Abstract
Chronic hepatitis C Virus (HCV) infection is one of the major causes of liver fibrosis and liver transplantation in the United States. Circulating microRNA (miRNA) in the blood are emerging as biomarkers for pathological conditions. In the present study, we performed a systematic screening approach to identify upregulated miRNAs in the plasma/serum of HCV infected patients with different stages of hepatic histological disease severity. We initially screened serum samples of HCV infected patients with fibrosis and compared with sera of healthy volunteers using serum miRNA array profiling, and identified a group of modulated miRNAs. Subsequent study demonstrated that miR-20a and miR-92a in HCV infected fibrosis patients sera were significantly upregulated when compared with that of healthy volunteers or non-HCV associated liver disease. We have also observed an increase of plasma miR-20a and miR-92a in acute and chronic HCV infected patients as compared to that of healthy volunteers. However, there was no correlation between the plasma/serum levels of any of these miRNAs with HCV viral loads. We next investigated longitudinal plasma samples from HCV infected patients. Our results suggested that miR-20a and miR-92a remained unaltered in HCV infected patients who progressed from acute to chronic infection. On the other hand, miR-92a expression was reduced in acute to resolved individuals. These data provide evidence that plasma/serum levels of miR-20a and miR-92a have potential as sensitive and cost effective biomarkers for early detection of HCV infection.
Conclusion
The circulating miR-20a may serve as a potential for predictive biomarker in HCV mediated fibrosis.
Keywords: Hepatitis C virus, microRNA, biomarkers
INTRODUCTION
Chronic HCV infection associated liver disease is an important public health problem worldwide. An estimated 200 million people worldwide and 4 million people in the United States are infected with HCV. In the United States, HCV genotypes 1a and 1b are predominant in patients with chronic infection (1). Approximately 20% chronically infected patients develop end-stage liver disease (2). Chronic HCV infection is characterized by high levels of inter-individual variation in disease progression (3). Although several chronically infected individuals never develop cirrhosis, but some may develop severe fibrosis. A number of cellular factors, demographic and clinical characteristics, as well as viral factors, have been associated with the development of HCV-related liver fibrosis (4).
MicroRNAs (miRNAs) are a class of small, single stranded non-coding RNA of 22 nucleotides with a characteristic hairpin secondary structure (5, 6). They regulate gene silencing either by targeting mRNA directly into degradation or by inhibiting translation. Aberrant expression of miRNAs has been linked to variety of cancers, including hepatocellular carcinoma (7, 8).
Several groups have reported the presence of miRNAs in human serum and plasma, called circulating miRNAs (9, 10). These miRNAs are not affected by endogenous RNases in the blood. In addition, circulating miRNAs display consistent profiles between healthy individuals and significantly altered levels in disease conditions (11, 12). These characteristics of circulating miRNAs established their potential value as biomarkers for changes in physiological and pathological conditions (12). For example, miR-25 and miR-223 are shown to be serum biomarkers for lung cancer (13), miR-184 for squamous cell carcinoma (14), and miR-92a for leukemia (15). Circulating miR-122 and miR-155 were identified as inflammation biomarker in different forms of liver injuries (16–19). miR-141 and miR-375 were the most promising markers correlated with prostate tumor progression (20). The circulating miRNAs can also be used to predict the clinical outcomes of non-small-cell lung cancer patients (21).
In our present study, circulating miRNAs, miR-20a and miR-92a, were identified as possible predictive biomarkers for HCV mediated liver disease. Our data showed that an increase in circulating miR-20a correlate with HCV mediated liver fibrosis severity, which may serve as predictor for liver disease progression. We have also observed that miR-20a and miR-92a are upregulated in acute and chronic stage of HCV infection. To our knowledge this is the first report describing a group of miRNAs upregulated in HCV infection which could be used as a potential predictive biomarker.
MATERIALS AND METHODS
Study design and patient samples
Our study was approved by Saint Louis University and Massachusetts General Hospital Institutional Review Board and written informed consent was obtained from all subjects. A total of 86 sera samples including 44 HCV infected patients with different stages of fibrosis, 20 non-HCV associated patients with liver fibrosis and 22 healthy volunteers were included in this study. The liver fibrosis stage was evaluated according to Batts and Ludwig scoring system in patients with chronic hepatitis C, including 33 (F0–F2) early stage and 11 (F3–F4) late stage fibrosis. We included plasma samples from patients with different clinical stages of HCV infection, including acute (4–24 weeks from time of exposure), chronic (33–163 weeks from time of exposure) and resolved samples from Massachusetts General Hospital, Harvard Medical School. All subjects in the control group had normal aminotransferase activities, no history of liver disease or alcohol abuse and were tested negative for HBV, HCV and HIV infections. Table 1 shows the characteristics of patients infected with HCV with different clinical stages and those with non-HCV associated liver disease were included in this study.
Table 1.
Patient Characteristics
| Plasma samples | Serum samples | ||||||
|---|---|---|---|---|---|---|---|
| Healthy volunteers | Patients with acute HCV | Patients with chronic hepatitis | Patients with resolved HCV infection | Healthy volunteers | Patients with HCV mediated fibrosis | Patients with Non-HCV associated liver disease | |
| Sample numbers | 10 | 29 | 18 | 11 | 22 | 44 | 20 |
| Age (years)* | 32±9 | 28±8 | 28±8 | 27±7 | 38±9 | 53±9 | 60±12 |
| Gender (Male/Female) | 5/5 | 10/19 | 9/9 | 1/10 | 17/5 | 29/15 | 13/7 |
| ALT (IU/L)† | ND | 128 (12–2299) | 116 (17–736) | 14 (9–48) | 14 (6–37) | 63 (18–391) | 49 (12–118) |
| AST (IU/L) † | ND | 75 (16–1935) | 46.5 (26–358) | 22 (13–43) | 19 (9–25) | 52.5 (16–520) | 44 (25–88) |
| HCV viral load (log10 copies/ml)# | N/A | 6.5±6.4 | 5.8±5.5 | UD | N/A | 6.6±6.2 | N/A |
| Stage of fibrosis | N/A | N/A | N/A | N/A | N/A | 33/11 | 9/11 |
denotes Mean ± SD;
denotes median (range);
denotes HCV viral load in log10 ± SEM; N/A denotes not applicable; ND, not done; UD, undetermined
Serum miRNA expression profiling
We performed Human Serum & Plasma miScript miRNA PCR Array (MIHS-106Z, Qiagen, CA, USA) that profiles the expression of 84 miRNAs detectable and differentially expressed in serum, plasma, and other bodily fluids. Human serum miScript miRNA PCR Array was used for miRNA profiling in serum samples (n=4 from each group) of healthy volunteers, early stage (F0–F2) and late stage (F3–F4) HCV infected fibrosis patients. In brief, RNA was reverse transcribed to cDNA using miScript Reverse Transcription kit (Qiagen) according to the manufacturer’s instructions. Real-time qPCR was performed using miScriptSYBR Green PCR kit (Qiagen) with the manufacturer provided miScript Universal primer. Array data was analyzed using free web based software http://pcrdataanalysis.sabiosciences.com/mirna/arrayanalysis.php and automatically perform all ΔΔCt fold change calculations.
miRNA-specific quantitative real-time RT-PCR
Total RNA was isolated from 200 μl of plasma/serum by miRVana PARIS kit (Ambion, Austin, TX), according to the manufacturer’s instructions. Synthetic spiked-in C.elegans miR-39 was added to the plasma/serum and cell culture supernatant samples prior to RNA extraction as an internal control. There is no consensus on the use of housekeeping miRNAs and it was reported that frequently used reference genes like U6 small nuclear RNA (RNU6B) and 5S ribosomal RNA are easily degraded in plasma/serum samples (22). In addition, a large variation of serum U6 levels was reported in several studies (23). We used TaqMan qRT-PCR assays to examine the expression of miRNAs in plasma/serum RNA of all samples. All reagents, primers and probe were purchased from Applied Biosystems. Real-time PCR was performed using an ABI 7500 Sequence Detection System and fold changes in gene expression were calculated using the 2−ΔΔCt method. The mean miRNA level of the three real-time quantitative PCR experiments was calculated for each case.
Cell culture and HCV infection
Immortalized human hepatocytes (IHH) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 100 U/ml of penicillin G, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 atmosphere. We have grown HCV genotypes 1a (clone H77) in IHH as previously described (24).
Statistical analysis
Data were analyzed by non-parametric tests using Wilcoxon test for comparison of paired samples, and Mann-Whitney U-test for two nonparametric groups. Receiver operating characteristic (ROC) curves were constructed and the area under the curve (AUC) was calculated to evaluate specificity and sensitivity of predictive value or feasibility of using serum/plasma miRNA as a marker for liver disease progression. A p-value of less than 0.05 was considered statistically significant. All statistical analyses were performed and graphs were generated using GraphPad Prism 5.0 (GraphPad Software Inc, California).
RESULTS
Profiling of serum miRNA levels in HCV infected patients with liver fibrosis
To generate a comprehensive set of miRNA expression profiles in HCV infected fibrosis patients, we analyzed expression of 84 miRNAs from 4 samples of each group- healthy volunteers (control group), HCV infected patients with early stage (F0–F2) liver fibrosis (Group 1) and late stage (F3–F4) liver fibrosis (Group 2). The miRNA expression profile was generated using Qiagen miRNA Array web based software. Clustering analysis revealed that HCV infected liver disease patients expressed distinct pattern of miRNAs (data not shown). 36 miRNAs were differentially expressed in HCV infected patients compared to healthy volunteers and normalized with spiked-in cel-miR-39. We chose the following six miRNAs, which displayed significant differences in fold change between HCV infected patients and healthy controls: miR-20a, miR-25, miR-27a, miR-92a, miR-148a, and miR-195 to validate miRNA array results by real time qPCR. We also observed that miR-574–3p expression level was similar in all categories of samples in miRNA array analysis. Since no significant difference was found in the levels of miR-574–3p between controls, non-HCV associated samples and HCV infected samples, and we used miR-574–3p as an additional endogenous control to normalize the samples in further analysis.
Among these dysregulated serum miRNAs, miR-21 and miR-122 were also upregulated in our array, which were recently reported to be highly expressed in HCV infected patients. Although there is some information about circulating serum miR-21 in chronic HCV, these reports are conflicting (25, 26). Further, miR-21 may be a non-specific biomarker for chronic HCV, since other studies have suggested its potential as biomarker for other cancers (12). miR-122 has also been shown to be differentially regulated in liver injury irrespective of etiology (16, 17, 27). We chose not to include these two miRNAs as predictors for HCV mediated liver disease progression.
Candidate miRNAs, miR-20a and miR-92a, are highly expressed in serum of HCV infected patients with liver fibrosis
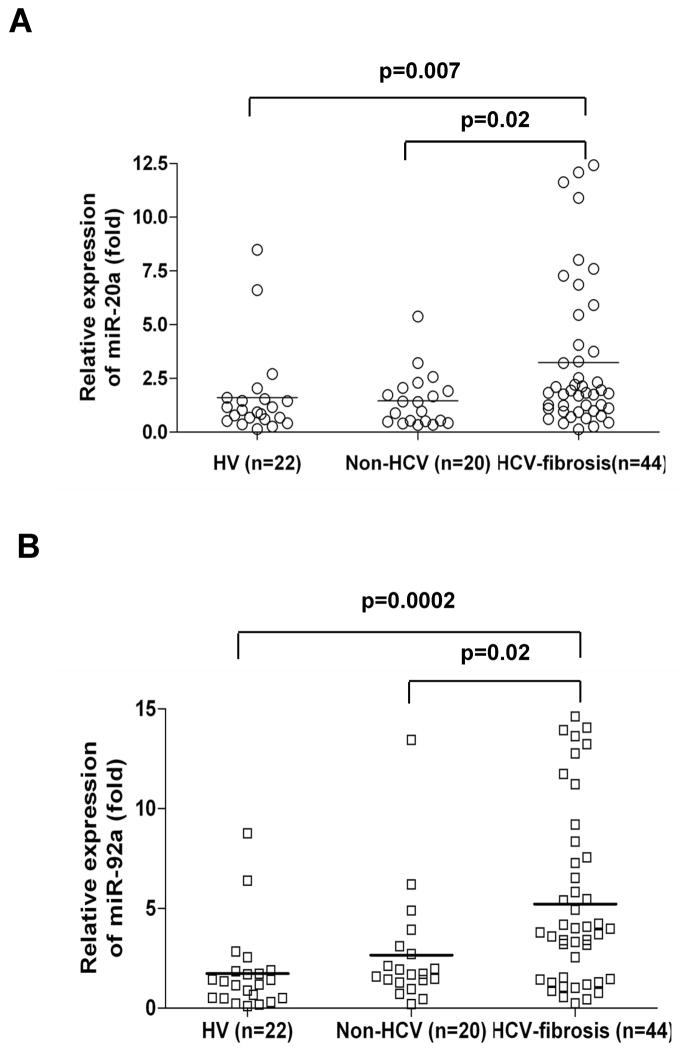
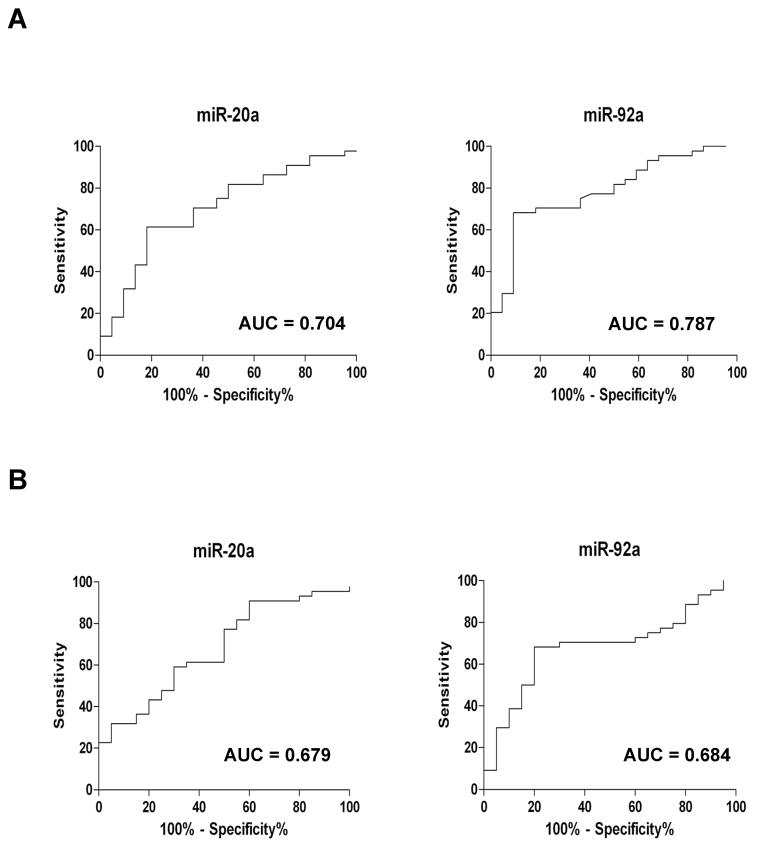
We further validated the six miRNAs individually in a small cohort of sera from healthy volunteers, non-HCV and HCV infected samples. After validation, we chose to examine the expression levels of miR-20a and miR-92a in subsequent studies. We observed that miR-20a and miR-92a expression were significantly upregulated in HCV infected patients with fibrosis as compared to healthy volunteers and non-HCV related liver disease patients with fibrosis (Fig. 1, panels A and B). We also observed that expression levels of miR-20a in serum is gradually increased from early stage to late stage fibrosis in HCV infected patients (Fig. 2, panel A). However, level of miR-92a was decreased in in late stage fibrosis in HCV infected patient sera (Fig. 2, panel B). We next determined the predictive value of these miRNAs in identifying the HCV mediated liver fibrosis progression in a cohort of 22 healthy controls, 20 non-HCV liver disease patients and 44 patients with HCV. The levels of two miRNAs in these serum samples were measured and ROC analysis was performed on individual miRNAs. miR-20a had AUC of 0.704±0.067 (95%CI=0.571–0.836) with a sensitivity of 61.4% and specificity of 81.8%, and miR-92a had AUC of 0.787±0.058 (95%CI=0.672–0.901) with sensitivity of 70.5% and specificity of 77.3% in separating the healthy controls from HCV infected patients (Fig. 3, panel A). Further, miR-20a displayed AUC of 0.679±0.070 (95%CI=0.542–0.817) with sensitivity of 61.4% and specificity of 65%, and miR-92a displayed AUC of 0.684±0.069 (95%CI=0.548–0.819) with sensitivity of 70.5% and specificity of 70% (Fig. 3, panel B) in separating non-HCV infected fibrosis patients from HCV infected fibrosis patients.
Fig. 1. Upregulation of serum levels of miR-20a and miR-92a.
Scatter plots of serum levels of (A) miR-20a and (B) miR-92a in healthy volunteers, HV (n=22), non-HCV associated liver fibrosis patients, Non-HCV (n=20) and HCV infected fibrosis patients, HCV-fibrosis (n=44). The line indicates the median value per group. Fold regulation are expressed as RQ based on 2^(−ΔΔCt) method. Mann-Whitney U test was used to determine statistical significance.
Fig. 2. Serum levels of miR-20a and miR-92a and histological liver disease severity in HCV infected fibrosis patients.
HCV infected fibrosis patients group was subdivided into early stage fibrosis (F0–F2), Early fibrosis (n=33) and late stage fibrosis (F3–F4), Late fibrosis (n=11). Scatter plots of serum levels of (A) miR-20a, (B) miR-92a in healthy volunteers, HV (n=22), HCV-early fibrosis (n=33) and HCV-late fibrosis (n=11). The line indicates the median value per group. Fold regulation are expressed as RQ based on 2^(−ΔΔCt) method. Mann-Whitney U test was used to determine statistical significance.
Fig. 3. Receiver operating characteristic (ROC) analysis using serum miR-20a and miR-92a for discriminating HCV infected fibrosis patients.
ROC curves with corresponding area under the ROC curve (AUC) for (A) miR-20a and miR-92a in discriminating HCV infected fibrosis patients from healthy volunteers and (B) miR-20a and miR-92a in discriminating HCV infected fibrosis patients from non-HCV infected liver fibrosis patients.
Expression level of miR-20a and miR-92a in HCV infected patient with different clinical status
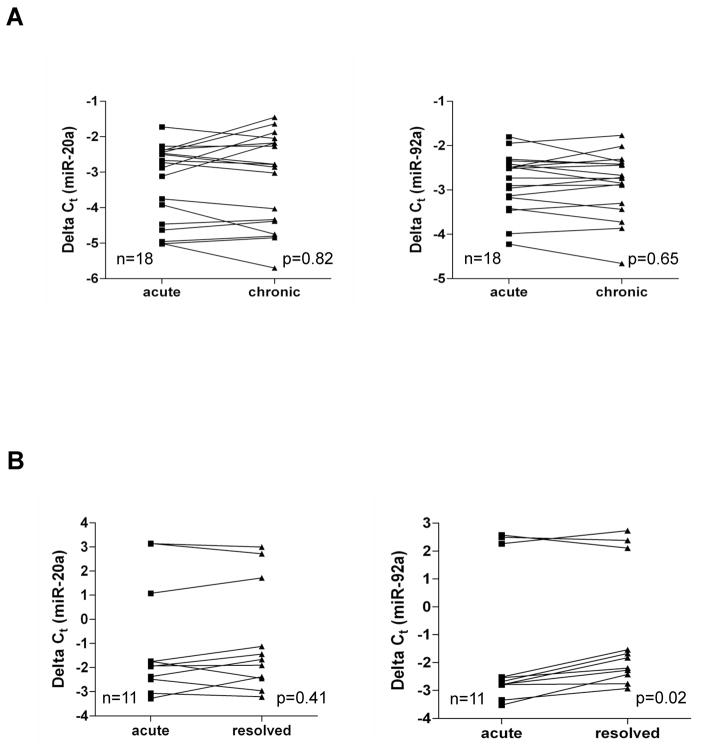
We next examined the expression level of these miRNAs in HCV infected patients with acute, chronic or resolved samples. The plasma levels of these two miRNAs in HCV infected patients during different stages of HCV infection (29 with acute hepatitis, 18 with chronic hepatitis and 11 with resolved infection) were analyzed. Our data indicated that in acute and chronic stage of HCV infection, miR-20a and miR-92a levels are significantly elevated when compared with healthy volunteers (Fig. 4, panels A and B). On the other hand, expression level of miR-20a and miR-92a declines in resolved infection. Thus, an increase in expression of miR-20a and miR-92a in plasma may correlate with early stage of HCV infection. We also performed ROC analysis to determine the predictive value of these miRNAs for detection of early stage of HCV infection. Our analysis showed that miR-20a had AUC of 0.883±0.058 (95%CI=0.769–0.996) with sensitivity of 89.6% and specificity of 80%, and miR-92a had AUC of 0.889±0.057 (95%CI=0.778–1.001) with a sensitivity of 89.6% and specificity of 90% in separating the healthy volunteers from acutely infected HCV patients (Fig. 5, panel A). miR-20a also displayed AUC of 0.983±0.019 (95%CI=0.944–1.022) with a sensitivity of 100% and specificity of 80%, and miR-92a had AUC of 0.989±0.015 (95%CI=0.960–1.018) with a sensitivity of 100% and specificity of 80% in separating healthy volunteers from chronic HCV infected patients with no fibrosis (Fig. 5, panel B). We further analyzed the longitudinal samples for status of plasma miRNAs in acute to chronic HCV infected patients (18 pairs). Our results suggested that both miR-20a and miR-92a levels in plasma remained unchanged in acute to chronic pairs of HCV infected patients (Fig. 6, panel A). Interestingly, when we analyzed the longitudinal samples of acute to resolved groups (11 pairs), only miR-92a expression is significantly reduced (Fig. 6, panel B). There is no significant change in miR-20a expression in acute to resolved group. While our longitudinal sample data is promising, larger sample group in future study will help to confirm the use of these miRNAs as potential biomarker in early stage of infection.
Fig. 4. Scatter plots of plasma levels of miR-20a and miR-92a in HCV infected patients with different clinical status.
Scatter plots of plasma levels of (A) miR-20a and (B) miR-92a in healthy volunteers (HV, n = 10), acutely infected HCV patients (acute, n = 29), HCV infected patients with chronic hepatitis (chronic, n = 18) and resolved HCV infected patients (resolved, n=11). The line indicates the median value per group. Fold regulation are expressed as RQ based on 2^(−ΔΔCt) method. Mann-Whitney U test was used to determine statistical significance.
Fig. 5. Receiver operating characteristic (ROC) analysis using plasma miR-20a and miR-92a for discrimination of early stage of HCV infection.
ROC curves with corresponding area under the ROC curve (AUC) for (A) miR-20a and miR-92a in discriminating acutely infected HCV patients from healthy controls, and (B) miR-20a and miR-92a in discriminating chronic infected HCV patients from healthy controls.
Fig. 6. Plasma levels of miR-20a and miR-92a in HCV infected longitudinal samples.
(A) Levels of miR-20a and miR-92a in 18 pairs of HCV infected patients that progressed from acute to chronic phase of infection. (B) Levels of miR-20a and miR-92a in 11 pairs of acute to resolved paired samples from HCV infected patients. p-value indicates statistical differences in miRNA level between paired samples determined by Wilcoxon matched pairs test.
Upregulation of candidate miRNAs in in vitro HCV infected culture supernatants
To further verify that these identified miRNAs are indeed upregulated from HCV infection, we determined their status in the HCV infected culture supernatants as compared to that of mock infected culture supernatants. We found that miR-20a and miR-92a were highly upregulated in HCV infected culture supernatants in comparison to that of mock infected control, whereas miR-574–3p expression was similar between mock treated and HCV infected culture supernatants (Fig. 7).
Fig. 7. Differential expression of miRNAs in supernatant of HCV infected cells.
Relative expression of miRNAs in mock infected and HCV infected culture supernatants at 6 days post infection. Data presented here as RQ based on 2^(−ΔΔCt) method and representative of two independent experiments.
DISCUSSION
The search for non-invasive biomarkers for diagnosis of diseases has become a rapidly growing area of clinical research (28). Unlike screening for large numbers of mRNAs, a small group of miRNAs or even one specific miRNA might be sufficient to differentiate patients from healthy individuals. In this report, we demonstrated that upregulation of selected miRNAs were associated with progression of liver fibrosis in HCV infected patients. We identified two miRNAs (miR-20a and miR-92a) in association with HCV infection and liver fibrosis. We also observed that expression levels of miR-20a and miR-92a follow an increasing trend in acute and chronic hepatitis. Interestingly, lack of significant differential pattern of plasma levels of miR-20a in acute to chronic hepatitis (longitudinal samples) but significantly elevated expression in fibrosis stage of HCV infection suggested that miR-20a may be a good predictive biomarker for HCV mediated liver disease progression.
miR-17–92 cluster is a proto-oncogenic cluster (also called oncomir-1) consisting of six miRNAs which include miR-20a and miR-92a (29). Increased expression of miR-20a was found in the plasma of chronic lymphocytic leukemia (CLL) patients (30) and in the serum of individuals with gastric cancer (31). Serum miR-92a level was increased in epithelial ovarian cancer (32). Circulating miR-92a level was also upregulated in patients with colorectal cancer (CRC), and advanced adenomas as compared to that of controls, suggesting that circulating miR-92a may serve as a biomarker on early detection of benign lesions before neoplastic formation of CRC (33). Apart from the oncogenic potential, miR17–92 cluster is involved in regulation of fibrosis in rodents and human liver (34). We observed that miR-92a is upregulated in acute and chronic HCV infected sera and reduced in resolved samples, suggesting its potential for early detection marker. Interestingly, plasma miR-92a expression was higher in acute to chronic HCV infected patients with highest AUC value. We also observed the presence of these miRNAs in HCV infected culture supernatants as compared to mock infected hepatocytes. Thus, we believe that HCV infection in hepatocytes enhances expression of these miRNAs, which in turn, releases in circulation. Future work is needed to understand the role of these miRNAs in HCV infection.
Chronic HCV infection is associated with liver fibrosis, and eventually develops end stage liver disease. The progression of liver fibrosis varies in HCV infected patients (3), therefore identifying a predictive biomarker will help in developing treatment strategy. Further, current follow-up for fibrosis is liver biopsy or measurement of liver stiffness, and these procedures have limitations. Therefore, minimally invasive serological marker may be a good alternative for assessment of liver disease progression. Our study demonstrated an upregulation of miR-20a in hepatitis C infected patients which positively correlate with progression of liver fibrosis. On the other hand, circulatory miR-92a level is inversely correlated with fibrosis stage. There are reports suggesting that miRNAs get secreted in the extracellular milieu in response to inflammation or injury to hepatocytes. miR-21 is positively correlated with HCV mediated fibrosis (35). miR-21 is known to target SMAD7 and thereby, enhances TGF-β mediated fibrosis. Increased levels of miR-21 in association with necroinflammation and drug induced liver injury are also reported (36, 37). While our manuscript is in preparation, Trebica et al (27) reported that circulating miR-122 levels is inversely correlated with fibrosis stages. The molecular mechanisms of HCV mediated liver fibrosis are different than that of NASH or NAFLD. Therefore, it is possible that the HCV specific circulatory miRNAs play a role in promotion of liver fibrosis. Indeed, further studies are necessary to elucidate the underlying mechanism.
In conclusion, our study demonstrated that miR-20a and miR-92a are expressed at higher levels in serum/plasma of patients with HCV infection as compared with healthy individuals, suggesting that these serum/plasma miRNAs may serve as potential biomarkers of the HCV infection. We have further demonstrated that serum miR-20a expression is elevated with early to late fibrosis stage of HCV infected patients, but not in non-HCV infected patients, suggesting its potential as a predictive biomarker for HCV mediated liver disease progression.
Acknowledgments
We like to thank Patricia Osmack for helping us with the serum samples. This work was supported by research grant DK081817 from the National Institutes of Health and Saint Louis University Liver Center Seed Grant.
References
- 1.Zein NN, Rakela J, Krawitt EL, Reddy KR, Tominaga T, Persing DH. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Collaborative Study Group. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]
- 2.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 3.Missiha SB, Ostrowski M, Heathcote EJ. Disease progression in chronic hepatitis C: modifiable and nonmodifiable factors. Gastroenterology. 2008;134:1699–1714. doi: 10.1053/j.gastro.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 4.Probst A, Dang T, Bochud M, Egger M, Negro F, Bochud PY. Role of hepatitis C virus genotype 3 in liver fibrosis progression--a systematic review and meta-analysis. J Viral Hepat. 2011;18:745–759. doi: 10.1111/j.1365-2893.2011.01481.x. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brase JC, Wuttig D, Kuner R, Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 18.Ding X, Ding J, Ning J, Yi F, Chen J, Zhao D, Zheng J, et al. Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Report. 2012;5:1428–1432. doi: 10.3892/mmr.2012.838. [DOI] [PubMed] [Google Scholar]
- 19.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, Beissbarth T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 22.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi R, Weiland M, Gao XH, Zhou L, Mi QS. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55:1640–1642. doi: 10.1002/hep.25558. [DOI] [PubMed] [Google Scholar]
- 24.Kanda T, Basu A, Steele R, Wakita T, Ryerse JS, Ray R, et al. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J Virol. 2006;80:4633–4639. doi: 10.1128/JVI.80.9.4633-4639.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 27.Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2012:00814–8. doi: 10.1016/j.jhep.2012.10.015. pii:S0168–8278. [DOI] [PubMed] [Google Scholar]
- 28.Mo MH, Chen L, Fu Y, Wang W, Fu SW. Cell-free Circulating miRNA Biomarkers in Cancer. J Cancer. 2012;3:432–448. doi: 10.7150/jca.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Haaften G, Agami R. Tumorigenicity of the miR-17–92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moussay E, Wang K, Cho JH, van Moer K, Pierson S, Paggetti J, Nazarov PV, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2011;108:6573–6578. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 33.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 34.Kodama T, Takehara T, Hikita H, Shimizu S, Shigekawa M, Tsunematsu H, Li W, et al. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. J Clin Invest. 2011;121:3343–3356. doi: 10.1172/JCI44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 37.Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]