Abstract
AUF1 is a family of four proteins generated by alternative pre-mRNA splicing that form high affinity complexes with AU-rich, mRNA-destabilizing sequences located within the 3′ untranslated regions of many labile mRNAs. While AUF1 binding is most frequently associated with accelerated mRNA decay, emerging examples have demonstrated roles as a mRNA stabilizer or even translational regulator for specific transcripts. In this review, we summarize recent advances in our understanding of mRNA recognition by AUF1 and the biochemical and functional consequences of these interactions. In addition, unique properties of individual AUF1 isoforms and the roles of these proteins in modulating expression of genes associated with inflammatory, neoplastic, and cardiac diseases are discussed. Finally, we describe mechanisms that regulate AUF1 expression in cells, and current knowledge of regulatory switches that modulate the cellular levels and/or activities of AUF1 isoforms through distinct protein post-translational modifications. This article is part of a Special Issue entitled: RNA Decay mechanisms.
Keywords: AU-rich element, RNA-binding protein, mRNA turnover, Gene regulation, Alternative splicing, RNA structure
1. Introduction
Gene expression is regulated through the cumulative activities of many cellular mechanisms. The control of mRNA turnover is a key regulatory step that plays an integral role in this process. The cytoplasmic level of any given mRNA is dictated by a complex interplay between the synthetic and processing rates of the transcript, countered by the decay mechanisms that eliminate specific transcripts from the cellular mRNA population (Fig. 1A). This relationship allows mRNA decay to influence gene expression in several ways. First, a stable mRNA accumulates to higher steady-state levels than an unstable transcript under conditions where synthetic rates are constant (Fig. 1B). This property is clearly demonstrated with ectopically expressed reporter mRNAs, where insertion of a cis-acting mRNA-destabilizing sequence alone is sufficient to significantly diminish mRNA levels [1,2]. Second, the stability of a mRNA impacts the speed with which transcript levels approach new steady states following changes in the synthetic rate [3]. While the abundance of any mRNA will increase or decrease in response to a corresponding change in its rate of transcription, unstable mRNAs will reach their new steady-state levels faster than stable transcripts (Fig. 1C). This principle contributes to the rapid induction of transcripts encoding inflammatory factors in response to pro-inflammatory stimuli, as these mRNAs tend to have very short half-lives (t1/2) [4]. A further benefit to rapid decay of mRNAs that encode inflammatory mediators, cell cycle proteins, and signaling molecules is that, following acute induction, they can be degraded to limit cellular complications of sustained expression [5]. Finally, the decay rates of specific mRNAs can be modulated, contributing to changes in their steady-state levels in the absence of or cooperatively with alterations in transcription rates [5,6]. The turnover kinetics of diverse mRNA subpopulations are sensitive to a variety of stimuli, including cellular responses to signaling molecules (e.g., steroid hormones, cytokines, inflammatory mediators, many others), infectious agents (e.g., lipopolysaccharide), and other cellular stresses.
Fig. 1.
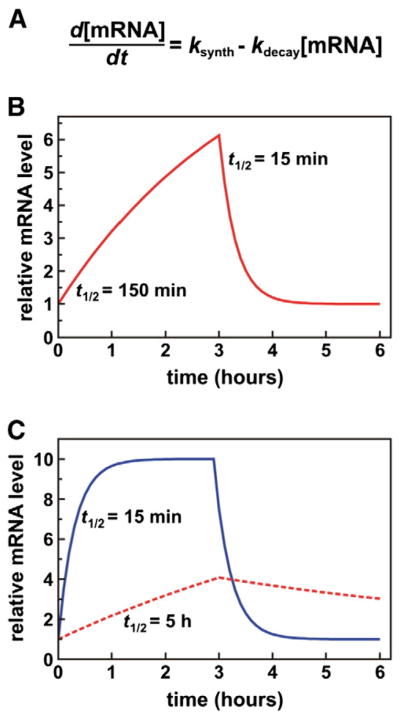
Contributions of mRNA turnover kinetics to the control of gene expression. (A) The impact of mRNA synthesis (ksynth) and degradation (kdecay) rates on mRNA concentrations as modeled by Hargrove and Schmidt [3], where synthesis behaves as a zero-order function (i.e., its rate is independent of the existing mRNA concentration), while decay follows first-order kinetics. Here, the mRNA half-life (t1/2) is given by ln2/kdecay. (B) Regulating mRNA levels by modulating decay rates. In this simulation, an mRNA with t1/2=15 min is stabilized 10-fold (yielding t1/2=150 min) at t=0, dramatically increasing mRNA levels without any change in the synthetic rate. At t=3 h, the mRNA half-life is returned to the original 15 min, resulting in a rapid decrease in mRNA level and restoration of the original mRNA steady state. (C) In these simulations, the RNA synthetic rate (ksynth) is modulated for both unstable (t1/2=15 min, solid blue line) and relatively stable (t1/2=5 h, dashed red line) mRNAs. At t=0 both mRNAs are present at equal concentrations (=1). However, at that point, ksynth for each mRNA is increased 10-fold. The unstable mRNA reaches its new steady state within approximately 1 h, while the rate of increase for the stable mRNA is much slower. The stable mRNA will eventually reach the same steady state, but will require approximately 20 h to do so. At t=3 h, ksynth is returned to its original value for both mRNAs. As expected, levels of the unstable mRNA decrease quickly to re-establish its original steady-state level, again within approximately one hour, while diminution of the stable mRNA occurs over a much longer time frame.
2. AU-rich elements
In mammalian cells, cytoplasmic decay rates vary dramatically among different mRNAs, with stable transcripts often present for many hours or days while labile mRNAs exhibit much shorter half-lives (t1/2=2 h or less) [5,6]. Decay rates are directed via cis-acting sequence elements contained within each mRNA. These RNA sequences serve as binding sites for trans-acting factors that may positively or negatively impact the mRNA degradation process.
The best characterized family of cis-acting mRNA stability determinants are the adenylate/uridylate-rich elements (AU-rich elements or AREs) frequently contained within the 3′-untranslated regions (3′UTRs) of mRNAs encoding cytokines/lymphokines, proto-oncogenes, inflammatory mediators and G protein-coupled receptors [4,7,8]. In general, the presence of an ARE accelerates mRNA turnover in cells, and bioinformatics surveys estimate that 5–8% of human genes contain ARE sequences [9]. Prototypical AREs contain AUUUA pentamers within a U-rich region and can range in size from 40 to 150 nucleotides [4]. Some examples contain one or two copies of the canonical pentamer in a U-rich region, while others are composed of overlapping arrays of pentameric repeats. Still other ARE sequences can accelerate mRNA decay despite lacking any AUUUA motifs [10].
AU-rich elements function by forming ribonucleoprotein (RNP) complexes with members of a set of RNA-binding factors collectively termed ARE-binding proteins (ARE-BPs). To date, over twenty ARE-BPs have been identified, although functional significance has only been defined for a subset [5,11,12]. The RNA-binding domains of ARE-BPs are varied and include RNA Recognition Motifs (RRMs), zinc fingers and K homology (KH)-domains. Some trans-factors can function as mRNA destabilizers when bound to an ARE-containing mRNA (e.g., AUF1, TTP, and KSRP), while others stabilize the transcript to which they are bound (e.g., HuR) [13–18]. Yet another subset of ARE-BPs represses translation of their mRNA targets (TIA-1, TIAR) [19,20]. However, since different ARE-BPs can target common RNA sequences [21–23] and may exhibit similar tissue distributions [24,25], it follows that ARE-BP binding to mRNA substrates may be subject to competitive and/or combinatorial influences. These relationships may be compounded further by variations in the subcellular localization of the targeted transcript and/or post-translational modifications of ARE-BPs [26].
This review focuses on the ARE-BP known as AUF1 (ARE/poly(U)-binding/degradation factor 1) and its functions in regulating mRNA decay. Biochemical characterization of AUF1-containing RNPs has revealed many features of RNA recognition, ancillary factor recruitment, and RNA conformational remodeling by this protein family. Finally, we will examine several physiological targets of AUF1 and the current state of knowledge regarding mechanisms that regulate AUF1 expression and function.
3. The AUF1 family of proteins
AUF1, also known as heterogeneous nuclear ribonucleoprotein (hnRNP) D, was originally identified as a cytosolic activity that accelerated decay of polysome-associated c-myc mRNA in vitro [13,27]. Subsequent purification and cloning identified a family of four proteins derived by alternative splicing of a common pre-mRNA that formed direct, high-affinity complexes with a variety of ARE substrates [28,29]. The inclusion or exclusion of exons 2 and/or 7, encoding 19 and 49 amino acid inserts near the N- and C-termini, respectively, is responsible for the differences between the isoforms (Fig. 2). Named according to their apparent molecular weights, the p45AUF1 isoform contains sequences encoded by both exon 2 and exon 7, p42AUF1 retains the exon 7-encoded domain and p40AUF1 the exon 2-encoded domain, while p37AUF1 lacks sequences from either differentially spliced exon. All four isoforms contain two tandemly arranged, non-identical RRM domains as well as an 8-amino acid glutamine-rich motif located C-terminal to RRM2 [14,28]. The RRM domains are required but not sufficient for high-affinity RNA binding [30]. All AUF1 proteins form stable dimers in solution and bind canonical ARE substrates with low- to mid-nanomolar affinity [30,31]. The sequence specificity of AUF1 binding is somewhat relaxed, as polyuridylate substrates lacking canonical AUUUA motifs also bind AUF1 with similar affinity [32,33]. Inclusion of the exon 2-encoded domain immediately N-terminal of RRM1 modestly inhibits RNA binding, as isoforms containing this sequence (p40AUF1 and p45AUF1) bind a model ARE substrate with approximately 3- to 5-fold lower affinity than their exon 2-deficient counterparts (p37AUF1 and p42AUF1, respectively) [28,31]. On extended RNA substrates, AUF1 dimers can bind sequentially to form oligomeric protein structures [32]. However, RNA-induced AUF1 oligomers are significantly more stable for the p42AUF1 and p45AUF1 isoforms, suggesting that sequences encoded by exon 7 enhance secondary binding events required to form these higher-order complexes [31].
Fig. 2.
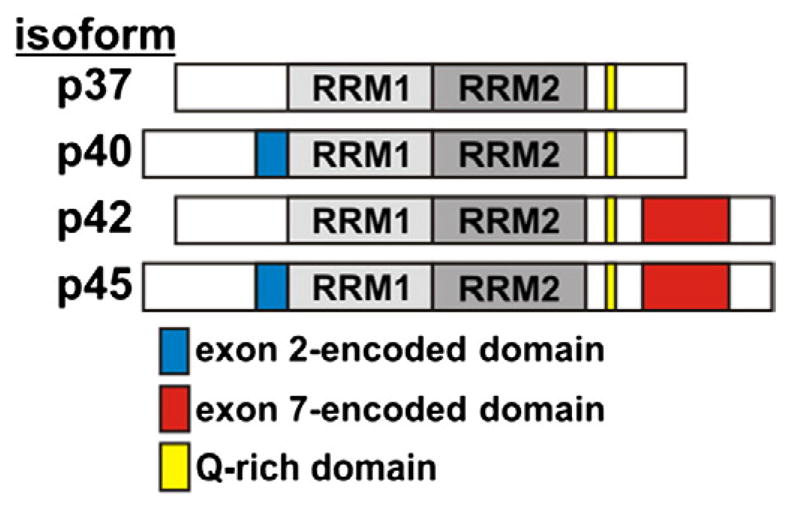
Domain organization of AUF1 proteins. The locations of peptide sequences encoded by alternatively spliced exons and the glutamine-rich (Q-rich) domain are shown flanking the tandem RNA Recognition Motifs (RRMs) common to all AUF1 isoforms.
In most cell types, p42AUF1 and p45AUF1 appear to be largely nuclear, while the smaller isoforms reside in both the nuclear and cytoplasmic compartments [14,34–36]. While the mechanical basis for this distribution remains unclear, several studies have identified potential biochemical mediators of AUF1 protein localization. For example, all isoforms contain a common 19-amino acid C-terminal domain that can bind the nuclear transport factor transportin 1 [37]. However, in an alternative model insertion of the exon 7-encoded domain inhibits nuclear import (p42AUF1 and p45AUF1), suggesting that their delivery to the nucleus may require co-transport with alternative nuclear cargoes [38]. Selected AUF1 isoforms can also form stable complexes with specific nuclear (scaffold attachment factor-β) or cytoplasmic (14-3-3σ) factors [35,39], which may further enrich concentrations of individual isoforms in these compartments. Finally, biochemical data indicate that each AUF1 isoform can form complexes with all others [38], suggesting that any AUF1 protein could be carried within a heterodimer or higher-order protein assembly to specific cellular locations. Together, these data suggest that the subcellular distributions of AUF1 isoforms may be maintained by a complex equilibrium involving diverse molecular determinants and protein-binding events, which could potentially be exploited to modulate AUF1 localization in response to cellular stresses or other signaling events. Finally, observations that specific AUF1 isoforms accumulate in nuclei portended functions beyond the cytoplasm. Strong evidence indicates that AUF1 is required for telomere maintenance, involving transcriptional activation of the telomerase reverse transcriptase (TERT) gene [40,41], and possibly direct interaction with telomeric repeat sequences [42,43]. While these activities indicate a broader role for AUF1 in the regulation of both genome maintenance and gene expression, they are beyond the scope of this review and hence not discussed further.
4. Mechanism of AUF1-induced mRNA decay
The biochemical linkage between the recognition of mRNA substrates by AUF1 and their targeting to ribonucleolytic activities remains incomplete, but data reported by a number of investigators have provided insights into potential mechanisms, and have identified many of the ancillary protein factors involved. ARE-containing transcripts are degraded via the deadenylation-dependent mRNA decay pathway [5]. In this pathway, 3′ → 5′ removal of the mRNA poly(A) tail by dedicated cellular deadenylase activities (e.g., Caf1, Pop2, and PARN) is the initial catalytic and rate-limiting step. As such, deadenylated mRNA decay intermediates are normally difficult to detect from cellular sources. Once the poly(A) tail has been removed, the body of the mRNA is rapidly degraded by nucleases of the exosome complex in the 3′ → 5′ direction, and/or by 5′-decapping and the Xrn1 5′ → 3′ exoribonuclease (reviewed in Refs. [5,44]). While binding of ARE-targeted mRNA-destabilizing proteins (e.g., AUF1, TTP, and KSRP) to mRNA substrates promotes transcript decay, none of these proteins exhibit intrinsic nucleolytic activity. Rather, these factors must recruit downstream components of the mRNA decay machinery to facilitate mRNA degradation [17,45–47].
The mRNA decay activity of AUF1 appears to be principally mediated through the isoforms normally found in the cytoplasm (p37AUF1 and p40AUF1) [48], although roles for the normally nuclear isoforms in the degradation of specific mRNA targets have been described [49,50]. Current models of AUF1 function suggest that it nucleates the formation of large multi-subunit complexes on ARE-containing transcripts (Fig. 3). The AUF1- and Signal Transduction-Regulated Complex (ASTRC) is composed of cap-dependent translation initiation factors and molecular heat-shock chaperone proteins [51]. The formation of the ASTRC complex is a three-step process that is initially mediated by the formation of AUF1 oligomers via sequential dimer binding to ARE-containing transcripts [32]. The second step is the recruitment of additional transacting proteins which may include eukaryotic translation initiation factor 4G (eIF4G), poly(A)-binding protein (PABP), heat shock proteins Hsp70 and Hsc70, as well as Hsp27 and potentially as yet unidentified factors [51–53]. Association of Hsp27, in particular, is associated with targeting ASTRC-associated mRNAs for decay, since ARE-directed mRNA turnover is dramatically slowed in Hsp27-deficient cells [51].
Fig. 3.
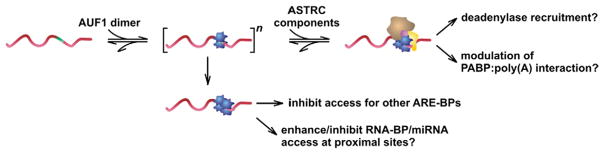
Functional consequences of AUF1 binding to ARE-containing mRNA substrates. An mRNA containing an ARE within its 3′UTR (green insert) is targeted by AUF1 dimers (blue), which may involve multiple rounds of protein recruitment leading to the formation of oligomeric AUF1 protein arrays on the mRNA substrate. In this example AUF1 complex formation is limited to two rounds (yielding an AUF1 tetramer) for clarity. Possible functional consequences of AUF1 oligomerization on mRNA substrates may include recruitment of ancillary protein components (ASTRC), steric obstruction of competing ARE-BPs, or modulation of trans-factor binding at adjacent sites by local RNA conformational remodeling. Biochemical and cellular evidence supporting individual mechanisms is described in the text.
The third and final step of AUF1-induced mRNA decay requires the acceleration of 3′-deadenylation. To date, few details have been resolved that define the specific role(s) that AUF1/ASTRC play in this process. Conceivably, the ASTRC complex may recruit specific deadenylase activities to the targeted mRNP and/or activate such enzymes already in place. The latter model is appealing since in cell-free mRNA decay reactions, partially purified AUF1 proteins can destabilize polysome-associated mRNA substrates without direct competition of PABP using exogenous poly(A) [13]. A third non-exclusive possibility is that ASTRC modifies the interaction of PABP (an ASTRC component) with the mRNA 3′-poly(A) tail, thus enhancing access for cytosolic deadenylase enzymes. Finally, components of the exosome, a large assembly containing 3′ → 5′ exoribonucleolytic and RNA helicase activities, have been identified in complexes containing AUF1 [45,46]. While direct recruitment of the exosome was once considered a potential model for initiating mRNA decay, recent data suggest that this complex is not a major contributor to mRNA 3′-deadenylation [54]. As such, it is more likely that exosome recruitment by AUF1-containing complexes serves to accelerate degradation of the mRNA body once deadenylation has been completed.
While targeting mRNA substrates for rapid degradation is considered the primary functional consequence of AUF1 binding, at least for the major cytoplasmic isoforms, studies from several groups also suggest that AUF1 can indirectly influence mRNA abundance or function by dynamically competing with other factors for RNA binding sites. This concept followed logically from observations that many proteins target AREs or similar RNA sequences [11]. Among ARE-BPs, AUF1 in particular exhibits several features that make it an ideal “generic competitor” for ARE target site occupancy: (i) the binding site size for an AUF1 dimer on an ARE substrate is approximately 34 nt, significantly larger than the length of RNA required for optimal binding of HuR (15 nt) or TTP (9 nt) [21,22,31], (ii) the sequence specificity of AUF1 binding is relatively relaxed among ARE-BPs with a general preference for polyuridylate stretches [32,55], and (iii) AUF1 binding is highly dynamic in solution, with dissociative half-times of 15 s or less [32]. These properties give AUF1 the ability to form high affinity complexes with a wide range of AREs and similar sequences, and occlude large sections of those RNA domains when bound. However, the dynamic nature of AUF1 binding should also permit rapid re-establishment of binding equilibria following changes in the concentrations or RNA-binding activities of AUF1 or any competing factors. For example, in vitro studies indicate that AUF1 and the mRNA-stabilizing protein HuR contact the same region of the androgen receptor mRNA [56]. Functional competition between AUF1 and HuR was observed in an intestinal crypt-derived cell model, where polyamine depletion enhanced HuR binding while simultaneously suppressing AUF1 binding to JunD mRNA, with the net effect of stabilizing the targeted transcript [57]. Another example of direct competition for a specific ARE was observed on c-myc mRNA, where AUF1 and TIAR compete for a common RNA target site [58]. By displacing the translational repressor TIAR, the net effect of AUF1 is to enhance mRNA translational efficiency in this context.
Although the biochemical properties of AUF1 listed above are ideal for an effective competitor of ARE binding, the size of ARE sequences found in many mRNAs (40–150 nt) remains large relative to the AUF1 binding footprint [4]. As such, ample opportunity exists for combinatorial as well as competitive relationships between the cellular ARE-BP population and specific ARE target sites. For example, AUF1 and HuR can bind to independent as well as overlapping sites on mRNAs encoding p21 and cyclin D1 [23]. More recently, an in-cell FRET study showed that AUF1 and HuR co-localize in both nuclear and cytoplasmic compartments, and observed AUF1/AUF1, HuR/HuR and even AUF1/HuR interactions consistent with proximal association on common RNA substrates [59]. This ability of mRNA-destabilizing and -stabilizing proteins to bind simultaneously and/or competitively to individual mRNA substrates could serve as a fine-tuning mechanism to regulate mRNA turnover rates, or could conceivably trigger alternative functional outcomes.
A final and unexpected mechanism by which AUF1 may function does not appear to require its RNA-binding activity; rather, p45AUF1 can enhance the affinity of the tandem zinc finger (TZF) domain of TTP for ARE targets. FRET-based assays show that the affinity of the TTP TZF domain for an 11-nt ARE substrate is enhanced by 5-fold if p45AUF1 is present in the binding reaction [60]. Since p45AUF1 can bind the TTP TZF domain in the absence of RNA [60] and AUF1 proteins do not form detectable complexes with RNA substrates this small [31], enhancement of TTP TZF:ARE binding by AUF1 likely results from allosteric consequences of the AUF1:TTP TZF interaction. The functional significance of this ternary p45AUF1:TTP TZF:ARE complex is currently unknown.
5. Reciprocal relationship between AUF1 binding and local RNA structure
Current data indicate that the RNA sequence requirements for AUF1 recruitment are relatively relaxed (described above). However, biochemical studies have demonstrated that AUF1 binding affinity is highly sensitive to RNA secondary structure. This was first observed in the context of the core ARE from TNFα mRNA, where multivalent cations stabilize a folded conformation that significantly inhibits association of p37AUF1 [33,61], supporting a model whereby AUF1 preferentially recognizes single-stranded domains of ARE targets. By contrast, Hsp70 binding to the TNFα ARE was less sensitive to cation-induced RNA folding, while HuR bound both folded and unfolded forms of this RNA substrate with comparable affinities [2]. These observations suggested that local RNA structures could modulate the equilibrium between a mRNA-destabilizing (AUF1) and mRNA-stabilizing (HuR) factor competing for a common ARE target. Consistent with this model, peripheral mutations that stabilized TNFα ARE folding inhibited decay of a reporter mRNA in cells, while compensatory mutations that weakened local ARE folding reversed this effect [2]. Although the tested TNFα ARE structures were limited to a highly conserved RNA hairpin within the ARE itself, it is likely that these models will also apply to ARE sequences involved in RNA duplexes or higher-order structures with RNA domains adjacent or even distal to the ARE.
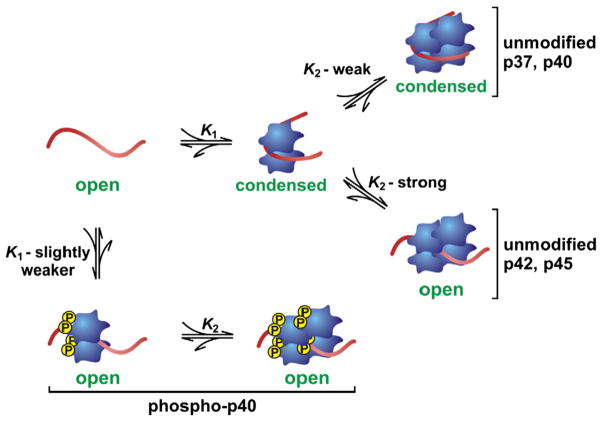
While the presence of local RNA secondary structure can significantly impact the affinity of AUF1 binding, assembly of AUF1 RNPs can also dramatically alter the local RNA conformational landscape. Using FRET-based in vitro RNA folding assays, all AUF1 isoforms have been shown to alter the local structure of model ARE substrates [31,61]. In all cases, the initial dimer binding event brings the 5′- and 3′-ends of the ARE target in close proximity, consistent with adoption of a condensed RNA conformation. However, the effect of the second AUF1 dimer binding event on local RNA structure is isoform-specific (Fig. 4). AUF1 isoforms that lack exon 7-encoded sequences (p37AUF1, p40AUF1) retain a condensed RNA conformation in the AUF1 tetramer RNP complex, while those containing the exon 7-encoded domain (p42AUF1, p45AUF1) induce extended conformations on RNA substrates [31]. It is possible that isoform-specific differences in this mechanical consequence of RNA binding could contribute to differential roles of AUF1 variants in mRNA fate. However, in addition to AUF1 isoform selection, protein post-translational modifications can also impact local RNA structure in AUF1 RNPs. In THP-1 monocytes, polysome-associated p40AUF1 is normally phosphorylated within its exon 2-encoded domain at residues Ser83 and Ser87. While this has a small but measureable inhibitory effect on the affinity of p40AUF1 binding to an ARE target, the presence of these phosphorylated residues maintains RNA substrates in elongated conformations, even at the first binding step [62].
Fig. 4.
Isoform- and phosphorylation-dependent control of local RNA structure by AUF1. Association of unphosphorylated AUF1 dimers induces structural condensation of ARE targets, regardless of the protein isoform, a conformation that is maintained following the secondary round of p37AUF1 or p40AUF1 dimer binding that generates AUF1 tetramer:ARE complexes (top). However, the K2 binding event that generates tetrameric p42AUF1 or p45AUF1 complexes on an RNA substrate induces an extended RNA conformation (center), similar to that observed following both initial and secondary complexes formed with p40AUF1 dimers phosphorylated at Ser83 and Ser87 (bottom).
The functional significance of local RNA structural remodeling by AUF1 remains unclear, but emerging models for combinatorial trans-factor binding events suggest some possibilities. At the simplest level, AUF1-induced conformational changes on an ARE-containing mRNA substrate could occlude or enhance access for other RNA-binding proteins or miRNAs targeting proximal binding sites. Evidence supporting roles for HuR in promoting or restricting miRNA access at local sites on mRNA has been proposed by several groups, and could conceivably involve similar mechanisms [63–65]. Alternatively, the unique conformations of specific AUF1:ARE RNPs may present specialized binding determinants for recruitment of ancillary protein factors including components of ASTRC that involve both protein (AUF1) and RNA recognition. This paradigm is similar to the recruitment of some coregulator complexes to gene promoters, which can involve recognition of both proteins and adjacent DNA sequences [66,67]. In any case, AUF1 isoform- and phosphorylation-dependent changes in the composition and architecture of these RNPs provide a diverse population of molecular determinants to direct downstream macromolecular association events.
6. Physiological targets of AUF1
AUF1 can regulate the decay kinetics of myriad transcripts including many encoding cell cycle regulators such as p16INK4a, p21WAF1/CIP1, and cyclin D1, early response genes, inflammatory mediators and cytokines such as TNFα and interleukin-1β (IL-1β), G protein-coupled receptors, and oncoproteins (reviewed in Refs. [68,69]). A common theme among these transcripts is that their expression must be tightly regulated in response to various cellular signals. AUF1 has predominantly been identified as a protein associated with destabilization of ARE-containing mRNAs. Consistent with this model, AUF1-null mice overexpress the pro-inflammatory cytokines TNFα and IL-1β owing to their impaired ability to rapidly degrade the transcripts encoding these proteins [70]. This inability to suppress TNFα and IL-1β production yields an endotoxin hypersensitivity phenotype, where challenge with endotoxin results in symptoms of severe septic shock including vascular hemorrhage, intravascular coagulation and high mortality [70]. In addition, AUF1-null mice develop chronic, spontaneous inflammatory dermatitis characterized by pruritus and excoriations [71]. However, in a system/transcript-dependent manner, evidence has also been presented indicating that AUF1 may stabilize and/or modulate the translational efficiency of specific ARE-containing mRNAs, and that this may also occur in an isoform-specific manner [55,72–74]. This functional flexibility allows AUF1 to coordinate several aspects of the inflammatory response. While the examples described above highlight its roles in suppressing expression of selected pro-inflammatory cytokines, AUF1 can also enhance production of anti-inflammatory signals. Specifically, in monocytes AUF1 promotes translation of the mRNA encoding transforming growth factor-β-activated kinase 1 (TAK1), a member of the NF-κB signaling pathway which is necessary for inducing expression of the anti-inflammatory cytokine IL-10 [75]. Furthermore, AUF1 binds ARE-like sequences in the 3′UTR of IL-10 mRNA and enhances its expression following stimulation with lipopolysaccharide (LPS). This effect is mediated specifically by the p40AUF1 isoform, and likely involves stabilization of the IL-10 transcript [76].
AUF1 also regulates the expression of many mRNAs whose products manipulate processes associated with tumor development, as these frequently include 3′UTR ARE-like sequences. Current evidence indicates that AUF1 may exert pro- and anti-tumorigenic roles in different cellular contexts. For example, transgenic mice that overexpress p37AUF1 develop undifferentiated sarcomas that progress to late stage and result in death, and concomitantly overexpress the cell cycle regulator cyclin D1 as well as the oncogenes c-myc and c-fos [77]. Recently, AUF1 was also implicated as a central driver of a paracrine signaling pathway from stromal fibroblasts that enhances proliferation and invasiveness of breast cancer cells. In this system, AUF1 targets and suppresses CDKN2A mRNA, thus decreasing expression of its encoded protein product, the cell cycle inhibitor p16INK4A [78]. In turn, diminished p16INK4A levels lead to increased secretion of stromal cell-derived factor 1 and matrix metalloproteinase-2, which stimulate the endothelial–mesenchymal transition in breast cancer cells. In a third example, splenic B cell follicles from AUF1 knockout mice show enhanced apoptosis associated with reduced expression of members of the Bcl-2 family of apoptotic inhibitors [79], suggesting an anti-apoptotic function for AUF1 in this case.
However, several studies have also implicated anti-tumorigenic roles for AUF1. For example, while Bcl-2 expression was suppressed by the loss of AUF1 in splenic B cells [79], other reports associate AUF1 binding to the ARE within the bcl-2 mRNA 3′UTR with suppression of this transcript [80,81]. Also, although cyclin D1 levels were elevated in mice that overexpress p37AUF1 [77], increased AUF1 binding decreases levels of cyclin D1 mRNA in UVC-treated HeLa cells [23]. Cyclin D1 expression was also suppressed in a non-small-cell lung cancer line following prostaglandin A2 treatment, involving selective induction of p45AUF1 which bound within the 3′UTR of cyclin D1 mRNA and accelerated its decay [49]. Finally, AUF1 binding destabilizes VEGF mRNA, decreasing the production of this powerful pro-angiogenic factor [82,83]. Together, these data suggest apparently contradictory functions for AUF1, promoting both pro- and anti-tumorigenic events in diverse model systems. However, it is likely that context-specific parameters including AUF1 dosage, isoform selectivity, and the availability and/or accessibility of protein co-factors and competing ARE-BPs all contribute to the distinct functional outcomes of AUF1 binding to specific mRNAs in individual cell types.
Beyond implicating AUF1 in mechanisms contributing to inflammatory diseases and cancer, increased expression of AUF1 has also been observed in selected cardiac pathologies. AUF1 protein levels are increased in failing human hearts, which smooth muscle cell models indicate may result from sustained exposure to β-adrenergic receptor agonists [84]. Since AUF1 binds and destabilizes β-adrenergic receptor mRNAs, enhanced AUF1 expression is thus an appealing model contributing to the agonist-mediated downregulation of β-adrenergic receptor signaling associated with heart failure. AUF1 expression is also enhanced in models of cardiac hypertrophy and stress including sustained JNK signaling, LPS, and phorbol ester treatment, where it directs accelerated decay of the mRNA encoding the regulatory subunit of protein phosphatase 2A, B56α [85]. Finally, AUF1 suppresses expression of sarco(endo)plasmic reticulum Ca2+-ATPase 2a (SERCA2a) in phorbol ester-stimulated neonatal rat ventricular myocytes, an event involving an as yet undefined phosphorylated form of AUF1 [86].
7. Regulation of AUF1
The expression and function of AUF1 can be regulated by several different means. While selected stimuli including estradiol, prostaglandin A2, and oxidative stress can modulate levels of one or more AUF1 isoforms in specific cell backgrounds, essentially no mechanistic details of their action are known [49,72,87]. However, AUF1 expression can also be regulated by post-transcriptional mechanisms that may integrate multiple mRNA decay pathways (described below) to collectively impact the cellular concentration of this ARE-BP. In addition, selected AUF1 isoforms may be post-translationally modified, including changes in phosphorylation, methylation, ubiquitination, or protein isomerization at specific sites. These events can alter the ability of AUF1 to bind ARE-containing transcripts and/or influence the decay rates of mRNA targets.
Two post-transcriptional mechanisms that control AUF1 expression exploit alternative splicing events that occur in the 3′UTR of AUF1 mRNA. While the AUF1 gene contains 10 exons, the translational termination codon is contained within exon 8. As a result, sequences encoded by exons 9 and 10 are exclusively contained within the mRNA 3′UTR. Furthermore, the introns linking these 3′UTR exons are not always excised [88], yielding a total of five AUF1 mRNA 3′UTR splice variants that are detectable in cytoplasmic RNA samples (Fig. 5). While the most abundant AUF1 mRNA variants normally lack sequences between exons 8 and 10, one regulatory event facilitated by alternative splicing in the AUF1 3′UTR operates on mRNAs that retain intron 9. A highly conserved ARE-like sequence within this intron suppresses expression of a luciferase reporter mRNA but also binds AUF1, suggesting that regulation of these mRNA variants may include a negative feedback loop [88]. However, 3′UTR variants where intron 9 was selectively spliced also reduced reporter gene expression. In these cases, the 107-nucleotide exon 9 is retained within the 3′UTR, forcing the exon9:exon10 junction to be positioned well over 50–55 nucleotides downstream of the translational termination codon, a configuration that activates mRNA degradation via the nonsense-mediated mRNA decay (NMD) pathway in mammalian cells [44]. Importantly, siRNA-mediated depletion of either Upf1 or Upf2, trans-acting factors involved in the NMD cascade, stabilized AUF1 mRNAs where intron 9 had been selectively spliced [89]. These data confirmed a role for NMD in degrading these mRNAs and provided the first evidence of a functional linkage between the NMD and ARE-directed mRNA decay pathways. Finally, beyond binding selected 3′UTR variants of its own mRNA, AUF1 can interact with transcripts encoding other ARE-BPs including HuR, TIA-1, TIAR, KSRP, and NF-90 [90], suggesting that relative levels of these factors may be regulated through extensive cross-talk networks.
Fig. 5.
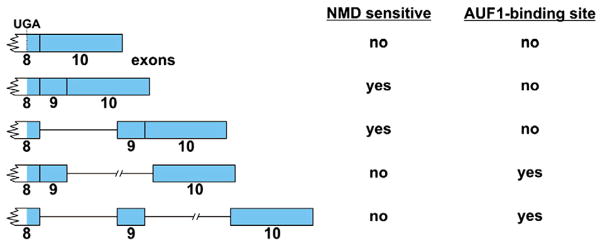
AUF1 mRNA 3′UTR variants generated by alternative pre-mRNA splicing. 3′UTR exon sequences are represented by blue boxes and retained introns by lines. The sensitivity of each mRNA variant to the NMD pathway and AUF1 recognition is indicated at right and discussed in the text.
AUF1 protein levels and function can also be regulated via specific protein modifications. Although AUF1 was originally identified as a phosphoprotein [14], the first defined phosphorylation events were observed on polysome-associated p40AUF1 at Ser83 and Ser87 (Fig. 4). In THP-1 monocytic leukemia cells, stimulation with the phorbol ester TPA dephosphorylated these residues concomitant with stabilization of candidate AUF1-binding mRNAs [34]. Since the principal biochemical consequence of AUF1 phosphorylation at these sites was to alter the conformation of AUF1-bound RNA ligands (described above), these data suggested that RNA allosteric mechanisms could link AUF1 binding to induction or obstruction of mRNA catabolic processes. However, while phosphorylation of p40AUF1 at Ser83 and Ser87 is associated with destabilization of ARE-containing transcripts, hyperphosphorylation of AUF1 by nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) correlates with stabilization of several AUF1 target mRNAs [91], although the precise locations of these AUF1 modifications have not yet been identified.
Beyond phosphorylation, additional post-translational modifications have also been identified on AUF1 proteins. One example is protein methylation within the C-terminal RGG (arginine-glycine-glycine) repeat motifs, specifically arginine dimethylation within the third RGG repeat, which is common to all four AUF1 protein isoforms [92]. This C-terminal domain is required for AUF1-directed suppression of VEGF expression in RAW-246.7 macrophages, although the biochemical basis for its contributions to mRNA decay remains unknown [82]. Interestingly, an arginine methyltransferase inhibitor reduced AUF1 levels in these cells [82], raising the intriguing possibility that this modification might contribute to the regulation of AUF1 protein stability. Cellular concentrations of AUF1 are also regulated by control of AUF1 degradation through the ubiquitin–proteasome pathway [53,93]. Both p37AUF1 and p40AUF1 can be poly-ubiquitinated, while inclusion of the exon 7-encoded domain in p42AUF1 and p45AUF1 appears to inhibit this modification [94]. More recent data have demonstrated that the heat shock protein Hsp27 (a component of the ASTRC complex) may bind directly to ARE-containing mRNA targets, interact with AUF1 and promote ubiquitination of the AUF1 protein [95]. Since Hsp27 phosphorylation by the p38MAPK pathway (and selected others) enhances its ability to promote ubiquitination of associated proteins, phospho-Hsp27-directed ubiquitination and consequent degradation of AUF1 provides an appealing mechanism to explain the stabilization of many ARE-containing mRNAs observed following MAPK activation. Ubiquitination of p45AUF1 and p42AUF1 is also promoted by the von Hippel–Lindau tumor suppressor gene product (pVHL), which suppresses AUF1 protein levels in pVHL-positive cell models under normoxic conditions [83]. pVHL also forms a stable complex with AUF1 on VEGF mRNA, which is rapidly degraded in these cells. Under hypoxia, pVHL dissociates from AUF1, leading to accumulation of AUF1 protein but also stabilization of VEGF mRNA [83]. In this context, pVHL not only regulates cellular AUF1 levels, but may also function as an important co-factor in mediating the mRNA-destabilizing activity of AUF1 on selected mRNA targets.
Finally, a novel mechanism that suppresses AUF1 function during eosinophil or T cell activation involves isomerization by the peptidyl-prolyl cis-trans isomerase Pin1. In resting cells, catalytically inactive, phosphorylated Pin1 can form complexes with AUF1, but does not perturb the ability of AUF1 to bind and promote degradation of GM-CSF mRNA [96,97]. Pin1 binds protein substrates using phospho-Ser-Pro or phospho-Thr-Pro motifs [98]. Since only p40AUF1 and p45AUF1 contain canonical Pin1 binding sites (phospho-Ser83-Pro84), other AUF1 isoforms are likely recruited to Pin1 via heterodimeric AUF1 complexes. However, leukocyte activation induces Pin1 dephosphorylation, which activates its proline isomerization activity [96,97]. The resulting conformational transition in AUF1 is likely responsible for the subsequent inhibition of its RNA-binding activity, and hence abrogating its ability to destabilize mRNA substrates.
8. Conclusion
AUF1 proteins bind to AREs and similar RNA sequences with high affinity, but the functional consequences of these events can vary widely. The best characterized outcome of AUF1:ARE complex formation is accelerated mRNA decay. Recent data indicate that this is an ordered but also a dynamic process requiring multiple factors in addition to the AUF1 protein and the ARE-containing substrate transcript. Alternatively, AUF1 binding may be associated with mRNA stabilization or regulation of translation in some cases. Functional outcomes are likely context dependent, and may involve competitive and/or collaborative relationships with other RNA-binding proteins or even miRISC complexes. Over the past decade, several groups have also made progress on the regulation of AUF1 function, mostly involving post-translational mechanisms that alter AUF1 protein stability or RNA-binding activity. However, these studies also prompt a host of new and intriguing questions. Precise roles of ASTRC or other components in mediating mRNA decay following complex assembly remain unresolved, as are the impacts of local RNA structural remodeling mediated by AUF1 and regulated by its phosphorylation status. Furthermore, little is known about the mechanisms that control AUF1 expression in many cellular contexts, including those that enhance AUF1 levels during heart failure or in models of cardiac hypertrophy. Similarly, many potential roles of dysregulated AUF1 expression or activity in tumor progression remain unresolved, despite the vast array of cancer-related mRNAs that are targeted by this factor. Finally, the enriched concentrations of AUF1 normally detected in nuclei indicate important but heretofore undefined functions in this organelle, and observations linking AUF1 to telomere maintenance suggest that these roles may include some that are uncoupled from the RNA-binding activity of this factor.
Acknowledgments
Research support for the Brewer and Wilson labs is provided by National Institutes of Health grants R01 CA052443 (to G.B.) and R01 CA102428 (to G.M.W.).
Footnotes
This article is part of a Special Issue entitled: RNA Decay mechanisms.
References
- 1.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hargrove JL, Schmidt FH. The role of mRNA and protein stability in gene expression. FASEB J. 1989;3:2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- 4.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 9.Halees AS, El-Badrawi R, Khabar KS. ARED organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 13.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 17.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 20.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 21.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 23.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouble A, Morello D. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene. 2000;19:5377–5384. doi: 10.1038/sj.onc.1203910. [DOI] [PubMed] [Google Scholar]
- 25.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 26.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 29.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 30.DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 31.Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem. 2010;285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 33.Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 34.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A+U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J Biol Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 35.Arao Y, Kuriyama R, Kayama F, Kato S. A nuclear matrix-associated factor, SAF-B, interacts with specific isoforms of AUF1/hnRNP D. Arch Biochem Biophys. 2000;380:228–236. doi: 10.1006/abbi.2000.1938. [DOI] [PubMed] [Google Scholar]
- 36.Inoue A, Arao Y, Omori A, Ichinose S, Nishio K, Yamamoto N, Kinoshita Y, Mita S. Identification of S1 proteins B2, C1 and D1 as AUF1 isoforms and their major role as heterogeneous nuclear ribonucleoprotein proteins. Biochem J. 2003;372:775–785. doi: 10.1042/BJ20021719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 39.He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pont AR, Sadri N, Hsiao SJ, Smith S, Schneider RJ. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol Cell. 2012;47:5–15. doi: 10.1016/j.molcel.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 43.Eversole A, Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol. 2000;20:5425–5432. doi: 10.1128/mcb.20.15.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 46.Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen TM, Hsu CH, Tsai SJ, Sun HS. AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay. Nucleic Acids Res. 2010;38:8061–8071. doi: 10.1093/nar/gkq717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, Turrin DK, De La Cruz MP, Pestka S, Brewer G. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 54.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 55.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J Biochem. 2012;151:423–437. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- 57.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 59.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedar VP, Zucconi BE, Wilson GM, Blackshear PJ. Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J Biol Chem. 2012;287:5459–5471. doi: 10.1074/jbc.M111.312652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson GM, Sutphen K, Moutafis M, Sinha S, Brewer G. Structural remodeling of an A+U-rich RNA element by cation or AUF1 binding. J Biol Chem. 2001;276:38400–38409. doi: 10.1074/jbc.M106509200. [DOI] [PubMed] [Google Scholar]
- 62.Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- 63.Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem. 2011;286:41442–41454. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 65.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holbert MA, Sikorski T, Carten J, Snowflack D, Hodawadekar S, Marmorstein R. The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting. J Biol Chem. 2007;282:36603–36613. doi: 10.1074/jbc.M705812200. [DOI] [PubMed] [Google Scholar]
- 67.Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J. Intrinsic nucleic acid-binding activity of chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell. 2012;47:228–241. doi: 10.1016/j.molcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 68.Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Front Biosci. 2011;16:2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sadri N, Schneider RJ. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J Invest Dermatol. 2009;129:657–670. doi: 10.1038/jid.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor alpha mRNA. J Biol Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. [DOI] [PubMed] [Google Scholar]
- 73.Paschoud S, Dogar AM, Kuntz C, Grisoni-Neupert B, Richman L, Kuhn LC. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, Pestka S. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol Cell Biol. 2011;31:602–615. doi: 10.1128/MCB.00835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J Interferon Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 78.Al-Ansari MM, Hendrayani SF, Shehata AI, Aboussekhra A. p16(INK4A) Represses the paracrine tumor-promoting effects of breast stromal fibroblasts. Oncogene. doi: 10.1038/onc.2012.270. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sadri N, Lu JY, Badura ML, Schneider RJ. AUF1 is involved in splenic follicular B cell maintenance. BMC Immunol. 2010;11:1. doi: 10.1186/1471-2172-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1) J Biol Chem. 2010;285:27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Nicolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 is a bcl-2 A+U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J Biol Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
- 82.Fellows A, Griffin ME, Petrella BL, Zhong L, Parvin-Nejad FP, Fava R, Morganelli P, Robey RB, Nichols RC. AUF1/hnRNP D represses expression of VEGF in macrophages. Mol Biol Cell. 2012;23:1414–1422. doi: 10.1091/mbc.E11-06-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xin H, Brown JA, Gong C, Fan H, Brewer G, Gnarra JR. Association of the von Hippel–Lindau protein with AUF1 and posttranscriptional regulation of VEGFA mRNA. Mol Cancer Res. 2012;10:108–120. doi: 10.1158/1541-7786.MCR-11-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pende A, Tremmel KD, DeMaria CT, Blaxall BC, Minobe WA, Sherman JA, Bisognano JD, Bristow MR, Brewer G, Port JD. Regulation of the mRNA-binding protein AUF1 by activation of the beta-adrenergic receptor signal transduction pathway. J Biol Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- 85.Glaser ND, Lukyanenko YO, Wang Y, Wilson GM, Rogers TB. JNK activation decreases PP2A regulatory subunit B56alpha expression and mRNA stability and increases AUF1 expression in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2006;291:H1183–H1192. doi: 10.1152/ajpheart.01162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blum JL, Samarel AM, Mestril R. Phosphorylation and binding of AUF1 to the 3′-untranslated region of cardiomyocyte SERCA2a mRNA. Am J Physiol Heart Circ Physiol. 2005;289:H2543–H2550. doi: 10.1152/ajpheart.00545.2005. [DOI] [PubMed] [Google Scholar]
- 87.Guo GE, Ma LW, Jiang B, Yi J, Tong TJ, Wang WG. Hydrogen peroxide induces p16(INK4a) through an AUF1-dependent manner. J Cell Biochem. 2010;109:1000–1005. doi: 10.1002/jcb.22474. [DOI] [PubMed] [Google Scholar]
- 88.Wilson GM, Sun Y, Sellers J, Lu H, Penkar N, Dillard G, Brewer G. Regulation of AUF1 expression via conserved alternatively spliced elements in the 3′ untranslated region. Mol Cell Biol. 1999;19:4056–4064. doi: 10.1128/mcb.19.6.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Banihashemi L, Wilson GM, Das N, Brewer G. Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay. Mol Cell Biol. 2006;26:8743–8754. doi: 10.1128/MCB.02251-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 92.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 93.Laroia G, Sarkar B, Schneider RJ. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc Natl Acad Sci U S A. 2002;99:1842–1846. doi: 10.1073/pnas.042575699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laroia G, Schneider RJ. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 2002;30:3052–3058. doi: 10.1093/nar/gkf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knapinska AM, Gratacos FM, Krause CD, Hernandez K, Jensen AG, Bradley JJ, Wu X, Pestka S, Brewer G. Chaperone Hsp27 modulates AUF1 proteolysis and AU-rich element-mediated mRNA degradation. Mol Cell Biol. 2011;31:1419–1431. doi: 10.1128/MCB.00907-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Esnault S, Shen ZJ, Whitesel E, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates granulocyte-macrophage colony-stimulating factor mRNA stability in T lymphocytes. J Immunol. 2006;177:6999–7006. doi: 10.4049/jimmunol.177.10.6999. [DOI] [PubMed] [Google Scholar]
- 97.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 98.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]