acid and 5-fluoro-4-methyl-3-aminobenzoic acid. Both compounds were chemically prepared from commercially available 2-fluoro-5-nitrobenzoic acid and 5-fluoro-4-methyl-3-nitrobenzoic acid, respectively, by catalytic hydrogenation (Figure S3). The ΔptmT/H mutant was pulse-fed with the compounds and the products were analyzed. The results showed that feeding experiments with 2-fluoro-5-aminobenzoic acid gave rise to fluorinated TM-025 and TM-026, designated as TM-025F and TM-026F (Figures 3F and 4). These two compounds gave quasi-molecular ions m/z 413 and 547, respectively, 18 atom mass unit (amu) higher than those of TM-025 and TM-026 (Figure S4). Further confirmation of the products was carried out by comparing their MS/MS fragmentation patterns with those of TM-025 and TM-026 (Figure S4) and by NMR analyses (Figures S9-S12). Whereas 2-fluoro-5-aminobenzoic acid was incorporated in relatively high yields, giving rise to 4–5 mg/L of each TM-025F and TM-026F, feeding experiments with 5-fluoro-4-methyl-3-aminobenzoic acid did not give any new products (Figure S2F).
Figure 4.
Chemical structures of pactamycin analogs.
To test the antimalarial activity of TM-025F and TM-026F, the compounds were subjected to an in vitro antimalarial activity assay. The compounds, along with TM-025, TM-026, and pactamycin were tested against chloroquine sensitive (D6) and multidrug-resistant (Dd2 and 7G8) strains of Plasmodium falciparum. The results showed that TM-025F and TM-026F maintain strong activity against malarial parasites with IC50 in low nM concentrations (Table 1). The results also revealed that fluorine atom substitution in the aminoacetophenone moiety of TM-025 and TM-026 does not significantly affect their antimalarial activity in vitro.
Table 1.
Antimalarial activity of pactamycin analogs.
| Compound | IC50 (nM) |
||
|---|---|---|---|
| D6 | Dd2 | 7G8 | |
| pactamycin | 4.9 | 3.9 | 4.2 |
| TM-025 | 7.7 | 10.5 | 7.1 |
| TM-026 | 11.5 | 14.0 | 9.1 |
| TM-025F | 12.3 | 16.8 | 12.4 |
| TM-026F | 26.5 | 39.1 | 24.9 |
| chloroquine | 10.6 | 114 | 89.5 |
D6, chloroquine-sensitive P. falciparum; Dd2 and 7G8, multidrug-resistant P. falciparum.
To examine if the fluorinated analogs retain the target selectivity of TM-025 and TM-026, antibacterial activity tests were performed against both Gram-positive (Mycobacterium smegmatis, Staphylococcus aureus, Bacillus subtilis) and Gram-negative (Pseudomonas aeruginosa and E. coli) bacteria in an agar diffusion assay. Whereas pactamycin is active against all bacteria tested, neither TM-025F nor TM-026F showed any significant activity at the concentrations used (Figure S13), suggesting their high selectivity for plasmodial cells, but not for bacteria.
In conclusion, we have used an efficient mutasynthetic strategy to generate fluorinated TM-025 and TM-026, two promising pactamycin analogs, in S. pactum. Similar to the parent compounds, the fluorinated analogs maintain excellent activity and selectivity toward chloroquine-sensitive and multidrug-resistant strains of P. falciparum. The results also provide insights into the biosynthesis of 3-aminobenzoic acid, which is relatively obscure, as well as substrate preferences of some of the enzymes in the pathway.
Supplementary Material
Acknowledgment
This work was funded by the general research funds from the OSU College of Pharmacy. This publication was made possible, in part, by the Mass Spectrometry Facilities and Service Core of the Environmental Health Sciences Center, Oregon State University, grant number P30 ES00210, National Institute of Environmental Health Sciences, NIH.
Footnotes
Supporting Information Available Experimental procedures, Table S1, Figures S1-S13. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bhuyan BK. Appl Microbiol. 1962;10:302–304. doi: 10.1128/am.10.4.302-304.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White FR. Cancer Chemother Rep. 1962;24:75–78. [PubMed] [Google Scholar]
- 3.Taber R, Rekosh D, Baltimore D. J Virol. 1971;8:395–401. doi: 10.1128/jvi.8.4.395-401.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otoguro K, Iwatsuki M, Ishiyama A, Namatame M, Nishihara-Tukashima A, Shibahara S, Kondo S, Yamada H, Omura S. J Antibiot (Tokyo) 2010;63:381–384. doi: 10.1038/ja.2010.50. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 6.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Mol Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 7.Hanessian S, Vakiti RR, Dorich S, Banerjee S, Lecomte F, DelValle JR, Zhang J, Deschenes-Simard B. Angew Chem Int Ed Engl. 2011;50:3497–3500. doi: 10.1002/anie.201008079. [DOI] [PubMed] [Google Scholar]
- 8.Hanessian S, Vakiti RR, Dorich S, Banerjee S, Deschenes-Simard B. J Org Chem. 2012;77:9458–9472. doi: 10.1021/jo301638z. [DOI] [PubMed] [Google Scholar]
- 9.Knapp S, Yu Y. Org Lett. 2007;9:1359–1362. doi: 10.1021/ol0702472. [DOI] [PubMed] [Google Scholar]
- 10.Malinowski JT, McCarver SJ, Johnson JS. Org Lett. 2012;14:2878–2881. doi: 10.1021/ol301140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Roongsawang N, Shirasaka N, Lu W, Flatt PM, Kasanah N, Miranda C, Mahmud T. Chembiochem. 2009;10:2253–2265. doi: 10.1002/cbic.200900339. [DOI] [PubMed] [Google Scholar]
- 12.Lu W, Roongsawang N, Mahmud T. Chem Biol. 2011;18:425–431. doi: 10.1016/j.chembiol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Chan KK, O'Hagan D. Methods Enzymol. 2012;516:219–235. doi: 10.1016/B978-0-12-394291-3.00003-4. [DOI] [PubMed] [Google Scholar]
- 14.Adams ES, Rinehart KL. J Antibiot (Tokyo) 1994;47:1456–1465. doi: 10.7164/antibiotics.47.1456. [DOI] [PubMed] [Google Scholar]
- 15.Rinehart KL, Jr, Potgieter M, Delaware DL, Seto H. J. Am. Chem. Soc. 1981;103:2099–2101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.