Abstract
To identify susceptibility loci for visceral leishmaniasis we undertook genome-wide association studies in two populations; 989 cases and 1089 controls from India, and 357 cases in 308 Brazilian families (1970 individuals). The HLA-DRB1-HLA-DQA1 locus was the only region to show strong evidence of association in both populations. Replication at this region was undertaken in a second Indian population comprising 941 cases and 990 controls, resulting in Pcombined=2.76×10−17 and OR(95%CI)=1.41(1.30-1.52) across the three cohorts at rs9271858. A conditional analysis provided evidence for multiple associations within the HLA-DRB1-HLA-DQA1 region, and a model in which risk differed between three groups of haplotypes better explained the signal and was significant in the Indian discovery and replication cohorts. In conclusion the HLA-DRB1-HLA-DQA1 HLA class II region contributes to visceral leishmaniasis susceptibility in India and Brazil, suggesting shared genetic risk factors for visceral leishmaniasis that cross the epidemiological divides of geography and parasite species.
Leishmania are protozoan parasites that live in macrophages. They are transmitted by sand flies and cause severe and debilitating cutaneous, as well as fatal visceral disease in subtropical/tropical regions of Old and New Worlds. Leishmaniasis is classified by WHO as one of the neglected tropical diseases. It affects 12 million people and there are an estimated 1.5 million new cases annually1. Of these, 500,000 are cases of potentially fatal visceral leishmaniasis caused by the Leishmania donovani complex, 90% of which occurs in three foci in India/Bangladesh/Nepal, Sudan, and Brazil. Skin-tests and lymphocyte responses indicate that only 1 in 5-10 infected individuals develop clinical disease2-4. The importance of host genetic factors is indicated by familial clustering5 and high sibling risk ratios6. However, human genetic studies undertaken to date (reviewed7-9) provide inconsistent results. As part of the Wellcome Trust Case Control Consortium 2 (WTCCC2) study of 15 complex disorders and traits, we report the first genome-wide association study (GWAS) of visceral leishmaniasis across major foci of disease caused by L. donovani in India and L. infantum chagasi in Brazil.
Subjects for the India discovery GWAS (Supplementary Table 1, online methods) were recruited from Bihar state in northeast India. Patients and controls were matched for self-reported age, sex, religion, caste and geographic region of recruitment. The Brazilian family-based sample was collected as part of the Belém Family Study and from study sites near Natal6,10,11 (Supplementary Table 1, online methods).
All individuals were genotyped at the Wellcome Trust Sanger Institute on the custom Illumina Human660W-Quad chip (online methods). After performing stringent quality control (QC) procedures12 (online methods) the Indian discovery dataset comprised 2078 individuals (989 cases and 1089 controls) genotyped at 526,731 SNPs and the Brazilian discovery dataset comprised of 1970 individuals (357 cases; 308 families) genotyped at 553,323 SNPs.
As ancestry differences and close relationships between case and control individuals can confound association studies, we used a variance components method (sometimes referred to as a mixed model), which models the pair-wise relatedness between individuals to account for population structure in the data13. This approach accounts for relatedness on different scales, from close relatives to distant ancestral structure, and it can thus be applied both to the case-control samples in India and the family data from Brazil. Similar models have been recently used in several other association studies14-18. An advantage of our implementation of the mixed model is that we are able to estimate the effect sizes on the log-odds scale. Using this method, the genomic inflation factor (λ) was 1.03 for the India analysis and 1.02 for the Brazil analysis, showing that it successfully handled any population structure or relatedness in the data. The mixed model approach was used for all analyses presented here, for both discovery and replication data and in further dissection of association signals.
Previous studies attempting to identify the genetic components of visceral leishmaniasis susceptibility have typically been small, underpowered, family studies or candidate gene studies19-23. We present the results from our discovery GWAS and replication data (see below) at these previously identified loci in Table 1. None of the loci previously found to be associated with visceral leishmaniasis consistently show an association with visceral leishmaniasis in our three datasets.
Table 1.
Signals of association at previously reported non-HLA loci for visceral leishmaniasis
| SNP | Chr | Locus | Position (bp) | Risk Allele |
Popb | Ref | This Study | Odds Ratio |
95% CI |
P- value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4674259 | 2q35 | CXCR2 | 218,699,250 | A | India | 19 | Indian Discovery | 0.92 | 0.81-1.04 | 0.195 |
| Indian Replicationc |
0.87 | 0.77-1.00 | 0.040 | |||||||
| Brazil Discovery | 1.24 | 1.04-1.49 | 0.017 | |||||||
| rs4672875a | 2q35 | CXCR1 | 218,729,630 | A | India | 19 | Indian Discovery | 1.05 | 0.89-1.23 | 0.582 |
| Indian Replication |
NA | NA | NA | |||||||
| Brazil Discovery | 0.82 | 0.65-1.05 | 0.104 | |||||||
| rs2276631 | 2q35 | SLC11A1 | 218,957,257 | A | Sudan | 23 | Indian Discovery | NA | NA | NA |
| Indian Replication |
0.96 | 0.80-1.15 | 0.633 | |||||||
| Brazil Discovery | NA | NA | NA | |||||||
| rs3731865 | 2q35 | SLC11A1 | 218,958,247 | G | Sudan | 23 | Indian Discovery | NA | NA | NA |
| Indian Replication |
0.99 | 0.82-1.19 | 0.933 | |||||||
| Brazil Discovery | NA | NA | NA | |||||||
| rs17860704 | 6q27 | DLL1-FAM120B | 170,444,354 | G | Sudan | 22 | Indian Discovery | NA | NA | NA |
| Indian Replication |
1.14 | 0.97-1.34 | 0.117 | |||||||
| Brazil Discovery | NA | NA | NA | |||||||
| rs159273 | 17q12 | CCL1 | 29,714,798 | C | Brazil | 21 | Indian Discovery | 1.05 | 0.95-1.15 | 0.362 |
| Indian Replication |
NA | NA | NA | |||||||
| Brazil Discovery | 1.15 | 0.86-1.56 | 0.334 | |||||||
| rs2063979 | 17q12 | CCL16 | 31,327,679 | A | Brazil | 21 | Indian Discovery | 0.99 | 0.87-1.13 | 0.913 |
| Indian Replication |
1.05 | 0.92-1.19 | 0.467 | |||||||
| Brazil Discovery | 0.92 | 0.77-1.10 | 0.335 | |||||||
| rs854680 | 17q12 | CCL16 | 31,333,164 | C | Brazil | 21 | Indian Discovery | 1.00 | 0.88-1.13 | 0.938 |
| Indian Replication |
1.07 | 0.94-1.22 | 0.277 | |||||||
| Brazil Discovery | 1.16 | 0.97-1.39 | 0.103 | |||||||
| rs228953 | 22q12 | IL2RB | 35,861,382 | G | Sudan | 20 | Indian Discovery | NA | NA | NA |
| Indian Replication |
0.97 | 0.85-1.10 | 0.615 | |||||||
| Brazil Discovery | NA | NA | NA |
Odd ratios are given for the previously reported risk allele. Positions are in NCBI human genome build 36 coordinates. ‘NA’ indicates that the SNP was not genotyped in this sample.
r2=1 (HapMap Utah residents with ancestry from northern and western Europe (CEU)) and 0.71 (HapMap Gujarati Indians in Houston, Texas (GIH)) with previously reported rs3138060.
Population in which the initial SNP association was reported.
The Indian replication sample reported in the table overlaps significantly with the Mehrotra et al (2011)19 case-control population in which rs4674259 was genotyped as part of a separate Sequenom panel.
Because different pathogen species are responsible for disease in India and Brazil, we first examined each GWAS separately. Our primary focus will be on regions showing association in both GWAS studies. This would be appropriate if similar host genetic factors affected susceptibility to the two different parasite species. Observing the same association in different populations, with different study design (population-based and family-based association, respectively) also adds to the robustness of potential findings.
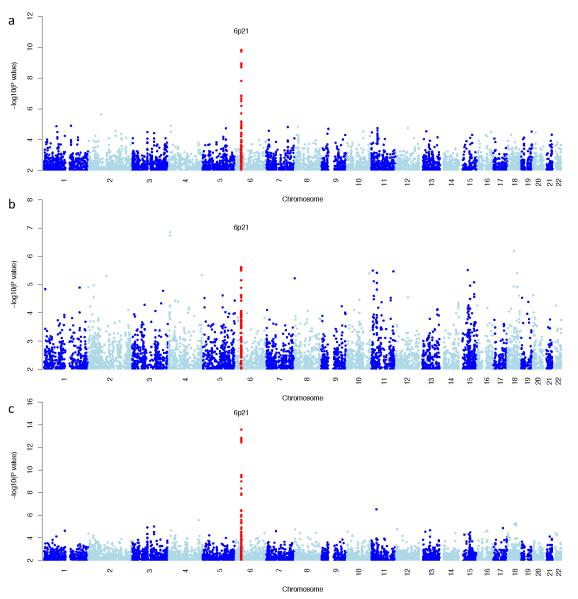
We combined the India and Brazil GWAS through a fixed effects meta-analysis (Fig. 1c). Two regions had a Pcombined <10−6, one of which was the HLA-DRB1-HLA-DQA1 class II region (Supplementary Table 4). Details of the regions showing association (P<10−5) in only one of the two studies are shown in (Fig 1, and Supplementary Tables 2, 3 and 4). Apart from the MHC, none of the association regions with P <10−5 in the separate India or Brazil analysis showed significant association in the combined analysis.
Figure 1.
Plot of genome-wide association results in the separate (a,b) and combined (c) discovery GWAS using a variance components method. SNPs in red show regions with replicated association to visceral leishmaniasis susceptibility. a) Indian discovery data at 526,731 SNPs b) Brazilian discovery data at 553,323 SNPs c) Plot of the meta-analysis genome-wide association results.
A second Indian cohort was used in a replication analysis of the findings from the two discovery GWAS. These samples were genotyped on the Immunochip 24, a purpose-designed Illumina chip, some content of which was chosen by WTCCC2 for deep replication of its GWAS findings. After QC (online methods), the Indian replication dataset comprised 1931 individuals (941 visceral leishmaniasis cases and 990 controls). Outside of the MHC, none of the regions that were associated at P<10−5 in either the separate Indian or Brazilian GWAS or the combined discovery meta-analysis replicated at P<0.1 in the Indian replication data (Supplementary Tables 2, 3 and 4). However, only four of the regions associated in the Brazil GWAS were available on the Immunochip.
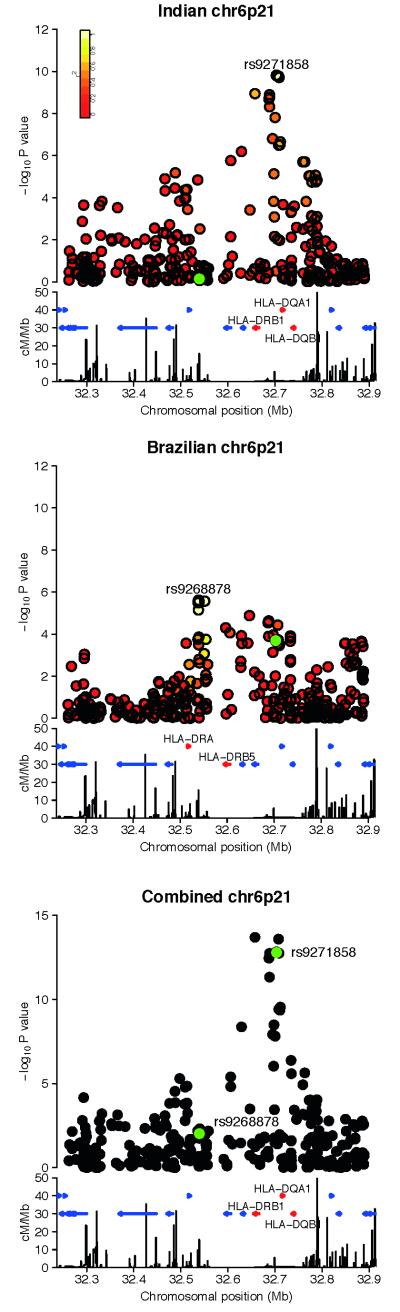
The MHC was the only region which replicated in all three cohorts (rs9271858, Pcombined=2.76×10−17 and OR(95%CI)=1.41(1.30-1.52)). Interestingly while there is shared signal at the MHC in both discovery GWAS, which clearly replicates, there are also differences between the India and Brazil results (Figure 2, Supplementary Table 4).
Figure 2.
Regional association plots of the signal at the MHC in the a) India discovery b) Brazil discovery c) Meta analysis of the discovery cohorts. The −log10 P values are shown on the upper part of each plot. SNPs are colored based on their r2 with the labeled hit SNP, calculated in the a) Indian controls b) Brazilian founders c) no r2 colouring. The SNP coloured in green is the hit SNP in the a) Brazil cohort b) India cohort c) India and Brazil cohorts. The bottom section of each plot shows the fine scale recombination rates estimated from individuals in the CEU HapMap population, and genes are marked as horizontal blue lines. Genes flanking the hit SNP are coloured red and are labeled.
In the India discovery GWAS, the two SNPs most associated with visceral leishmaniasis in the MHC were in strong LD (rs9271842 and rs9271858, r2=1 in Indian controls). Only one of these SNPs was present in the replication data (rs9271858), therefore for convenience we focus below on this SNP, and refer to it as the top SNP. This SNP is also significant in the Brazil data (PBrazil=2.04×10−4), but conversely the top MHC signal in the Brazil data (rs9268878) was not significant in the India data (PIndia=0.71, PReplication=0.58 at rs4428528, r2=1). The top MHC SNP signal in Brazil is 163.6 Kb upstream of the India signal, and is in almost complete linkage equilibrium with the top India SNP (r2= 0.06 in the Brazil founders). Further replication in a Brazilian dataset is required to assess this additional MHC signal: it could be a population or parasite species-specific association, or alternatively a false-positive finding.
In the Indian discovery cohort, when conditioning on the top SNP from the Indian discovery GWAS (rs9271858), there was still residual association signal at the MHC; two highly correlated SNPs both had P<10−3 (rs9271252 and rs9271255, r2=1 in Indian controls). For convenience, because it is typed in the discovery and replication samples, we focus in what follows on one of these, rs9271255, with a P value of 8.79×10−4 in the conditional analysis. This SNP is not highly correlated with rs9271858 (r2=0.21 in Indian controls). When conditioning on both rs9271858 and rs9271255, no association signal remains with P <10−3. In the India replication data, when conditioning on rs9271858, rs9271255 is still significant (P=5.72×10−3). This conditional analysis was repeated in the Brazil discovery data; when conditioning on rs9271858, rs9271255 was significant with a P value of 1.91×10−5.
We turn now to a more detailed examination of this replicated MHC signal (at rs9271858 and rs9271255). We focus on the Indian data, for which the Indian replication cohort is from a comparable population and can therefore be used to replicate cohort specific association signals. To further explore the Indian MHC signal, we estimated phase for the SNP genotypes for all individuals in the Indian discovery data. We subsequently fitted a model to the phased haplotypes with different risk parameters for the four haplotypes defined by the phased genotypes at the two aforementioned SNPs, rs9271858 and rs9271255. After additional exploration of the model (Supplementary Results), it was apparent that combining two of the four haplotypes, to give a model with three risk classes, fitted the data as well as the model with four risk classes (Supplementary Figure 1), and also gave a better fit than the two-SNP model (Supplementary Results).
At rs9271858 and rs9271255 respectively, the haplotype with highest risk is defined by the alleles GA and the haplotype with intermediate risk by AA, with the haplotypes AG and GG being protective relative to the other two haplotypes, and having statistically indistinguishable risk. This model with three risk classes had P=1.65×10−11 in the Indian discovery data, and where the protective haplotypes are set as the baseline with OR=1, the intermediate haplotype OR(95%CI)=1.28(1.09-1.50) and the highest risk haplotype OR(95%CI)=1.72(1.48-2.00). This analysis replicated in the Immunochip data: the model with three risk classes was significant at P= 4.78×10−6, where the intermediate haplotype OR(95%CI)=1.19(1.00-1.40) and the highest risk haplotype OR(95%CI)=1.50(1.27-1.76).
Our results establish that common polymorphisms in the HLA-DRB1-HLA-DQA1 region of the MHC are genetic risk factors for visceral leishmaniasis, and importantly appear to cross the epidemiological divides of geography and parasite species. For GWAS associations, the top SNPs are not necessarily the causal variants, but are expected to be correlated with the functional variants. There are SNPs in the MHC which are in moderate LD with rs9271858 (r2 ~ 0.2) and are located up to 500kb away (e.g. rs408359, r2=0.19 calculated in the Indian controls). This extensive linkage disequilibrium is common in the MHC, and makes it particularly difficult to assess possible functional candidates within the region of association25.
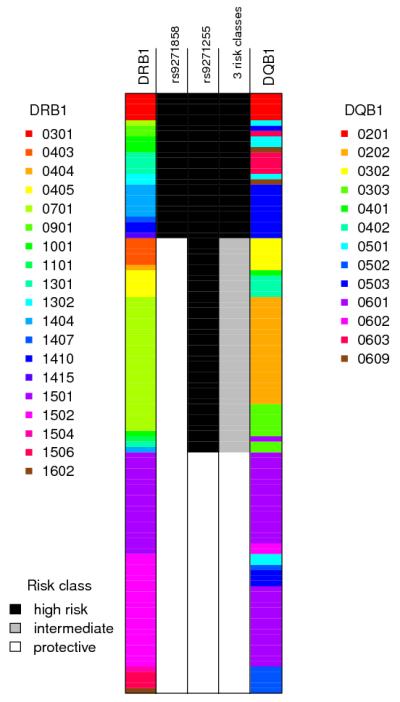
One possibility is that the association signal could be driven by functional variation in the adjacent highly polymorphic classical HLA class II genes, DRB1 and DQB1, each of which is a natural immunological candidate. The classical alleles at these loci were genotyped in a subset of individuals in both of the discovery cohorts, and phased (statistically) onto the haplotypes estimated from all the SNP data (see Supplementary Results). In this phased data set, chromosomes carrying the protective haplotype at rs9271858 and rs9271255 always carried one of the three classical DRB1 alleles 15, 16 and 01, and vice versa (Figure 3, Supplementary Table 5). We note that the DRB*01 and *16 alleles are very rare in the Indian data (Supplementary Table 5), so we have limited power to assess their possible role. This correlation was also perfect in the 142 phased Brazil discovery sample chromosomes (Online Methods) where all of the chromosomes carrying the protective haplotype defined by rs9271858 and rs9271255 also carried one of DRB1 *15, *16 or *01 (Supplementary Table 5, Supplementary Figure 2). This lends support to the possibility that the association signal could be driven by classical HLA alleles, and the biological importance of endogenous processing and presentation of antigen from infected macrophages and dendritic cells to CD4 T cells that drive the immune response in visceral leishmaniasis26,27. Nevertheless, additional research will be needed to establish whether or not these DRB1 alleles are themselves functionally involved in protection against visceral leishmaniasis, or merely correlated with an as-yet uncharacterised functional variant. Further details of the HLA analysis are presented in the Supplementary.
Figure 3.
Schematic representation of the HLA and SNP phased Indian discovery haplotypes in the MHC region. Each row represents an individual haplotype, the three middle columns show different risk models as described in the text, with the shading denoting which class in the model that haplotype is in: column 2 shows the classification of haplotypes based solely on the allele (risk or protective) present at the top SNP rs9271858; column 3 shows the classification of haplotypes based solely on the allele (risk or protective) present at the top SNP in the conditional analysis, rs9271255; column 4 shows the three risk groups as described in the text, which can also be defined as the allele combinations (risk-risk, protective-risk, protective-protective) at SNPs rs9271858 and rs9271255 that most parsimoniously capture their phase-known haplotype associations with disease (see text and Supplementary Figure 1). In India (N=3732), the frequencies of the 2-SNP haplotypes are A_A 0.301, A_G 0.310, G_A 0.372, G_G 0.017. In Brazil (N=1026), the frequencies of the 2-SNP haplotypes are A_A 0.406, A_G 0.097, G_A 0.390, G_G 0.105. The two flanking columns show the HLA alleles at DRB1 and DQB1 and are colour-coded as shown in the legends. The protective risk class correlates perfectly with haplotypes carrying DRB1 *15, *16 and *01. This analysis used the subset of individuals in the Indian discovery data for whom classical HLA alleles were available, and the plot above shows only the 112 haplotypes for which the posterior probability of the estimated phase at both DRB1 and DQB1 was >0.9. Note the haplotype carrying DRB1*0101 is rare and not shown in the plot as it did not occur on a haplotype with a successfully phased DQB1 allele.
Previous studies of HLA associations in human leishmaniasis have provided confusing and often contradictory results generally compromised by small sample size and lack of power (reviewed9). One small study did find protection associated with DRB1*15/*16 for visceral leishmaniasis caused by Old World L. infantum28, the same parasite species that causes visceral leishmaniasis in Brazil29. Protection has also been associated with DR2 (=DRB1*15/*16) in small studies of cutaneous and mucocutaneous forms of leishmaniasis caused by L. mexicana30 and L. braziliensis31,32, respectively.
There have been relatively few successful GWAS studies of infectious disease susceptibility33-36. Our GWAS has identified HLA class II region polymorphisms as significant genetic risk factors for visceral leishmaniasis, with the risk associated with the high-risk haplotype being high by GWAS standards: OR(95%CI) estimated from replication data of 1.50(1.27-1.76). Importantly, genetic variation in the MHC has not been found to be associated with all infectious disease. Our observations focus attention on determining the precise nature of HLA class II variation in determining visceral leishmaniasis susceptibility and offer the potential to contribute to development of novel strategies for disease control.
ONLINE METHODS
Samples
Supplementary Table 1 provides characteristics of samples. In India, discovery and replication sets comprised cases and controls from villages located within a radius of ~100km from the city of Muzaffarpur covering the districts of Muzaffarpur, Vaishali, Samastipur, Saran, Sheohar, East Champaran and Sitamarhi in Bihar State in northeast India. Further epidemiological and demographic details relating to the study site are described elsewhere37. An annual incidence rate of 2.49 clinical visceral leishmaniasis cases/1,000 persons has been reported in the region37. L. donovani sensu strictu (zymodeme MON-2) was confirmed as the causative agent of visceral leishmaniasis in the study region, in accordance with other reports on clinical isolates from kala-azar patients in the state of Bihar38-41. The controls had no history of visceral leishmaniasis or a family history of visceral leishmaniasis among first-, second-, or third-degree relatives. Informed written consent in Hindi was obtained from all participating individuals and from parents of children under 18 years old. Approval for the study was provided by the Ethical Committee of the Institute of Medical Sciences, Banaras Hindu University, Varanasi, India.
From northeast Brazil, 64 families were collected during 1991-1994 as part of the Belém Family Study6,42. These families were ascertained from medical records of the Fundacão National de Saude in the States of Para, Maranhão, Piaui based on data from the 1983-85 and 1993-94 epidemics. A further 244 families were collected from study sites near Natal where transmission of the parasite is focal and transient, allowing identification of neighbourhoods with ongoing or recent transmission. These families were ascertained from medical records of the Fundacão National de Saude in Rio Grande do Norte. All families were of equivalent socioeconomic status. The causative agent of visceral leishmaniasis was confirmed as L. infantum chagasi in a subset (~10% over 4 States) of visceral leishmaniasis patients6. Further clinical, epidemiological, family structure and demographic information relating to the families are described elsewhere6,10,21,42,43.
Ethical approval for the Belem Family Study was obtained originally from the local ethics committee at the Instituto Evandro Chagas, Belém, Para, Brazil6,42. Approval for continued use of the Belem Family Study samples, and for collection and use of the samples from Natal, has been granted from the local Institutional Review Board at the Universidade Federal do Rio Grande do Norte (CEP-UFRN 94-2004), nationally from the Comissão Nacional de Ética em Pesquisa (CONEP: 11019), and from the Ministerios Cencia e Tecnologia for approval to ship samples out of Brazil (portaria 617; 28 September 2005). Informed written consent for sample collection was obtained from adults, and from parents of children <18 years old. The Brazilian populations studied are long-term (>200 years) admixtures of Caucasian, Negroid and Native Indian ethnic backgrounds, as confirmed in recent analysis of the Natal families11.
Individuals from both India and Brazil were classified as affected when they had been diagnosed with clinical visceral leishmaniasis that responded positively to specific anti-leishmanial treatment. Data on sub-clinical disease or asymptomatic infections were not included. Diagnosis of clinical visceral leishmaniasis was made on the basis of clinical, parasitological (splenic aspirates in India; bone marrow aspirates in Brazil) and serological criteria as described elsewhere6,10,21,42,44-46.
DNA sample preparation
For India non-invasive buccal swabs were collected and genomic DNA prepared in the Cambridge laboratory. For Brazil, blood was collected by venepuncture from all available family members. Genomic DNA was prepared directly from blood in Natal10, or from Epstein-Barr virus-transformed B cells cultured from fresh or cryopreserved peripheral blood mononuclear cells in Cambridge21. Genomic DNA for all cases was shipped to the Sanger Institute, Cambridge. Quality was validated using the Sequenom iPLEX assay designed to genotype 4 gender SNPs and 26 SNPs present on the Illumina Beadchips. DNA concentrations were quantified using a PicoGreen assay (Invitrogen) and an aliquot assayed by agarose gel electrophoresis. A DNA sample was considered to pass quality control if the DNA concentration was greater than or equal to 50 ng/μl, the DNA was not degraded, the gender assignment from the iPLEX assay matched that provided in the patient data manifest and genotypes were obtained for at least two thirds of the SNPs on the iPLEX.
Genotyping
Indian discovery cases and controls and the Brazil families were genotyped at the Sanger Institute on the Illumina Infinium platform using the Human660-Quad, a custom chip designed by WTCCC2 and comprising Human550 supplemented with 60,000 additional probes that were intended to allow the genotyping of common CNVs from the Structural Variation Consortium47. Replication genotyping for India was carried out at the Sanger Institute using the Illumina Immunochip, a custom chip designed by WTCCC2 and the Immunochip Consortium and comprising 196,524 SNPs. For both chips, bead intensity data was processed and normalized for each sample in BeadStudio; data for successfully genotyped samples was extracted and genotypes called using Illuminus.
Quality controls
SNPs. SNPs were excluded if the Fisher information for the allele frequency was not close to unity (information <0.98) or if the minor allele frequency (MAF) was very low (defined as <0.01%), or if the missingness >0.02, or for extreme departures from Hardy-Weinberg equilibrium (HWE P-value <1×10−20). After applying the above filters in the Indian discovery 526,731 autosomal SNPs remained for further analysis. For the Brazilian families, the above filters left 553,323 autosomal SNPs. Of the two discovery cohorts, 521,134 overlapping SNPs passed QC in both cohorts. Cluster plots of the hit SNPs described in this study are shown in Supplementary Figure 3.
Samples
For quality control of samples typed on the Indian and Brazilian Illumina Human660 quad data, a Bayesian clustering method was used to infer and exclude outlying individuals on the basis of call rate, heterozygosity and signal intensity12,48. To remove signal intensity outliers observed for raw intensity data, the difference between the A channel intensity and the B channel intensity was averaged over all SNPs on autosomes for each sample. A similar approach was used taking intensity measures from the A channel on the non-pseudo autosomal X chromosomes to identify outliers and infer gender for all Indian and Brazilian samples. Samples were removed if their inferred gender was discordant with the recorded gender after cross-checking with original database entries, or if less than 90% of the SNPs typed by Sequenom on entry to sample handing (see above) agreed with the genome-wide data. Duplicated individuals were also excluded. For the Indian discovery sample a total of 121/2199 individuals were removed following these quality control checks. For the Indian replication sample 111/2042 individuals were removed. For the Brazilian family study, additional quality control checks were carried out in order to verify (and if necessary adjust) the expected pedigree relationships between the individuals. Genome-wide identity by descent (IBD) sharing probabilities for all pairs of individuals were estimated using a subset of 11,177 autosomal markers using the software PLINK49. These estimates were used to assist in pedigree checking and resulted in 49 additional post quality control individuals being removed due to unresolved pedigree relationship discrepancies. In total, for the Brazilian family study, 189/2159 individuals were removed.
Statistical analyses
We carried out association analyses using a novel13 variance components method (a linear mixed model, similar to Kang et al.17). The linear mixed model explicitly accounts for correlations in individuals’ phenotypes due to their relatedness through specification of the phenotypic variance/covariance matrix in terms of parameters representing underlying genetic and environmental components of variance (estimated during the model fitting procedure) and pairwise relatedness measures (kinship coefficients), estimated prior to model fitting on the basis of the genome-wide genotype data. In this analysis we used a standard linear mixed model
where Y=(y1,…,yn)T is a vector of responses (coded 1/0 for case/control) on n subjects, X=(xik) is the n × K matrix of predictor values for any variables to be modelled as fixed effects (including variables representing covariates and genotypes at any SNPs currently under test), β=(β1, … βK)T are regression coefficients (to be estimated) representing the linear effects of the predictors on the response, and Q and ε are random effects assigned the distributions and ε~N(0, σe2I), where and are parameters (to be estimated) representing the genetic and environmental components of variance respectively, I is the n × n identity matrix and Φ is the n × n matrix of pairwise kinship coefficients.
Although use of this linear mixed model was originally proposed for pedigrees with known relationships50-53, recently this approach has gained popularity for use with samples of unknown or uncertain relationship13,17,18,54, including apparently unrelated samples who may nevertheless display distant levels of common ancestry. For this purpose, the pairwise kinship coefficients modelling either close or distant relatedness are estimated (prior to fitting the linear mixed model) on the basis of genome-wide genotype data, rather than being fixed at known theoretical values.
Computational considerations have led to the development of several faster approximations for constructing tests of the fixed effects of interest in the above linear mixed model15-17,53,55,56. These approximate tests have been implemented in various software packages including EMMAX, TASSEL, FaST-LMM and GenABEL. For the analyses presented here, we used our own implementation of the linear mixed model developed for a previous study13 that has the advantage, in common with the recently-developed GEMMA package57 of fitting the full (rather than an approximate) model, which in principal can lead to a small increase in power, depending on the underlying true model. Our implementation also allows a transformation between the parameters of the linear model shown above and the logistic model (which is the usual model for case/control data), allowing the convenient estimation of effects on the log odds scale and the generation of resulting odds ratio estimates. We confirmed that the Indian discovery results did not change significantly after the inclusion of the first 10 PCs in the linear mixed model (data not shown).
Combined P-values across Indian (discovery and replication) and Brazilian datasets were calculated in R using an inverse variance fixed effects meta-analysis.
In addition, for the Brazilian family study we verified our results using two complementary alternative methods implemented in the software packages FBAT58 and ROADTRIPS59. Results were comparable across the different methods (data not shown).
HLA typing
The HLA DRB1 and DQB1 typing of 100 selected Indian discovery individuals was performed using a sequencing-based method60. To select these individuals, SNPs in the class II region DRA-QB2 (32,514,320 to 32,840,188 NCBI Build 36) were first pruned in PLINK49 to generate a set of 78 SNPs in approximate linkage equilibrium (r2<0.8) and with MAF>0.1. LD blocks across these 78 SNPs were then visualised using Haploview61 and the top 59 haplotype tagging SNPs were selected and used to generate haplotypes for all 1866 unrelated individuals in fastPHASE62. The subset of 100 individuals selected for sequence-based HLA typing represented the 135 most common 59-SNP haplotypes in the region. Automated sequencing was carried out on ABI Prism 3730 or 3730xl Genetic Analysers (Applied Biosystems; Foster City, CA, USA) and HLA-DRB1 analysis was carried out using ASSIGNV4.0.1.36 (Conexio Genomics; Applecross, WA, Australia). Where possible, individuals were assigned DRB1 or DQB1 specificities to the 4-digit level63, i.e. to a specific HLA protein encoded on each chromosome (for example, DRB1*1501, where *1501 is a specific functional DRB1 protein). Where this was not possible, individuals were assigned to a 2-digit level allele group, e.g. HLA-DRB1*15 indicates an allele group for which all specific functional DRB1 proteins share a common ancestry.
HLA-DRB1 and DQB1 typing was further performed in a subset of 71 unrelated individuals from the Brazilian families using the same sequence-based method as described above for the Indian samples.
PHASING the SNP and HLA data
SNPs in the MHC region and HLA-DRB1 and -DQB1 alleles were phased using IMPUTE264 and PHASE65. The Indian discovery and Brazilian replication cohorts were treated separately throughout the phasing pipeline.
For the Indian discovery data, IMPUTE264 was used to phase all the SNPs on chromosome 6 for 1866 individuals, using HapMap3 populations as the haploid reference panel. For the Brazilian data 5,667 SNPs in the MHC (20,733,613- 41,984,313bp) were phased. The phasing of the Brazilian data was performed in two stages, firstly with an unrelated set of 498 parents and secondly an unrelated set of 268 offspring, totalling 766 individuals. The trio relationships were then used to check the accuracy of both SNP and HLA allele phasing.
Treating the IMPUTE2 SNP phasing as fixed, PHASE65 was then used to phase the HLA alleles typed at HLA-DRB1 and HLA-DQB1 onto HLA region SNP haplotypes, selecting only haplotypes phased at probability >0.9. PHASE65 was used to phase the multi-allelic HLA-DRB1 and HLA-DQB1 alleles onto the known SNP haplotypes, specifying known phase at all of the SNPs and the PHASE options –MR and -d1. The HLA alleles were given positions: - DRB1 was set at 32,597,662bp, between rs28756238 and rs41546317 and HLA-DQB1 was set at 32,735,635bp between rs28724231 and rs1063355. Both HLA-DRB1 and HLA-DQB1 alleles were phased in the Indian data, and HLA-DRB1 was phased in the Brazilian data.
After QC of the PHASE data, in the Indian discovery set there was a total of 3,732 SNP haplotypes, 154 with HLA-DRB1 alleles and 153 with HLA-DQB1 alleles. Of these, 112 were phased to 4-digit level for both DRB1 and DQB1. In the Brazilian dataset there was a total of 1,481 SNP haplotypes, 142 of which were phased with 2 digit HLA-DRB1 alleles.
Haplotype analyses
Haplotype associations were investigated by applying two methods; a conditional analysis (see Results) and a Bayesian approach. Both methods reached the same result, each fitting a three-haplotype model of risk.
GENECLUSTER, which adopts a Bayesian approach66 was used to look for primary and secondary association signals at known and putative SNPs. This method analyses the genealogy of the case-control sample to find evidence for causal mutation(s). It uses the genealogy of a reference haplotype panel (in this case the HapMap3 GIH haplotypes) to approximate the genealogy of the case-control sample at positions across the region by clustering the case-control haplotypes under the leaves of the reference genealogy. Placing a disease mutation on a branch of the reference genealogy segregates the haplotypes at the leaves into two different risk groups: those case-control haplotypes that fall under the disease mutation (and therefore carries the mutation) and those that do not. For a given mutation in the tree, GENECLUSTER carries out a Bayesian test of association. The model can also be extended to two disease mutations in the genealogy. For this data, the genealogical tree was estimated on chromosome 6 at 32,678,817 bp. The log10(Bayes Factor) for the 2-mutation (three haplotype) model was 7.36, compared to log10(Bayes Factor) 5.42 for a single mutation (two haplotype) model.
Following the linear mixed model conditional analysis, we phased the SNP data for all individuals in the India discovery data and found that the two SNPs associated in the conditional analysis (rs9271858 and rs9271255) tag the haplotypes from the GENECLUSTER analysis. The first mutation perfectly correlated with the protective haplotype and the second mutation separated the risk halotype into two, which are well tagged by the two SNP intermediate and risk haplotypes (r2 protective=1, intermediate=0.49, risk=0.60) (overall correlation 0.86).
Supplementary Material
ACKNOWLEDGEMENTS
The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z). We thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, M.L. Perez, C.R. Stribling, S. Taylor and A. Wilk of the Wellcome Trust Sanger Institute’s Sample and Genotyping Facilities for technical assistance. We thank D. Davison for making available his program ‘Shellfish’ for calculating principal components in large genetic datasets. CCAS is supported by a Wellcome Trust Fellowship (097364/Z/11/Z). HJC is supported by a Wellcome Senior Fellowship in Basic Biomedical Science (087436/Z/10/Z). PD is supported in part by a Royal Society Wolfson Merit Award and work was supported in part by WTCHG core grants 090532/Z/09/Z and 075491/Z/04/B. Collection of samples and epidemiological data, sample preparation, and sequence-based HLA typing was supported by grants from The Wellcome Trust (074196/Z/04/Z and 085475/Z/08/Z to JMB, SS, SMBJ and MEW) and the National Institutes of Health [Tropical Medicine Research Centre award P50AI074321 to SS in India; Tropical Medicine Research Centre award P50 AI-30639 to Dr. Edgar M. Carvalho and SMBJ in Brazil; R01 AI076233 (MEW, JMB); R01 AI048822 (MEW, SMBJ, JMB)]. Special thanks to all subjects who contributed samples, and clinicians and field staff in India and Brazil who helped with recruitment of study subjects.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Alvar J, et al. Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucheton B, et al. The interplay between environmental and host factors during an outbreak of visceral leishmaniasis in eastern Sudan. Microbes Infect. 2002;4:1449–1457. doi: 10.1016/s1286-4579(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 3.Sacks DL, Lal SL, Shrivastava SN, Blackwell JM, Neva FA. An analysis of T cell responsiveness in Indian Kala-azar. J. Immunol. 1987;138:908–913. [PubMed] [Google Scholar]
- 4.Ho M, Siongok TK, Lyerly WH, Smith DH. Prevalence and disease spectrum in a new focus of visceral leishmaniasis in Kenya. Trans R. Soc. Trop. Med. Hyg. 1982;76:741–746. doi: 10.1016/0035-9203(82)90095-5. [DOI] [PubMed] [Google Scholar]
- 5.Cabello PH, Lima AM, Azevedo ES, Kriger H. Familial aggregation of Leishmnaia chagasi infection in northeastern Brazil. Am. J. Trop. Med. Hyg. 1995;52:364–365. doi: 10.4269/ajtmh.1995.52.364. [DOI] [PubMed] [Google Scholar]
- 6.Peacock CS, et al. Genetic epidemiology of visceral leishmaniasis in northeastern Brazil. Genet Epidemiol. 2001;20:383–396. doi: 10.1002/gepi.8. [DOI] [PubMed] [Google Scholar]
- 7.Burgner D, Jamieson SE, Blackwell JM. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell JM, et al. Genetics and visceral leishmaniasis: of mice and man. Parasite Immunol. 2009;31:254–266. doi: 10.1111/j.1365-3024.2009.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeronimo SM, et al. Genetic predisposition to self-curing infection with the protozoan Leishmania chagasi: a genomewide scan. J Infect Dis. 2007;196:1261–9. doi: 10.1086/521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettinger NA, et al. Genetic admixture in Brazilians exposed to infection with Leishmania chagasi. Ann Hum Genet. 2009;73:304–13. doi: 10.1111/j.1469-1809.2009.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellenguez C, Strange A, Freeman C, Donnelly P, Spencer CC. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astle W, Balding DJ. Population Structure and Cryptic Relatedness in Genetic Association Studies. Statistical Science. 2009;24:451–471. [Google Scholar]
- 15.Lippert C, et al. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–5. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–60. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amin N, van Duijn CM, Aulchenko YS. A genomic background based method for association analysis in related individuals. PLoS ONE. 2007;2:e1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrotra S, et al. Genetic and functional evaluation of the role of CXCR1 and CXCR2 in susceptibility to visceral leishmaniasis in north-east India. BMC Med Genet. 2011;12:162. doi: 10.1186/1471-2350-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bucheton B, et al. Identification of a novel G245R polymorphism in the IL-2 receptor beta membrane proximal domain associated with human visceral leishmaniasis. Genes Immun. 2007;8:79–83. doi: 10.1038/sj.gene.6364355. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson SE, et al. Genome-wide scan for visceral leishmaniasis susceptibility genes in Brazil. Genes Immun. 2007;8:84–90. doi: 10.1038/sj.gene.6364357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fakiola M, et al. Genetic and functional evidence implicating DLL1 as the gene that influences susceptibility to visceral leishmaniasis at chromosome 6q27. J Infect Dis. 2011;204:467–77. doi: 10.1093/infdis/jir284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamed HS, et al. SLC11A1 (formerly NRAMP1) and susceptibility to visceral leishmaniasis in The Sudan. Eur J Hum Genet. 2004;12:66–74. doi: 10.1038/sj.ejhg.5201089. [DOI] [PubMed] [Google Scholar]
- 24.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bakker PI, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundar S, Reed SG, Sharma S, Mehrotra A, Murray HW. Circulating T helper 1 (Th1) cell- and Th2 cell-associated cytokines in Indian patients with visceral leishmaniasis. Am J Trop Med Hyg. 1997;56:522–5. doi: 10.4269/ajtmh.1997.56.522. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho EM, et al. Immunologic markers of clinical evolution in children recently infected with Leishmania donovani chagasi. J Infect Dis. 1992;165:535–40. doi: 10.1093/infdis/165.3.535. [DOI] [PubMed] [Google Scholar]
- 28.Meddeb-Garnaoui A, et al. Association analysis of HLA-class II and class III gene polymorphisms in the susceptibility to mediterranean visceral leishmaniasis. Hum Immunol. 2001;62:509–517. doi: 10.1016/s0198-8859(01)00237-3. [DOI] [PubMed] [Google Scholar]
- 29.Kuhls K, et al. Comparative microsatellite typing of new world leishmania infantum reveals low heterogeneity among populations and its recent old world origin. PLoS Negl Trop Dis. 2011;5:e1155. doi: 10.1371/journal.pntd.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivo-Diaz A, et al. Role of HLA class II alleles in susceptibility to and protection from localized cutaneous leishmaniasis. Hum Immunol. 2004;65:255–61. doi: 10.1016/j.humimm.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Cabrera M, et al. Polymorphism in TNF genes associated with mucocutaneous leishmaniasis. J. Exp. Med. 1995;182:1259–1264. doi: 10.1084/jem.182.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petzl-Erler ML, Belich MP, Queiroz-Telles F. Association of mucosal leishmaniasis with HLA. Human Immunology. 1991;32:254–260. doi: 10.1016/0198-8859(91)90088-q. [DOI] [PubMed] [Google Scholar]
- 33.Jallow M, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thye T, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khor CC, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet. 2011;43:1139–41. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 37.Singh SP, Reddy DC, Mishra RN, Sundar S. Knowledge, attitude, and practices related to Kala-azar in a rural area of Bihar state, India. Am J Trop Med Hyg. 2006;75:505–8. [PubMed] [Google Scholar]
- 38.Manna M, Majumder HK, Sundar S, Bhaduri AN. The molecular characterization of clinical isolates from Indian Kala-azar patients by MLEE and RAPD-PCR. Med Sci Monit. 2005;11:BR220–7. [PubMed] [Google Scholar]
- 39.Chatterjee M, Manna M, Bhaduri AN, Sarkar D. Recent kala-azar cases in India: isozyme profiles of Leishmania parasites. Indian J Med Res. 1995;102:165–72. [PubMed] [Google Scholar]
- 40.Thakur CP, Dedet JP, Narain S, Pratlong F. Leishmania species, drug unresponsiveness and visceral leishmaniasis in Bihar, India. Trans R Soc Trop Med Hyg. 2001;95:187–9. doi: 10.1016/s0035-9203(01)90160-9. [DOI] [PubMed] [Google Scholar]
- 41.Sundar S, et al. Resistance to treatment in Kala-azar: speciation of isolates from northeast India. Am J Trop Med Hyg. 2001;65:193–6. doi: 10.4269/ajtmh.2001.65.193. [DOI] [PubMed] [Google Scholar]
- 42.Blackwell JM, et al. Immunogenetics of leishmanial and mycobacterial infections: the Belem Family Study. Philos Trans R Soc Lond B Biol Sci. 1997;352:1331–45. doi: 10.1098/rstb.1997.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeronimo SM, et al. An emerging peri-urban pattern of infection with Leishmania chagasi, the protozoan causing visceral leishmaniasis in northeast Brazil. Scand J Infect Dis. 2004;36:443–9. doi: 10.1080/00365540410020451. [DOI] [PubMed] [Google Scholar]
- 44.Khalil EA, Zijlstra EE, Kager PA, El Hassan AM. Epidemiology and clinical manifestations of Leishmania donovani infection in two villages in an endemic area in eastern Sudan. Trop Med Int Health. 2002;7:35–44. doi: 10.1046/j.1365-3156.2002.00832.x. [DOI] [PubMed] [Google Scholar]
- 45.Fakiola M, et al. Classification and regression tree and spatial analyses reveal geographic heterogeneity in genome wide linkage study of Indian visceral leishmaniasis. PLoS One. 2010;5:e15807. doi: 10.1371/journal.pone.0015807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zijlstra EE, el-Hassan AM, Ismael A, Ghalib HW. Endemic kala-azar in eastern Sudan: a longitudinal study on the incidence of clinical and subclinical infection and post-kala-azar dermal leishmaniasis. Am J Trop Med Hyg. 1994;51:826–36. doi: 10.4269/ajtmh.1994.51.826. [DOI] [PubMed] [Google Scholar]
- 47.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edin. 1918;52:399–433. [Google Scholar]
- 51.Henderson CR. Estimation of variance and covariance components. Biometrics. 1953;9:226–252. [Google Scholar]
- 52.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–8. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 55.Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–85. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svishcheva GR, Axenovich TI, Belonogova NM, van Duijn CM, Aulchenko YS. Rapid variance components-based method for whole-genome association analysis. Nat Genet. 2012;44:1166–70. doi: 10.1038/ng.2410. [DOI] [PubMed] [Google Scholar]
- 57.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horvath S, Wei E, Xu X, Palmer LJ, Baur M. Family-based association test method: age of onset traits and covariates. Genet Epidemiol. 2001;21(Suppl 1):S403–8. [PubMed] [Google Scholar]
- 59.Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet. 2010;86:172–84. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sayer D, et al. HLA-DRB1 DNA sequencing based typing: an approach suitable for high throughput typing including unrelated bone marrow registry donors. Tissue Antigens. 2001;57:46–54. doi: 10.1034/j.1399-0039.2001.057001046.x. [DOI] [PubMed] [Google Scholar]
- 61.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 62.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marsh SG, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su Z, Cardin N, Donnelly P, Marchini J. A Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association studies. Statistical Science. 2009;24:430–450. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.