Abstract
The genes and pathways that govern the functions and expansion of hematopoietic stem cells (HSC) are not completely understood. In the current study, we investigated the roles of serine/threonine Pim kinases in hematopoiesis in mice. We generated PIM1 transgenic mice (Pim1-Tx) over-expressing human PIM1 driven by vav hematopoietic promoter/regulatory elements. Compared to wild-type littermates, Pim1-Tx mice showed enhanced hematopoiesis as demonstrated by increased numbers of Lin−Sca-1+c-Kit+ (LSK) hematopoietic stem/progenitor cells and cobblestone area forming cells, higher BrdU incorporation in long-term HSC population, and a better ability to reconstitute lethally irradiated mice. We then extended our study using Pim1−/−, Pim2−/−, Pim3−/− single knockout (KO) mice. HSCs from Pim1−/− KO mice showed impaired long-term hematopoietic repopulating capacity in secondary and competitive transplantations. Interestingly, these defects were not observed in HSCs from Pim2−/− or Pim3−/− KO mice. Limiting dilution competitive transplantation assay estimated that the frequency of LSKCD34− HSCs was reduced by ~28-fold in Pim1−/− KO mice compared to wild-type littermates. Mechanistic studies demonstrated an important role of Pim1 kinase in regulating HSC cell proliferation and survival. Finally, our PCR array and confirmatory RT-PCR studies identified several genes including Lef-1, Pax5 and Gata1 in HSCs that were affected by Pim1 deletion. Our data provide the first direct evidence for the important role of Pim1 kinase in the regulation of HSCs. Our study also dissects out the relative role of individual Pim kinase in HSC functions and regulation.
Keywords: Serine/threonine kinase, Pim kinase, Hematopoietic stem cells, Hematopoietic stem cell transplantation, Apoptosis, Proliferation, Transgenic mouse, Knockout mouse
INTRODUCTION
The fate of hematopoietic stem cells (HSCs) including their survival, self-renewal, expansion and differentiation is regulated by both extrinsic and intrinsic factors. Several molecules and pathways important in HSC regulation have been identified, including homeobox protein B4, Slug transcription factors, and the morphogenic signaling molecules [1–3]. However, despite significant advances in our understanding of HSC biology over the last several decades, the detailed molecular mechanisms and pathways regulating HSC functions are not completely known, and many more genes and molecular pathways remain to be discovered.
Pim (proviral insertion in murine lymphomas) protein kinases are a small family of constitutively active, highly conserved oncogenic serine/threonine kinases that has 3 members: Pim1, Pim2 and Pim3 [4–7]. Pim kinases share > 60% of homology at amino acid level, and many of their functions overlap with each other, notably in lymphomagenesis. For instance, EμMyc-EμPim2 double transgenic mice develop B cell lymphoid tumors similar to those arising in EμMyc-EμPim1 transgenic mice [8]. Additionally, Pim3 can compensate for the loss of Pim1 and Pim2 in MuLV-induced lymphomagenesis. Pim kinases are aberrantly expressed in many types of cancers including prostate cancer, colon cancer, and lymphoid leukemia/lymphoma [9–15], and Pim kinase inhibitors are being developed for the treatment of cancer [16].
Beside oncogenic potential, Pim kinases are involved in normal cellular functions as well, including the regulation of early B lymphopoiesis [17], the self-renewal of mouse embryonic stem cells [18], and myocardial regeneration [19]. The roles of Pim kinases in hematopoiesis are not well understood. PIM1 is highly expressed in human fetal hematopoietic tissues such as the liver and spleen [10], and Pim1 kinase is a key target for HOXA9, a homeoprotein important in hematopoiesis [20]. In vitro studies also suggested an important role of Pim kinase in protecting hematopoietic cells from apoptosis [21] and in enhancing growth factor- independent survival in myeloid cells [22, 23]. A more recent study from Grundler, et al. [24] suggested that Pim1 regulates the CXCR4 chemokine receptor expression in HSCs. However, a detailed analysis of the roles of individual Pim kinase in hematopoiesis using stringent serial transplantation and competitive limiting dilution transplantation is still lacking. Furthermore, it will be important to fully understand the roles of Pim kinases in hematopoiesis before initiating testing Pim inhibitors in clinical settings.
In the current study, we performed detailed HSC functional analyses using PIM1 transgenic mice (Pim1-Tx) and Pim single knockout (KO) mice. Our data demonstrated an important role of Pim1 kinase in the regulation of HSCs.
MATERIALS AND METHODS
Mice
Pim1-Tx mice
Pim1-Tx mice were generated by microinjection of a construct containing entire human PIM1 coding sequence into FVB/J zygotes (details described in supplementary materials). All our studies were performed in accordance withMedical University of South Carolina Institutional Animal Care and Use Committee approved-procedures.
Pim single KO mice and Pim1−/−Pim2−/−Pim3−/− triple KO (TKO) mice
Pim1−/−/2−/− and Pim2−/−/3−/− double KO mice were generated by Mikkers, et al. [25], and were kindly provided by Drs Paul B. Rothman (Johns Hopkins University) and Anton Berns (Netherlands Cancer Institute). Pim1−/−, Pim2−/− and Pim3−/−KO mice in this study were generated by systematically breeding wild-type (WT) FVB/J mice with the Pim double KO mice. Pim TKO mice were generated by crossbreeding Pim1 heterozygous (Pim1−/+) Pim2−/−Pim3−/− mice.
FVB/J mice
FVB/J mice as transplant recipients were purchased from the Jackson Laboratory.
Flow cytometry analysis
To determine the percentage and absolute number of Lin−Sca-1+c-Kit+ (LSK) cells, the mice were sacrificed and bones (2 femurs and 2 tibias per mouse) and spleen were harvested. Red blood cells (RBC) were depleted using ACK lysis buffer. Total bone marrow (BM) mononuclear cells and splenocytes were counted. The cells were then stained with biotin-conjugated anti-CD3, anti-CD4, anti-CD5, anti-CD8a, anti-CD11b, anti-B220, anti-Gr-1, anti-TER-119; APC-conjugated c-Kit; and PE-conjugated Sca-1 antibodies, followedby staining with streptavidin-PE-Cy7 or streptavidin-FITC (all these antibodies were purchased from BD Pharmingen, CA).
To determine the surface expression of CXCR4, the cells were stained with rabbit anti-mouse CXCR4 antibody (Abcam) followed by Alexa Flour 488-conjugated goat anti-rabbit antibody (Invitrogen).
To determine HSC cell proliferation and apoptosis, multicolor flow cytometry was used. Briefly, 3,000 to 6,000 sorted LSK cells were cultured in StemSpan SFEM medium (StemCell Technologies) supplemented with 100 ng/mL of murine stem cell factor (SCF), Thrombopoietin (TPO), and Flt3 (all from Invitrogen). Cells were counted every 48 hrs and stained with APC-H7-conjugated c-Kit, PE-conjugated Sca-1, and biotin-conjugated lineage-specific antibodies followed by streptavidin-Brilliant-violet-605 (Biolegend). The cells were then stained with Aqua Live/dead fixable dye (Invitrogen) followed by fixation and permeabilization using BD Cytofix/Cytoperm kit. The fixed cells were subsequently stained with PE-Cy7-conjugated Ki67 antibody (BD Pharmingen) and FITC-conjugated caspase 3 antibody (BD Pharmingen). Flow cytometry was performed on BD LSRFortessa and analyzed with FlowJo software.
Colony forming Unit (CFU) and cobblestone area forming cell (CAFC) assay
CFU assays were performed in complete M3434 methylcellulose medium (Stem Cell Technologies) following the manufacturer’s instructions. The CAFC assay was performed as previously described [26]. The frequencies of CAFCs were determined at week 2 and week 5 using L-Calc software (StemCell Technologies).
In vivo BrdU incorporation assay
In vivo BrdU incorporation was performed as described [27, 28] with minor modifications. Briefly, mice were intraperitoneally injected with 2 doses (8 and 2 hours before sacrifice) of bromodeoxyuridine (BrdU, BD Pharmingen) at 50 μg/gram body weight. BM cells were then isolated and enriched for Lin− cell population using lineage cell depletion kit (Miltenyi Biotec). At least 1.5×106 Lin− BM cells were labeled with PE-conjugated-Sca-1 (D7), APC-conjugated-c-Kit (2B1), PerCP-eFluor 710-conjugated-CD135 (eBioscience), and eFluor 450-conjugated CD34 (eBioscience), followed by fixation and staining with FITC–conjugated BrdU antibody (BrdU Flow Kit, BD Pharmingen).
Primary, secondary, competitive and limiting dilution hematopoietic stem cell transplantation (HCT)
For primary HCT, BM cells were isolated from male Pim1-Tx mice, Pim single KO mice, or wild-type (WT) littermates. RBC-depleted BM cells were injected (cell doses were indicated in the text) via tail-vein to lethally irradiated (11Gy) female FVB/J recipient mice. Animal survival was monitored daily. To determine hematological recovery, peripheral blood was collected from transplant recipient mice by retro-orbital sampling under anesthesia condition. Whole blood cell counts were measured using a Beckman Hemogram counter. Male donor cell engraftment was determined as described below using quantitative PCR for sex-determining region Y (Zfy1).
For secondary HCT, BM cells were obtained from primary transplanted recipient mice at 4 months post transplantation, and 1×107 BM cells/recipient were injected into lethally irradiated female FVB/J mice. Male donor cell engraftment was measured.
For competitive repopulation assay, 5×105 male BM donor cells from Pim1-Tx mice, Pim single KO mice, or WT controls were mixed with 2×105 female competitive BM cells from FVB/J mice, and transplanted into lethally irradiated female FVB/J mice.
For limiting dilution competitive transplantation assay, we used a previously described method [29] with minor modifications. Briefly, sorted LSKCD34− male donor cells at doses of 15, 45 and 150 cells were mixed with 1.5×105 female competitor BM cells and injected into lethally irradiated female FVB/J recipients. The frequencies of HSCs were calculated using ELDA program as described [30].
PCR-based male donor cell engraftment analysis
Male donor cell engraftment in female transplant recipients was determined as described [27, 31, 32]. Briefly, genomic DNAs were extracted from RBC-lysed peripheral blood cells or BM cells using the DNANeasy Kit (QIAGEN), and further purified using Ethanol precipitation method. Twenty ng of genomic DNA were mixed with SYBR Green PCR master mix reagents (Bio-Rad) and real-time (RT)-PCR was performed. Donor cell engraftment was estimated by percentage of male DNA calculated from the standard curve by PCR for sex-determining region Y (Zfy1) [27]. Bcl-2: 5′-AAGCTGTCACAGAGGGGCTA and 5′-CAGGCTGGAAGGAGAAGATG and Actin: 5′-TGTTACCAACTGGGACGACA and 5′-ACCTGGGTCATCTTTTCACG were used as reference genes.
Homing assay
Homing assay was performed using sorted Lin−Sca-l+ male donor cells [33]. The cells were injected into lethally irradiated female recipients (60,000 cells/recipient). The homing efficiency of donor male cells to BM and the spleen at 12 hours post transplantation was determined by PCR quantification of male DNA as described.
Hematopoiesis PCR array and RT- PCR analysis
Commercially available hematopoietic stem cells and hematopoiesis PCR array (PAMM-054A, Qiagen) was performed as per the manufacturer’s instructions. Briefly, LSKcells were sorted from WT, Pim1-Tx, Pim1−/−, or TKO mice (3–6 mice per group). RNAs were isolated using RNeasy plus Mini Kit (Qiagen) and converted to cDNA by RT2 First Strand Kit (Qiagen). The cDNA samples were mixed with RT2 qPCR master mixes and a total of 84 hematopoiesis-specific genes were analyzed. Expression of mRNA for each gene was normalized against hprt, hsp90ab1, gapdh, and actin β endogenous control genes. Gene expression level in Pim1-Tx and KO mice was calculated using the 2−ΔΔCt value against WT mice.
Confirmatory RT-PCR on selected genes was performed using RNA samples isolated from LSK cells. The following primers were used: Actb: 5′-GATCTGGCACCACACCTTCT and 5′-GGGGTGTTGAAGGTCTCAAA; Il6st: 5′-CATGCTTTCAGGCTTTCCTC and 5′-CCATACATGAAGTGCCATGC; Lef1: 5′-TCACTGTCAGGCGACACTTC and 5′-TGAGGCTTCACGTGCATTAG; Gata1: 5′-TGTCCTCACCATCAGATTCCA and 5′-TCCCTCCATACTGTTGAGCAG; Trim10: 5′-CAACTGGAGGGGTTAGACGG and 5′-GGCACTTTCTGGTTTCACATCT; Pax5: 5′-AACTTGCCCATCAAGGTGTC and 5′-GGCTTGATGCTTCCTGTCTC.
Statistical Analysis
The values were reported as Mean ± SEM of multiple experiments or Mean ± SD from a representative experiment. Differences were analyzed by Student’s t test or as indicated. p< 0.05 was regarded as significant.
RESULTS
Generation of vav-hPIM transgenic (Pim1-Tx) mice
Pim1 Tx mice over-expressing murine Pim1 driven by Eμ immunoglobulin promoter were previously reported [34]. In that model, Pim1 over-expression was restricted to the lymphoid lineage, which limits the investigation of the role of Pim1 in HSCs. We generated Pim1-Tx mice bearing full-length human PIM1 sequence encoding a 33 kDa PIM1 protein under the control of vav hematopoietic regulatory elements and SV40 sequences (Fig. S1A). Consistent with hematopoietic-specific expression pattern driven by vav regulatory elements, our Pim1-Tx mice expressed human PIM1 mRNA in all hematopoietic tissues including spleen, bone marrow and thymus, but not in non-hematopoietic tissues such as kidney (Fig. S1B). Pim1-Tx mice had an increased expression of Pim1 protein as demonstrated by western blot analysis using Pim1 antibody (19F7) that recognizes both human- and mouse-origin of Pim1 (Fig. S1C).
We investigated the relative expression level of hPIM1 in various hematopoietic cell subsets, including LSK HSCs, B220+ B cells, and Gr-1+ granulocytes using qRT-PCR. Human PIM1 is expressed in all these cell subsets, and the expression levels were comparable between LSK HSCs and B220+ B cells. However, hPIM1 expression was ~4-fold lower in Gr-1+ granulocytes (Fig. 1A). This is consistent with previous observation that luciferase transgene expression in vav-tTA–driven luciferase Tx mice was much higher in B lymphoid cells than in myeloid cells [35].
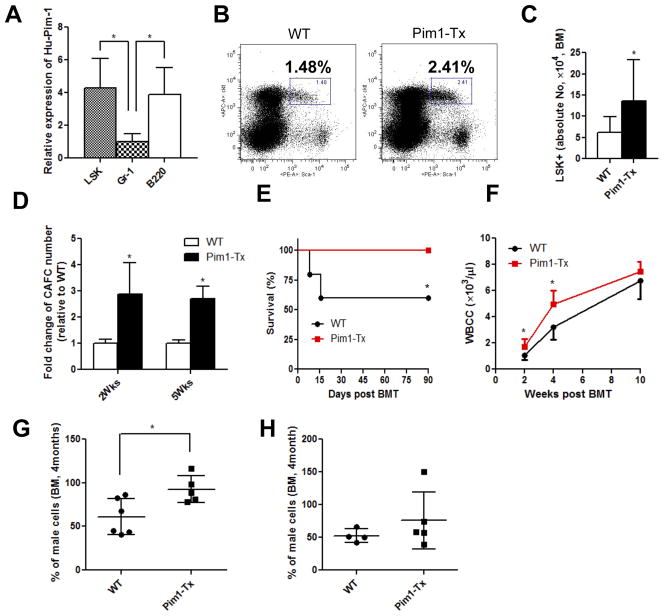
Figure 1. Increased hematopoiesis in Pim1-Tx mice. (A) Quantitative RT-PCR of human PIM1 in various hematopoietic cell subsets.
BM cells isolated from Pim1-Tx mice were sorted to obtain Lin−Scal-1+c-Kit+ (LSK) cells, Gr-1+ granulocytes and B220+ B lymphocytes. RNAs were isolated and quantitative RT-PCR performed to analyze Hu-PIM1 expression (*p < 0.05, data are representative of 2 independent experiments). (B) Increased BM LSK HSPCs in Pim1-Tx mice. BM cells from Pim1-Tx or WT littermates were depleted of Lin+ cells using murine lineage cell depletion kit. The Lin− cells were then stained with Scal-1 and c-Kit antibodies. Representative flow cytometry dot plots were shown. (C) The absolute LSK cell numbers (from 2 femurs and 2 tibias per mouse) in Pim1-Tx mice and WT littermates (n=12, *p < 0.05). (D) Increased CAFCs in Pim1-Tx mice. Cobblestone forming assay were performed and CAFC numbers were calculated as described (*p<0.05, data are representative of 2 independent experiments). (E) Improved survival in recipients transplanted with Pim1-Tx BM cells. FVB/J mice were lethally irradiated (11 Gy) and injected via tail vein with total BM cells (1×105 cells/recipient mouse) isolated from Pim1-Tx mice or WT littermates. Animal survival was monitored daily (n=5/group, *p<0.05). (F) Enhanced hematological recovery in lethally irradiated transplant recipient mice receiving Pim1-Tx BM cells. The BM transplanted mice were bled at 2, 4 and 10 wks post transplant. WBC counts were measured by Beckman Hemogram counter (n=5, *p<0.05). (G) Enhanced engraftment of Pim1-Tx BM cells in competitive BM transplantation assay. 5×105 total BM cells from male Pim1-Tx or male WT littermates, along with 2×105 WT female competitor BM cells, were transplanted into lethally irradiated FVB/J female mice. Recipient mice were sacrificed at 4 months post transplant and percentage of male donor engraftment in BM was estimated by RT-PCR as decried in the Methods and Materials (*p<0.05). (H) No significant increase in engraftment in Pim1-Tx BM secondary transplanted recipient mice. The primary transplant recipient mice as described in Fig. 1F were sacrificed at 4 months post transplant and BM cells harvested and injected into lethally irradiated female FVB/J mice (1×107/recipient mouse). Male donor cell engraftment in BM was analyzed at 4 months post secondary transplantation.
We have followed Pim1-Tx mice and WT littermate progenies for two years. Consistent with the oncogenic potential of Pim1 kinase, we found that approximately 10% of Pim1-Tx mice developed acute lymphoblastic leukemia/lymphoma between 141 and 435 days of life while WT littermates were leukemia free in entire lifetime (data not shown). Interestingly, none of Pim1-Tx mice developed myeloid lineage leukemia. The exact cause for the absence of myeloid malignancies in our Pim1-Tx mice remains to be investigated, but may in part due to the relatively low expression of hPIM1 in mature myeloid cells like granulocytes in these mice [35].
Increased hematopoietic stem/progenitor cell number in Pim1-Tx mice
The relatively high level of hPIM1 expression in LSK cells of Pim1-Tx mice allowed us to determine the roles of Pim1 kinase in hematopoiesis. LSK cells represent a heterogeneous, but virtually all hematopoietic stem/progenitor cells (HSPCs) in mice. We first evaluated the percentage and absolute number of LSK cells in both young (4 months) and aged (1.5 yrs) Pim1-Tx mice that were free of leukemia/lymphoma. Compared to WT littermates, both young and aged Pim1-Tx mice had increased percentage and absolute number of LSK cells in the BM. Representative flow cytometry dot plots showed ~2-fold increase in the percentage LSK cells in Pim1-Tx mice (Fig. 1B). The total LSK cell number (from 2 femurs and 2 tibias of each mouse) was 13.6± 9.8×104 in Pim1-Tx mice, compared to 6.2±3.2×104 in age-matched WT littermates (Fig. 1C, p < 0.05). This increase in the absolute number of LSK cells in Pim1-Tx mice was largely due to the increased percentage of LSK cells. The total numbers of BM mononuclear cells were quite similar between Pim1-Tx mice and WT littermates at all ages (data not shown).
The body weights of Pim1-Tx mice were similar to those of WT littermates. However, Pim1-Tx mice had an increased spleen weight in aged (1.5 yrs old) mice (Fig. S2A). Further analysis indicated that the absolute number of LSK cells in the spleen of Pim1-Tx mice was also significantly increased (Fig. S2B, p<0.05).
We measured peripheral white blood cell count, red blood cell count, hemoglobin concentration, and platelet count in Pim1-Tx mice, and observed no significant differences between Pim1-Tx mice and WT littermates (data not shown). Detailed cell subset analysis also did not show significant differences in peripheral Gr-1+ granulocytes, B220+ B cells or CD3+ T cells between Pim1-Tx and WT littermates (Fig. S3A). Furthermore, we examined the BM tissue by microscopy following H&E staining and did not observe evidences indicative of myelodysplastic or myeloproliferative changes in the Pim1-Tx mice we examined. Morphologically, the BM section of Pim1-Tx mice that were free of leukemia/lymphoma was very similar to that of WT littermates (Fig. S3B). These results suggest that over-expression of Pim1 expands HSPCs without affecting their differentiation.
Enhanced hematopoietic functions in Pim1-Tx mice
We performed cobblestone-area-forming-cell (CAFC) assay, an in vitro functional assay of HSPCs [26]. We observed a > 2-fold increase in 2-week and 5-week CAFC in Pim1-Tx BM cells (Fig. 1D, p<0.05), suggesting that Pim1 overexpression leads to enhanced clonogenic activity and expansion of BM HSPCs.
HSCs are characterized by their abilities to self-renew and to differentiate into all lineages of hematopoietic cells. These hallmarks of HSC functions are best demonstrated using transplantation model in which HSCs reconstitute lethally irradiated recipients. We thus transplanted lethally irradiated (11 Gy) FVB/J mice with BM cells harvested from Pim1-Tx mice or WT littermates (1×105 cells/recipient mouse). This cell dose only rescues about half of lethally irradiated mice, thus allowing us to determine the protective effects of Pim1-Tx HSCs. As showed in Fig. 1E, about 40% of mice transplanted with WT BM cells died within 2 weeks post-transplantation. In contrast, all the mice transplanted with BM cells from Pim1-Tx mice survived. Additionally, the mice transplanted with Pim1-Tx cells had significantly higher peripheral WBC counts, especially during the early stage following transplant (Fig. 1F).
To determine the repopulating capacity of Pim1-Tx HSCs, we carried out competitive BM transplantation experiments. We transplanted 5×105 BM cells from male Pim1-Tx or WT littermates, along with 2×105 of competitive female FVB/J BM cells, into lethally irradiated female FVB/J recipients. Male donor cell engraftment in BM at 4 months post transplant was estimated by quantitative PCR measuring the sex-determining region Y (Zfy1) [31, 32]. As shown in Fig. 1G, recipients transplanted with Pim1-Tx cells had a higher percentage of donor-derived male cells compared to those transplanted with WT cells (p<0.05), indicating that Pim1-Tx BM cells are more efficient and competent in reconstituting the hematopoietic system than WT cells.
To investigate the self-renewal and long-term repopulating ability of Pim1-Tx HSCs, we performed secondary transplantation using BM cells from the primary competitive transplant recipient mice. Male donor cell engraftment was measured in BM at 4 months after the secondary transplant. No difference was observed in donor cell contribution between Pim1-Tx and WT secondary transplant recipients (Fig. 1H). We also performed limiting dilution competitive transplantation using sorted LSKCD34− cells and compared the donor cell contribution at 6 weeks and 16 weeks post transplantation. At 6 weeks post transplantation, we estimated the frequency of active HSCs in Pim1-Tx was 1 in 29.9, compared to 1 in 47.3 in WT littermate controls (Fig. S4A). However, at 4 months post transplantation, the difference in the estimated frequency of active HSCs between Pim1-Tx and WT littermates disappeared (Fig. S4B). These data imply that overexpression of Pim1 kinase may expand HSC population initially, followed by HSC exhaustion.
Decreased hematopoietic functions in Pim1−/− KO mice
To further confirm the functional role of Pim1 on hematopoiesis and to dissect out the relative contribution of individual Pim kinase in hematopoiesis, we extended our studies using Pim1−/−, Pim2−/−, and Pim3−/− mice. We first performed in vitro BM CFU assay and compared CFUs in Pim1−/−, Pim2−/−, Pim3−/−, and WT mice. As shown in Fig. 2A, the BM CFUs-GEMM in Pim1−/− mice was reduced by ~50% in comparison to WT BM cells. In contrast, no difference was observed in BM CFUs-GEMM between Pim2−/− or Pim3−/− and WT mice.
Figure 2. Decreased hematopoiesis in Pim1−/− mice, but not in Pim2−/− or Pim3−/− mice.
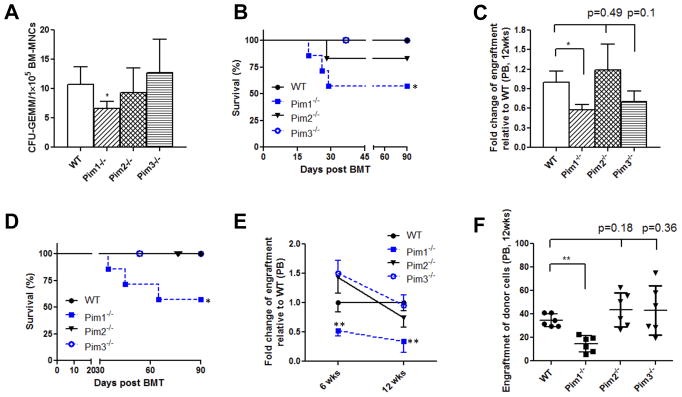
(A) Lower frequency of BM CFU-GEMMs in Pim1−/− mice, but not in Pim2−/− or Pim3−/− mice. BM cells were plated in MethocultR GF 3434 and CFU-GEMMs were determined at 12–14 days (*p<0.05, data are representative of 3 independent experiments). (B) Reduced animal survival in primary non-competitive BM transplant mice receiving Pim1−/− BM cells. Lethally irradiated female FVB/J mice were injected via tail vein with BM cells obtained from male WT, Pim1−/−, Pim2−/−, or Pim3−/− mice (5×105 cells/recipient mouse). Animal survival was monitored daily (n=7/group, *p<0.05). (C) Reduced donor cell engraftment in primary non-competitive transplant mice receiving Pim1−/− BM cells. Peripheral blood samples from animals in Fig. 2B were analyzed for donor cell engraftment as described (n= 4 for Pim1−/− mice and n= 6 for other genotypes, *p<0.05). (D) Reduced animal survival in secondary BM transplant mice receiving Pim1−/− BM cells. The primary transplant recipient mice as described in Fig. 2C were sacrificed at 4 months post transplant and BM cells harvested and injected into lethally irradiated female FVB/J mice (1×107/recipient mouse). Animal survival was monitored daily (n=6/group). (E) Reduced donor cell engraftment in secondary transplant mice receiving Pim1−/− BM cells. Secondary transplant recipient mice were sacrificed at 4 months post transplant and male donor cell engraftment in BM was analyzed (n=6/group, n=3 for Pim1−/− in Fig. 2E, *p<0.05, **p<0.01). (F) Reduced donor engraftment in competitive BM transplant mice receiving Pim1−/− BM cells. Competitive BM transplantation was performed as described using WT, Pim1−/−, Pim2−/−, or Pim3−/− BM cells. Male donor engraftment in peripheral blood at 12 weeks post transplant was analyzed (n=7, **p<0.01).
We next compared the ability of BM cells from Pim1−/−, Pim2−/−, Pim3−/− mice to reconstitute hematopoiesis in primary transplant recipient mice. In this series of BM transplantation experiments, we injected 5×105 male cells/recipient to ensure that enough control mice survive the transplantation. As shown in Fig. 2B, the lethally irradiated mice transplanted with WT BM cells all survived throughout the experiments. In contrast, around 50% of the lethally irradiated mice receiving Pim1−/− BM cells died within 6 weeks after transplant. The survival of transplant mice receiving Pim2−/− or Pim3−/− BM cells was comparable to those receiving WT BM cells. We also measured peripheral blood donor male cell engraftment in survived recipients. Male donor cell engraftment in Pim1−/− BM transplanted recipients was significantly lower than that of WT BM transplant recipient mice. Pim2−/− and Pim3−/− BM cells showed no impairment in donor cell reconstitution (Fig. 2C).
To further define the role of Pim kinases on HSC self-renewal and reconstitution, we performed secondary BM transplantation (Fig. 2D–2E). We harvested BM cells from primary BM transplanted mice (Fig. 2C) at 4 months post transplantation, and injected equal number of total BM cells into lethally irradiated female recipients. Again, ~50% of secondary transplant mice receiving Pim1−/− BM cells died within 10 weeks post transplantation (Fig. 2D). None of WT, Pim2−/− or Pim3−/− secondary transplanted mice died. The male donor cell engraftment in the peripheral blood of Pim1−/− secondary transplant recipient mice was also significantly lower than that in Pim2−/−, Pim3−/− or WT recipients at both 6 weeks and 12 weeks post transplantation (Fig. 2E, p<0.01). We further performed competitive transplant assays, and found that Pim1−/− BM cells were less competitive and less efficient in repopulating the lethally irradiated hosts compared to WT, Pim2−/− or Pim3−/− BM cells (Fig. 2F). These results demonstrated that the self-renewal potential and the long-term repopulating capacity of HSCs were impaired in Pim1−/− mice while preserved in Pim2−/− and Pim3−/− mice.
Reduced frequency of Long-term HSCs in Pim1−/− mice
We performed competitive repopulating unit (CRU) assays with limiting dilutions of sorted LSKCD34 cells to quantify the frequency of active HSCs in Pim1−/− mice. Various doses of sorted male LSKCD34 cells (15, 45 and 150 cells) from Pim1−/− mice or WT controls were mixed with competitor female BM cells, and transplanted into lethally irradiated female FVB/J mice. Peripheral blood samples were collected at 6 weeks and 4 months post transplantation, and analyzed for donor engraftment. Scatter plots demonstrated a dramatically reduced male donor engraftment in recipients transplanted with Pim1−/− LSKCD34 cells (Fig. 3A). Poisson statistical analysis at 6 weeks post transplant showed a 7-fold reduction in CRUs in Pim1−/− KO mice (CRUs: 1 in 19.1 in WT mice; 1 in 146.5 in Pim1−/− mice) (Fig. S5A–C). This reduction in CRUs in Pim1−/− KO mice increased to 28-fold when estimated at 4 months post transplantation (CRUs: 1 in 11.7 in WT mice and 1 in 328 in Pim1−/− mice) (Fig. 3B and Fig. S5D, n= 48).
Figure 3. Reduced competitive repopulating units (CRU) in Pim1−/− mice.
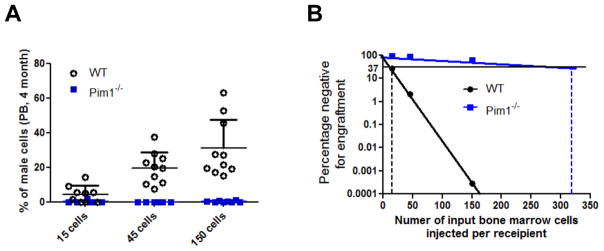
Scatter plot (A) and limiting dilution plot (B) were shown. Various doses of male LSK+CD34− HSCs (15, 45, and 150 cells/recipient mouse) were mixed with 1.5×105 WT female FVB BM cells and transplanted into lethally irradiated female FVB mice. Male donor engraftment in peripheral blood was estimated at 4 months post transplant. Poisson statistical analysis of limiting-dilution was performed to allow estimation of CRUs in each group (n = 28 for WT and n= 20 for Pim1−/− mice). The plot shows the percentage of recipient mice containing less than 1% donor male cells in the peripheral blood at 4 months after transplantation versus the number of cells injected per mouse. The horizontal line indicates the point at which 37% of the transplanted mice are non-engrafted in each group; the CRU frequency is estimated at the point where 37% of the mice are non-engrafted, by conventional Poisson statistical methods and limiting-dilution analysis. The vertical dashed lines highlight the CRU frequencies in Pim1−/− and WT mice.
Minimal effects of Pim1 kinase on CXCR4 expression and HSC homing to BM
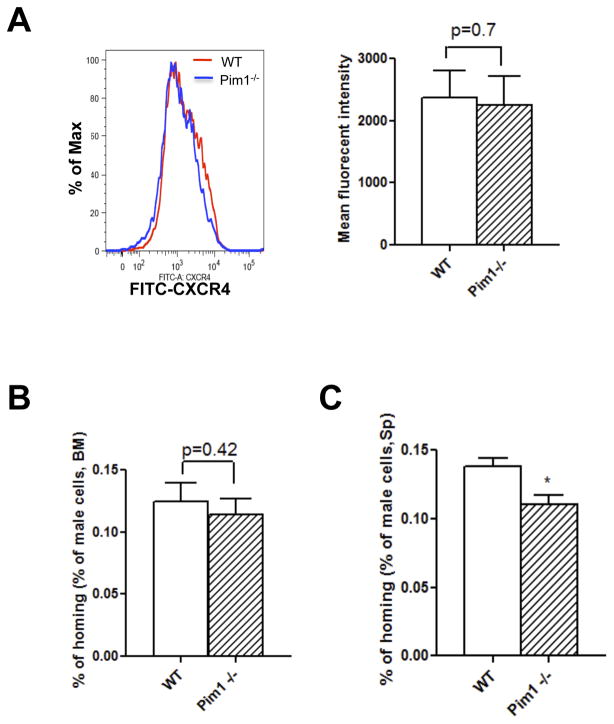
Previous study suggested that Pim1−/− mice were defective in CXCR4 expression and in HSC homing [24]. We first measured CXCR4 expression in Lin− BM cells. We did not observe differences in the percentage of CXCR4 positive cells, nor in the CXCR4-FITC fluorescence intensity between WT and Pim1−/− mice (Fig. 4A). We also performed in vivo HSC homing assays with sorted Lin−Sca-l+ BM cells from male WT or Pim1−/− mice. The homing of male Lin−Sca-l+ BM cells into BM and spleen was determined at 12 hrs post transplantation using PCR analysis. The homing efficiency at 12 hrs post transplant was estimated at ~0.13% in WT BM cells, which is comparable to that reported by other groups [33]. We did not observe any differences in HSC homing to BM between WT and Pim1−/− Lin−Sca-1+ cells (Fig. 4B). There was a slight reduction (~20%) in homing to the spleen with Pim1−/− HSCs compared to WT HSCs (Fig. 4C). These data suggested that Pim1 has minimal effects on BM CXCR4 level and on HSC homing.
Figure 4. Minimal effects of Pim1 kinase on BM CXCR4 expression and HSC homing.
(A) Comparable surface CXCR4 level between WT and Pim1−/− BM cells. Lin− BM cells obtained from WT or Pim1−/− mice were stained with CXCR4 antibody and analyzed by flow cytometry. Representative CXCR4 histogram (left panel) and mean fluorescence intensity (right panel) were shown (n=5). (B) and (C) Minimal effect of Pim1 kinase on HSC homing to bone marrow and spleen. 60,000 sorted Lin−Sca-1+ cells obtained from male WT or Pim1−/− mice were injected intravenously into lethally irradiated female mice. The recipient mice were sacrificed at 12 hours post transplantation, and BM and spleen were isolated. The percentage of male donor HSC homing to BM (B) and spleen (C) was calculated (n=3, *p<0.05).
Increased HSC proliferation and cell survival by Pim1 kinase
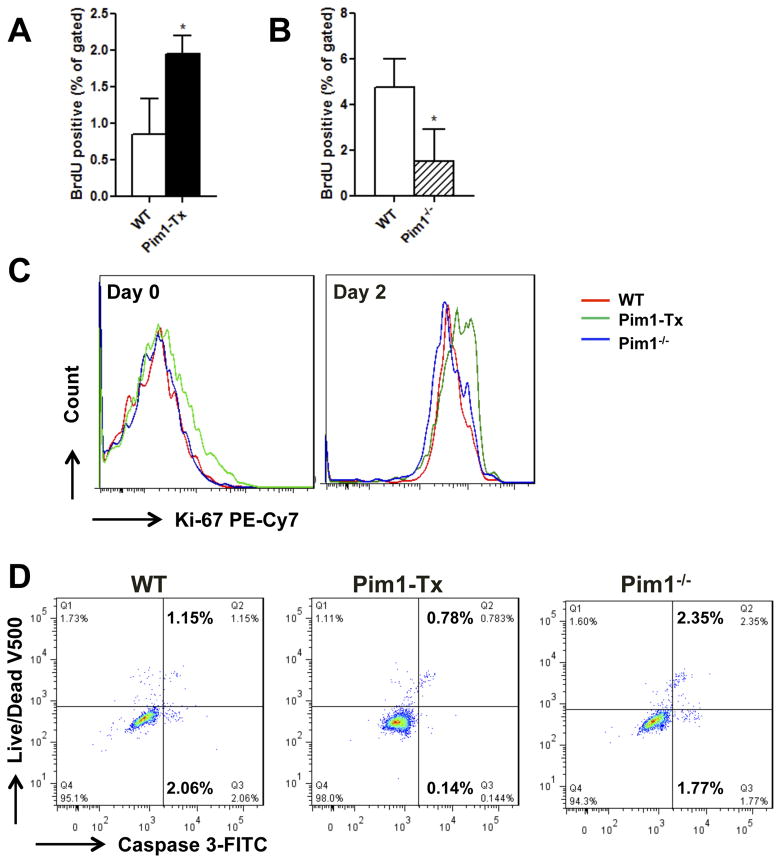
We performed additional studies to understand the mechanisms through which Pim1 kinase promotes hematopoiesis. We reasoned that Pim1 kinase might regulate HSC cell proliferation and survival. To test this hypothesis, we first quantified in vivo BrdU incorporation in long-term HSCs (LT-HSCs defined as Lin−Sca-1+c-Kit+CD34−CD135−), Short-term HSCs (ST-HSCs defined as Lin−Sca-1+c-Kit+CD34+CD135−) and multi-potent progenitors (MPPs defined as Lin−Sca-1+c-Kit+CD34+CD135+) [27, 36, 37] (Fig. S6). Compared to WT littermates, LT-HSCs from Pim1-Tx mice had significantly higher in vivo BrdU labeling (Fig. 5A). Interestingly, no differences in BrdU labeling were observed in ST-HSCs or MPPs between WT and Pim1-Tx mice (data not shown). We also measured BrdU labeling in Pim1−/− mice. Consistent with our hypothesis, BrdU incorporation in LT-HSCs was significantly reduced in Pim1−/− mice compared to that in WT littermates (Fig. 5B).
Figure 5. Enhanced HSC proliferation and cell survival in Pim1-Tx mice.
(A) Increased in vivo BrdU incorporation in LT-HSCs of Pim1-Tx mice. WT or Pim1-Tx mice were injected with 2 doses of BrdU. The mice were then sacrificed and BM cells harvested and labeled with antibodies as described in Methods and Materials. BrdU-positive cells in the LT-HSC (LSK+CD34−CD135−) population were analyzed (n=3, *P <0.05). (B) Decreased in vivo BrdU incorporation in LT-HSCs of Pim1−/− mice (p<0.05). (C) Increased Ki67 labeling in Pim1-Tx LSK cells. Sorted LSK+ cells from WT, Pim1-Tx or Pim1−/− mice were cultured in StemSpan media supplemented with SCF, TPO-1 and Flt3 (100 ng/ml for each) for 2 days. The cells were then stained with PE-Cy7-conjugated Ki67 antibody, Aqua Live/dead fixable dye, FITC-conjuagated Caspase 3 antibody, and analyzed with flow cytometry. Representative Ki67 labelings on freshly isolated LSK cells (left panel) and LSK cells that have been cultured for 2 days (right panel) were shown. (D) Increased cell survival in Pim1-Tx LSK cells. Sorted LSK+ cells from WT, Pim1-Tx or Pim1−/− mice were cultured for 2 days and stained with Aqua Live/dead fixable dye and Caspase 3 antibody as described (data are representative of 3 independent experiments).
Ki67 is a commonly used marker for cell proliferation. We thus measured Ki67 labeling in freshly isolated LSK cells and LSK cells that had been cultured ex vivo for 2 days in the presence of SCF, TPO and Flt3 (Fig. 5C). Compared to WT or Pim1−/− mice, both freshly isolated and cultured LSK cells from Pim1-Tx mice showed an increase in Ki67 labeling as demonstrated by the shift towards right in the Ki67 fluorescence intensity. Our results support an important role of Pim1 kinase in promoting HSC cell proliferation.
To determine the role of Pim1 kinase in HSC cell survival, LSK cells cultured ex vivo were stained with Live/Dead dye and Caspase 3 antibody. As shown in Fig. 5D, in comparison to WT mice, LSKs from Pim1-Tx mice showed an reduction in both Live/Dead dye+Caspase 3+ cell population and Live/Dead dye−Caspase 3+ cell population. Pim−/− LSK cells showed an increase in cell death compared to WT LSK cells (Fig. 5D). Similarly, freshly isolated LSK cells from Pim1-Tx mice showed a ~2-fold reduction in apoptotic cell death (Fig. S8B).
Given the increase in cell proliferation and decrease in apoptotic cell death observed with Pim1-Tx LSK cells, one would expect Pim1-Tx LSK cells should expand more rapidly and Pim−/− LSK cells slower in ex vivo cultures. Indeed, at day 4 of ex vivo culture, the LSK cells from Pim1-Tx, WT and Pim1−/− mice expanded at 19.3±0.6 fold, 14.3±0.7 fold and 4.1±0.3 fold, respectively (Fig. S7). These data suggest that the hematopoietic phenotypes we observed in Pim1-Tx and Pim1−/− mice are likely related to the effects of Pim1 kinase on HSC cell proliferation and survival.
Hematopoiesis-associated genes affected by Pim1 kinase
To identify potential downstream target genes of Pim1 kinase in HSCs, we isolated mRNAs from purified BM LSK cells of Pim1-Tx, WT, Pim1−/−, and Pim TKO mice and performed the Hematopoietic Stem Cells and Hematopoiesis PCR Array (Qiagen). This commercially available PCR array allows for simultaneous analysis of a total of 84 genes important in hematopoiesis. We have performed 3 independent sets of experiments and were able to reliably amplify around 70 genes. Six genes showed differential expression pattern in Pim1-Tx vs. Pim1−/− and Pim TKO LSK cells. We then performed confirmatory RT-PCR on these six genes. We found that the expression levels of Lef1 and Pax5 were upregulated in Pim1−/− and Pim TKO LSK cells. Expression of Gata1 was down-regulated by ~2-fold in Pim1−/− and Pim TKO LSK cells (Fig. 6).
Figure 6. Hematopoiesis-associated genes affected by Pim1 deletion.
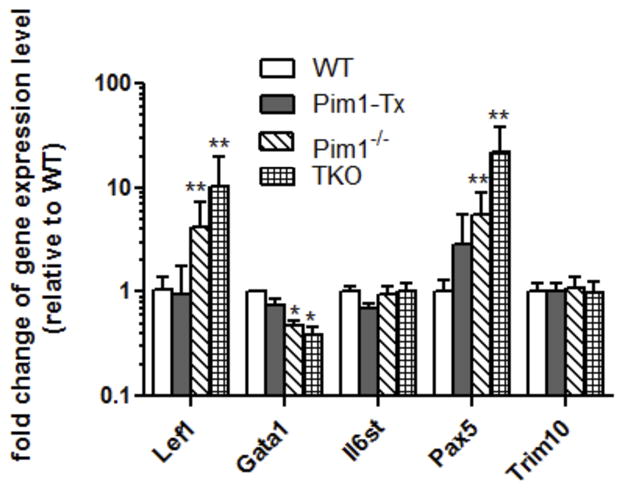
BM LSK cells were isolated from the WT, Pim1-Tx, Pim1−/− and Pim TKO mice (3–6 mice/group) and RNA isolated and quantitative RT-PCR array performed. (n=3. *p<0.05, **p<0.01).
Different response pattern to cytokines/growth factors in Pim1 versus Pim2 and Pim3
To further validate and understand the differences of Pim1 versus Pim2 and Pim3 in hematopoiesis, we measured their gene expression level in response to several cytokines/growth factors that are important in HSC proliferation and expansion. We cultured Lin− BM cells with SCF, IL-3, IL-6, TPO, Flt3 alone or in combination for 24 hrs. As shown in Fig. S9, Pim1 mRNA was upregulated by IL-3, IL-6 and TPO. In contrast, Pim2 and Pim3 mRNA expression was not changed with these cytokines. These data suggested that Pim1 might be associated with signal pathway(s) that is different from Pim2 or Pim3, thus having different roles in hematopoiesis.
DISCUSSION
In the current study, we utilized transgenic and knockout mouse models to investigate the roles of Pim1 kinase in hematopoiesis. We demonstrated that Pim1 kinase regulates the number, proliferation, self-renewal, and repopulating capacity of murine HSCs. Our study was the first to unequivocally demonstrate a role for Pim1 kinase in HSC regulation. Pim1 transgenic mice driven by Eμ-immunoglobulin promoter and Pim knockout mice were previously reported by others [25, 34]. The use of Eμ immunoglobulin enhancer likely restricts the Pim1 transgene expression to lymphoid lineage. There were no data described in those publications reporting any changes in HSCs in those mice. It is unknown if the authors had examined the HSC population and observed no differences or if the HSC population was simply not investigated in those mice. In our Pim1-Tx mice, we used vav-hematopoietic regulatory elements, which allow for the enforced expression of Pim1 kinase in HSCs and the evaluation of its roles in hematopoiesis.
In contrast to the overlapping potentials between Pim1, Pim2 and Pim3 in lymphomagenesis, we find that Pim1 kinase has a unique role in hematopoiesis. The function of Pim1 in HSCs is distinct from Pim2 or Pim3 and cannot be compensated by other isoforms. Our data also show that IL-3, IL-6 and SCF up-regulate gene expression of Pim1, but not Pim2 or Pim3 in HSCs. These findings suggest that Pim1 in HSCs is likely associated with unique downstream pathways different from those of Pim2 or Pim3.
The hematopoietic phenotypes were much more pronounced in Pim1−/− mice than in Pim1-Tx mice. For instance, the frequency of HSCs was reduced by more than 28-fold in Pim1−/− mice. In contrast, the changes were subtle in Pim1-Tx mice; the numbers of LSK cells, CAFCs, and the HSC frequency were only increased by 0.5–2 fold in Pim1-Tx mice. The modest hematopoietic phenotype seen in our Pim1-Tx mice may in part be attributed to the relatively low level (~0.5–2 fold) of overexpression of Pim1. We used human PIM1 cDNA instead of genomic DNA for our transgene vector construction, which may lead to relatively low level of transgene expression. Another possibility is that the vav-regulatory elements [38] does not permit overexpression of hPIM1 transgene in osteoblasts or mesenchymal stem cells (stromal cells) that constitute the marrow niche microenvironment and support hematopioesis [39], thus diminishing the overall contribution of Pim1 in hematopoiesis.
Contradictory to the observation by Grundler, et al. [24], we did not find differences in CXCR4 level between WT and Pim1−/− BM cells in our mouse model system. No defects in HSC homing to bone marrow were observed with Pim1−/− BM cells. A slight decrease in Pim1−/− HSC homing to spleen was noted, but this decrease was minimal and could not be solely attributed to the dramatic impairment in hematopoiesis we observed with Pim1−/− mice. The reasons for the discrepancy between our findings and those previously reported are unclear, but it could be related to the different methodology used to measure HSC homing.
We found that Pim1 kinase plays an important role in HSC cell proliferation and survival. Our findings are consistent with previous observations in cancer cells. Pim1 can promote cell-cycle progression and cell proliferation by phosphorylating and activating cell cycle regulators (i.e., phosphatases Cdc25A and Cdc25C) [40, 41]. Pim1 can reduce apoptosis by phosphorylating and inhibiting the proapoptotic BH3 protein BAD [42, 43]. More recently, Pim kinase was found to be important in the regulation of energy metabolism and cell growth [44]. It is very likely that Pim1 exerts diverse biological functions on HSCs. The proliferative effects of Pim1 kinase on HSCs can expand HSC population as we demonstrated in Pim1-Tx mice. On the other hand, this proliferative effect may cause stem cell exhaustion as we observed in Pim1-Tx secondary transplant mice (Fig. 1H) and the diminishing HSC frequency with time as we saw in our Pim1-Tx mice (Fig. S4).
We identified several potential genes affected by the deletion of Pim1 kinase in HSCs, including Lef-1, Pax5 and Gata1. Lef-1 is a transcription mediator critical in Wnt signaling. Lef-1 regulates cell cycle and growth through genes such as cyclin D1 and c-myc [45], and plays a major role in T-cell development, B-cell proliferation, and pro-B cell apoptosis [46]. Lef-1 gene expression was up-regulated in Pim1−/− LSK cells and in Pim TKO LSK cells (Fig. 6). It remains to be determined if this up-regulation of Lef-1 in Pim1-deleted LSK cells reflects a compensatory mechanism of HSC or if Lef-1 is negatively regulated by Pim1 kinase. Pax5 is a transcription factor important for B cell development [47]. Overexpression of Pax5 was found to cause marked inhibition of myelopoiesis [48]. Gata1 is a key regulator of erythroid and megakaryocytic homeostasis [49]. Pim1−/− mice and Pim TKO mice exhibit erythrocyte microcytosis and thrombocytopenia ([50] and our unpublished data). It is unclear if the erythrocyte microcytosis and thrombocytopenia seen in these mice are caused by the down-regulation of Gata1 gene. Additional work is needed to more clearly define the mechanism by which Pim1 kinase affects these genes.
In conclusion, our study demonstrated a novel, previously unknown role of Pim kinases in the regulation of HSCs. In addition, our findings support the notion that oncogenes are important not only in tumorigenesis, but also in normal cell development. Identifying the roles of Pim1 kinase in hematopoiesis is an important first step in developing safe and effective therapeutic agents for the treatment of cancers.
Supplementary Material
(A) The construct used for pronuclear injection. The entire coding DNA sequence of human PIM1 was cloned into vav-regulatory elements and SV40 sequences. (B) Human PIM1 mRNA expression. Human PIM1 mRNA expression was expressed in all hematopoietic tissue but not in non-hematopoietic tissue. Spleen, thymus, bone marrow, and kidney tissues were harvested from WT control mice and Pim1-Tx mice. Messenger RNA was prepared and qRT-PCR performed. The samples were then run on 2% agarose gel. (C) Pim kinase protein expression. Pim1 was over-expressed in the splenocytes of Pim1-Tx mice. Cell lysates were prepared from the splenocytes of Pim1-Tx and WT littermate and were subject to immunoblot analysis using 19F7 Pim1 antibody, which recognizes both human- and mouse-origin of Pim1.
(A) Increased spleen weight and size in Pim1-Tx mice. Pim1-Tx mice and WT littermates (1.5 yrs old) were sacrificed and spleen was harvested and weighed. Left panel: three representative mice per group were shown (scale bar indicates 1cm). Right panel: average spleen weight in Pim1-Tx mice and WT littermates (n=11, *p<0.05). (B) Increased LSK HSPCs in spleen in Pim1-Tx mice. Red-blood cell- depleted splenocytes from Pim1-Tx mice and WT littermates were stained with Lineage-specific, Scal-1, and c-Kit antibodies and analyzed on flow cytometry. Total number of LSK HSPCs (calculated by total BM mononuclear cells × %LSK) was shown (n=12, *p < 0.05).
(A) Peripheral blood cell subset analysis. Peripheral blood cells were stained with Gr-1, B220, and CD3 antibody. The absolute numbers of these cell subsets were calculated as described in the Materials and Methods (n=8). (B) Bone marrow morphology. Bone were harvested from Pim1-Tx mice or wild-type control mice, fixed, sectioned, stained with H/E and examined under microscope (representative of 5 mice in each group, scale bar indicates 20μm). Pim1-Tx mice showed no evidences of myelodysplastic/myeloproliferative disorder.
(A) CRUs measured at 6 weeks post transplant. (B) CRUs measured at 4 months post transplant. Various doses of male LSK+CD34− HSCs (15, 45, and 150 cells/recipient mouse) were mixed with 1.5×105 WT female FVB BM cells and transplanted into lethally irradiated female FVB mice. Male donor engraftment was estimated as described at 6 weeks (A: peripheral blood samples; n=29 for WT and n=27 for Pim1-Tx) and 4 months (B: BM samples; n=29 for WT and n=24 for Pim1-Tx) post transplant. Poisson statistical analysis of limiting-dilution was performed to allow for estimation of CRUs in each group. The plot shows the percentage of recipient mice containing less than 1% donor male cells at 6 weeks and at 4 months post transplantation versus the number of cells injected per mouse.
Limiting-dilution assay was performed as described using Pim1−/− and WT mice. Panels A-C are blood samples collected at 6 weeks post transplant (A: scatter plot; B: CRU frequencies; C: limiting dilution plot). Panel D is BM samples collected at 4 months post transplant and shown the CRU frequency in Pim1−/− and WT mice.
BM Lin−cells were labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for the long-term HSCs (LT-HSCs, Lin−Sca-1+c-Kit+CD34−CD135−), Short-term HSCs (ST-HSCs, Lin−Sca-1+c-Kit+CD34+CD135−) and multipotent progenitors (MPPs, Lin−Sca-1+c-Kit+CD34+CD135+). BrdU incorporation was analyzed for LT-HSCs.
Sorted LSK cells (6,000/well) from WT, Pim1-Tx and Pim1−/− mice were cultured in StemSpan media supplemented with SCF, TPO-1 and Flt3 (100 ng/ml for each) for 4 days. Total cell numbers were counted and averaged from 3–4 wells/group (data are representative of 4 independent experiments).
BM Lin− cells were enriched by Lineage cell depletion Kit (Miltenyi Biotec) from WT, Pim1-Tx and Pim1−/− mice. The cells were then stained with biotin-labeled lineage specific antibodies, PE-conjugated Scal-1, and APC-H7-conjugated c-Kit followed by streptavidin-Brilliant-violet-605 and Aqua Live/Dead dye. The cells were then fixed and subsequently stained with PE-Cy7-conjugated Ki-67 antibody and FITC-conjugated Caspase 3 antibody. All the cells were gated on LSK population.
BM Lin− cells isolated from C56B/6 mice were cultured in DMEM medium supplemented with 20% heat-inactive FBS and 100 ng/ml of SCF, IL-3, IL-6, TPO, Flt3 alone or in combination for 24 hrs. Cells were harvested and mRNA isolated for RT-PCR analysis of Pim1 (A), Pim2 (B), and Pim3 (C) gene expression.
Acknowledgments
We thank Dr. Robert Stuart, Director of our Hematological Malignancies and BMT program for valuable discussion and critical reading of the manuscript. We thank Dr. Jerry M. Adams, Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia for the kind gift of vav-hCD4 vector; and Dr. Peter D. Aplan, Genetics Branch, CCR, NCI, Bethesda MD for advice on developing the construct for generating transgenic mice. We thank Richard Peppler and Haiqun Zeng at the HCC Flow Cytometry Core for performing flow cytometry analysis. This work is supported by MUSC Hollings Cancer Center Startup Fund, Hollings Cancer Center ACS IRG (YK), ASCO Conquer Cancer Foundation Career Development Award (YK), NIH 1K08HL 103780-01A1 (YK), and NIH 3P30CA138313-01S3. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agents.
Footnotes
Author contributions:
Y-W.L generated the vav-Pim1-Tx mice. N.A. performed all experiments. S.M. genotyped the mice. J.N.K assisted in flow cytometry experiments. Y.W provided stromal cells for CAFC experiments. Z.L. and A.S.K participated in research design. N.A. and Y.K. designed research and wrote the paper.
DISCLOSURE
The authors declare no competing financial interests.
References
- 1.Zhang XB, Beard BC, Beebe K, Storer B, Humphries RK, Kiem HP. Differential effects of HOXB4 on nonhuman primate short- and long-term repopulating cells. PLoS Med. 2006;3 (5):e173. doi: 10.1371/journal.pmed.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, Shao L, Bai H, Wang ZZ, Wu WS. Slug deficiency enhances self-renewal of hematopoietic stem cells during hematopoietic regeneration. Blood. 2010;115(9):1709–1717. doi: 10.1182/blood-2009-07-232934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell C, Risueno RM, Salati S, Guezguez B, Bhatia M. Signal control of hematopoietic stem cell fate: Wnt, Notch, and Hedgehog as the usual suspects. Curr Opin Hematol. 2008;15 (4):319–325. doi: 10.1097/MOH.0b013e328303b9df. [DOI] [PubMed] [Google Scholar]
- 4.Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–1015. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, van Wezenbeek P, Melief C, Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 6.Breuer ML, Cuypers HT, Berns A. Evidence for the involvement of pim-2, a new common proviral insertion site, in progression of lymphomas. EMBO J. 1989;8(3):743–748. doi: 10.1002/j.1460-2075.1989.tb03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32(1):153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 8.Allen JD, Verhoeven E, Domen J, van der Valk M, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15(10):1133–1141. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Bhattacharya N, Weaver M, Petersen K, Meyer M, Gapter L, Magnuson NS. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci. 2001;2(3):167–179. [PubMed] [Google Scholar]
- 10.Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci U S A. 1989;86(22):8857–8861. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YW, Beharry ZM, Hill EG, Song JH, Wang W, Xia Z, Zhang Z, Aplan PD, Aster JC, Smith CD, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010;115(4):824–833. doi: 10.1182/blood-2009-07-233445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen WW, Chan DC, Donald C, Lilly MB, Kraft AS. Pim family kinases enhance tumor growth of prostate cancer cells. Mol Cancer Res. 2005;3(8):443–451. doi: 10.1158/1541-7786.MCR-05-0007. [DOI] [PubMed] [Google Scholar]
- 13.Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114(19):4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popivanova BK, Li YY, Zheng H, Omura K, Fujii C, Tsuneyama K, Mukaida N. Proto-oncogene, Pim-3 with serine/threonine kinase activity, is aberrantly expressed in human colon cancer cells and can prevent Bad-mediated apoptosis. Cancer Sci. 2007;98(3):321–328. doi: 10.1111/j.1349-7006.2007.00390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66(13):6741–6747. doi: 10.1158/0008-5472.CAN-05-4272. [DOI] [PubMed] [Google Scholar]
- 16.Alvarado Y, Giles FJ, Swords RT. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5(1):81–96. doi: 10.1586/ehm.11.69. [DOI] [PubMed] [Google Scholar]
- 17.Domen J, van der Lugt NM, Acton D, Laird PW, Linders K, Berns A. Pim-1 levels determine the size of early B lymphoid compartments in bone marrow. J Exp Med. 1993;178(5):1665–1673. doi: 10.1084/jem.178.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aksoy I, Sakabedoyan C, Bourillot PY, Malashicheva AB, Mancip J, Knoblauch K, Afanassieff M, Savatier P. Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases Pim-1 and Pim-3. Stem Cells. 2007;25(12):2996–3004. doi: 10.1634/stemcells.2007-0066. [DOI] [PubMed] [Google Scholar]
- 19.Fischer KM, Cottage CT, Wu W, Din S, Gude NA, Avitabile D, Quijada P, Collins BL, Fransioli J, Sussman MA. Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing Pim-1 kinase. Circulation. 2009;120(21):2077–2087. doi: 10.1161/CIRCULATIONAHA.109.884403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu YL, Passegue E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109(11):4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pircher TJ, Zhao S, Geiger JN, Joneja B, Wojchowski DM. Pim-1 kinase protects hematopoietic FDC cells from genotoxin-induced death. Oncogene. 2000;19(32):3684–3692. doi: 10.1038/sj.onc.1203684. [DOI] [PubMed] [Google Scholar]
- 22.Nosaka T, Kitamura T. Pim-1 expression is sufficient to induce cytokine independence in murine hematopoietic cells, but is dispensable for BCR-ABL-mediated transformation. Exp Hematol. 2002;30(7):697–702. doi: 10.1016/s0301-472x(02)00808-1. [DOI] [PubMed] [Google Scholar]
- 23.Lilly M, Kraft A. Enforced expression of the Mr 33,000 Pim-1 kinase enhances factor-independent survival and inhibits apoptosis in murine myeloid cells. Cancer Res. 1997;57 (23):5348–5355. [PubMed] [Google Scholar]
- 24.Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, Pogacic V, Villa A, Ehret S, Berridge G, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med. 2009;206(9):1957–1970. doi: 10.1084/jem.20082074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24(13):6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Os RP, Dethmers-Ausema B, de Haan G. In vitro assays for cobblestone area-forming cells, LTC-IC, and CFU-C. Methods Mol Biol. 2008;430:143–157. doi: 10.1007/978-1-59745-182-6_10. [DOI] [PubMed] [Google Scholar]
- 27.Merchant A, Joseph G, Wang Q, Brennan S, Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood. 2010;115(12):2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson SM, Torrice CD, Bell JF, Monahan KB, Jiang Q, Wang Y, Ramsey MR, Jin J, Wong KK, Su L, et al. Mitigation of hematologic radiation toxicity in mice through pharmacological quiescence induced by CDK4/6 inhibition. J Clin Invest. 2010;120(7):2528–2536. doi: 10.1172/JCI41402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himburg HA, Muramoto GG, Daher P, Meadows SK, Russell JL, Doan P, Chi JT, Salter AB, Lento WE, Reya T, et al. Pleiotrophin regulates the expansion and regeneration of hematopoietic stem cells. Nat Med. 2010;16(4):475–482. doi: 10.1038/nm.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 31.An N, Kang Y. Using quantitative real-time PCR to determine donor cell engraftment in a competitive murine bone marrow transplantation model. Journal of Visualized Experiments. 2013 doi: 10.3791/50193. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrne P, Huang W, Wallace VM, Shean MK, Zhang Z, Zhong Q, Theodossiou C, Blakesley H, Kolls JK, Schwarzenberger P. Chimerism analysis in sex-mismatched murine transplantation using quantitative real-time PCR. Biotechniques. 2002;32(2):279–280. 282–274, 286. [PubMed] [Google Scholar]
- 33.Colvin GA, Lambert JF, Dooner MS, Cerny J, Quesenberry PJ. Murine allogeneic in vivo stem cell homing. J Cell Physiol. 2007;211(2):386–391. doi: 10.1002/jcp.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56(4):673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim WI, Wiesner SM, Largaespada DA. Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol. 2007;35(8):1231–1239. doi: 10.1016/j.exphem.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15(4):659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105(7):2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 38.Ogilvy S, Elefanty AG, Visvader J, Bath ML, Harris AW, Adams JM. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood. 1998;91(2):419–430. [PubMed] [Google Scholar]
- 39.Shen Y, Nilsson SK. Bone, microenvironment and hematopoiesis. Curr Opin Hematol. 2012;19 (4):250–255. doi: 10.1097/MOH.0b013e328353c714. [DOI] [PubMed] [Google Scholar]
- 40.Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274(26):18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 41.Bachmann M, Kosan C, Xing PX, Montenarh M, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38(3):430–443. doi: 10.1016/j.biocel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571(1–3):43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 43.Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beharry Z, Mahajan S, Zemskova M, Lin YW, Tholanikunnel BG, Xia Z, Smith CD, Kraft AS. The Pim protein kinases regulate energy metabolism and cell growth. Proc Natl Acad Sci U S A. 2011;108(2):528–533. doi: 10.1073/pnas.1013214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96(10):5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reya T, O’Riordan M, Okamura R, Devaney E, Willert K, Nusse R, Grosschedl R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13(1):15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 47.Schebesta M, Heavey B, Busslinger M. Transcriptional control of B-cell development. Curr Opin Immunol. 2002;14(2):216–223. doi: 10.1016/s0952-7915(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 48.Sekine R, Kitamura T, Tsuji T, Tojo A. Efficient retroviral transduction of human B-lymphoid and myeloid progenitors: marked inhibition of their growth by the Pax5 transgene. Int J Hematol. 2008;87(4):351–362. doi: 10.1007/s12185-008-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu R, Yamamoto M. Contribution of GATA1 dysfunction to multi-step leukemogenesis. Cancer Sci. 2012;103(12):2039–2044. doi: 10.1111/cas.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laird PW, van der Lugt NM, Clarke A, Domen J, Linders K, McWhir J, Berns A, Hooper M. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21(20):4750–4755. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The construct used for pronuclear injection. The entire coding DNA sequence of human PIM1 was cloned into vav-regulatory elements and SV40 sequences. (B) Human PIM1 mRNA expression. Human PIM1 mRNA expression was expressed in all hematopoietic tissue but not in non-hematopoietic tissue. Spleen, thymus, bone marrow, and kidney tissues were harvested from WT control mice and Pim1-Tx mice. Messenger RNA was prepared and qRT-PCR performed. The samples were then run on 2% agarose gel. (C) Pim kinase protein expression. Pim1 was over-expressed in the splenocytes of Pim1-Tx mice. Cell lysates were prepared from the splenocytes of Pim1-Tx and WT littermate and were subject to immunoblot analysis using 19F7 Pim1 antibody, which recognizes both human- and mouse-origin of Pim1.
(A) Increased spleen weight and size in Pim1-Tx mice. Pim1-Tx mice and WT littermates (1.5 yrs old) were sacrificed and spleen was harvested and weighed. Left panel: three representative mice per group were shown (scale bar indicates 1cm). Right panel: average spleen weight in Pim1-Tx mice and WT littermates (n=11, *p<0.05). (B) Increased LSK HSPCs in spleen in Pim1-Tx mice. Red-blood cell- depleted splenocytes from Pim1-Tx mice and WT littermates were stained with Lineage-specific, Scal-1, and c-Kit antibodies and analyzed on flow cytometry. Total number of LSK HSPCs (calculated by total BM mononuclear cells × %LSK) was shown (n=12, *p < 0.05).
(A) Peripheral blood cell subset analysis. Peripheral blood cells were stained with Gr-1, B220, and CD3 antibody. The absolute numbers of these cell subsets were calculated as described in the Materials and Methods (n=8). (B) Bone marrow morphology. Bone were harvested from Pim1-Tx mice or wild-type control mice, fixed, sectioned, stained with H/E and examined under microscope (representative of 5 mice in each group, scale bar indicates 20μm). Pim1-Tx mice showed no evidences of myelodysplastic/myeloproliferative disorder.
(A) CRUs measured at 6 weeks post transplant. (B) CRUs measured at 4 months post transplant. Various doses of male LSK+CD34− HSCs (15, 45, and 150 cells/recipient mouse) were mixed with 1.5×105 WT female FVB BM cells and transplanted into lethally irradiated female FVB mice. Male donor engraftment was estimated as described at 6 weeks (A: peripheral blood samples; n=29 for WT and n=27 for Pim1-Tx) and 4 months (B: BM samples; n=29 for WT and n=24 for Pim1-Tx) post transplant. Poisson statistical analysis of limiting-dilution was performed to allow for estimation of CRUs in each group. The plot shows the percentage of recipient mice containing less than 1% donor male cells at 6 weeks and at 4 months post transplantation versus the number of cells injected per mouse.
Limiting-dilution assay was performed as described using Pim1−/− and WT mice. Panels A-C are blood samples collected at 6 weeks post transplant (A: scatter plot; B: CRU frequencies; C: limiting dilution plot). Panel D is BM samples collected at 4 months post transplant and shown the CRU frequency in Pim1−/− and WT mice.
BM Lin−cells were labeled with fluorescent-labeled antibodies and analyzed by flow cytometry for the long-term HSCs (LT-HSCs, Lin−Sca-1+c-Kit+CD34−CD135−), Short-term HSCs (ST-HSCs, Lin−Sca-1+c-Kit+CD34+CD135−) and multipotent progenitors (MPPs, Lin−Sca-1+c-Kit+CD34+CD135+). BrdU incorporation was analyzed for LT-HSCs.
Sorted LSK cells (6,000/well) from WT, Pim1-Tx and Pim1−/− mice were cultured in StemSpan media supplemented with SCF, TPO-1 and Flt3 (100 ng/ml for each) for 4 days. Total cell numbers were counted and averaged from 3–4 wells/group (data are representative of 4 independent experiments).
BM Lin− cells were enriched by Lineage cell depletion Kit (Miltenyi Biotec) from WT, Pim1-Tx and Pim1−/− mice. The cells were then stained with biotin-labeled lineage specific antibodies, PE-conjugated Scal-1, and APC-H7-conjugated c-Kit followed by streptavidin-Brilliant-violet-605 and Aqua Live/Dead dye. The cells were then fixed and subsequently stained with PE-Cy7-conjugated Ki-67 antibody and FITC-conjugated Caspase 3 antibody. All the cells were gated on LSK population.
BM Lin− cells isolated from C56B/6 mice were cultured in DMEM medium supplemented with 20% heat-inactive FBS and 100 ng/ml of SCF, IL-3, IL-6, TPO, Flt3 alone or in combination for 24 hrs. Cells were harvested and mRNA isolated for RT-PCR analysis of Pim1 (A), Pim2 (B), and Pim3 (C) gene expression.