Summary
Most herpes simplex virus 2 (HSV-2) reactivations in humans are subclinical and associated with rapid expansion and containment of virus. Previous studies have shown CD8+ T cells persist in genital skin and mucosa at the dermal-epidermal junction (DEJ), the portal of neuronal release of reactivating virus, for prolonged time periods after lesions are cleared1,2. The phenotype and function of this persistent CD8+ T-cell population remain unknown. Here, using cell type-specific laser capture microdissection, transcriptional profiling and T-cell receptor beta (TCRβ) genotyping on sequential genital skin biopsies, we show CD8αα+ T cells are the dominant resident population of DEJ CD8+ T cells that persist at the site of prior HSV-2 reactivation. CD8αα+ T cells located at the DEJ lack chemokine receptor expression required for lymphocyte egress and recirculation, express gene signatures of T-cell activation and antiviral activity, and produce cytolytic granules during clinical and virological quiescent time periods. Sequencing of the TCRβ chain repertoire revealed that the DEJ CD8αα+ T cells are oligoclonal with diverse usage of TCR VB genes, which differ from those commonly described for MAIT and NKT cells. Dominant clonotypes overlapped among multiple recurrence episodes over a period of two and a half years. Episodes of rapid asymptomatic HSV-2 containment were also associated with a high CD8 effector-to-target ratio and focal enrichment of CD8αα+ T cells. These studies indicate DEJ CD8αα+ T cells are tissue resident cells that appear to play a fundamental role in immune surveillance and in initial containment of HSV-2 reactivation in human peripheral tissue. Elicitation of CD8αα+ T cells might be a critical component for developing effective vaccines against skin and mucosal infections.
Clinical studies have shown that 50-80% of HSV reactivations are subclinical and of short duration (<6 hours)3-5, and that CD8+ T cells not only infiltrate selectively to the site of viral reactivation, but also persist locally at DEJ for months after resolution of herpes lesions1,2. To define the phenotype, function and diversity of these DEJ CD8+ T cells during virological quiescence as well as early containment of reactivation, we used cell type-specific laser capture microdissection (LCM) to identify and select individual CD8+ T cells from the DEJ (DEJ CD8), dermis near blood vessels (BV CD8), HSV-2-affected area, and contralateral HSV-2-unaffected genital tissue (Control CD8) (Supp Fig. 1A&B), to examine the persistent nature, antiviral signature and TCR repertoire of these cells from sequential skin biopsies. The purity of captured CD8 cells was validated by the abundance of CD8A and the absence of CD4 gene expression (Supp Fig. 1C). Detailed immunofluorescent staining demonstrated that DEJ-localized CD8+ T cells expressed CD3ε and the T-cell receptor (TCR) β, but not TCRγδ (Supp Fig. 2). These cells did not express co-receptors and markers for dendritic cells (DC), natural killer (NK) cells, and nonconventional T cells, such as NK T cells and mucosal associated invariant T (MAIT) cells (Supp Fig. 3). Thus, the CD8+ T cells that persist at the DEJ are CD8+ TCRαβ T cells.
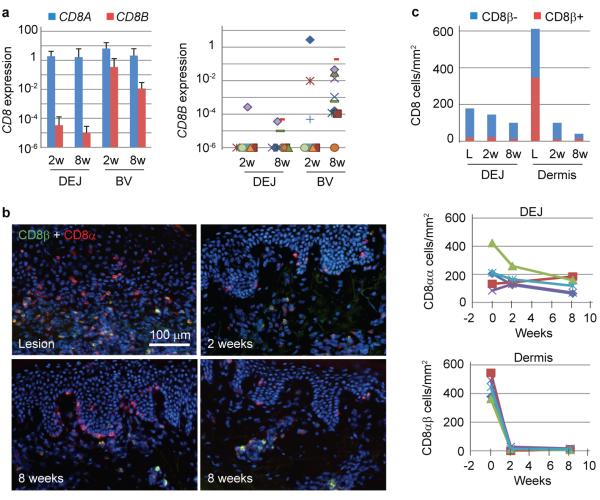
Because tissue-based CD8+ T cells and memory precursors have been shown to express varied levels of CD8α and CD8β6,7, we measured gene expression of these co-receptors by quantitative TaqMan PCR. DEJ CD8 cells almost exclusively expressed CD8A mRNA. CD8B was detected in only 1/8 and 3/10 subjects biopsied at 2 and 8 weeks post-healing from the HSV-2 affected area, respectively. In contrast, BV CD8 cells from the same biopsies expressed high levels of CD8B transcripts in 9/10 patients (Fig. 1A). Cell surface expression of CD8α and CD8β chains circumstantiated these transcriptional patterns (Fig. 1B&C). CD8α+β− cells predominated at the DEJ from the time of active lesion to 2 and 8 weeks post-healing, ranging from 85% to 91% of the total DEJ CD8α+ cells, and the number of these cells remained relatively stable. In contrast, CD8α+β+ cells localized mainly in the dermis during HSV-2 ulceration and rapidly diminished after completely healing, resulted in a 98% reduction in cell density during the first 2 weeks post-healing at a rate (169 cells/mm2/week) almost 10 times higher than that of CD8α+β− cells at the DEJ (17.5 cells/mm2/week) for the same period. When detected in either 2 or 8 week post-healed biopsy tissue, CD8α+β+ cells were mainly located near the blood vessels. These data indicate, at both the RNA and protein level, CD8+ T cells that selectively persist and preferentially localize at the DEJ have a CD8αα homodimer phenotype.
Figure 1. CD8αα+, but not CD8αβ+, T cells persist at the DEJ in human HSV-2 infection.
a, Average and individual expression of CD8A and CD8B genes in DEJ CD8 and BV CD8 cells laser captured from tissues of 2 (n = 8) and 8 (n = 10) week post-healed (wph) biopsies, relative to ACTB. b, Differential localization of CD8αα+ (red) to the DEJ and CD8αβ+ (yellow) to the dermis in active lesions, 2 wph and 8 wph tissue (high and low density areas). f, Quantitation of CD8αα+ and CD8αβ+ T cells at DEJ and dermis (upper) over time. Individual and average (light blue line) kinetics of DEJ CD8αα+ cells (middle) and dermal CD8αβ+ T cells (lower) over time (n = 4). Lesions: week 0.
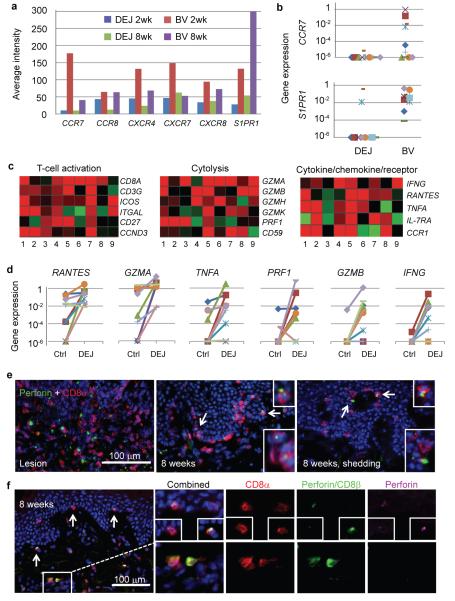
To investigate the potential mechanism for persistence of CD8αα+ T cells at the DEJ, we compared the transcriptional profiles of DEJ CD8 versus BV CD8 cells isolated from the same tissue biopsy obtained at 2 and 8 weeks post-healing. Gene expression of chemokine receptors and lymphocyte-trafficking receptors was significantly different between DEJ CD8 and BV CD8 cells. We observed a down-regulation of CCR7, CCR8, CXCR4, CXCR6, CXCR7 and S1PR1 transcripts in DEJ CD8 compared to BV CD8 cells (Fig. 2A). Using TaqMan PCR, we found no detectable CCR7 gene expression in DEJ CD8 cells from 8 week post-healed tissue in 9/10 subjects; an expression pattern also observed for S1PR1, which was undetectable in 8/10 subjects (Fig. 2B). Comparatively, high levels of CCR7 and S1PR1expression were detected in BV CD8 cells. It has been shown that CCR7 is required for guiding lymphocytes exiting peripheral tissue and S1PR1 functions in lymphocyte recirculation and trafficking in the blood8-11. The absence of transcriptional activity of both genes in DEJ CD8 cells suggests that a potential mechanism for CD8αα+ T cells persistence at the DEJ is a lack of chemotactic requirements for exit.
Figure 2. Tissue residence and effector function of DEJ CD8αα+ T cells in post-healed tissue.
a, Down-regulation of chemokine receptor expression in DEJ CD8 as compared to BV CD8 cells. Data are average signal intensities on the Illumina Beadarray (n=5). b, CCR7 and S1PR1 expression by QPCR. One subject per symbol. c, Activated effector signatures of DEJ CD8 cells. Pair-wised comparison was performed between DEJ CD8 and control CD8 cells from the same individual at the 8wph time point (n = 9). Red: up-regulated, green: down-regulated. d, QPCR analysis of antiviral gene expression (n = 9). e, Perforin protein expression in DEJ CD8 cells of active lesion (left) and 8 wph with (right) and without (middle) virus shedding. f, Perforin protein expression in DEJ CD8αα+ T cells but not BV CD8αβ+ T cells. Perforin: yellow granules (both pink and green); CD8α: red; CD8β: green. Arrows denote enlarged area of perforin expression.
We also compared expression profiles of genes critical for T-cell activation and antiviral and cytolytic function between captured DEJ CD8 and Control CD8 cells from 8 week post-healed biopsies12, a time of presumptive virological quiescence. Up-regulation of genes for TCR co-receptors (CD8A and CD3G) and co-stimulation (ICOS, and ITGAL known as LFA-1A), G1/S cell cycle transition (CD27 and CCND3), cytolytic activity (PRF1, GZMA, GZMB, GZMK and GZMH), and cytokines/chemokines and receptors (IFNG, TNFA, RANTES and IL-7RA) suggests that DEJ CD8 cells were actively engaging and eliminating infection (Fig. 2C). We performed quantitative PCR on a subset of these genes to further delineate the expression patterns (Fig. 2D). RANTES and GZMA expression was up-regulated in DEJ CD8 cells from all 10 subjects. GZMB and IFNG were induced in 6/10 and 7/10 subjects, respectively, a significantly higher proportion compared to contralateral control samples (1/9 and 2/9, respectively) (p<0.01, Fisher exact test). Perforin protein expression was also detected in DEJ CD8 cells at 8 weeks post-healing (Fig. 2E); no perforin granules were detected in the contralateral Control CD8 cells (data not shown). The strongest perforin granule expression at the 8 week post-healing time point was seen in DEJ cells from patients who had evidence of subclinical HSV-2 shedding (Fig. 2E). Co-staining of biopsy tissue showed that perforin granules were detected in DEJ CD8α+β− cells (Fig. 2F upper panels) but not in BV CD8α+β+ cells (Fig. 2F lower panels). The expression of cytolytic granules and mRNA patterns of T-cell activation and antiviral responses indicate that DEJ CD8αα+ T cells are programmed and regulated to respond to virus infection.
We investigated the role CD8αα+ T cells play in the early containment of HSV-2 reactivation during episodes of subclinical recurrence. HSV-2 DNA was detected in tissue sections in two of the ten 8 week post-healed biopsies. DNA levels were significantly lower than those during a genital lesion (<20 copies/5×104 cells vs. 106 copies/5×104 cells, respectively) and the biopsies concurrently displayed no histologic evidence of ulceration or epithelial cell necrosis, indicating an episode of spontaneous asymptomatic shedding controlled by the host immune system1,13,14. Immunofluorescence staining revealed a punctate distribution of HSV-2 antigen in several keratinocytes dispersed in the epidermis (Fig. 3A). CD8+ T cells and HSV-2-infected cells formed clusters in which 2 to 4 CD8+ T cells were in direct contact with a virally infected cell (Fig. 3B). This high effector-to-target ratio during asymptomatic shedding is in contrast to that which occurs during clinical episodes of recurrent genital herpes (Fig. 3C). In situ Qdot-pMHC multimer staining identified HSV-2 specific CD8+ T cells present in these shedding biopsies (Supp Fig. 4) along with increased numbers of perforin granules (Fig. 3D), providing evidence of antigen-specific early containment of virus infection. Furthermore, high-density CD8αα+ T cells were found in foci at the DEJ and there was increased recruitment of CD8αβ+ cells to the DEJ (Fig. 3E). Again, we detected perforin granule expression in CD8α+β− T cells, suggesting these CD8αα+ T cells are cytolytically active during early host antiviral control (Supp Fig. 5). These studies suggest that CD8αα+ T cells are important effectors in early immune containment.
Figure 3. DEJ CD8 cells in early containment during asymptomatic HSV-2 reactivation.
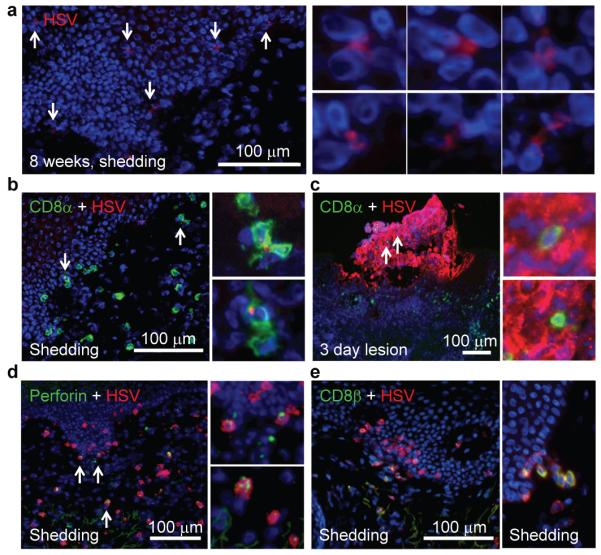
a, HSV-2 antigen expression in keratinocytes during asymptomatic HSV-2 reactivation. Arrows denote infected cells. b, CD8 effector-to-target ratio during asymptomatic (b) and lesional (c) HSV-2 reactivation. d, High level of CD8α+ T cells expressing perforin granules in during asymptomatic shedding. Arrows denote enlarged area. e, Focal accumulation of CD8αα+ T cells (red, left) and recruitment of CD8αβ+ T cells (yellow, right) to the DEJ.
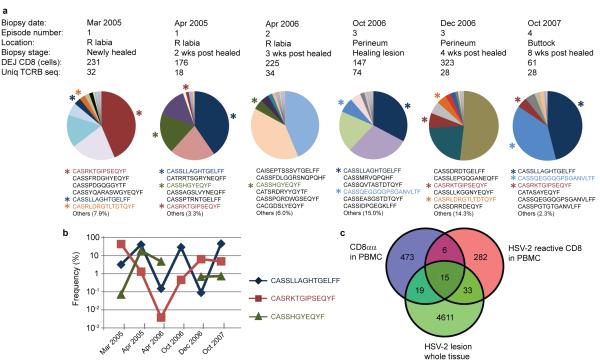
In order to further understand the persistence, diversity and specificity of CD8αα+ T cells at the DEJ, we analyzed the amino acid sequence in the hypervariable complementarity-determining region 3 (CDR3) of the T-cell receptor β chain (TCRB) in DEJ CD8 cells using high-throughput sequencing assays15. We collected DEJ CD8 cells via LCM from six biopsies obtained during a two and a half year time period from an individual with long-standing recurrent HSV-2 who participated in a series of trials involving sequential skin biopsies during and after HSV-2 reactivation (Fig. 4A). The sequence pattern of DEJ CD8αα+ T cells indicated an oligoclonal immune profile, with the top six most abundant sequences making up 85% to 98% of the TCRB repertoire in each DEJ CD8 sample (Fig. 4A & Supp. Table 1). Twelve TCRB CDR3 sequences were common among the six tissue biopsies (Fig. 4A, Supp. Table 2); two sequences were detected in all six biopsies and one was present in five biopsies over the 2.5 year time period (Fig. 4B). Moreover, both the abundant sequences and the 12 commonly shared sequences (Supp Table 1&2) involved a diverse utilization of VB gene families; e.g., 18 VB genes were found among the 27 unique TCRB sequences that predominated in DEJ CD8αα+ T cells. Interestingly, evidence of constrained VB gene usage as described in CD8αα+ MAIT cells was not seen16,17; Vβ2 and Vβ13 families were excluded in DEJ CD8 cells.
Figure 4. Characteristics of DEJ CD8 cell TCRβ repertoire.
a, Oligoclonal distribution and dominant CDR3 sequences of the DEJ CD8 cell TCRβ repertoire. DEJ CD8 cells were laser captured from six biopsies obtained during and after multiple episodes of herpes recurrence from one individual. The pie chart illustrates the distribution of the unique CDR3 sequences detected in the DEJ CD8 TCRβ repertoire from each biopsy. Amino acid (aa) sequences of the top six clones are listed in the order of abundance. Shared aa sequences between biopsies are highlighted in the same color as pie charts. Each color represents one clone. b, Frequencies of the three most commonly shared DEJ CD8 clones. c, Venn diagram of overlapping TCRβ repertoires among 3 T-cell populations from the same individual: tissue infiltrating T cells from a healing lesion biopsy, blood PBMC-derived HSV-2 reactive CD8+ T cells and PBMC-derived CD8αα+ T cells. Clone tracking was performed at the nucleotide level.
To examine the antigen specificity of DEJ CD8αα+ T cells, we sequenced TCRB from tissue infiltrating T cells during lesion healing, PBMC-derived CD8αα+ T cells, and HSV-2 reactive CD8+ T cells isolated directly ex vivo from PBMCs from the same individual18. Fifteen TCRB CDR3 sequences were shared by all three T-cell populations (Fig. 4C & Supp Table 3), and one of these sequences, CASRLDRGTLTDTQYF, was also detected in DEJ CD8 cells from two separate biopsies 21 months apart from distinct HSV-2 recurrences (3/2005, newly healed of episode #1; and 12/2006, 4 weeks post-healing of episode #3; Fig. 4A). These results provide evidence that DEJ CD8 cells are comprised of a CD8αα+ T-cell population that responds to HSV-2 infection in genital tissue. The sequence CASRLDRGTLTDTQYF and the two commonly shared sequences in all six DEJ CD8 samples, CASSLLAGHTGELFF and CASRKTGIPSEQYF, were abundant in the lesion-healing biopsy sample. These sequences were ranked #2, #6 and #12 in frequency among 4,678 unique TCR sequences detected, further supporting the important role of DEJ CD8αα+ T cells in response to HSV-2 antigen in tissue.
Our studies demonstrate the ability of dissecting cell type-specific immune responses in situ in humans and provide several novel insights into the tissue resident immunity in genital skin and mucosa. Accumulating evidence indicates that antigen-specific CD8+ T cells retain long-term residence at the peripheral site of infection, exhibiting superior protection over their blood counterparts against re-infection of invading pathogens1,19-25. Our human studies have shown that CD8+ T cells persisting at the DEJ site of prior HSV-2 lesions possess a CD8αα homodimer phenotype. The persistence of CD8αα+ T cells contrasts with the rapid depletion of CD8αβ+ T cells after lesion healing. These CD8αα+ T cells lack chemotactic signals, such as CCR7 and S1PR1, distinguishing them from circulating CD8αβ+ T cells.
The detailed characterization of cell type-specific markers and TCR VB gene diversity indicate the CD8αα+ T cells described in this study are not innate or semi-innate T cells, rather, they are components of the adaptive immune response retained in the barrier tissue as sentinel cells. CD8αα homodimer expression has been associated with high-affinity antiviral effector T cells and hypothesized to preserve optimal effector cells at the portal of pathogen entry for long-lived mucosal memory7,26. Whether DEJ CD8αα+ T cells are directed at a unique class of viral antigens is unknown and is of major interest.
The unique anatomic localization of DEJ CD8αα+ T cells contiguous to the neural gateway of reactivating virus suggested these cells might recognize and contain HSV infection. While functional relevance remains to be proven, the activated gene signature, oligoclonal immune profile and HSV-2 specific T-cell response at sites of prior reactivation support the notion that DEJ CD8αα+ T cells might mediate a rapid and effective peripheral immunity to control frequent HSV antigen exposure in human skin and genital tract. The prompt CD8 antiviral response at the site of virus release during asymptomatic HSV reactivation is in sharp contrast to the delayed CD8+ T-cell infiltration during a lesion-forming herpes recurrence, in which CD8+ T-cell infiltration occurs 2 to 3 days after initial infection, following NK and CD4+ T-cell arrival27,28. Enhancement of the quantity and the function of tissue resident CD8αα+ T cells might potentially be a mechanism for improved immunotherapeutic treatment and for prevention of HSV-2 reactivation in humans. Similarly, the immunological role of these persisting T cells in other chronic infections of the skin and genital tract, such as HIV, should also be explored.
Methods
Human biopsies
The study protocol, biopsy procedure and informed consent were approved by the University of Washington Institutional Review Board. Enrolled subjects have culture-proven symptomatic genital HSV-2 infection and were all HIV-1/2 seronegative. All genital skin biopsies were collected from HSV-2 seropositive immunocompetent adults at the time of active lesion and then at 2 and 8 weeks after complete lesion healing, and from contralateral unaffected (control) genital skin1,2,12. Tissue biopsies were snap frozen in OCT within 12 hours of collection and stored at −80°C until processing. Peripheral blood was collected at the same time points.
Immunofluorescence staining
Detection of T-cell markers, viral antigen and HSV-2 specific CD8+ T cells in genital tissue was performed as described1,2. Tyramide Signal Amplification (TSA) (Invitrogen) was applied for the first antibody in dual staining, followed by incubation with CD8α–AF647. For triple staining, sequential TSA was performed by applying first antibody to perforin and then antibody to CD8β, followed by incubation with CD8α–AF647. Antibodies used in this study are listed in Supplementary Table 4.
CD8-specific laser capture microdissection (LCM)
Frozen tissue was sectioned into 8 μm slices and placed on a polyethylene naphthalate (PEN) membrane slide (Carl Zeiss, Germany). Tissue sections were fixed in 100% ethanol, incubated with CD8α–AF647 on ice for 5 min, washed in PBS, and dried with ethanol (75%, 95% and 100%) and xylene. CD8+ cells were identified and laser microdissected using either a PALM MicroBeam instrument (Carl Zeiss) or an Arcturus Veritas microdissection system (Molecular Devices). About 100 CD8+ cells were laser captured from each sample for transcriptional analysis, unless otherwise specified.
RNA extraction, cDNA amplification and hybridization to Illumina beadarrays
Total RNA from LCM captured CD8 cells from each group were extracted using Arcturus PicoPure RNA Isolation kits ((Life Technologies), and quantified by a NanoDrop 1000 Spectrophotometer. One nanogram of total RNA was then used to generate and amplify cDNA through whole transcriptome amplification using WT-Ovation Pico RNA Amplification System (NuGEN). The cDNA were biotin labeled following a protocol from NuGEN and labeled cDNA (750 ng) were then used to hybridize to Illumina HumanRef8_v3 beadarrays in the Shared Resources at Fred Hutchison Cancer Center.
Bead array data analysis
Detailed analysis has been described previously12. Briefly, raw data were imported to GenomeStudio (V2010.3, Illumina) for initial quality control. Normalized Data were then exported to R and differentially expressed genes between DEJ CD8 and Control CD8 or between DEJ CD8 and BV CD8 were selected. The differentially expressed genes were further analyzed using an unsupervised hierarchical clustering method and using SpotFire DecisionSite for functional genomics (Version 9.1.2). Enriched functional categories and network analysis for the set of differentially expressed genes were performed using Ingenuity Pathway Analysis (IPA 8.8). The GoMiner program was used to annotate all the 18,401 genes on Illumina Human Ref8_v3 bead arrays.
Microarray data accession number
Complete array data can be accessed under GEO accession numbers GSE39625 and GSE44975.
TaqMan real-time PCR
TaqMan PCR assays were performed using primer/probe sets obtained from Applied Biosystem (Supplementary Table 5). Relative expression to ACTB is shown for each gene. HSV-2 DNA detection in tissue was described previously1,29.
DNA extraction and amplification
Genomic DNA from laser capture microdissected CD8+ T cells was extracted using Arcturus PicoPure DNA Extraction kits (Life Technologies). Recovered DNA was whole genome amplified through multiple displacement amplification using REPLI-g Mini kit (Qiagen).
TCRB sequencing
TCR beta chain sequencing was performed at Adaptive Biotechnologies using the ImmunoSEQ platform15. All TCR beta chains were PCR amplified from 400 ng of DNA from each sample. The library was sequenced using Illumina HiSeq and the resulting reads were processed using and uploaded onto the ImmunoSeq Analyzer secure relational database.
Purification of HSV reactive CD8+ T cells from PBMC ex vivo
We obtained bulk HSV-2 reactive CD8 T cells as described previously18 and adapted the method for HSV-2. HSV-2-infected HeLa cell debris was added to cultured peripheral blood mononuclear cell (PBMC)-derived dendritic cells, which were then incubated with negatively selected CD8 T cells from the same subject and stained with 7-AAD for viability assessment and with anti-CD8α, anti-CD3, and anti-CD137 conjugated to fluorochromes. The cells were sorted on a FACSAria III instrument and live, CD3+, CD8α+, and CD137bright cells were selected. Approximately 5,000 directly sorted cells were submitted for TCRβ sequencing.
Purification, sorting and deep sequencing of CD8αα+ T cells from PBMC
CD8αα T cells were purified from PBMC and labeled with antibodies of CD8α Pacific Blue, CD8β APC, CD3 APC-Cy7, αβ TCR Alexa 488, and CD4 PE-Cy7. The cells were sorted using BD FACS Aria. CD8αα T cells, described as CD3+αβ TCR+CD4−CD8α+CD8β−, were collected and expanded using a rapid expansion protocol30 in the presence of 50 IU/ml of recombinant human IL-2 and 1 ng/ml of recombinant human IL-7 (rhuIL-7). A total of 890,000 CD8α+β−CD4− T cells (98% pure) were collected and submitted to Adaptive Biotechnologies (Seattle, WA) for TCRβ deep sequencing.
Supplementary Material
Acknowledgements
We thank M. Huang, H. Xie, J. Vazquez and D. McDonald for technical assistance. M. Prlic and C. Desmarais for discussion and reading the manuscript. M. Miner for editing. We also thank our study participants and S. Kuntz and M. Stern for clinical assistance. This work was supported by grants from the National Institutes of Health (R37AI042528, R01AI04252815, P01AI030731 and R56AI093746) and the James B. Pendleton Charitable Trust.
Footnotes
Author Contributions. J.Z. and L.C. conceived the study and wrote the manuscript. J.Z. and T.P. developed the technology and analyzed and interpreted the data. K.P., A.S.K., A.K., L.J. and K.D. performed the experiments. D.M.K. isolated HSV-2 reactive CD8+ T cells and peptide epitope used in the study. C.J. and A.W. directed human biopsy studies. H.R. contributed to TCR data analysis and interpretation. All authors contributed to the discussion.
Author Information. Reprints and permission information are available at www.nature.com/reprints. The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Zhu J, et al. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J Exp Med. 2007;204:595–603. doi: 10.1084/jem.20061792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, et al. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med. 2009;15:886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wald A, Zeh J, Selke S, Ashley RL, Corey L. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med. 1995;333:770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- 4.Wald A, et al. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N Engl J Med. 2000;342:844–850. doi: 10.1056/NEJM200003233421203. [DOI] [PubMed] [Google Scholar]
- 5.Mark KE, et al. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J Infect Dis. 2008;198:1141–1149. doi: 10.1086/591913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayday A, Theodoridis E, Ramsburg E, Shires J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nature Immunol. 2001;2:997–1003. doi: 10.1038/ni1101-997. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, et al. Mucosal memory CD8(+) T cells are selected in the periphery by an MHC class I molecule. Nature Immunol. 2011;12:1086–1095. doi: 10.1038/ni.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 9.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nature Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 10.Debes GF, et al. Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues. Nature Immunol. 2005;6:889–894. doi: 10.1038/ni1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi H, Flavell RA. Cutting edge: regulation of T cell trafficking and primary immune responses by sphingosine 1-phosphate receptor 1. J Immunol. 2005;174:2485–2488. doi: 10.4049/jimmunol.174.5.2485. [DOI] [PubMed] [Google Scholar]
- 12.Peng T, et al. An effector phenotype of CD8+ T cells at the junction epithelium during clinical quiescence of herpes simplex virus 2 infection. J Virol. 2012;86:10587–10596. doi: 10.1128/JVI.01237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiffer JT, et al. Mucosal host immune response predicts the severity and duration of herpes simplex virus-2 genital tract shedding episodes. Proc. Natl Acad. Sci. USA. 2010;107:18973–18978. doi: 10.1073/pnas.1006614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiffer JT, et al. Frequent release of low amounts of herpes simplex virus from neurons: results of a mathematical model. Sci. Transl Med. 2009;1:7ra16. doi: 10.1126/scitranslmed.3000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bourhis L, et al. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 2011;32:212–218. doi: 10.1016/j.it.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Jing L, et al. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin. Invest. 2012;122:654–673. doi: 10.1172/JCI60556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark RA, et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci. Transl Med. 2012;4:117ra7. doi: 10.1126/scitranslmed.3003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nature Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 21.Gebhardt T, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477:216–219. doi: 10.1038/nature10339. [DOI] [PubMed] [Google Scholar]
- 22.Jiang X, et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity. 2003;18:593–603. doi: 10.1016/s1074-7613(03)00112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purwar R, et al. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PloS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc. Natl Acad. Sci. USA. 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trautmann L, et al. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham AL, Turner RR, Miller AC, Para MF, Merigan TC. Evolution of recurrent herpes simplex lesions. An immunohistologic study. J Clin. Invest. 1985;75:226–233. doi: 10.1172/JCI111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magaret A, Wald A, Huang ML, Selke S, Corey L. Optimizing PCR Positivity Criterion for Detection of Herpes Simplex Virus DNA on Skin and Mucosa. Journal of clinical immunology. 2007;45:1618–1620. doi: 10.1128/JCM.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddell SR, et al. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.