Abstract
Filamentous fungus Fusarium poae is a worldwide cause of the economically important disease Fusarium head blight of cereal grains. The fungus is itself commonly infected with a bisegmented dsRNA virus from the family Partitiviridae. For this study, we determined the structure of partitivirus Fusarium poae virus 1 (FpV1) to a resolution of 5.6 Å or better by electron cryomicroscopy and three-dimensional image reconstruction. The main structural features of FpV1 are consistent with those of two other fungal partitiviruses for which high-resolution structures have been recently reported. These shared features include a 120-subunit T=1 capsid comprising 60 quasisymmetrical capsid protein dimers with both shell and protruding domains. Distinguishing features are evident throughout the FpV1 capsid, however, consistent with its more massive subunits and its greater phylogenetic divergence relative to the other two structurally characterized partitiviruses. These results broaden our understanding of conserved and variable elements of partitivirus structure, as well as that of picobirnavirus, and support the suggestion that a phylogenetic subcluster of partitiviruses closely related to FpV1 should constitute a separate taxonomic genus.
Keywords: Capsid protein, Cryoelectron microscopy, dsRNA virus, Electron cryomicroscopy, Fungal virus, Virus structure
1. Introduction
Viruses in the family Partitiviridae have bisegmented dsRNA genomes, with each segment packaged in a separate virus particle. They persistently infect their plant, fungus, or protozoan hosts, and routinely transmit by intracellular means such as cell division and cell-cell fusion (Ghabrial et al., 2005, 2008). In plants, they generally are not associated with disease and hence are also called cryptoviruses. Plant partitiviruses are grouped in two genera, Alphacryptovirus and Betacryptovirus, though genome sequences indicate that members of the genus Alphacryptovirus are phylogenetically diverse (Boccardo and Candresse, 2005; Crawford et al., 2006; Ghabrial et al., 2008; Willenborg et al., 2009). No sequences are available for the genus Betacryptovirus. Fungal partitiviruses are grouped in one genus, Partitivirus, though genome sequences indicate that members of this genus, too, are phylogenetically diverse (Boccardo and Candresse, 2005; Crawford et al., 2006; Ghabrial et al., 2008; Willenborg et al., 2009). Protozoan partitiviruses are so far restricted to the genus Cryspovirus (Nibert et al., 2009) and infect species of the apicomplexan genus Cryptosporidium, including the human pathogens C. parvum and hominis (Khramtsov et al., 1997; Green et al., 1999; Leoni et al., 2003).
There are four recognized families of dsRNA viruses that have mono- or bisegmented genomes enclosed by icosahedral protein capsids: Totiviridae (monosegmented; T=1 capsid; fungus, protozoan, and possibly arthropod hosts), Birnaviridae (bisegmented; T=13 capsid; vertebrate and arthropod hosts), Partitiviridae (see above; T=1 capsid), and Picobirnaviridae (bisegmented; T=1 capsid; vertebrate hosts). Partitiviruses encompass the smallest of these genomes, ranging in total lengths from 3.2 to 4.4 kbp (Ghabrial et al., 2005, 2008). The only picobirnavirus genome for which a full-length sequence has been reported falls within this same range, at 4.3 kbp (Wakuda et al., 2005). In comparison, the smallest reported toti- and birnavirus genomes are 4.6 and 5.9 kbp, respectively (Fauquet et al., 2005). The two genome segments of partitiviruses respectively encode the viral RNA-dependent RNA polymerase (RdRp) and the viral capsid protein (CP). Some partitiviruses additionally package one or more satellite segments of dsRNA that are likely not essential for replication (Oh and Hillman, 1995; Compel et al., 1998; Kim et al., 2005).
The first three-dimensional (3D) structures of partitivirus particles have been recently determined by transmission electron cryomicroscopy (cryoTEM) and 3D image reconstruction as well as by X-ray crystallography (Ochoa et al., 2008; Pan et al., 2009; Tang et al., 2010). These structures are for two distantly related members of the genus Partitivirus that can co-infect the saprophytic, filamentous fungus Penicillium stoloniferum: Penicillium stoloniferum viruses F and S (PsV-F and -S) (Bozarth et al., 1971; Buck and Kempson-Jones, 1973, 1974; Kim et al., 2003, 2005). The structures have revealed a number of distinctive features, including quasisymmetrical CP dimers, 60 of which together form the T=1 capsid; domain swapping between the two subunits within the shell domain of each CP dimer; prominent surface arches, one from each dimer, formed by protruding domains; and the occurrence of diamond-shaped CP tetramers (dimers of dimers) as probable assembly intermediates. Most of these features have also been recently observed in the X-ray crystal structure of the rabbit picobirnavirus (raPBV) capsid (Duquerroy et al., 2009). Thus, given their structural similarities and despite their different host ranges, partiti- and picobirnaviruses appear likely to share a distinct evolutionary lineage relative to other dsRNA viruses.
In the current study, we advanced structural characterizations of the family Partitiviridae by determining the cryoTEM structure of another fungus-infecting member, Fusarium poae virus 1 (FpV1), from the genus Partitivirus (Fekete et al., 1995; Compel et al., 1998). This virus was originally named FUPO-1 (Compel et al., 1998), but was renamed FpV1 for better consistency with exisiting nomenclature for other partitiviruses (Ghabrial et al., 2005). The phytopathogenic fungal host Fusarium poae is a causal agent of Fusarium head blight, or scab, of cereal grains worldwide and is of considerable importance because it not only reduces crop yields but also produces mycotoxins that pose threats to humans and other animals that consume the grains from infected plants (Stenglein, 1999).
PsV-F and -S are distantly related within the same phylogenetic subcluster, whereas FpV1 belongs to a distinct subcluster and is therefore more distantly related to PsV-F and -S than those are to each other (Boccardo and Candresse, 2005; Crawford et al., 2006; Ghabrial et al., 2008; Willenborg et al., 2009) (Table 1). The fact that the level of sequence identity among the CPs of PsV-F, PsV-S, and FpV1 is quite low (pairwise identity scores between FpV1 and PsV-F or -S are respectively 13% or 14%, as compared to 19% between PsV-F and -S, in alignments with T-Coffee (Poirot et al., 2003) generated for this study) led us to expect that a structure determination for FpV1 virions may reveal unique variations on the recently described architectural designs of PsV-F and -S (Ochoa et al., 2008; Pan et al., 2009; Tang et al., 2010). In addition, the CP of FpV1 is substantially (almost 50%) larger than those of PsV-F and -S (Table 1), suggesting that its structure may include other distinctive features with specific functions.
Table 1.
Recognized viruses in the genus Partitivirus with sequenced genomic RNA(s)a
| Subclusterb | Virus name (abbrev.)c | dsRNA1 (bp) | dsRNA2 (bp) | RdRp (aa, kDa) | CP (aa, kDa) |
|---|---|---|---|---|---|
| 1 | AoV | 1754 | 1555 | 539, 62 | 433, 47 |
| DdV1 | 1787 | 1585 | 539, 62 | 436, 48 | |
| DdV2 | 1781 | 1611 | 539, 62 | 442, 50 | |
| FsV1 | 1645 | 1445 | 519, 60 | 413, 44 | |
| GaV-MS1 | 1782 | 1586 | 539, 61 | 443, 47 | |
| OPV1 | 1744 | 1567 | 539, 63 | 430, 46 | |
| PsV-F | 1677 | 1500 | 538, 62 | 420, 47 | |
| PsV-S | 1754 | 1582 | 539, 62 | 434, 47 | |
| 2 | AhV | 2180 | 2135 | 665, 78 | 652, 74 |
| CrV1 | 2207 | 2305 | 663, 77 | 661, 73 | |
| FpV1 | 2203 | 2185 | 673, 78 | 637, 70 | |
| HaV | 2325 | nd | 734, 87 | nd | |
| HmV | 2247 | nd | 706, 83 | nd | |
| PoV1 | 2296 | 2223 | 706, 82 | 636, 71 | |
| RhsV-717 | 2363 | 2206 | 730, 86 | 683, 76 | |
| RnV1 | 2299 | 2279 | 709, 84 | 686, 77 |
Viruses recognized as species by the International Committee on Taxonomy of Viruses
As defined in previous papers (see text).
Virus names and Genbank numbers: AoV, Aspergillus ochraceous virus (EU118277 and EU118278); DdV1, Discula destructiva virus 1 (AF316992 and AF316993); DdV2, Discula destructiva virus 2 (AY033436 and AY033437); FsV1, Fusarium solani virus 1 (D55668 and D55669); GaRV-MS1, Gremmeniella abietina RNA virus MS1 (AY089993 and AY089994); OPV1, Ophiostoma partitivirus 1 (AM087202 and AM087203); PsV-F, Penicillium stoloniferum virus F (AY738336 and AY738337); PsV-S, Penicillium stoloniferm virus S (AY156521 and AY156522); AhV, Atkinsonella hypoxylon virus (L39125 and L39126); CrV1, Ceratocystis resinifera virus 1 (AY603051 and AY603052); FpV1, Fusarium poae virus 1 (AF015924 and AF047013); HaV, Heterobasidion annosum virus (AAL79540); HmV, Helicobasidium mompa virus (BAD32677); PoV1, Pleurotus ostreatus virus 1 (AY533036 and AY533038); RhsV-717, Rhizoctonia solani virus 717 (AF133290 and AF133291); and RnV1, Rosellinia necatrix virus 1 (AB113347 and AB113348). Viruses for which 3D structures have been determined are bolded.
2. Materials and methods
2.1. Fungal culture and purification of FpV1 virions
A culture of F. poae strain A-11 (Compel et al., 1998) was kindly supplied by László Hornok (University of Agricultural Sciences, Gödöllo, Hungary). This fungus was then maintained in the lab on slants of potato dextrose agar containing 0.5% yeast extract. For purification of FpV1 virions, a protocol was devised from one previously reported for Penicillium chrysogenum virus (Jiang and Ghabrial, 2004). F. poae was grown in a stationary culture containing potato dextrose broth supplemented with 0.5% yeast extract. Mycelium was harvested from a 7-day culture by straining through miracloth in a Buchner funnel and homogenized in 0.1 M sodium phosphate, pH 7.4, containing 0.2 M KCl and 0.5% mercaptoethanol in a Waring blender. The buffer was used at a rate of 3 ml/g of wet mycelium. The homogenate was clarified by emulsification with an equal volume of chloroform and the emulsion was broken by centrifugation at 5,000 rpm in a Beckman JA-14 rotor. The upper aqueous layer was then subjected to two cycles of differential centrifugation with all high speed pellets suspended in the 0.1 M phosphate buffer, pH 7.4. The first cycle consisted of ultracentrifugation for 2.5 h at 27,000 rpm in a Beckman Type-30 rotor followed by centrifugation at 10,000 rpm for 10 min in a Beckman JA-20 rotor. The resultant supernatant was then centrifuged at 40,000 rpm for 1.5 h in a Beckman 50Ti rotor. These second high-speed pellets were resuspended in the same phosphate buffer and centrifuged at 10,000 rpm for 10 min in a Beckman JA-20 rotor. Final purification was performed by rate zonal centrifugation in sucrose gradients (100–400 mg/ml) made in 0.1 M sodium phosphate, pH 7.4. The gradients were centrifuged at 24,000 rpm for 2.5 h in a Beckman SW28 rotor and the UV-absorbing band was collected by puncturing the tube with a syringe. This harvested sample was diluted 1:1 with the sodium phosphate buffer and centrifuged at 40,000 rpm in a Beckman 50Ti rotor. The final pellet was suspended in buffer A (0.05 M Tris-HCl, pH 7.6, containing 150 mM NaCl and 5 mM EDTA), and stored at 4°C for short periods before analysis.
2.2. CryoTEM and 3D image reconstruction of FpV1
Small aliquots (~2–3 μl) of purified FpV1 virions were vitrified, and cryoTEM image data were recorded essentially as described for earlier work on PsV-S (Ochoa et al., 2008). For FpV1, a total of 240 micrographs were recorded in an FEI Polara microscope on a 4K2 Gatan Ultrascan CCD camera at a nominal magnification of 76,900, yielding an effective pixel size of 1.95 Å. All FpV1 images were acquired with the automated LEGINON system (Suloway et al., 2005) from specimens maintained at liquid nitrogen temperatures. Image data were recorded under low-dose conditions (~15 e/Å2), with 200 keV electrons, at underfocus settings ranging from 1.0 to 2.6 μm. Only those micrographs that exhibited minimal specimen drift and astigmatism were selected for further processing.
The program RobEM (http://cryoem.ucsd.edu/programDocs/runRobEM.txt) was used to extract individual particle images and to estimate the defocus level of each micrograph. A total of 16,522 particle images were boxed, each 2712 pixels in size. Image processing was carried out similarly as for both PsV-F and PsV-S (Tang et al., 2010). Briefly, the current version of AUTO3DEM (Yan et al., 2007b) was used to perform initial steps in constructing a model by the random method (Yan et al., 2007a) and then to refine it to about 8-Å resolution. The subroutines P3DR and PO2R were next used as stand-alone applications to refine and improve the map manually while adjusting different combinations of inverse temperature factor (Havelka et al., 1995) and amplitude-weighting schemes (Bowman et al., 2002) as described (Tang et al., 2010). The final cryoTEM map of FpV1 has been deposited in the EMBL-EBI Electron Microscopy Database (ID code EMD-5171).
2.3. Uses of structure data for other viruses
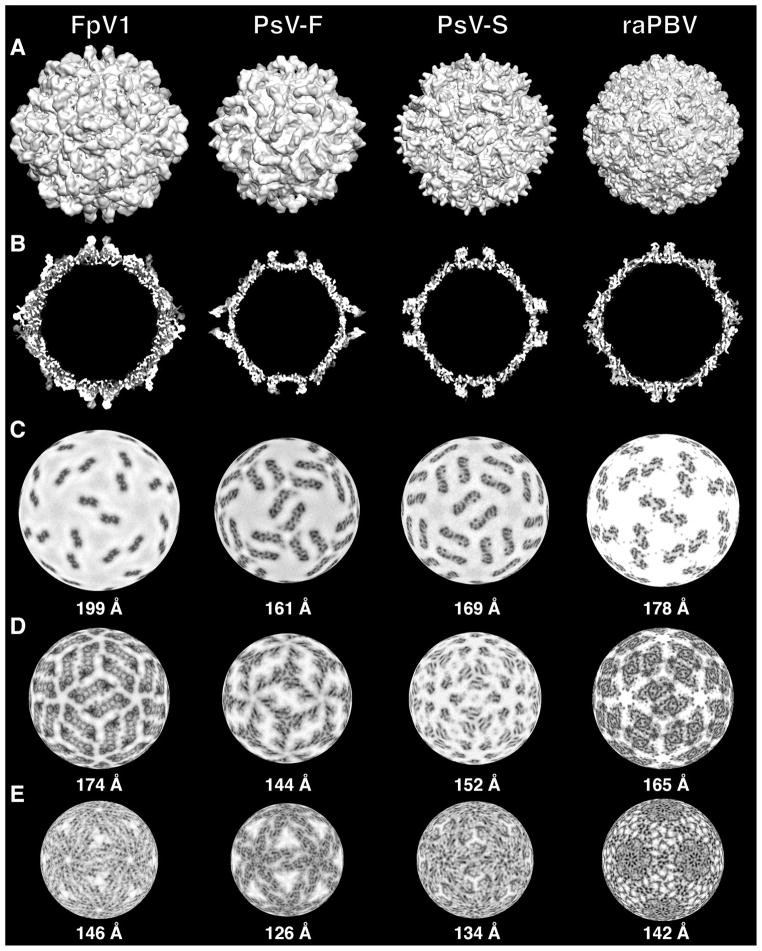
For use in Figs. 3 and 5, cryoTEM maps of PsV-F and -S capsids were available from recent work by some of the current authors but can also be obtained from the EMBL-EBI Electron Microscopy Database (http://www.ebi.ac.uk/pdbe/emdb/) (ID codes EMD-5161 and EMD-5163, respectively). For use in Fig. 5, full capsid coordinates of raPBV were obtained from the VIPER database (http://viperdb.scripps.edu/) (Reddy et al., 2001) and used in the EMAN program module pdb2mrc (Ludtke et al., 1999) to generate a simulated 3D density map at 8-Å resolution. This map was then displayed and analyzed with the programs UCSF Chimera (Goddard et al., 2007) and RobEM.
Fig. 3.
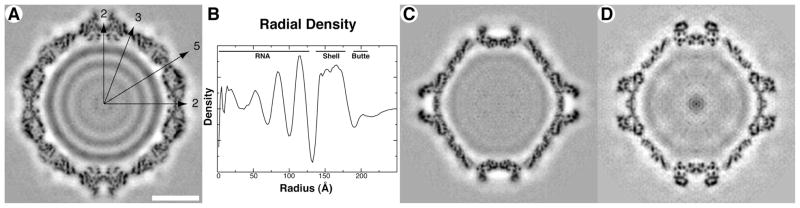
Cryo-reconstruction of FpV1. (A) Equatorial section with density values coded in grayscale (lowest density in white). Icosahedral symmetry axes (2, 3, 5) are labeled in the upper right quadrant. Scale bar, 100 Å. (B) Radial density plot. Evident layers in FpV1 are labeled at top. (C, D) Same as for panel A except for partitiviruses PsV-F (C) and PsV-S (D).
Fig. 5.
Comparisons of FpV1, PsV-F, PsV-S, and raPBV capsids. CryoTEM maps of the partitiviruses and a simulated density map of raPBV were used to generate the images (see Materials and methods). (A) Surface views of maps all rendered at 8-Å resolution. (B) Threshold-rendered equatorial sections. (C–E) Density-coded radial sections at the indicated radii through the capsid regions of each virus. The different radii were chosen to allow side-by- side comparison of morphologically analogous regions of the different capsids: protruding domains (C), upper part of the shell domains (D), and lower part of the shell domains (E).
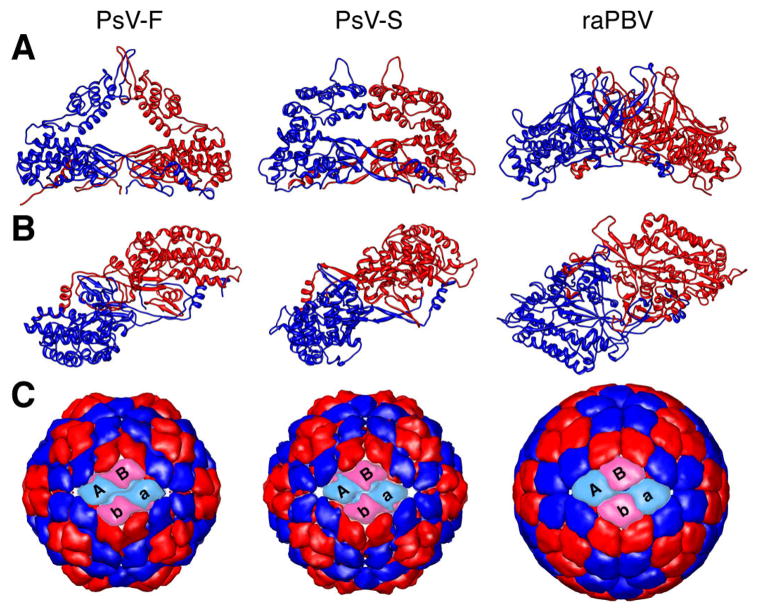
For use in Fig. 6A and B, atomic coordinates for the CP dimers of PsV-F and PsV-S were available from recent work by some of the current authors but can also be obtained from the RCSB Protein Data Bank (www.rcsb.org/pdb/) (ID codes PBD-3ES5 and PDB-3IYM, respectively). Atomic coordinates for the CP dimer of raPBV were obtained from the RCSB Protein Data Bank (ID code PDB-2VF1). For use in Fig. 6C, we generated models of the PsV-F, PsV-S, and raPBV capsids at 8-Å resolution using only the Cα atoms within the shell domain of each CP subunit and rendering those with the MULTISCALE MODELS module from UCSF Chimera. The rendered domains included aa residues 42–191 and 314–420 of PsV-F CP, and aa residues 39–171 and 339–434 of PsV-S CP. For raPBV, only aa residues at radii lower than 163 Å were rendered.
Fig. 6.
Further comparisons of CP dimers in PsV-F, PsV-S, and raPBV. (A) Side views of the backbone trace of the CP dimer of each virus (CPA, blue; CPB, red), which occupies the asymmetric unit of the T=1 icosahedron. (B) Same as in panel A, but rotated by 90° (horizontal axis) to place the protruding domains closest to the viewer. Each dimer is also rotated by 37° counterclockwise on the page to match the angular orientation of the left front dimer in panel C. (C) Outside view of the capsid of each virus, constructed from only the Cα atoms of the CP shell domains (see Materials and methods). The two front CP dimers in each capsid are shown in cyan and pink instead of blue and red. The respective front dimers are labeled A–B and a–b.
3. Results and discussion
3.1. Purified FpV1 virions
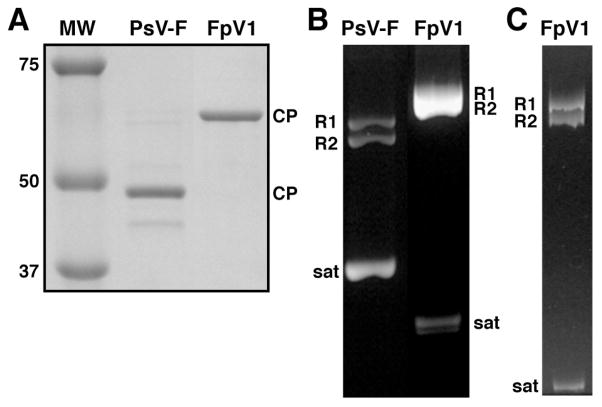
Large quantities of purified FpV1 virions were obtained from the F. poae culture by differential centrifugation and gradient sedimentation. SDS/polyacrylamide-gel electrophoresis of the virion sample showed a single, major protein band, with a relative molecular weight near 65,000 (Fig. 1A), consistent with a molecular mass of 70 kDa expected for FpV1 CP based on its reported sequence. Agarose-gel electrophoresis of RNAs in the virion sample showed a major, broad band with a slower mobility than those of the genomic dsRNAs of PsV-F (1500- and 1677-bp), which were run as markers in an adjacent lane (Fig. 1B). When a smaller sample of virion RNAs was run on a polyacrylamide gel, the major RNA band more clearly separated into a doublet (Fig. 1C), consistent with the two similarly sized genomic dsRNAs of FpV1 (2185- and 2203-bp). Two closely migrating, smaller RNAs were also present in our preparation of FpV1 virions (Fig. 1B, C). These RNAs exhibited a faster mobility than the 677-bp satellite dsRNA of PsV-F (Fig. 1B) and are consistent with the presence of an ~550-bp satellite dsRNA previously observed in virion preparations from this same F. poae strain by Compel et al. (1998).
Fig. 1.
Protein and RNA gels of purified PsV-F and FpV1 virions. (A) SDS/polyacrylamide gel stained with Coomassie blue. Molecular weight (MW) markers were run in one lane as shown, with approximate masses in kDa labeled at left. Positions of the PsV-F (middle lane) or FpV1 (right lane) CP are labeled at right. (B) Agarose gel stained with ethidium bromide. Positions of the PsV-F (left lane) or FpV1 (right lane) genomic dsRNA1 (R1), genomic dsRNA2 (R2), or satellite RNA (sat) are labeled. (C) Nondenaturing 5% polyacrylamide gel stained with ethidium bromide. Labeling as in panel B.
3.2. Electron cryomicrographs of FpV1 virions
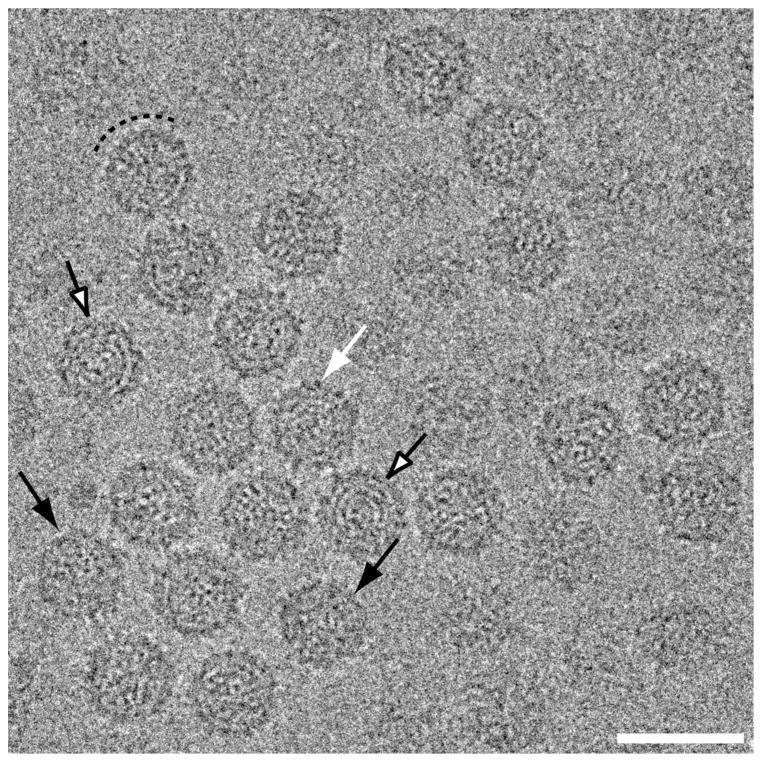
Transmission electron micrographs were recorded from unstained, vitrified samples of purified FpV1 virions (see Materials and methods). These cryomicrographs showed fields of regularly sized, intact particles with diameters near 400 Å (Fig. 2). The projected profiles of individual particles are largely round, though some particles exhibit a more angular profile (Fig. 2, black arrows). The only clear surface features are short (~15-Å) projections that are visible on most particles upon closer inspection (Fig. 2, white arrow). In some particles, the capsid has a beaded appearance suggestive of the presence of morphological capsomers or subunits (Fig. 2, dashed semi-circle). A few particles show RNA fingerprints in the central regions (Fig. 2, open arrows).
Fig. 2.
Transmission electron micrograph of vitrified sample of purified FpV1 virions. Most particles exhibit circular profiles, but some are more angular (black arrows). Short surface projections are visible on some particles (white arrow). Many capsid regions have a beaded appearance suggestive of morphological capsomers (dashed arc), and some particles exhibit RNA fingerprints in the central regions (open arrows). Scale bar, 500 Å.
3.3. CryoTEM reconstruction of FpV1 virions
Using the same basic procedures as had worked well with PsV-S and -F images in our other recent study (Tang et al., 2010), we obtained a novel cryoTEM reconstruction (cryo-reconstruction) from >16,000 particle images of FpV1 virions, first to about 8-Å resolution and ultimately to an estimated resolution of 5.6 Å based on a conservative, Fourier-shell correlation score of 0.5 (van Heel and Schatz, 2005). Given the striking appearances of structural features in the final reconstruction, especially the ridges and troughs of α-helical elements (see below), however, we conclude that its effective resolution approaches 5 Å.
An equatorial section view of the FpV1 cryo-reconstruction highlights the approximately circular profile of the capsid outer surface (Fig. 3A). The outermost and innermost diameters of the capsid are near 420 Å and 270 Å, respectively. The beaded appearance of the capsid is very evident and explained by thinned areas between the CP subunits near the icosahedral fivefold (I5), threefold (I3), and twofold (I2) symmetry axes. Projections, all ~25 Å in height, flank the I2 axes and represent structures analogous to the surface arches of PsV-F and -S (see below). Without these projections, the outermost diameter of the capsid is near 370 Å. Punctate and linear features within the capsid region of the reconstructed density map appear likely to represent secondary-structure elements including α-helices and β-sheets. Three well-defined, regularly spaced rings of density, with an average center-to-center distance of ~30 Å and attributable to genomic RNA, are evident in the particle interior. A radially averaged density plot provides another perspective on the radial distribution of the capsid and RNA features within the FpV1 virions (Fig. 3B).
3.4. 3D structure of FpV1 capsid
The outer surface topography of the FpV1 capsid is quite convoluted (Fig. 4A). Deep depressions occur at each of the icosahedral symmetry axes, with the broadest pits encompassing the I5 and I3 axes. Each I2 axis is crossed by a long and narrow, linear canyon, with capsid structures rising steeply on both sides. A second long and narrow, linear canyon, somewhat shallower than the preceding one, is found on the distal side of each of these elevated regions and runs nearly parallel to the I2-crossing canyon. All of these depressions interconnect to create a more-or-less continuous network on the capsid surface and circumscribe the elevated regions as discrete morphological elements.
Fig. 4.
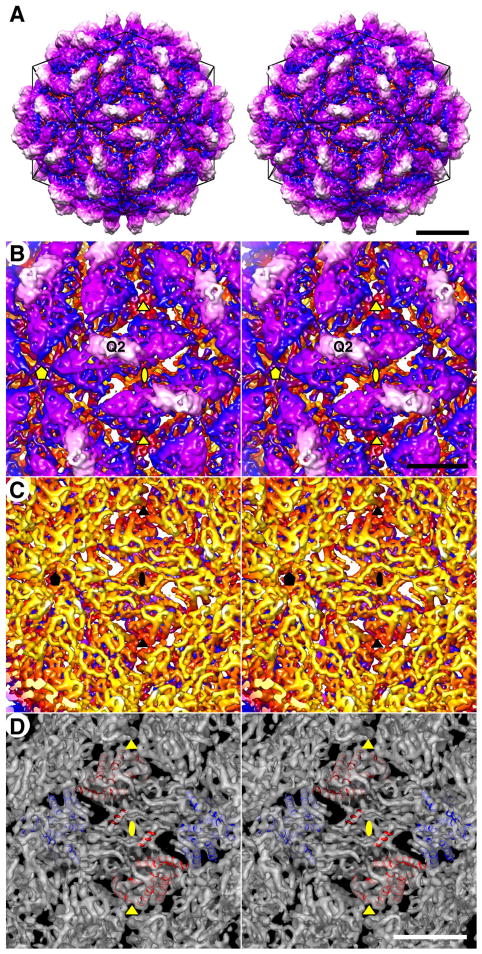
FpV1 capsid structure. (A) Stereo view of the outer surface along an I2 axis. A model icosahedron is superimposed (black lines). The capsid is radially depth-cued, from yellow (innermost); through rust, blue, and purple; to white (outermost) (also applies to panels B and C). The density threshold for this display is 1σ. Scale bar, 100 Å. (B) Magnified stereo view of the outer surface. The density threshold for this display is 2 σ to reveal additional features of the capsid densities (also applies to panels C and D). Encompassed I2, I3, and I5 axes are respectively marked by ellipse, triangle, and pentagon symbols (also applies to panels C and D), and one Q2 axis is also labeled. Scale bar, 50 Å (also applies to panel C). (C) Stereo view of the inner surface. (D) Stereo view of the inner surface with modeled helices for the CP dimers in two adjacent asymmetric units (CPA, blue; CPB, red). Scale bar, 50 Å.
The elevated regions cover the remainder of the capsid surface, number 60 in total, and are identical to one another. As implied above, each elevated region runs parallel alongside one of the I2-spanning canyons and at lower radii has the profile of an elongated parallelogram or oval (Fig. 4A, B; blue and purple shading, respectively). At these lower radii each elevation has the appearance of a broader “mesa”, which is then topped by a further-projecting, centrally located “butte” (Fig. 4A, B; white shading). The long axes of the mesas and buttes adopt different angles relative to the line between adjacent I5 axes in the views in Fig. 4A and B: about a 15° angle for the mesa vs. about a −15° angle for the butte. Notably, each elevation appears to be strongly twofold symmetrical, with the butte spanning the quasi-twofold (Q2) axis. These 60 dimeric elevations of FpV1 are therefore consistent with the capsid structure and organization of partitiviruses PsV-F and -S (Ochoa et al., 2008; Pan et al., 2009; Tang et al., 2010), with the elevated complexes representing 60 quasisymmetrical CP dimers and the buttes being analogous to the dyadic arches of the two other partitiviruses.
Inside views of the FpV1 capsid reveal the inner surface topography to be smoother than the outer (Fig. 4C). However, deep pits do occur at each of the icosahedral symmetry axes, as better visualized in the equatorial section (see Fig. 3A). Especially in the interior surface view, many ridged, tubular density features are visible within the capsid map, consistent with modeling as α-helices (Fig. 4D). Moreover, such modeling allows one to recognize that these inner regions of the capsid, directly underlying the two halves of the dyadic elevations, are also related by a Q2 axis (Fig. 4C, D). These findings thus further contribute to interpreting the FpV1 capsid as comprising 60 quasisymmetrical CP dimers.
Each of the CP dimers in FpV1 contains two quasi-equivalent types of CP subunit. These are designated subunit CPA, which lies nearer the I5 axis (Fig. 4D, blue helices), and subunit CPB, which lies nearer the I3 axis (Fig. 4D, red helices), according to the terminology adopted for other dsRNA viruses (Castón et al., 1997; Grimes et al., 1998). The ~5-Å resolution limit of the current map is insufficient to allow us to define the connectivity of the different secondary-structure elements unambiguously. Hence, we are unable yet to report a de novo chain trace for any significant stretch of the CP backbone to make reliable comparisons with other viruses.
One other notable aspect of the FpV1 capsid organization is that a CP dimer runs along both sides of each I2-spanning canyon and in an antiparallel orientation to each other (Fig. 4B, D). Together, they define a twofold-symmetrical, compact, diamond-shaped tetramer. These tetramers represent likely assembly intermediates, 30 of which would associate to form the full FpV1 capsid, as previously suggested for PsV-F and -S (Pan et al., 2009; Tang et al., 2010).
3.5. Further comparisons of FpV1 with PsV-F and -S
The limited resolution of the FpV1 cryo-reconstruction and lack of a backbone trace for the CP restricts comparisons with the CP structures of PsV-F and -S, yet some other comparisons of these three partitiviruses are informative. Consistent with its substantially more massive CP subunits, the FpV1 capsid appears to have a much greater average thickness and a slightly larger diameter than either of the other capsids (Fig. 3; Fig. 5A, B). The FpV1 capsid also has a much rounder profile, giving it the appearance of being more fully “stuffed”. FpV1 has prominent surface projections analogous to, yet distinct from, the arches in PsV-F and -S (Fig. 3; Fig. 5A, B). Indeed, the projections from the two subunits in each CP dimer in FpV1 do not interact in a manner that produces a passageway and hence are better described as “buttes” instead of “arches”. In all three partitiviruses, these dimeric outermost regions adopt unique angles relative to the line between adjacent I5 vertices: about −15° in FpV1 vs. about 66° in PsV-F and 37° in PsV-S (Fig. 5A, C). Despite such differences, the shell domains of the CP subunits in FpV1 are aligned almost identically to those in PsV-F and -S, as readily seen in radial sections through the shell domains of each virus (Fig. 5D, E). In these sections, the dimeric shell regions in all three viruses adopt a 35–40° angle relative to the line between adjacent I5 vertices. The shell domain in each also appears to be predominantly formed from complex bundles of α-helical elements, consistent with secondary-structure predictions indicating high α-helical content in each and also consistent with the strongly α-helical make-ups of other dsRNA viruses including members of the families Reo- and Totiviridae (Grimes et al., 1998; Naitow et al., 2002). Thin “rods” of density appear to extend from the capsid into the outer ring of RNA density in several locations in the FpV1 cryo-reconstruction. These extensions are less regular and less well defined than those reported for PsV-F and -S (Pan et al., 2009; Tang et al., 2010), and in the absence of a chain trace for FpV1 CP, we cannot yet assign these densities to the CP N-terminus as done for the two other viruses.
Curiously, the RNA rings in FpV1 have similar spacings to those in PsV-F and -S (25–35 Å), but the densities of the two outer rings in FpV1 are much greater than those in the other two viruses (Fig. 3A, C, D). As highlighted in Table 1, the two genome segments of FpV1 differ in length by less than 1% (Compel et al., 1998), whereas those of PsV-F and -S differ by about 12% and 11%, respectively (Kim et al., 2003, 2005). Since partitiviruses are known to package their segments into separate virus particles, and given that all partitivirus cryo-reconstructions to date have included a mixture of images of both particle types, the preceding numbers indicate that FpV1 should have less variation in the length of RNA packaged per particle than do PsV-F and -S. This greater consistency of packaging density would in turn be expected to reinforce the RNA ring densities at particular radii more strongly in FpV1 than in PsV-F and -S, as observed in Fig. 3. However, both FpV1 and PsV-F additionally package satellite RNAs (see Fig. 1B, C), which may complicate this simple explanation.
3.6. Partitivirus phylogenetics and taxonomy
Previous phylogenetic analyses have indicated that there are least two distinguishable subclusters of viruses within the current genus Partitivirus (Boccardo and Candresse, 2005; Crawford et al., 2006; Ghabrial et al., 2008; Willenborg et al., 2009). Characterized members of one subcluster, including PsV-F and -S, have CPs and RdRps sized 44–50 and 60–63 kDa, respectively, whereas characterized members of the other subcluster, including AhV and FpV1, have CPs and RdRps sized 70–77 and 77–87 kDa, respectively (Table 1). The genome segments of the two subclusters are also differently sized, consistent with their different encoded protein sizes: 1445–1787 and 2135–2363 bp for genome segments of the PsV-S/PsV-F and the AhV/FpV1 clusters, respectively (Table 1). Structural evidence in this study corroborates that the FpV1 capsid possesses several distinct features relative to those of PsV-F and -S. In taxonomic regards, therefore, we favor dividing the genus Partitivirus into two new genera (perhaps Alpha- and Betapartitivirus) to reflect these two subclusters, each of which is represented by at least eight formally recognized species (Ghabrial et al., 2008). The FpV1 structure described here would thereby be the first 3D structure reported for a member of its new genus.
3.7. Comparisons of partitivirus and picobirnavirus structures
Partiti- and picobirnaviruses both have bisegmented dsRNA genomes and small (<450-Å-diameter) capsids with 120-subunit T=1 symmetry. Picobirnaviruses have recently been grouped in a separate taxonomic family because of a variety of differences from partitiviruses including (i) infection of vertebrate hosts; (ii) probable capacity for effective extracellular transmission; (iii) probable copackaging of both genome segments in the infectious virion; (iv) possession of a second open reading frame (ORF) in the CP-encoding genome segment; (v) autoproteolytic maturation of the CP; and (vi) phylogenetically distinct CP and RdRp sequences (Wakuda et al., 2005; Duquerroy et al., 2009). Recent structure determinations for members of both families (Duquerroy et al., 2009; Pan et al., 2009; Tang et al., 2010; this study) allow for comparisons of their capsid architectures as described below.
The various structural views in Fig. 5 show a number of similarities between the three structurally characterized partitiviruses and the one structurally characterized picobirnavirus. The capsid diameter and thickness of raPBV are within the ranges defined by the partitiviruses (Fig. 5A, B). The protruding domains in raPBV do not form arches but yet are quite similar in size and position to the “buttes” of FpV1 (Fig. 5A–C), even adopting a similar angle (about -15°) relative to the line between adjacent I5 vertices. In addition, the raPBV shell domains have essentially the same alignment and angle (about 37°) as do those in the three partitiviruses (Fig. 5D, E). Simplified displays of the PsV-F, PsV-S, and raPBV capsids at 8-Å resolution, constructed from the backbone traces of only the Cα atoms of the shell domains, graphically demonstrate their shared organizations (Fig. 6C). Thus, though unique details with regard to specific densities are evident in each of the four viruses, they overall appear to share very similar architectures, especially in the shell domains. Greater divergence is evident in the protruding domains, though not in ways that clearly differentiate the two families. Notably, the protruding domain of FpV1 appears generally more similar to that of raPBV than to that of either PsV-F or PsV-S.
One general architectural difference that can be identified among these four viruses regards the pattern of contacts between the CP shell domains across the I2 axes of each capsid. In the original, 7.3-Å structure of PsV-S, densities were tentatively assigned to CPA vs. CPB subunits such that contacts across the I2 axes were interpreted to be CPB:CPB only (Ochoa et al., 2008). With the subsequent PsV-F crystal structure and identification of domain swapping between CPA and CPB, however, it became clear that the swapped regions mediate contacts across the I2 axes and are in fact CPA:CPA only (Pan et al., 2009). Because of the limited resolution of the cryo-reconstruction, we cannot yet interpret this architectural detail for FpV1. Nevertheless, in the raPBV crystal structure, in addition to CPA:CPA contacts from the swapped regions, there are CPB:CPB contacts across the I2 axes (Duquerroy et al., 2009). Thus, this varying pattern of I2 contacts between shell domains is a basic difference between at least some partiti- and picobirnaviruses. The additional CPB:CPB contacts observed in raPBV may contribute to stabilizing the virion during its extracellular spread between host cells or host individuals. Notably, however, the protruding domains of all four viruses are centered on the Q2 axis of the CP dimer and do not interact across the I2 axis with the protruding domains from the adjacent CP dimer within the diamond-shaped tetramer.
A detailed comparison of the protein folds of the partiti- and picobirnavirus CP dimers, which is the subject of an ongoing investigation, shows that these viruses share a capsid organization and CP fold that are distinct in several respects from those of other dsRNA viruses (Tao et al., manuscript in preparation). Thus, they likely have a shared evolutionary lineage and might even warrant grouping in a separate taxonomic order. Partiti- and picobirnaviruses exhibit a number of basic biological differences between them, however, which leads us to support their continued assignment to separate taxonomic families.
4. Conclusions
The 3D structure reported here for a third fungal partitivirus, FpV1, broadens our understanding of conserved and variable structural elements among partiti- and picobirnaviruses and supports the suggestion that a phylogenetic subcluster of partitiviruses closely related to FpV1 should constitute a separate taxonomic genus. Ongoing studies of partitiviruses from other genera or phylogenetic subclusters, including viruses obtained from plant and protozoan hosts, will lead us towards a fuller appreciation of the structural diversity within this family and of the evolutionary path(s) between partiti-, picobirna- and other dsRNA viruses.
The roles of the protruding domains in partitivirus biology remain a mystery. In picobirnaviruses, these domains appear likely to function in binding to cell-surface receptors and are also recognized by host antibodies (Duquerroy et al., 2009). Given that partitiviruses lack the capacity for efficient extracellular spread, however, a surface-receptor-binding function for their protruding “arches” and “buttes” appears very unlikely. Although the protruding domains certainly contribute to stabilizing the CP dimers by providing additional A:B contacts above the shell domain, they seem to be overly elaborate structures to fulfill simply that role. This raises a number of questions. For example, are partitiviruses capable of nonpersistent transmission by invertebrate vectors that have yet to be discovered, and might the protruding domains have a function in vector attachment for such transmission? Also, might the protruding domains interact with one or more host factors relating to localization inside the cell during partitivirus multiplication? Partitivirus virions are thought to accumulate in large, cytoplasmic aggregates enclosed in vesicles (Border et al., 1972). The protruding domains might therefore play a role in forming the virion aggregates or promoting envelopment by membranes. Such sequestration might enhance virus survival and maintenance of persistent infection since dsRNA that may be released from free virions is a potent inducer of fungal host defenses (Segers et al., 2007).
The partitivirus RdRp has not been clearly identified in any of the icosahedrally averaged virion structures to date, as would be expected if there is only one RdRp molecule per particle (Buck and Kempson-Jones, 1974). To ensure efficient packaging, the RdRp appears likely to be noncovalently bound either (i) to the inner surface of the capsid as well as to the genome segment or (ii) to only the genome segment. Anchoring of RdRp molecules specifically near transcript-exit sites in the capsids of other dsRNA viruses has been previously demonstrated or discussed (Prasad et al., 1996; Castón et al., 1997; Zhang et al., 2003) and has also been proposed for partitiviruses (Ochoa et al., 2008; Pan et al., 2009; Tang et al., 2010). Pores through the shell (e.g., at the I5 and I3 axes; see Fig. 6) are logical candidates to be sites for plus-strand RNA exit during partitivirus transcription. Hence, the partitivirus RdRp seems likely to be anchored near at least one of these pores in each virus particle. It also remains possible that transcripts are first directed into the particle interior and only later released. However, if this were true, there would still be the question of which sites in the shell serve as transcript exit points, as well as other important questions such as how these exit sites are “discovered” by the interior transcripts and how entangling of transcript and genome segment might be avoided in this case. Future studies, such as by electron cryotomography, may provide a means to resolve the position(s) of the partitivirus RdRp molecule(s).
Acknowledgments
We thank R.S. Sinkovits and N.H. Olson for expert advice on ways to enhance the computer processing and imaging aspects of this study. We also thank Y.J. Tao (Rice University) for helpful comments on the manuscript. This work was supported by NIH grants R37 GM033050 and 1S10 RR020016 to T.S.B. and by Dept. of Agriculture National Research Initiative Competitive Research Program grant 2001-35319-10010 to S.A.G. The San Diego Supercomputer Center provided access to TeraGrid computing, and support from the University of California-San Diego and the Agouron Foundation to T.S.B. were used to establish and equip cryoTEM facilities at the University of California-San Diego.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boccardo G, Candresse T. Complete sequence of the RNA1 of an isolate of White clover cryptic virus 1, type species of the genus Alphacryptovirus. Arch Virol. 2005;150:399–402. doi: 10.1007/s00705-004-0422-8. [DOI] [PubMed] [Google Scholar]
- Border DJ, Buck KW, Chain EB, Kempson-Jones GF, Lhoas P, Ratti G. Viruses of Penicillium and Aspergillus species. Biochem J. 1972;127:4–6. doi: 10.1042/bj1270004p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol. 2002;76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozarth RF, Wood HA, Mandelbrot A. The Penicillium stoloniferum virus complex: two similar double-stranded RNA virus-like particles in a single cell. Virology. 1971;45:516–523. doi: 10.1016/0042-6822(71)90352-7. [DOI] [PubMed] [Google Scholar]
- Buck KW, Kempson-Jones GF. Biophysical properties of Penicillium stoloniferum virus S. J. Gen. Virol. 1973;18:223–235. doi: 10.1099/0022-1317-18-3-223. [DOI] [PubMed] [Google Scholar]
- Buck KW, Kempson-Jones GF. Capsid polypeptides of two viruses isolated from Penicillium stoloniferum. J Gen Virol. 1974;22:441–445. [Google Scholar]
- Castón JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compel P, Papp I, Bibo M, Fekete C, Hornok L. Genetic relationships and genome organization of double-stranded RNA elements of Fusarium poae. Virus Genes. 1998;18:49–56. doi: 10.1023/a:1008069318838. [DOI] [PubMed] [Google Scholar]
- Crawford LJ, Osman TA, Booy FP, Coutts RH, Brasier CM, Buck KW. Molecular characterization of a partitivirus from Ophiostoma himal-ulmi. Virus Genes. 2006;33:33–39. doi: 10.1007/s11262-005-0028-6. [DOI] [PubMed] [Google Scholar]
- Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Eighth Report of the International Committee for the Taxonomy of Viruses. Elsevier/Academic Press; San Diego: 2005. Virus Taxonomy. [Google Scholar]
- Fekete C, Giczey G, Papp I, Szabó L, Hornok L. High-frequency occurrence of virus-like particles with double-stranded RNA genome in Fusarium poae. FEMS Microbiol Lett. 1995;13:295–299. doi: 10.1111/j.1574-6968.1995.tb07790.x. [DOI] [PubMed] [Google Scholar]
- Ghabrial SA, Buck KW, Hillman BI, Milne RG. Partitiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; San Diego: 2005. pp. 581–590. [Google Scholar]
- Ghabrial SA, Ochoa WF, Baker TS, Nibert ML. Partitiviruses: general features. In: Mahy BWJ, van Regenmortel MHV, editors. Encyclopedia of Virology. 3. Vol. 4. Elsevier/Academic Press; San Diego: 2008. pp. 68–75. [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Green J, Gallimore CI, Clewley JP, Brown DWG. Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch Virol. 1999;144:2457–2465. doi: 10.1007/s007050050658. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Ziéntara S, Mertens PP, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Havelka WA, Henderson R, Oesterhelt D. Three-dimensional structure of halorhodopsin at 7 Å resolution. J Mol Biol. 1995;247:726–738. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- Jiang D, Ghabrial SA. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J Gen Virol. 2004;85:2111–2121. doi: 10.1099/vir.0.79842-0. [DOI] [PubMed] [Google Scholar]
- Khramtsov NV, Woods KM, Nesterenko MV, Dykstra CC, Upton SJ. Virus-like, double-stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol Microbiol. 1997;26:289–300. doi: 10.1046/j.1365-2958.1997.5721933.x. [DOI] [PubMed] [Google Scholar]
- Kim JW, Choi EY, Lee JI. Genome organization and expression of the Penicillium stoloniferum virus F. Virus Genes. 2005;31:175–183. doi: 10.1007/s11262-005-1793-y. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim SY, Kim KM. Genome organization and expression of the Penicillium stoloniferum virus S. Virus Genes. 2003;27:249–256. doi: 10.1023/a:1026343831909. [DOI] [PubMed] [Google Scholar]
- Leoni F, Gallimore CI, Green J, McLauchlin J. Molecular epidemiological analysis of Cryptosporidium isolates from humans and animals by using a heteroduplex mobility assay and nucleic acid sequencing based on a small double-stranded RNA element. J Clin Microbiol. 2003;41:981–992. doi: 10.1128/JCM.41.3.981-992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Woods KM, Upton SJ, Ghabrial SA. Cryspovirus: a new genus of protozoan viruses in the family Partitiviridae. Arch Virol. 2009;154:1959–1965. doi: 10.1007/s00705-009-0513-7. [DOI] [PubMed] [Google Scholar]
- Ochoa WF, Havens WM, Sinkovits RS, Nibert ML, Ghabrial SA, Baker TS. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure. 2008;16:776–786. doi: 10.1016/j.str.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Hillman BI. Genome organization of a partitivirus from the filamentous ascomycete Atkinsonella hypoxylon. J Gen Virol. 1995;76:1461–1470. doi: 10.1099/0022-1317-76-6-1461. [DOI] [PubMed] [Google Scholar]
- Pan J, Dong L, Lin L, Ochoa WF, Sinkovits RS, Havens WM, Nibert ML, Baker TS, Ghabrial SA, Tao YJ. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci USA. 2009;106:4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot O, O’Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Rothnagel R, Zeng CQY, Jakana J, Lawton JA, Chiu W, Estes MK. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Natarajan P, Okerberg B, Li K, Damodaran KV, Morton RT, Brooks CL, 3rd, Johnson JE. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J Virol. 2001;75:11943–11947. doi: 10.1128/JVI.75.24.11943-11947.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein SA. Fusarium poae: a pathogen that needs more attention. J Plant Path. 2009;91:25–36. [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang J, Pan J, Havens WM, Ochoa WF, Guu TSY, Ghabrial SA, Nibert ML, Tao YJ, Baker TS. Backbone trace of partitivirus capsid protein from electron cryomicroscopy and homology modeling. Biophys J. 2010 doi: 10.1016/j.bpj.2010.04.058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods. 2005;126:165–169. doi: 10.1016/j.jviromet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Willenborg J, Menzel W, Vetten HJ, Maiss E. Molecular characterization of two alphacryptovirus dsRNAs isolated from Daucus carota. Arch Virol. 2009;154:541–543. doi: 10.1007/s00705-009-0314-z. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM—an automated and high-throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]