Abstract
Thiol peroxidases (Tpxs) are dimeric 2-Cys peroxiredoxins from bacteria that preferentially reduce alkyl hydroperoxides. Catalysis requires two conserved residues, the peroxidatic cysteine and the resolving cysteine, which are located in helix α2 and helix α3, respectively. The partial unraveling of helices α2 and α3 during catalysis allows for the formation of an intramolecular disulfide between these two residues. Here we present three structures of Escherichia coli Tpx representing the fully folded (FF, peroxide binding site intact), locally unfolded (LU, disulfide bond), and partially locally unfolded (PLU, transitional state) conformations. We also compare known Tpx crystal structures and analyze the sequence-conservation patterns among nearly 300 Tpx sequences. Twelve fully conserved Tpx-specific residues cluster at the active site and dimer interface, and an additional 37 highly conserved residues are mostly located in a cradle providing the environment for helix α2. Using the structures determined here as representative FF, transitional, and LU Tpx conformations, we describe in detail the structural changes associated with catalysis in the Tpx subfamily. Key insights include the description of a conserved hydrophobic collar around the active site, a set of conserved packing interactions between helices α2 and α3 that allow the local unfolding of α2 to trigger the partial unfolding of α3, a conserved dimer interface that anchors the ends of helices α2 and α3 to stabilize the active site during structural transitions, and a conserved set of residues constituting a cradle that stabilizes the two discrete conformations of helix α2 involved in catalysis. The involvement of the dimer interface in stabilizing active-site folding and in forming the hydrophobic collar implies that Tpx is an obligate homodimer and explains the high conservation of interface residues.
Keywords: thiol peroxidase, Tpx, peroxiredoxin, antioxidant, structural transitions
INTRODUCTION
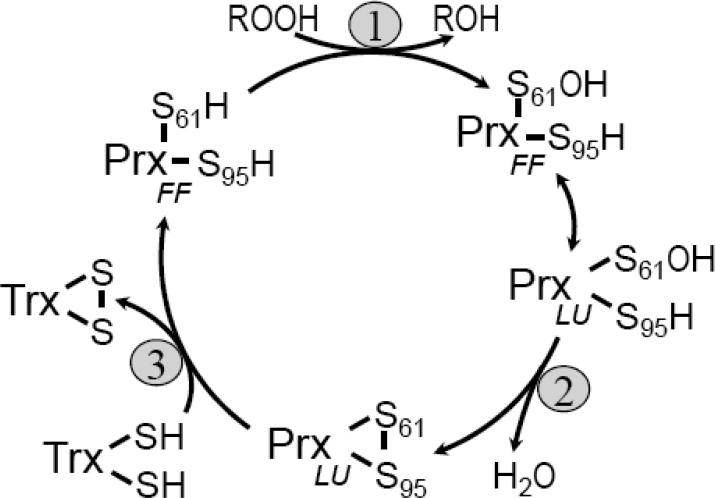
Peroxiredoxins (Prxs) are a ubiquitous family of peroxidases that reduce hydrogen peroxide (H2O2) and alkyl hydroperoxides with varying affinity1, and in some cases, peroxynitrite2,3. Prxs share a common fold, active site and catalytic cycle that uses a conserved Cys residue, called the peroxidatic Cys (CP), to attack the peroxide substrate (Figure 1). The CP residue is located in the first turn of α-helix 2 (α2) and sits in the bottom of the active-site pocket surrounded by the three other residues conserved in all Prxs, a Pro, a Thr and an Arg4. In all 2-Cys Prxs, a second Cys, called the resolving Cys (CR) is also required. Catalysis involves three chemical steps, (1) peroxidation, (2) resolution, and (3) recycling, and a local unfolding of the active site that allows disulfide bond formation between the CP and CR side chains (Figure 1); the two active site conformations are referred to as fully folded (FF; reduced form with a peroxide binding site) and locally unfolded (LU; disulfide form).
Figure 1.
Catalytic cycle of the Tpx subfamily. The three main chemical steps universal to the peroxiredoxin catalytic cycle, (1) peroxidation, (2) resolution and (3) recycling are shown along with the local unfolding step (double headed arrow) required for the resolution reaction. Peroxidation forms a Cys-sulfenic acid (S61-OH). Specific to the Tpx subfamily, the disulfide bond formed in the resolution step is intramolecular and thioredoxin (Trx) has been shown to be the most potent reductant8,10,13,16,17. S61 and S95 are the sulfur atoms of the peroxidatic and resolving cysteine residues, respectively, using EcTpx residue numbering. The fully folded (FF) and locally unfolded (LU) conformations are indicated.
Prxs can be organized into five subfamilies based on sequence similarity: Prx1, Prx6, Prx5, Tpx and BCP5. The Tpx (thiol peroxidase) subfamily consists of 2-Cys Prxs found in Gram-positive and Gram-negative eubacterial species6,7. The original description of a Tpx8 as well as a later Esherichia coli proteomic study9 suggested a periplasmic localization based on its release after osmotic shock. However, the lack of a signal sequence for export, immunoblots of Campylobacter jejuni cytoplasmic and periplasmic fractions10, and the mixed-disulfide formation between E. coli cytoplasmic thioredoxin mutants and Tpx upon peroxide challenge have led to the proposal that Tpx is primarily a cytoplasmic protein that is anomalously released during osmotic shock11. E. coli Tpx (EcTpx) is a potent reductant of alkyl hydroperoxides, having a ~200-fold preference for alkyl hydroperoxides over H2O28,12,13. With the exception of in vitro studies with C. jejuni Tpx10, other Tpxs react with alkyl hydroperoxides14,15,16,17, suggesting that in bacteria Tpx serves as the fundamental player in the removal of lipid hydroperoxides produced by oxidative stress.
The first structural view of a Tpx subfamily member was the LU conformation of EcTpx18. From this original structure, unique features of the Tpx subfamily were seen to be the position of CR within helix α3 and a short N-terminal extension that folds into a β-hairpin. Although only residues from a single chain are directly involved in catalytic chemistry, all characterized Tpxs are homodimers in solution12,17,19 and have an A-type dimer interface20.
Until now, six Tpx crystal structures have been determined (Table 1). Of these, three are in the FF conformation and three are in the LU conformation; two of these structures represent the FF and LU states of Mycobacterium tuberculosis Tpx (MtTpx) (Table 1). Additionally, NMR structures of the FF and LU states of Bacillus subtilis Tpx (BsTpx) have been published21. Nevertheless, no in-depth analysis of the structural transition between the FF and LU conformations has been published. Here we present crystal structures of EcTpx and two EcTpx mutants that reveal the FF, LU and a partially locally unfolded (PLU) state of EcTpx. We combine these three structures with an analysis of sequence-conservation patterns among nearly 300 Tpx sequences and comparisons with the previously determined structures to provide a detailed analysis of the structural changes associated with catalysis in the Tpx subfamily.
Table 1.
Deposited structures of Tpxs.
| Source and Conformationa | Resolution (Å) | Mutation | Sequence Identityb | PDB code | Reference |
|---|---|---|---|---|---|
| EcTpxLU | 1.8 | 100% | 3HVS | this study | |
| EcTpxFFc | 1.75 | CP61S | 100% | 3HVV | this study |
| EcTpxLUd | 2.1 | C82S, CR95S | 100% | 3HVX | this study |
| EcTpxLU | 2.2 | 100% | 1QXH | Choi et al., 200318 | |
| HiTpxLU | 1.9 | 63% | 1Q98 | NYSGXRCg | |
| MtTpxLUe | 1.75 | 51% | 1XVQ | Rho et al., 200617 | |
| MtTpxFF | 2.1 | CP60S | 51% | 1Y25 | Stehr et al., 200631 |
| SpTpxFF | 2.3 | 47% | 1PSQ | NYSGXRCg | |
| BsTpxLUf | NMR | 41% | 2JSY | Lu et al., 200821 | |
| BsTpxFFf | NMR | 41% | 2JSZ | Lu et al., 200821 | |
| AaTpxFF | 1.85 | 39% | 2YZH | RSGIg |
Organism abbreviations are: Ec = Escherichia coli; Hi = Haemophilus influenzae; Mt = Mycobacterium tuberculosis; Sp = Streptococcus pneumoniae; Bs = Bacillus subtilis; Aa = Aquifex aeolicus. The conformation of the active site is indicated as either fully folded (FF) or locally unfolded (LU). The LU conformation is locked in place by the formation of a disulfide bond between the CP and CR residues.
Sequence identity relative to EcTpx.
The fully folded conformation in EcTpx is distorted (see text).
The CP-CP disulfide in this structure creates a partial locally unfolded active site (see text).
The CP-CR disulfide in this structure has been broken by radiation.
The deposited structure is a monomer and no oligomerization information is given.
Structures deposited by the New York SGX Research Center for Structural Genomics (NYSGXRC) or RIKEN Structural Genomics/Proteomics Initiative (RSGI) structural genomics group without primary citation.
RESULTS AND DISCUSSION
In this work, the structures of three constructs of E. coli Tpx have been determined: wild type (EcTpx), a CP61S mutant (EcTpxC61S), and a double C82S, CR95S mutant (EcTpxC82,95S) at 1.8 Å, 1.75 Å, and 2.1 Å resolution, respectively (Table 2). All three structures have high-quality electron density for the ordered parts of the molecule (Figure 2) and adopt the standard Prx fold, including the Tpx-specific N-terminal antiparallel β-hairpin (Figure 3). These structures show three distinct conformations for the active site: EcTpx has an intramolecular CP-CR disulfide and is in the LU conformation, EcTpxC61S adopts the FF conformation but with a slightly distorted CP-loop, and EcTpxC82,95S presents a novel, PLU conformation with an intermolecular CP-CP disulfide bond. The features of these structures already fully described in publications of Tpx structures will not be discussed here.
Table 2.
Data collection and refinement statistics.
| EcTpx | EcTpxC61S | EcTpxC82,95S | EcTpxLAB1h | |
|---|---|---|---|---|
| PDB code | 3HVS | 3HVV | 3HVX | 3I43 |
| A. Data Collectiona | ||||
| Space group | P212121 | P3121 | P21 | P212121 |
| a (Å) | 38.83 | 59.54 | 44.72 | 38.83 |
| b (Å) | 64.00 | 59.54 | 123.36 | 64.00 |
| c (Å) | 137.95 | 73.32 | 61.93 | 137.95 |
| β (°) | 90 | 90 | 103.01 | 90 |
| Resolution limits (Å) | 69-1.8 (1.83-1.80) | 42-1.75 (1.84-1.75) | 62-2.1 (2.21-2.10) | 37-2.8 (2.95-2.8) |
| Unique observations | 28676 (1995) | 13577 (1403) | 32466 (2229) | 8854 (1276) |
| Multiplicity | 4.0 (3.8) | 7.7 (4.5) | 2.7 (2.1) | 2.9 (2.9) |
| Average I/σ | 11.7 (4.3) | 26.5 (3.2) | 10.7 (3.3) | 7.0 (2.1) |
| Completeness (%) | 97.2 (91.8) | 92.8 (63.1) | 96.6 (90.7) | 98.0 (99.3) |
| Rmeasb (%) | 8.9 (54.9) | 6.0 (50.9) | 7.8 (28.6) | 4.4 (12.6) |
| Rpimc (%) | 10.0 (52.0)e | 2.0 (22.0) | 5.0 (18.0) | 6.7 (24.0) |
| B. Refinement | ||||
| Rcryst (%) / Rfree (%) | 14.6 / 21.0f | 13.6 / 19.5g | 16.2 / 24.8g | 19.8 / 21.9f,h |
| Molecules in AU | 2 | 1 | 4 | |
| Number of protein residues | 334 | 167 | 668 | |
| Number of water molecules | 603 | 293 | 690 | |
| Total number of atoms | 3122 | 1591 | 5689 | |
| rmsd bond lengths (Å) | 0.023 | 0.023 | 0.019 | |
| rmsd bond angles (°) | 1.9 | 1.8 | 1.6 | |
| < B > protein (Å2) | 19.2 | 21.4 | 27.0 | |
| < B > solvent (Å2) | 37.7 | 23.4 | 18.2 | |
| Ramachandran plotd | ||||
| Favored (%) | 97.9 | 98.2 | 97.3 | |
| Outliers (%) | 0.0 | 0.0 | 0.3 |
Numbers in parentheses correspond to values in the highest resolution bin.
Rmeas is the multiplicity-weighted merging R-factor51.
Rpim is the precision-indicating multiplicity-weighted merging R-factor52.
Ramachandran plot generated using Molprobity41.
RmrgdF values51.
Ten percent of the data were set aside for Rfree calculations.
Five percent of the data were set aside for Rfree calculations.
Data sets LAB1 through LAB8 are of similar quality and have comparable statistics. As only rigid-body refinement against the same unit cell as EcTpx was done, refinement statistics are the same as for EcTpx.
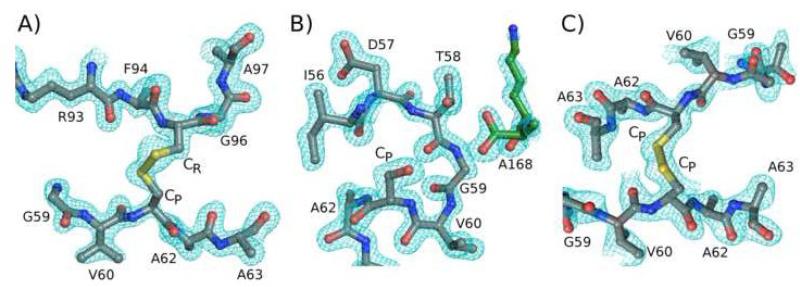
Figure 2.
Electron density quality and active site structures. A) 2Fo-Fc electron density (cyan) is shown for the disulfide bond in chain A of EcTpx. Peptide segments Gly59 - Ala63 and Arg93 - Ala97 (except, for clarity, the side chain of Phe94) are shown. The contour level is 1 ρrms. B) Electron density as in panel A is shown for the FF active site in EcTpxC61S. The peptide segments containing residues Val60 - Ala63 and Lys167 - Ala168 (of a symmetry mate, green) are shown; Ser61 is labeled as CP. The contour level is 2 ρrms. C) Electron density as in panel A is shown for the intermolecular CP-CP disulfide bond of EcTpxC82,95S. Residues Gly59 - Ala63 of chains C and D are shown; the contour level is 0.8 ρrms. In all panels, structures are colored by atom with C=grey, N=blue, O=red and S=yellow, and the CP and CR residues are indicated.
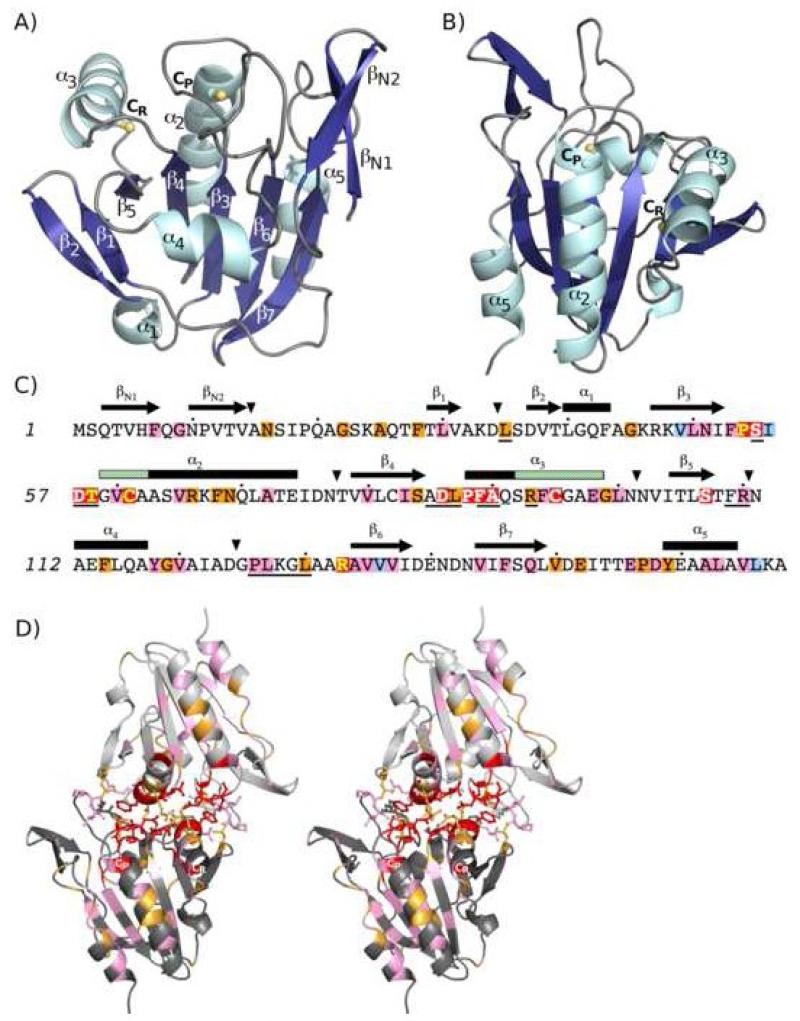
Figure 3.
Structure and sequence conservation within the Tpx subfamily. A) The structure of EcTpxC61S, a representative TpxFF, with only one chain of the dimer is shown. α-helices (cyan) and β-strands (blue) are labeled according to the common fold of all Prxs. Two features characteristic of the Tpx subfamily are the position of CR in helix α3 and an N-terminal extension containing a β-hairpin (βN1 and βN2). The CP and CR residues are shown as ball and stick with the Cys sulfur atom (or Ser61 oxygen atom) in yellow. B) The structure shown in panel A is rotated ~180° around the vertical axis to give the approximate views used in Figures 3D, 5 and 7. C) Tpx-specific sequence conservation is mapped onto the sequence of EcTpx. Background coloring is by residue conservation within the Tpx subfamily: red for 100% conserved, orange for > 90% conserved, pink for > 90% conserved between two amino acids and pale blue for conserved hydrophobic residues (see Table 3). Yellow text is used for the four residues conserved in all Prxs (Pro54, Thr58, CP61 and Arg133) and white text is used for those residues 100% conserved in Tpx sequences only. Secondary structure elements for the FF conformation are shown above the sequence, rectangles for 310-helices (α1) and α-helices, arrows for β-strands and triangles for insertions/deletions. The structure elements that locally unfold for disulfide formation (the first four residues of helix α2 and the last 7 residues of helix α3) are indicated by green hash marks in the secondary structure of α2 and α3. Solid lines underneath the sequence indicate residues that have > 5 Å2 surface area buried at the dimer interface. The conservation pattern found here is based on many more sequences than previous analyses17,18. D) Stereoview of the EcTpxC61S dimer, generated by crystal symmetry, shown looking down the two-fold axis. The two chains are dark grey and pale grey and the 17 residues that form the dimer interface (underlined in panel C) are shown as sticks. Coloring of the 100%, > 90%, and > 90% between two amino acids conserved residues (as in panel C) highlights their positions at the dimer interface, active site pocket, and the α2-cradle. The asymmetric conformations for Arg93 are visible, with each chain showing one conformation.
Overall Structures
EcTpx, the LU disulfide form
The two chains (one dimer) of EcTpx in the asymmetric unit are similar with a rmsd of 0.46 Å for 167 Cα atoms. The backbones of both chains are well-defined except for two regions in chain B, the turn between βN1 and βN2 and the loop containing CR. These segments also have the greatest Cα-rmsd between chains A and B. The greater order for these segments in chain A is due to crystal packing, suggesting chain B is more representative of the level of mobility present in solution. Surprisingly, while the CP-CR disulfide in chain A has clear connective electron density (Sγ-Sγ distance of 2.1 Å) (Figure 2A), the CP-CR disulfide is open in chain B (Sγ-Sγ distance of 3.3 Å). To determine if the broken disulfide was caused by synchrotron radiation damage22,23,24,25, a series of eight consecutive, in-house data sets were collected. The electron density for the chain B CP-CR disulfide bond was strong in the first data set and weakened over time, proving it is intact in a fresh crystal and sensitive to radiation-induced opening. Interestingly, the disulfides in the two chains adopt different conformations, with CP χ1 values of -66° versus 169°, and CR χ1 values of -65° versus 45° for chains A and B, respectively.
The structure of the LU disulfide conformation of EcTpx was previously determined at 2.2 Å resolution in a different crystal form (referred to here by the PDB code, 1QXH)18. The 1QXH structure has no notable differences with EcTpx (rmsd of ~0.7 Å for 159 Cα atoms). Different crystal packing and high B-factors in both structures contribute to the largest differences (up to 2.5 Å), which occur in the CP- and CR-loops. 1QXH also has one dimer in the asymmetric unit, and although nothing is packed on the disulfides, the two chains have distinct disulfide conformations, similar to what is seen in EcTpx.
EcTpxC61S, the FF form
EcTpxC61S, mimicking the reduced state, has one molecule in the asymmetric unit with a well-ordered main chain and clear electron density for residues 2 through 168. The protein adopts the FF conformation (Figure 3A and B), but it is slightly distorted as the crystal packing places the C-terminal carboxylate from Ala168 of a symmetry mate in the active site, hydrogen bonded to Thr58 and Arg133 (Figure 2B). Although the terminal carboxylate could mimic peroxide binding as has been seen for other Prxs with bound benzoate4,26,27,28,29, ethanediol30, or acetate31, neither oxygen atom is close enough to the CP to approximate the binding of substrate.
EcTpxC82,95S, the PLU form
The four chains in the asymmetric unit of EcTpxC82,95S are highly similar (rmsd of 0.3 to 0.5 Å for 167 Cα atoms) with well-ordered backbones except for residues 95 through 100 in all chains. The largest rmsds (up to 2.5 Å) occur in three regions of the protein which also have the highest B-factors: the turn between βN1 and βN2, the CR-loop (residues 95 through 100), and the C-terminal helix. An unexpected feature of the EcTpxC82,95S structure is a non-native intermolecular CP-CP disulfide bond formed between chains A and B and between chains C and D. Sensitivity of this mutant to undergo a non-physiologically relevant intermolecular oxidation is consistent with previous behavior12. The two CP-CP disulfides adopt the same conformation with one CP having a χ1 of ~ 180° (chains A and D) and the other having a χ1 of ~ -60° (chains B and C) (Figure 2C). A partial local unfolding of α2 (residues 59 through 61) and α3 (residues 96 through 98) accompanies the formation of the intermolecular disulfide.
In EcTpxC82,95S, the position of Ser82 matches that of Cys82 seen in EcTpx and EcTpxC61S. Although the χ1 angle of Ser82 is ~160° and of Cys82 is ~ -70°, the backbone position in β-strand 4 (β4) does not change. The lack of a special role for this residue is consistent with activity assays12,32 and it will not be discussed further.
Sequence conservation in the Tpx subfamily
To help identify residues important for Tpx function, we have generated a conservation pattern based on an alignment of 273 identified Tpx sequences (Figure 3C). All of the sequences are from bacteria and have identities as low as 28% relative to EcTpx. There are 12 residues 100% conserved, 20 residues greater than 90% conserved, 30 residues greater than 90% conserved as one of two amino acids and four residues maintained as a hydrophobic in all sequences. The roles of these 66 highly conserved residues are given in Table 3.
Table 3.
Roles for conserved residues.
| Residue | Conservationa | Roleb | Residue | Conservationa | Roleb |
|---|---|---|---|---|---|
| F7 | >90% w/ K | collar | A90 | 100% | dimer (~10 Å2) |
| G9 | >90% w/ K | structure | anchor (CP-, CR-loop) | ||
| N16 | >90% | structure | R93 | >90% | dimer (~70 Å2)c |
| G22 | >90% | structure | anchor (CR-loop) | ||
| A25 | >90% | structure | F94 | >90% w/ W | FF to LU |
| F28 | >90% | structure | CR95 | 100% | catalysis |
| L30 | >90% w/ V | structure | E98 | >90% w/ A | FF to LU |
| L35 | >90% | collar | G99 | >90% | FF to LU |
| dimer (~55 Å2) | L100 | >90% w/ I | FF to LU | ||
| G45 | >90% | structure | S107 | 100% | anchor (CP-loop) |
| V49 | hydrophobic | cradle | R110 | >90% w/ K | dimer (~70 Å2) |
| L50 | >90% w/ I | structure | F114 | >90% | structure |
| N51 | >90% w/ S | cradle | Y118 | >90% w/ F | structure |
| F53 | >90% w/ V | cradle | G119 | >90% | structure |
| FF to LU | V120 | > 90% w/ L | structure | ||
| P54 | 100% | catalysis | P126 | >90% w/ gap | dimer (~60 Å2) |
| S55 | 100% | dimer (~20 Å2) | L127 | >90% w/ gap | dimer (~20 Å2) |
| anchor (CP-loop) | L130 | >90% | dimer (~55 Å2) | ||
| I56 | hydrophobic | FF to LU | collar | ||
| D57 | 100% | dimer (~40 Å2) | R133 | 100% | catalysis |
| anchor (CP-, CR-loop) | FF to LU | ||||
| T58 | 100% | catalysis | A134 | >90% w/ S | structure |
| dimer (~15 Å2) | V135 | >90% w/ I | cradle | ||
| V60 | >90% w/ T | FF to LU | V136 | hydrophobic | structure |
| CP61 | 100% | catalysis | V137 | >90% w/ I | cradle |
| V65 | >90% w/ T | FF to LU | V144 | >90% w/ I | structure |
| R66 | >90% | FF to LU | F146 | >90% w/ H | cradle |
| F68 | >90% | FF to LU | Q148 | >90% w/ E | cradle |
| N69 | >90% | FF to LU | V150 | >90% | cradle |
| A72 | >90% w/ V | FF to LU | E152 | >90% | structure |
| V80 | >90% w/ I | cradle | E156 | >90% w/ H | cradle |
| I83 | >90% w/ V | structure | FF to LU | ||
| S84 | >90% | anchor (CP-loop) | P157 | >90% | cradle |
| D86 | 100% | dimer (~20 Å2) | D158 | >90% w/ N | cradle |
| anchor (CP-loop) | Y159 | >90% | cradle | ||
| L87 | >90% | dimer (~ 80 Å2) | FF to LU | ||
| P88 | 100% | anchor (CR-loop) | A162 | >90% w/ V | cradle |
| F89 | 100% | dimer (~ 80 Å2) | L163 | >90% w/ I | cradle |
| anchor (CR-loop) | V165 | >90% w/ A | cradle | ||
| collar | L166 | hydrophobic | cradle |
The substitution pattern used to define a conserved hydrophobic residue includes residues Ala, Phe, Gly, Ile, Leu, Met, Pro, Val, Trp, or Tyr. For residues conserved more than 90% as one of two amino acids, the identity of the second amino acid is given.
All of the well-conserved residues play one or more of the following six roles: dimer - packing at the dimer interface (buried surface area in parentheses); cradle - forming part of the cradle; FF to LU - changing conformation associated with catalysis; anchor - anchoring the ends of α2 or α3 (region anchored in parentheses); collar - forming the hydrophobic collar of the active site; catalysis - activating CP; structure - maintaining general protein structure.
Buried surface area is for the “in” conformation of Arg93.
From a broad scope, the connection of conservation with catalysis can be seen by considering the Prx structure as built around two hydrophobic cores, one on each face of the β-sheet. Looking at Figure 3B, the front side of the sheet is the side involved in catalysis with α2 laying centrally in a cradle with helices α3 and α5 as the right- and left-hand walls and four β-strands as the base. The backside core is involved primarily in folding and stability, but not catalysis.
Of the highly conserved residues, 17 are on the non-catalytic side and are for general protein structure (Table 3). The remaining 49 highly conserved residues are on the catalytic side (including the active site) or at the dimer interface (Figure 3D). Of the 12 residues 100% conserved, Pro54, Thr58, Cys61 and Arg133 are conserved among all Prxs and are directly in the peroxidatic active site4; Cys95 (the CR residue) is required for disulfide formation (step 2 of Figure 1), and remarkably the remaining seven are all at the dimer interface adjacent to the active site (see below). As can be seen in Figure 3D, most of the remaining 37 highly conserved residues are involved in the packing of α2 in its cradle. Notable is the sidedness of the conservation of helices α2, α5, and C-terminus of α3 and of strands β4, β3, β6, and β7, visible as an alternating pattern, showing it is specifically residues pointing into the cradle which are well conserved.
The Tpx dimer interface is highly conserved
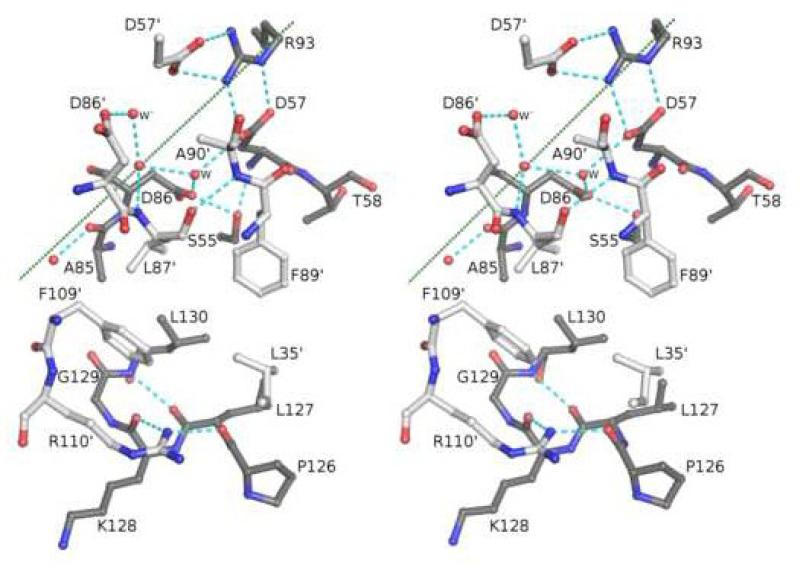
In the three structures determined here, 17 residues are directly involved in the A-type dimer interface20, giving a total surface area burial of ~1400 Å2 for each structure. Building on the description by Choi et al.18, the dimer interface of EcTpx includes three sets of interactions (Figure 4; prime denotes residues from the other monomer): (1) the side chain of Arg110‘ bridges the periphery of the interface by hydrogen bonding to the backbone oxygen of Pro126 and Lys128, creating a hydrogen-bonding network that also includes the backbone oxygen of Leu35‘ and the carboxylate of Asp37‘; (2) a large hydrophobic cluster containing the side chains of residues Leu35‘, Leu87‘, Phe89‘, Phe109‘, Ser55 (Cβ), Thr58 (Cγ), Ala85, Leu127, Gly129 and Leu130; and (3) asymmetric polar interactions at the two-fold axis involving hydrogen bonds formed between Asp57/Asp57‘ and Arg93/Arg93‘, and extending to include Ser55/Ser55‘ (Oγ), the carboxylate of Asp86, the backbone nitrogen of Leu87‘, and three waters (Figure 4), one of which is on the two-fold axis. Interaction sets 1 and 2 occur twice due to the two-fold symmetry.
Figure 4.
Packing interactions at the dimer interface. Stereoview of half of the EcTpx dimer interface with the two chains colored dark grey (chain A) and light grey (chain B, residue numbers contain a prime). One of every residue buried at the dimer interface is shown with the exception of two copies for residues Asp57, Asp86, and the water labeled W. Nearby residues not buried at the dimer interface, such as Asp37, are not shown. Of the three waters conserved at the dimer interface (red spheres), one is on the two-fold axis. Hydrogen bonding interactions are indicated by cyan dashes. The two-fold axis is marked by a dotted green line. Since the side chain of Arg93 is on the two-fold, Arg93‘ (not shown) cannot simultaneously occupy the symmetry-related position; this creates an asymmetric hydrogen bonding network.
Mapping the Tpx-specific conservation pattern onto the Tpx structure shows a striking cluster of the 100% conserved residues at the dimer interface (Figure 3D). Of the 17 interface residues, six are 100% conserved, four are greater than 90% conserved, and three are greater than 90% conserved between two amino acids, and among the published structures, three highly ordered waters are also conserved. The residues involved in the Arg110 hydrogen-bonding network (Figure 4) are the least-well conserved. This makes sense both because the backbone hydrogen bonding seen in this network does not require side-chain conservation and because the interaction is at the periphery of the interface.
The asymmetric hydrogen-bonding network involving the well-conserved residues Asp57 and Arg93 is not EcTpx specific but is observed in all of the Tpx crystal structures. The asymmetry is obligatory because the two-fold symmetry cannot be maintained without the overlap of the Arg93 side chains. We expect that the asymmetry is dynamic in that within a given dimer, the Arg93 and Arg93‘ side chains will share time in the “in” position bridging the Asp57/Asp57‘ side chains. The Arg93 side chain in the “out” position adopts a variety of positions in the Tpx structures, suggesting that the “in” position alone may be the reason for the high conservation of Arg93. Finally, the “in” and “out” conformations of the Arg93 residue can be propogated into altered backbone conformations and thus influence the conformation of α3 in that chain.
Whereas only residues from a single chain are directly involved in catalytic chemistry, the dimer interface appears to be intimately linked with activity. Over half of the surface area buried at the dimer interface involves residues in the CP-loop (residues 55 through 58) and residues associated with α3 (residues 85 through 93). Despite this intimate link, the dimer interactions remain remarkably unchanged during catalysis, consistent with solution studies showing Tpx dimerization independent of oxidation state12,17. The nine known Tpx crystal structures (Table 1) vary over ~20° in the monomer-monomer orientation due to sequence differences and a ~10° shift associated with the FF to LU transition. The Tpx dimer interface appears to be unique as it contains a significant twist (~30°) compared to other Prx A-type dimers20 and does not dissociate with the FF to LU transition like Prx118,33. Despite the unique features and the apparent importance of the Tpx dimer interface in catalysis, a similar surface area is buried upon dimerization as with other A-type dimers (~1300 to 1700 Å2 for Tpxs compared to ~1300 Å2 for Prx133 and ~1800 Å2 for Prx520).
Structural transitions during Tpx catalysis
Using the multiple crystal structures of Tpx subfamily members in the FF and LU conformations (Table 1), along with the Tpx-specific residue-conservation pattern, we can characterize the structural changes common to catalysis throughout the Tpx subfamily. As an unexpected addition, EcTpxC82,95S provides a snapshot of an intermediate conformation between the FF and LU. Due to their lower precision, the two NMR structures (Table 1) are not used in the following comparative structure overlays; however, the specific structural transitions we describe here are completely consistent with the more broadly defined conformational changes and heterogeneity observed in the solution studies21. For the remainder of this analysis, structures will be identified by the active-site conformation as a subscript, TpxFF for fully folded and TpxLU for locally unfolded. The partially locally unfolded EcTpxC82,95S structure will be referred to as EcTpxPLU. Also, for clarity, residue numbering is based on E. coli Tpx (Figure 3C).
TpxFF
The TpxFF conformation is similar among the nine chains present in the four TpxFF crystal structures (Table 1). β1 through β7 overlay well and have relatively low B-factors as expected for the central core of a protein (Figure 5A). The two regions of higher mobility are the loops at the C-terminal ends of α2 and α3, regions that are also the sites of insertions/deletions in Tpxs (Figure 3C).
Figure 5.
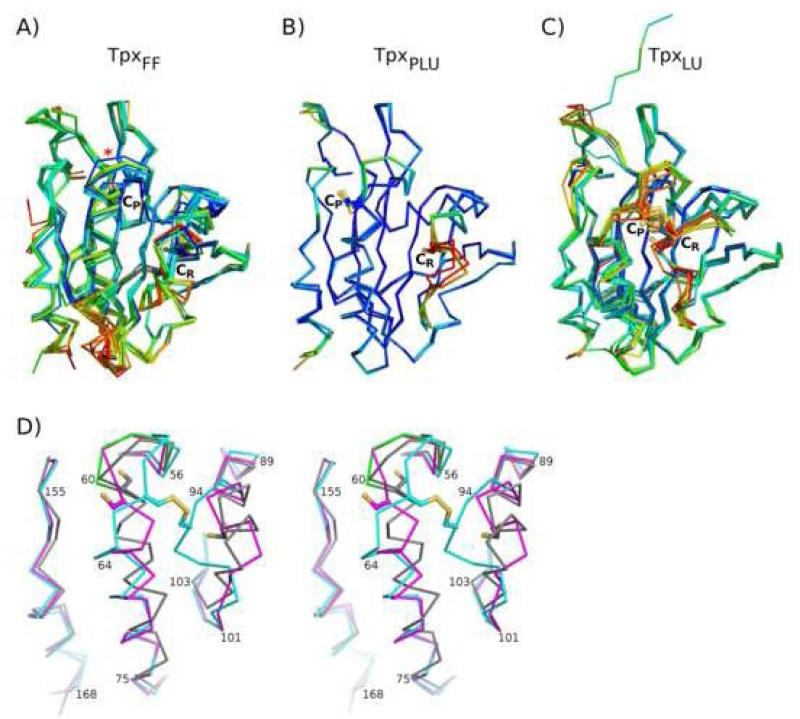
The FF, PLU and LU conformations. Shown are the overlays of the Tpx structures in the A) FF (TpxFF), B) PLU (CP-CP disulfide, TpxPLU) and C) LU (CP-CR disulfide, TpxLU) conformations. In panel A, the distorted CP-loop of the EcTpxFF structure is marked by an asterisk. In panel C, a notable difference is seen for MtTpxLU (PDB code 1XVQ), which has an extended N-terminus resulting from a crystallization artifact17. Each chain is colored by B-factor with red indicating higher values. The CP and CR residues are labeled and shown as ball and stick with sulfur atoms (or oxygen for Ser mutants) colored yellow. D) Stereoview overlay of the representative TpxFF (composite EcTpxFF, grey), TpxPLU (EcTpxPLU, magenta), and TpxLU (EcTpxLU, cyan) structures. The distorted CP-loop in EcTpxFF (green) is included to show its similarity to the PLU structure. The side chains of CP and CR residues are shown as in panels A through C.
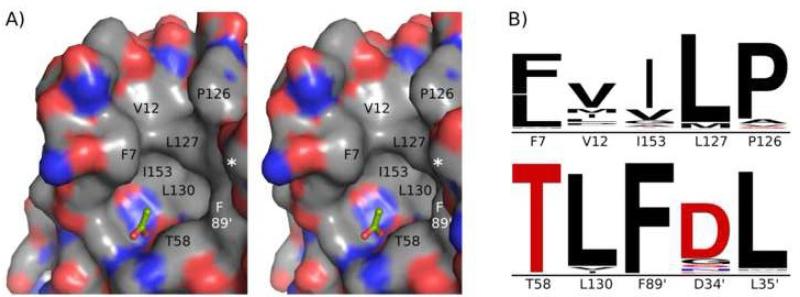
In all of the TpxFF structures, the active-site pocket (containing CP61) is well-ordered and surrounded by an extended hydrophobic collar involving ten residues (Figure 6). The hydrophobic collar is more extensive than was predicted from previous modeling of EcTpxFF18 and its non-polar nature is very well conserved among Tpxs, suggesting a potential importance of these residues in forming a substrate binding cleft recognizing alkyl hydroperoxide substrates and/or a protein docking surface. That the hydrophobic collar involves residues from the N-terminal β-hairpin provides a reason for the conservation of the hairpin in the Tpxs. Additionally, the participation of residues from both chains in the hydrophobic collar implies that although the chemistry of catalysis only requires residues from one chain, Tpx is an obligate homodimer.
Figure 6.
The hydrophobic collar of the Tpx active site. A) Stereoview of the composite EcTpxFF active-site surface. The active-site pocket containing the CP, Pro54, Thr58, and Arg133, is surrounded by a hydrophobic collar that tailors the substrate specificity of Tpxs to alkyl hydroperoxides. A bound acetate molecule (green sticks) modeled from pdb code 1Y25, mimics substrate binding and shows the methyl group pointing toward the collar. Collar residues are labeled with the asterisk representing Asp34‘ and Leu35‘ surfaces making up the right-hand wall of the collar. Asp34‘, Leu35‘, and Phe89‘ (white text) are from the second chain. Coloring is by atom with C=grey, N=blue, O=red and S=yellow. B) Sequence conservation for residues that form the hydrophobic collar. The relative size of the letters indicates conservation and all are plotted on the same scale. Hydrophobic residues are colored black. The nominally polar side chains of Asp34‘ and Thr58 contribute carbon atoms to the collar, and the importance of only the Cα and Cβ atoms for Asp34‘ is consistent with its lower sequence conservation.
In terms of active site conformation, one TpxFF structure differs from the rest, having residues 58 through 60 of the CP-loop shifted slightly (asterisk in Figure 5A), which causes α2 to begin one residue later. This is the EcTpxFF structure, and the distortion is related to the crystal packing interaction described earlier involving Ala168. This distortion shows that the FF active site retains flexibility that may facilitate catalysis, but it also makes the structure less useful for comparative studies. To provide an EcTpxFF structure with the more relevant CP-loop conformation, we have generated a composite structure by adjusting the positions of residues 56 through 61 based on equivalent residues from 2YZH, the next highest resolution FF structure (see Methods). This composite structure provides a relevant active-site conformation and is of greater utility in the comparative studies presented here and for future modeling studies such as drug design.
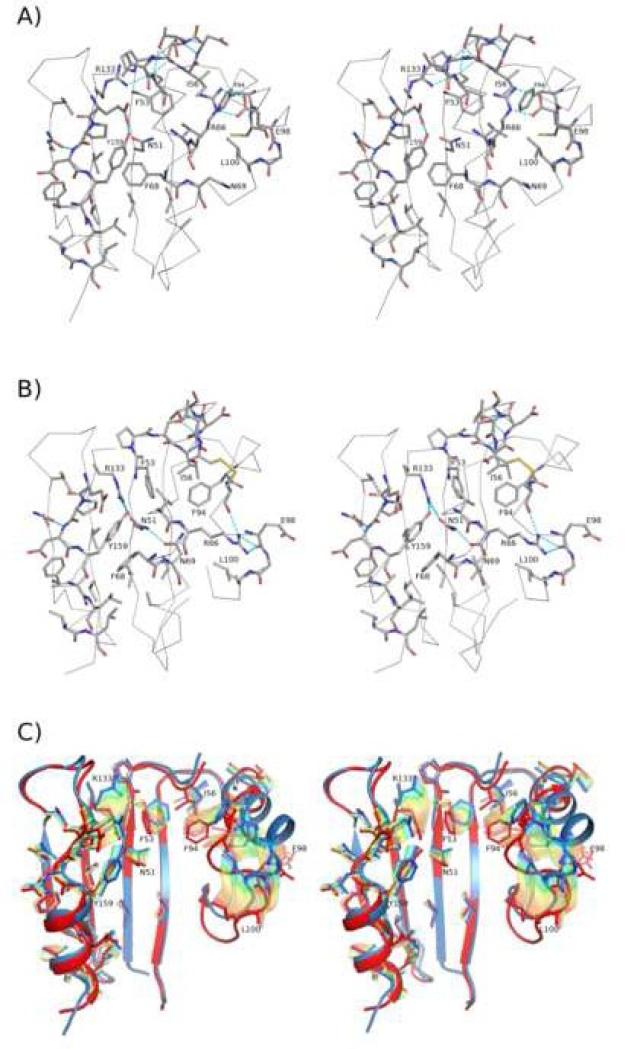
Taking the composite EcTpxFF structure as a representative model for the TpxFF conformation and combining it with sequence-conservation patterns, we can describe the Tpx-specific interactions that stabilize this conformation. In the FF conformation, α2 (residues 58 through 71) sits in the cradle lined by highly conserved residues from β4, β3, and β6 at the base, and α3 and α5 on the right- and left-hand sides (Figures 3D and 7A). Additional well-conserved residues from β7 (Phe145, Gln147 and Val149) do not contribute directly to the cradle but pack against and stabilize α5 with hydrophobic contacts and hydrogen bonds. The discrete position of α2 in the cradle of the TpxFF conformation is maintained by both hydrophobic and hydrophilic interactions involving the side chains of highly conserved residues. The α2-residues Val65, Phe68, and Ala72 pack on a large, hydrophobic surface created by conserved non-polar cradle residues (Val49, Phe53, Ile56, Val80, Phe94, Leu100, Val135, Val137, Ala162, Leu163, and Leu166). Two prominent polar links between α2 and α3 are hydrogen bonds between the side chains of Arg66 and Glu98, and between Asn69 and the backbone oxygen of Asn102 and Val103. Other notable hydrogen bonds in the cradle are formed between the side chains of Glu156 and Tyr159, between Arg133 and the CP, and between Asn51, a buried water and the backbone oxygen of Ser64.
Figure 7.
Catalysis-associated structural transitions in the Tpx subfamily. A) Stereoview of the composite EcTpxFF. Conserved residues (Table 3) in α2 and the cradle are shown as sticks and colored by atom with C=grey, O=red, N=blue, and S=yellow. The Oγ of Ser61 (CP) is colored yellow. Hydrogen bonds involving the side chains of conserved residues are indicated by cyan dashes. Select residues are labeled. B) Stereoview of EcTpxLU oriented and shown as in panel A. C) Stereoview of the interpolated conformational pathway between the TpxFF and TpxLU conformations. The structure is oriented as in A) but with α2 removed. The starting (FF, blue) and ending (LU, red) conformations of the side chains (sticks) and backbone (cartoon) are shown, and the side chains for the interpolated intermediate conformations are shown in semi-transparent rainbow colors (blue to red).
TpxLU
The TpxLU conformation is similar among the seven chains seen in the four TpxLU crystal structures (Table 1). Again, β1 through β7 overlay well and have low B-factors. For TpxLU, higher mobility is seen in four places: 1) the loop at the N-terminal β-hairpin, 2) the CP-loop, 3) the CR-loop, and 4) the loop before α5 (Figure 5C). The CP-CR disulfide has variable conformations and high B-factors suggesting it is highly flexible, consistent with the decreased number of NOEs in the solution structure of BsTpxLU21.
Using the EcTpxLU structure to represent the TpxLU conformation, we can describe the Tpx-specific features that stabilize this conformation of the protein. In the LU conformation, α2 (residues 63 through 71) sits in a discrete position in the cradle, tilted ~20° and twisted ~20° from its position in the FF conformation (Figure 3D). It is positioned by both hydrophobic and hydrophilic interactions with the cradle residues (Figure 7B). Compared to the FF conformation, α2-residues Val65, Phe68 and Ala72 have moved into new positions that are still buried, requiring a rearrangement of the hydrophobic core that is most notable for residues Phe53 and Phe94 which shift into the space vacated by the unraveling of the first turn of α2. In addition, new hydrogen bonds between the side chains of Arg66 and the backbone oxygen of CR95 and Glu98 stabilize the LU CR-loop. A second rearranged hydrogen-bonding network is created around the newly buried Asn69 by the side chains of residues Asn51, Arg133, and Tyr159.
TpxPLU
The TpxPLU conformation is represented by the four chains from EcTpxC82,95S. The structures are similar and interestingly show increased mobility not for the CP-loop but instead for the CR-loop (Figure 5B). The TpxPLU conformation is a hybrid of TpxFF and TpxLU; whereas the conformations of the CP-loop (residues 56 through 59) and α3 (residues 87 through 96) are similar to that in TpxFF, the position of the loop after α3 (beginning at residue 100) and the tilted and twisted conformation of α2 (beginning at residue 63) closely resemble TpxLU (Figure 5D). Due to the conformation of α2 in TpxPLU, the cradle-packing interactions are more similar to those in TpxLU. Interestingly, the conformation of the CP-loop through residue 59 matches that of the distorted CP-loop in EcTpxC61S rather than the canonical FF loop position, supporting the conclusion that the distortion reflects relevant active-site flexibility. The hybrid PLU structure implies that in terms of cradle packing, the Tpx sequences lock into one of two distinct conformations (FF and LU) rather than adopting a continuum of conformations.
Structural events associated with catalysis in the Tpx subfamily
The distinct grouping of the TpxFF and TpxLU structures and the high conservation of side chains involved in the transition show that among Tpxs, universally conserved changes in the structure accompany catalysis. Using the composite EcTpxFF and EcTpxLU structures as the two endpoints and the EcTpxPLU structures for insight into the transition process, we can describe the structural events associated with catalysis in the Tpx subfamily (Figure 7C).
Starting in the stable FF conformation, α2 is packed in the cradle with the active-site pocket in a conformation that can bind and turn over the alkyl hydroperoxide substrate (step 1 in Figure 1). Although the active site must be properly folded for activation of the CP residue, the CP-loop and N-terminus of α2 are flexible and dynamically sample partially unfolded conformations. Substrate reduction results in the formation of CP-sulfenic acid, and although the equilibrium constant may shift, the structure remains in dynamic equilibrium between the FF and LU conformations.
Based on insights from the PLU conformation, the local unfolding of the first turn of α2 is linked to the destabilization of α3; the motion in α2 moves the side chain of Arg66 and disrupts its hydrogen bonds with Glu98. This and other changes in packing interactions between α2 and α3 destabilize the C-terminal end of α3, increasing its tendency to unravel. In the presence of the CP-sulfenic acid, the enhanced unfolding of the C-terminal end of α3 exposes the CR residue, allowing it to react with the OH of the CP-sulfenic acid to form the CP-CR disulfide bond and covalently trap the protein in the LU conformation (step 2 in Figure 1). For the known Tpx structures, this transition involves the unfolding of residues 56 through 62 (the CP-loop and first turn of α2) and residues 94 through 103 (the last two turns of α3 and the CR-loop), and is accompanied by a tilt and a twist of the entire α2 helix through residue 75 (Figure 5D).
When α2 tilts and twists to adopt the stable LU conformation, the highly conserved side chains along the inside of α2 (Val65, Phe68, Asn69 and Ala72) are shifted and the cradle must repack to maintain good interactions. In terms of hydrophobic packing, the Val-rich hydrophobic patch remains largely unchanged in the two conformations, serving as a greasy, featureless surface that Phe68 and Ala72 can slide along and pack well against in both the FF and LU conformations. A new hydrophobic pocket created by the rotation of Phe53, Ile56 and Phe94 is however, the new packing surface for Val65. The new positions of these residues fill the space vacated by the CP. The rearrangements of the hydrophobic interactions are accompanied by changes in the hydrogen-bonding networks, largely due to the repositioning of Asn69 to face the base of the cradle rather than α3. Associated with the introduction of this buried polar group, a new hydrogen-bonding network is generated by the repositioning of Asn51, Arg133, Glu156, and Tyr159, with Arg133 replacing the buried water that was hydrogen bonded to Asn51 (Figure 7B and C). Lastly, new hydrogen bonds and hydrophobic packing stabilize the new packing between α2 and α3: Arg66 forms new hydrogen bonds with the backbone oxygen of CR and Glu98, and Leu100 repacks against α2 one turn lower than in the FF conformation.
The N-termini of the two regions that locally unfold are anchored to the structure by interactions of the highly conserved side chains Ser55, Asp57, Asp86, Ser107, Pro88, Phe89, Ala90, and Arg93 (Table 3). Interestingly, these anchoring residues that help prevent excessive unfolding of the active site region are also residues buried at the dimer interface. We conclude that the dimer interface not only contributes residues to the hydrophobic collar involved in substrate binding but is also required for proper formation of the FF active site and stabilization of the LU conformation. This intimate connection between the dimer interface and the catalytically required unfolding/refolding transition explains the strong conservation of the interface and underscores that Tpxs are obligate homodimers.
MATERIALS AND METHODS
Protein Purification and Crystallization
Wild type EcTpx, EcTpxC61S, and the double mutant EcTpxC82,95S were purified as previously described12 and stored in 5 mM potassium phosphate buffer, pH 7. All crystals were grown at 4 °C using the hanging-drop vapor-diffusion method. For EcTpx, 1 μL of reservoir solution containing 20% (w/v) PEG 8000, 0.1 M phosphate-citrate pH 4.2 and 0.2 M sodium chloride (Emerald Wizard Screen I #31) was mixed with 1 μL of protein at 10 mg/mL. Rod-shaped crystals grew to a final size of 0.05 × 0.05 × 0.2 mm3 in one week. Crystals were transferred to an artificial mother liquor like the reservoir but with 30% (w/v) PEG 8000, then pulled through oil and flash-frozen in liquid nitrogen for data collection.
For EcTpxC61S, 1 μL of reservoir solution containing 22% (w/v) PEG 8000 and 0.05 M potassium phosphate (optimization of Hampton Crystal Screen I #42) was mixed with 1 μL of protein at 3.4 mg/mL. Box-shaped crystals grew to a final size of 0.15 × 0.15 × 0.15 mm3 in two weeks. Crystals were flash-frozen in liquid nitrogen after serial transfer (1 min each) to solutions containing the reservoir mixed with increasing glycerol concentrations ranging from 5 to 20%.
For EcTpxC82,95S, 2 μL of reservoir solution containing 15% (w/v) PEG 8000, 0.1 M Tris pH 7.0 and 0.2 M magnesium chloride (optimization of Emerald Wizard Screen II #43) was mixed with 1 μL of protein at 6.4 mg/mL. Box-shaped crystals grew to final size of 0.05 × 0.1 × 0.05 mm3 in one week. Crystals were pulled through oil and flash-frozen in liquid nitrogen for data collection.
Data Collection
Data were collected using beamlines 5.0.1 or 5.0.3 at the Advanced Light Source (ALS, Lawrence Berkeley National Laboratory). Data sets for EcTpxC61S and EcTpxC82,95S were processed and scaled using iMosflm v1.0.034 and SCALA35 and EcTpx data were processed and scaled using the HKL suite of programs36. Final statistics showed the merged data sets were usable to 1.8 Å, 1.75 Å and 2.1 Å for EcTpx, EcTpxC61S and EcTpxC82,95S, respectively (Table 2).
Radiation damage test data sets were collected on a single EcTpx crystal using an in-house source (Raxis IV detector and Cu Kα radiation from a Rigaku RU300 rotating anode generator running at 50 kV, 100 mA; Δϕ = 1°, 80 20 min images). Eight successive data sets (EcTpxLAB1 through EcTpxLAB8) were collected on the same wedge of data and were processed and scaled using iMosflm and SCALA. Final statistics showed that the merged data sets were all usable to 2.8 Å and of similar quality (Table 2).
Structure Determination and Refinement
The structures of EcTpx, EcTpxC61S and EcTpxC82,95S were solved by molecular replacement in CCP437 with the program MOLREP38 using data from 15 to 4 Å resolution. All refinements were done using Coot39 and REFMAC40; during iterative manual rebuilding, water molecules were added in Coot using standard criteria (> 1 ρrms intensity in the 2Fo-Fc map, > 2.4 Å distance from nearest contact, no B-factors > 80 Å2). For the final rounds of refinement, B-factor and peptide-planarity weights were optimized and riding hydrogen atoms were added. No non-crystallographic symmetry restraints were applied and Molprobity41 was used to monitor geometry throughout refinement. Final refinement statistics for each model are in Table 2.
For EcTpx, the search model was a single chain from a published 2.2 Å structure of EcTpx (chain A of PDB code 1QXH)18. Rigid body refinement yielded an R of 40.9% and further restrained refinement at 1.8 Å resolution resulted in an R/Rfree = 28.8% / 32.5%. The resulting electron density maps allowed us to build C-terminal residues 159 through 168 and add two citrate molecules. Possible alternate side-chain conformations were visible in the electron density for residues 17, 160 and 165 of chain A and residues 7-10 and 78 of chain B but were not modeled. An alternate conformation for the disulfide bond in chain B was modeled based on EcTpxLAB1 data (see below) and the occupancy set to zero. The final R/Rfree values were 14.5% / 20.8%.
EcTpxC61S was solved using chain A of the above EcTpx structure as a search model. Rigid-body refinement yielded an R of 45.3% and restrained refinement extending to 1.75 Å resolution resulted in an R/Rfree of 33.6% / 49.6%. Clear electron density guided the rebuilding of residues 56 through 75 and 89 through 103 to change the locally unfolded conformation to the fully folded one and further refinement yielded an R/Rfree value of 30.2% / 35.4%. TLS refinement defining the entire chain as one TLS group dropped both R and Rfree by 2%. Possible alternate conformations were observed in the electron density for residues 67, 92, 93 and 160; only residue 93 was modeled with an alternate conformation. The final R/Rfree values were 13.6% / 19.5%. A composite structure was generated to show the more physiologically relevant conformation of the CP-loop. For this, residues 56 through 61 were remodeled based on the corresponding residues in chain A of PDB code 2YZH with the geometry being optimized in Coot. The deposited coordinate file contains both the conformation determined from electron density and the alternate conformation predicted for the composite structure. The alternate loop conformation has occupancy set to 0 as it is not actually present in this crystal form.
EcTpxC82,95S was solved using EcTpxC61S as a search model. Rigid body refinement yielded an R of 43.5% and restrained refinement extending to 2.1 Å resolution resulted in an R/Rfree of 29.7% / 38.4%. Residues 59 through 75 and 92 through 103 in all chains required manual rebuilding due to conformational changes associated with an intermolecular disulfide bond formed between the peroxidatic cysteines of two chains. TLS refinement with each chain defined as a TLS group dropped both R and Rfree by 1%. The final R/Rfree values were 16.2% / 24.8%. The higher Rfree value for this structure (~25% versus ~20%) and the slightly larger R/R free gap are reasonable considering its greater disorder (<B>protein ~27 Å2 versus 20 Å2) and the less extensive modeling of the ordered water (approximately half as many waters modeled per chain) due to the lower resolution of the data.
In each structure, water molecules were sorted by electron density in the final 2Fo-Fc map using an in-house program, with water 1 having the strongest density. Also, TLSanl42 was used to incoporate the TLS motions into the B-factors reported in the PDB files of EcTpxC61S and EcTpxC82,95S. DSSP43 was used to determine secondary structure and accessible surface area, and the Yale Morph server44 was used to interpolate the linear pathway between the EcTpxC61S and EcTpx structures.
Data sets testing radiation damage
Each of the eight in-house data sets were isomorphous with EcTpx and were refined with the same unit cell as the 1.8 Å EcTpx structure. Only rigid-body refinement of the starting model (the final EcTpx structure) was pursued so as to minimize model bias in the 2.8 Å refinement; this refinement yielded final R/Rfree values of 19.8% / 21.9% for EcTpxLAB1 and R/Rfree values of 19.2% / 22.0% for EcTpxLAB8. The first data set shows clear 2Fo-Fc density across the disulfide in chain B at 1.4 ρrms. Over the course of the eight data sets, the density for the disulfide gradually decreases to an intensity of 0.8 ρrms. To model the closed disulfide in chain B, the χ1 values of CP and CR were shifted to fit the side chain to the density and the geometry was optimized in Coot. This modification decreased the Sγ-Sγ distance to 2.2 Å. This closed disulfide conformation for chain B was added as an alternate conformation with zero occupancy in the EcTpx structure.
Sequence and structure comparisons
BLAST45 was used to find homologs in the Tpx subfamily of Prxs. The EcTpx sequence was used to search the nonredundant database on April 5th, 2009 using a BLOSUM62 scoring matrix and an expect score cutoff of 10-10; this cutoff was chosen based on the occurrence below this level of Prxs from other subfamilies. Sequences were filtered by the presence of CP and CR residues in the conserved positions. Of the 285 unique sequences identified in the initial search, eleven did not contain a CR in the conserved position. These may be 1-Cys Tpxs and were omitted (GI numbers: 83856669, 120436526, 89516394, 124259572, 187250433, 218520895, 108759791, 145702330, and 148979871). Sequence alignments were performed with ClustalW46 and after manual adjustment were analyzed for conservation patterns with the assistance of Consurf v3.047 and Weblogo48. Among the identified Tpx sequences, the sequence for Helicobacter pylori (GI: 54111572) unexpectedly did not conserve the Prx-wide conserved residue Thr58. Since it is not known if the structure is able to compenstate for this apparent deviation from the conserved Prx active site, this sequence was also omitted, leaving 273 sequences for the comparisons.
Structural overlays were performed with a locally written program using a pair wise distance cutoff of 3 Å49. To calculate differences in the interfacial angles of the dimers, dimers were superimposed using a matrix based on only one of the monomers. The angle required to superimpose the non-overlaid monomer was classified as the change in interfacial angle.
Molecular graphics were created using Pymol50 and figures were prepared using GIMP.
ACKNOWLEDGEMENTS
The authors would like to thank Laura Baker for protein preparation. This work was supported by a grant from the National Institutes of Health to L.B.P. with a subcontract to P.A.K. (R01 GM50389). The authors acknowledge the Proteins and Nucleic Acids Core facility of the Environmental Health Sciences Center at Oregon State University (NIEHS grant P30 ES00210).
ABBREVIATIONS
- Prx
peroxiredoxin
- Tpx
thiol peroxidase
- EcTpx
thiol peroxidase from Escherichia coli
- EcTpxC61S
C61S mutant of EcTpx
- EcTpxC82,95S
C82S and C95S mutant of EcTpx
- FF
fully folded active site
- LU
locally unfolded active site
- PLU
partially locally unfolded active site
- CP
peroxidatic cysteine
- CR
resolving cysteine
- TpxFF
fully folded Tpx
- TpxPLU
partially locally unfolded Tpx
- TpxLU
locally unfolded Tpx
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Bank accession numbers
The coordinates and structure factors have been deposited in the Protein Data Bank as 3HVS for EcTpx, 3HVV for EcTpxC61S, 3HVX for EcTpxC82,95S, and 3I43 for EcTpxLAB1.
References
- 1.Wood ZA, Schroder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 2.Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 3.Flohé L, Budde H, Hofmann B. Peroxiredoxins in antioxidant defense and redox regulation. Biofactors. 2003;19:3–10. doi: 10.1002/biof.5520190102. [DOI] [PubMed] [Google Scholar]
- 4.Karplus PA, Hall A. Structural survey of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- 5.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 7.Wan XY, Zhou Y, Yan ZY, Wang HL, Hou YD, Jin DY. Scavengase p20: a novel family of bacterial antioxidant enzymes. FEBS Lett. 1997;407:32–36. doi: 10.1016/s0014-5793(97)00302-5. [DOI] [PubMed] [Google Scholar]
- 8.Cha MK, Kim HK, Kim IH. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 9.Link AJ, Robison K, Church GM. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- 10.Atack JM, Harvey P, Jones MA, Kelly DJ. The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J Bacteriol. 2008;190:5279–5290. doi: 10.1128/JB.00100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao K. Subcellular localization and in vivo oxidation-reduction kinetics of thiol peroxidase in Escherichia coli. FEMS Microbiol Lett. 2008;289:41–45. doi: 10.1111/j.1574-6968.2008.01372.x. [DOI] [PubMed] [Google Scholar]
- 12.Baker LM, Poole LB. Catalytic mechanism of thiol peroxidase from Escherichia coli. Sulfenic acid formation and overoxidation of essential CYS61. J Biol Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha MK, Kim WC, Lim CJ, Kim K, Kim IH. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J Biol Chem. 2004;279:8769–8778. doi: 10.1074/jbc.M312388200. [DOI] [PubMed] [Google Scholar]
- 14.Dubbs JM, Mongkolsuk S. Peroxiredoxins in bacterial antioxidant defense. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 143–193. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger T, Budde H, Flohé L, Menge U, Singh M, Trujillo M, Radi R. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;423:182–191. doi: 10.1016/j.abb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 16.Olczak AA, Seyler RW, Jr., Olson JW, Maier RJ. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect Immun. 2003;71:580–583. doi: 10.1128/IAI.71.1.580-583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rho BS, Hung LW, Holton JM, Vigil D, Kim SI, Park MS, Terwilliger TC, Pedelacq JD. Functional and structural characterization of a thiol peroxidase from Mycobacterium tuberculosis. J Mol Biol. 2006;361:850–863. doi: 10.1016/j.jmb.2006.05.076. [DOI] [PubMed] [Google Scholar]
- 18.Choi J, Choi S, Cha MK, Kim IH, Shin W. Crystal structure of Escherichia coli thiol peroxidase in the oxidized state: insights into intramolecular disulfide formation and substrate binding in atypical 2-Cys peroxiredoxins. J Biol Chem. 2003;278:49478–49486. doi: 10.1074/jbc.M309015200. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Wan XY, Wang HL, Yan ZY, Hou YD, Jin DY. Bacterial scavengase p20 is structurally and functionally related to peroxiredoxins. Biochem Biophys Res Commun. 1997;233:848–852. doi: 10.1006/bbrc.1997.6564. [DOI] [PubMed] [Google Scholar]
- 20.Sarma GN, Nickel C, Rahlfs S, Fischer M, Becker K, Karplus PA. Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1021–1034. doi: 10.1016/j.jmb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Yang F, Li Y, Zhang X, Xia B, Jin C. Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase. Biochem Biophys Res Commun. 2008;373:414–418. doi: 10.1016/j.bbrc.2008.06.051. [DOI] [PubMed] [Google Scholar]
- 22.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr D Biol Crystallogr. 2000;56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 23.Ravelli RB, Garman EF. Radiation damage in macromolecular cryocrystallography. Curr Opin Struct Biol. 2006;16:624–629. doi: 10.1016/j.sbi.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Ravelli RB, McSweeney SM. The ‘fingerprint’ that X-rays can leave on structures. Structure. 2000;8:315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 25.Weik M, Ravelli RB, Kryger G, McSweeney S, Raves ML, Harel M, Gros P, Silman I, Kroon J, Sussman JL. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Declercq JP, Evrard C, Clippe A, Stricht DV, Bernard A, Knoops B. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 A resolution. J Mol Biol. 2001;311:751–759. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- 27.Smeets A, Loumaye E, Clippe A, Rees JF, Knoops B, Declercq JP. The crystal structure of the C45S mutant of annelid Arenicola marina peroxiredoxin 6 supports its assignment to the mechanistically typical 2-Cys subfamily without any formation of toroid-shaped decamers. Protein Sci. 2008;17:700–710. doi: 10.1110/ps.073399308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evrard C, Capron A, Marchand C, Clippe A, Wattiez R, Soumillion P, Knoops B, Declercq JP. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J Mol Biol. 2004;337:1079–1090. doi: 10.1016/j.jmb.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Evrard C, Smeets A, Knoops B, Declercq JP. Crystal Structure of the C47S Mutant of Human Peroxiredoxin 5. J of Chem Cryst. 2004;34:553–558. [Google Scholar]
- 30.Nakamura T, Yamamoto T, Inoue T, Matsumura H, Kobayashi A, Hagihara Y, Uegaki K, Ataka M, Kai Y, Ishikawa K. Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1. Proteins. 2006;62:822–826. doi: 10.1002/prot.20796. [DOI] [PubMed] [Google Scholar]
- 31.Stehr M, Hecht HJ, Jager T, Flohe L, Singh M. Structure of the inactive variant C60S of Mycobacterium tuberculosis thiol peroxidase. Acta Crystallogr D Biol Crystallogr. 2006;62:563–567. doi: 10.1107/S0907444906008249. [DOI] [PubMed] [Google Scholar]
- 32.Trujillo M, Mauri P, Benazzi L, Comini M, De Palma A, Flohe L, Radi R, Stehr M, Singh M, Ursini F, Jaeger T. The mycobacterial thioredoxin peroxidase can act as a one-cysteine peroxiredoxin. J Biol Chem. 2006;281:20555–20566. doi: 10.1074/jbc.M601008200. [DOI] [PubMed] [Google Scholar]
- 33.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 34.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 35.Evans PR. Recent advances in phasing. Proceedings of the CCP4 study weekend. 1997:97–102. [Google Scholar]
- 36.Otwinowski Z, Minor W. Carter CWJ, Sweet RM, editors. Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology. 1997. pp. 307–26. [DOI] [PubMed]
- 37.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howlin B, Butler SA, Moss DS, Harris GW, Driessen HPC. TLSANL: TLS parameter-analysis program for segmented anisotropic refinement of macromolecular structures. J of appl cryst. 1993;26:622–624. [Google Scholar]
- 43.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 44.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–75. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rozwarski DA, Gronenborn AM, Clore GM, Bazan JF, Bohm A, Wlodawer A, Hatada M, Karplus PA. Structural comparisons among the short-chain helical cytokines. Structure. 1994;2:159–173. doi: 10.1016/s0969-2126(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 51.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 52.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Cryst. 1997;30:203–205. [Google Scholar]