SUMMARY
RIP1 and RIP3 kinases are central players in TNF-induced programmed necrosis. Here, we report that the RIP homotypic interaction motifs (RHIMs) of RIP1 and RIP3 mediate the assembly of heterodimeric filamentous structures. The fibrils exhibit classical characteristics of β-amyloids, as shown by Thioflavin T (ThT) and Congo red (CR) binding, circular dichroism, infrared spectroscopy, X-ray diffraction, and solid-state NMR. Structured amyloid cores are mapped in RIP1 and RIP3 that are flanked by regions of mobility. The endogenous RIP1/RIP3 complex isolated from necrotic cells binds ThT, is ultrastable, and has a fibrillar core structure, whereas necrosis is partially inhibited by ThT, CR, and another amyloid dye, HBX. Mutations in the RHIMs of RIP1 and RIP3 that are defective in the interaction compromise cluster formation, kinase activation, and programmed necrosis in vivo. The current study provides insight into the structural changes that occur when RIP kinases are triggered to execute different signaling outcomes and expands the realm of amyloids to complex formation and signaling.
INTRODUCTION
Recent studies have implicated the intracellular signaling kinase RIP1 as a key switch of cell fate regulation. Depending on the cellular context, RIP1 controls whether the pleiotropic cytokine TNF induces NF-κB activation, apoptosis, or programmed necrosis (Moquin and Chan, 2010). The E3 ligases cIAP1/2 and LUBAC ubiquitinate RIP1 in the TNFR1 signaling complex (Walczak, 2011). Polyubiquitinated RIP1 then engages downstream adaptors such as NEMO to activate IKK to promote NF-κB transcriptional activity, leading to cell survival, proliferation, and differentiation (Walczak, 2011). When RIP1 ubiquitination is blocked by removal of the E3 ligases cIAP1 and cIAP2 through genetic ablation, RNA interference (RNAi) knockdown, or inhibitor of apoptosis (IAP) antagonists, RIP1 forms a secondary complex in the cytosol with Fas-associated death domain (FADD) and caspase-8—termed the Ripoptosome—to initiate apoptotic cell death (Feoktistova et al., 2011; Tenev et al., 2011; Wang et al., 2008). Active caspase-8 within the Ripoptosome cleaves and inactivates RIP1 (Chan et al., 2003; Lin et al., 1999) and RIP3 (Feng et al., 2007). When caspases are inhibited by pharmacological inhibitors or under certain physiological conditions such as viral infections, RIP1 and RIP3 form the necrosome to initiate a third pathway known as programmed necrosis or necroptosis (Cho et al., 2009; He et al., 2009; Zhang et al., 2009).
The understanding of programmed necrosis is still unfolding. Whereas it was originally thought to be associated with nonspecific cellular damages, genetic experiments in mice clearly show that caspase-8-mediated cleavage and inactivation of RIP1 and RIP3 is critical for preventing extensive necrosis during embryonic development in order to ensure proper clonal expansion of lymphocytes and to prevent extensive necrosis and inflammation in skin and intestinal epithelium (Kaiser et al., 2011; Oberst et al., 2011; Welz et al., 2011; Zhang et al., 2011). In addition to caspase inhibition, assembly of the RIP1/RIP3 necrosome also requires intact RIP1 and RIP3 kinase activity (Cho et al., 2009). Recent studies identified MLKL, a kinase-like protein, as a substrate of the RIP3 kinase (Sun et al., 2012; Zhao et al., 2012).
The structural basis for the association between RIP1 and RIP3 within the necrosome is poorly understood. Both RIP1 and RIP3 contain Ser/Thr kinase domains (KDs) at their N-termini, and RIP1 also has a death domain (DD) at its C terminus for recruitment to the TNF receptor signaling complex (Stanger et al., 1995; Sun et al., 1999; Yu et al., 1999) and for formation of the Ripoptosome (Feoktistova et al., 2011; Tenev et al., 2011; Wang et al., 2008) (Figure 1A). Unique segments of homologous sequences in RIP1 and RIP3 (RIP homotypic interaction motifs, RHIMs) (Figures 1A and 1B) were shown to mediate their interaction (Sun et al., 2002), which is crucial for the induction of programmed necrosis (Cho et al., 2009). The RHIM is found in a growing number of signaling adaptors with crucial functions in cell death and innate immunity (Moquin and Chan, 2010). For instance, macrophage necrosis induced through TLR-3/4 requires RHIM-mediated interaction between the adaptor TRIF and RIP3 (He et al., 2011). Similarly, RHIM-mediated interaction between the intracellular DNA sensor DAI and RIP3 causes necrosis of cells infected with murine cytomegalovirus (Upton et al., 2012).
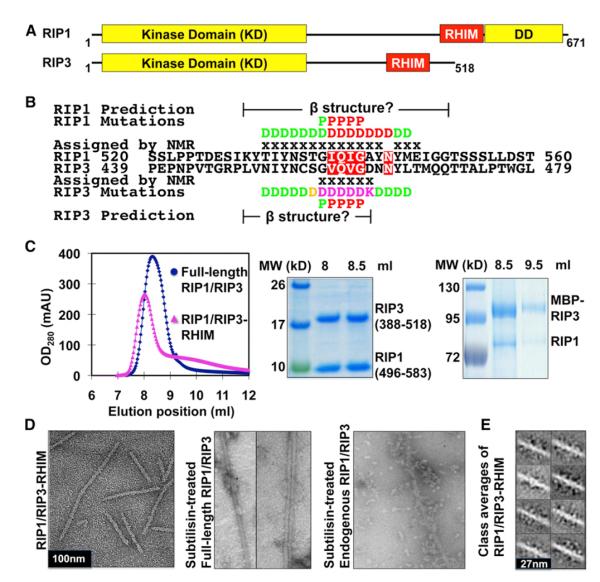
Figure 1. RIP1 and RIP3 Form a Filamentous Complex In Vitro and in Cells.
(A) Domain organization of human RIP1 and RIP3.
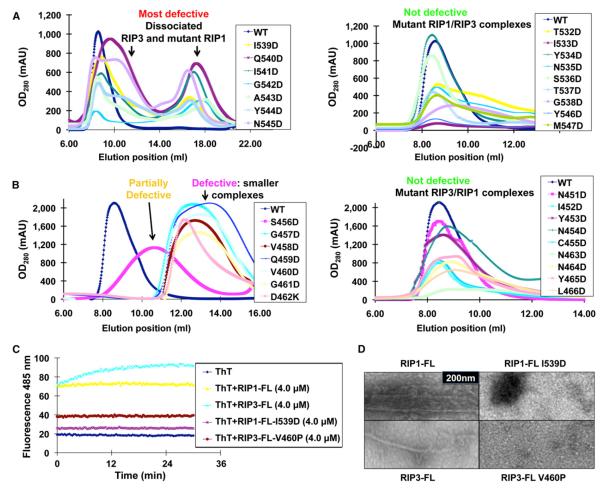
(B) Sequence alignment of RIP1 and RIP3 around the core RHIMs. Residues in RIP1 and RIP3 that are conserved across different species (Figure S1) are highlighted in red. Regions predicted to be in β sheet conformations are shown. Residues assigned by solid-state NMR are marked with “x.” Summary of mutagenesis results are displayed, with red indicating most defective mutants showing complete or partial dissociation between RIP1 and RIP3, magenta indicating defective mutants showing smaller complexes, orange indicating partially defective mutants with size of the complex between that of WT and defective mutants, and green indicating nondefective mutants.
(C) Coexpressed full-length and truncated RIP1/RIP3 complexes. Left, superimposed gel filtration profiles. Right, SDS-PAGE of the fractions.
(D) EM images of the RIP1/RIP3 complex.
(E) Representative class averages of the RIP1/RIP3-RHIM fibrils.
See also Figure S1.
Here, we show that RIP1 and RIP3 form an amyloid structure through their RHIMs and that this heterodimeric amyloid structure is a functional signaling complex that mediates programmed necrosis. Our results not only provide insights into the mechanism of RIP1 and RIP3 kinase activation but also further expand the realm of amyloid structures to normal physiological functions beyond those associated with human diseases (Eisenberg and Jucker, 2012).
RESULTS
The RIP1/RIP3 Complex Forms Filamentous Structures In Vitro and in Cells
The exact boundaries of RHIMs are unclear, but the sequence conservation is centered around the I(V)QI(V)G motif (Figure 1B and Figure S1A available online). We coexpressed RHIMs of RIP1 (residues 496–583) and His-tagged RIP3 (residues 388–518) (RIP1/3-RHIM) using regions that were previously shown to be sufficient for complex formation (Sun et al., 2002). The RIP1/3-RHIM complex copurified from Ni-affinity chromatography eluted around the void position of a Superdex 200 10/300 GL gel filtration column, which is much larger than the expected molecular mass of a heterodimer (Sun et al., 2002) (Figure 1C). We then coexpressed full-length RIP1 and RIP3 (RIP1/3-FL) in insect cells, which similarly coeluted around the void position of the gel filtration column (Figure 1C).
We investigated why RIP1/3-RHIM and RIP1/3-FL could form such large complexes. Secondary structure predictions suggested that the region between the KD and the DD in RIP1 (~residues 300–560) and the region C-terminal to the KD in RIP3 (~residues 300-end) are mostly unstructured random coils (Rost et al., 2004). The only exceptions are short segments of sequences around the I(V)QI(V)G motif, which show propensities for β strands (Figure 1B). Because amyloids are fibrous protein aggregates composed of cross-β cores, we asked whether RHIMs mediate assembly of amyloid-like fibrils.
We used electron microscopy (EM) to visualize the structures of the RIP1/RIP3 complexes. Consistent with our hypothesis, EM of negatively stained RIP1/3-RHIM revealed filamentous structures (Figure 1D). The fibrils exhibit a similar width of ~11–12 nm but vary in length. Class averages show substantial structural variability, but closer inspection suggests that the core of the complex, ~8 nm, may be ordered and that the variability may be mostly due to flexible extensions (Figure 1E). EM of negatively stained RIP1/3-FL showed mostly aggregates. We reasoned that the KDs and DD in full-length RIP1 and RIP3 might mediate additional interactions and mask the central amyloidal fibril architecture. Upon limited proteolysis to remove the flanking domains by subtilisin, the same enzyme used in the nuclear magnetic resonance (NMR) sample preparation (see section “Amyloid Core of the RIP1/RIP3 Complex”), clear fibril structures were revealed for RIP1/3-FL, similar to RIP1/3-RHIM (Figure 1D). When endogenous RIP1/RIP3 complex from HT-29 cells immunoprecipitated with anti-RIP1 antibody was treated with subtilisin and negatively stained, it showed mostly short and sometimes long filamentous structures under EM only upon necrosis induction with TNF, zVAD-fmk, and the IAP antagonist LBW242, but not before induction (Figures 1D and S1B).
The RIP1/RIP3 Complex Exhibits Classical Characteristics of Amyloid Fibrils
Amyloids are classically characterized using aromatic, cross-β binding dyes such as Thioflavin T (ThT) (LeVine, 1999) and Congo red (CR) (Klunk et al., 1989). To determine whether the RIP1/RIP3 fibrils are indeed amyloidal, we first characterized purified recombinant RIP1/RIP3 complexes in vitro. We added ThT to either RIP1/3-RHIM or RIP1/3-FL and measured its fluorescence after excitation at 430 nm. In comparison to ThT alone, RIP1/3-RHIM and RIP1/3-FL caused ThT to display an emission peak at ~485 nm with concomitant increase in fluorescence intensity (Figures 2A, 2B, and S2A). Similarly, CR showed a characteristic red shift from an absorption maximum of ~470 nm to ~540 nm upon addition of either RIP1/3-RHIM or RIP1/3-FL (Figures 2C and S2B) (Klunk et al., 1989). Therefore, both the ThT- and the CR-binding assays confirmed that the RIP1/RIP3 complex is amyloidal.
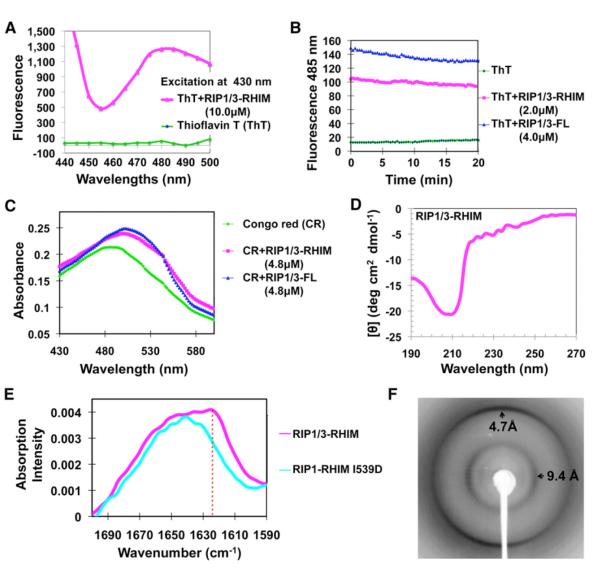
Figure 2. The RIP1/RIP3 Complex Is Amyloidal.
(A) Fluorescence emission spectra of ThT in the absence (green) and presence (magenta) of the RIP1/RIP3-RHIM complex.
(B) Both the RIP1/RIP3-RHIM and the full-length RIP1/RIP3 complex bind ThT.
(C) Absorption spectra of CR in the absence (green) and presence of either RIP1/RIP3-RHIM (magenta) or full-length RIP1/RIP3 complex (blue).
(D) Circular dichroism spectrum of the RIP1/RIP3-RHIM complex.
(E) Superimposed Fourier transform infrared spectra of RIP/RIP3-RHIM (magenta) and the I539D mutant of RIP1 (cyan). Only the WT RIP1/RIP3 complexes, not the RHIM mutant, showed the amide I’ maxima at 1,623 cm−1 (dashed vertical red line), which is characteristic of β-amyloid.
(F) An X-ray diffraction image of partially aligned RIP1/RIP3 fibrils. The arrows indicate equatorial and meridional reflections at 9.4 Å and 4.7 Å resolutions, respectively.
See also Figure S2.
We then used circular dichroism (CD) to estimate the secondary structure content of RIP1/3-RHIM, which revealed a prominent negative peak at ~210 nm (Figure 2D). Analysis of the spectrum with DICHROWEB (Whitmore and Wallace, 2004) indicated that the sample is a mixture of β sheet and random coil structures. Fourier transform infrared spectroscopy (FTIR) has been well established to distinguish cross-β amyloid fibrils from native β sheet proteins in that the former shows amide I’ maxima between 1,610 cm−1 and 1,630 cm−1 in wavenumbers, which is smaller than native β sheet proteins (Zandomeneghi et al., 2004). This difference has been attributed to smaller β sheet twist angles and larger numbers of β strands in amyloidal aggregates (Zandomeneghi et al., 2004). Indeed, RIP1/3-RHIM showed a prominent amide I’ absorbance maximum at 1,623 cm−1 (Figure 2E), which is consistent with the amyloidal nature of the complex. The RIP1-RHIM mutant I539D, which is no longer amyloidal (see section “Mutations of RIP1 and RIP3 Weaken Filament Formation”), did not exhibit this characteristic absorption.
Amyloids are characterized by cross-β quaternary structures in which the β sheets are parallel to the fiber axis, and the extended β strands lie approximately perpendicular to the axis. This arrangement produces characteristic diffraction patterns (Sunde et al., 1997). We partially aligned the RIP1/3-RHIM complex and obtained its diffraction pattern by using Cu radiation (Figure 2F). Orthogonal diffractions at 4.7 Å and 9.4 Å resolutions were observed, corresponding to the inter-β strand spacing along the meridional axis (parallel to the fibril axis) and the inter-β sheet stacking distance along the equatorial direction (perpendicular to the fibril axis), respectively. These data established unequivocally the cross-β amyloid core of the RIP1/RIP3 complex.
The Amyloid Core of the RIP1/RIP3 Complex
Secondary structure prediction suggests that the amyloid core sequences of RIP1 and RIP3 are much shorter than either the full-length or the truncated coexpression construct we used for obtaining the RIP1/RIP3 complex. To further map this interaction, we generated a series of coexpression constructs tagging either RIP1 or RIP3 with a His-MBP tag. His-tag pull-down experiments of the coexpression constructs showed that the interaction was retained, even when RIP1 was only 31 residues (525–555) and RIP3 was only 19 residues (446–464) in length (Figures 3A, S3A, and S3B).
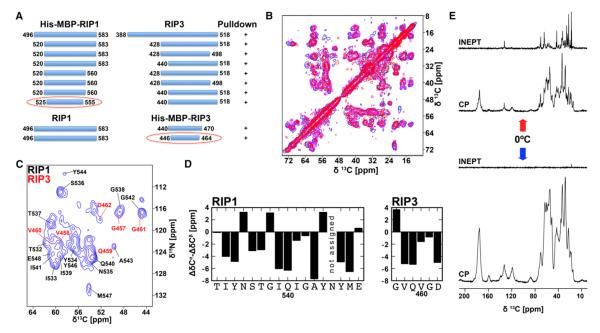
Figure 3. The Amyloid Core of the RIP1/RIP3 Complex.
(A) Mapping the interaction between RIP1 and RIP3 by using coexpression and His-tag pull-down. The shortest constructs that retained interaction are circled in red.
(B) Overlay of 2D DARR 13C-13C solid-state NMR spectra of the RIP1 (residues 496–583)/RIP3 (residues 388–518) complex (blue), its subtilisin-digested counterpart (magenta), and the RIP1 (residues 496–583)/RIP3 (residues 446–518) complex (red).
(C) 2D 15N-13C NCA solid-state NMR spectrum of subtilisin-digested RIP1/RIP3 complex. Site-specific assignments are indicated for RIP1 (black) and RIP3 (red).
(D) Plot of the difference in secondary chemical shift between Cα and Cβ, indicative of secondary structures (only ΔδCα for Gly).
(E) 13C 1D NMR spectra of the RIP1 (residues 496–583)/RIP3 (residues 388–518) complex. At above 0°C (top), the line width is generally narrower, and the INEPT pulse sequence, which is sensitive to relatively dynamic domains of the sample, gives an intense spectrum. At below 0°C (bottom), most of the dynamics are arrested, leading to no signal in the INEPT and an intense and broad cross-polarization (CP) spectrum, which is sensitive to the static domains of the sample.
See also Figure S3.
To further elucidate the core size and the secondary structure of the RIP1/RIP3 complex, we used solid-state NMR. Because sample conditions may influence amyloid structures, we collected 2D DARR 13C-13C correlation spectra at ~10 °C from three different coexpressed constructs, the RIP1 (residues 496–583)/ RIP3 (residues 388–518) complex, its subtilisin-digested counterpart, and the RIP1 (residues 496–583)/RIP3 (residues 446–518) complex. The overlay of these spectra showed very strong correspondence (Figure 3B), indicating that the size and the structure of the RIP1/RIP3 amyloid core are robust with respect to construct lengths and details of the preparation.
To obtain sequence-specific secondary structure information, we recorded high-resolution 13C-13C dipolar recoupling enhanced by amplitude modulation (DREAM) and dipolar-assisted rotational resonance (DARR) spectra; 15N-13C NCO, NCA, NcaCB, and NcoCX 2D spectra; and NCACX and NCACB 3D spectra on the subtilisin-digested complex at ~10 °C (Figures S3C, S3D). Using the mapped amyloid core sequences (Figures 1B and 3A) as targets, the majority of the peaks in the NCA spectrum were assigned, with 16 residues of RIP1 (T532–Y544 and Y546–E548) and 6 consecutive residues of RIP3 (G457–D462) (Figures 3C and 3D). Some of the residues were assigned with high confidence due to the nondegenerate 15N and 13C peaks in the three-dimensional (3D) spectra with additional support from Cα-Cα-1 cross-peaks in the two-dimensional (2D) DARR spectrum. All assignments are consistent with the RIP1/RIP3 core sequences. Negative and positive values of the difference in secondary chemical shifts between Cα and Cβ (i.e., ΔδCα-ΔδCβ or δCα for Gly) are indicative of β sheet and α-helical conformations, respectively. Most of the residues in the amyloid core are compatible with β sheet conformations, and the 4 central RHIM residues show consecutive negative shift differences (Figures 3D and S3E).
The spectra recorded with cross-polarization (CP)-based solid-state NMR above 0°C can be explained by ~37 resolved residues. Most of the residues outside the β-amyloid core were unobserved and either too dynamic or disordered to be visible in these spectra. Fast molecular dynamics can lead to partial or complete averaging of dipolar spin couplings that are essential for CP to work. When 1H-13C insensitive nuclei enhanced by polarization transfer (INEPT)—a sequence that only works when dipolar couplings are absent—was utilized, we observed intense and narrow spectra (Figure 3E). This indicated that there are very dynamic domains outside the amyloid core. This observation is consistent with the small core size observed from class averages of EM images (Figure 1E). At lower temperatures, these dynamics slowed down so that more domains in the sample could be detected with CP, but not with INEPT (Figure 3E).
Cross-Polymerization of the RIP1/RIP3 Complex
The sequence conservation at the core of the RHIMs of RIP1 and RIP3 prompted us to ask whether RIP1 or RIP3 alone could also form amyloid fibrils. Expression of His-Sumo-RIP1 (residues 496–583) or His-Sumo-RIP3 (residues 388–518) alone, followed by cleavage of the respective His-Sumo tag, showed that RIP1-RHIM or RIP3-RHIM eluted around the void position from a gel filtration column (Figure 4A). The RIP1 and RIP3 CD spectra showed similar secondary structures as the RIP1/RIP3 complex, and ThT fluorescence and CR absorption confirmed their amyloidal structures (Figure S4). EM of negatively stained RIP1 and RIP3 samples revealed that the homocomplexes are able to form fibrils by themselves as well (Figure 4B). However, these fibrils appeared to be less regular and shorter than those of the RIP1/RIP3 heterocomplex (Figure 1D).
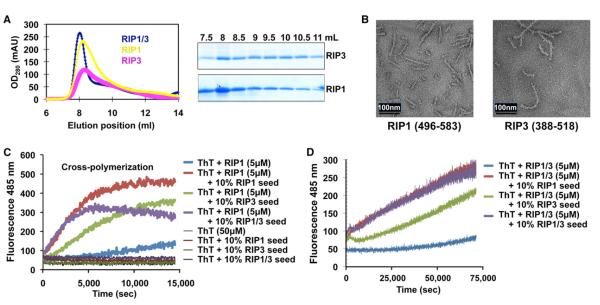
Figure 4. Cross-Polymerization and Mutagenesis of the RIP1/RIP3 Interaction.
(A) Left, superimposed gel filtration profiles of the RHIM fragments of RIP1 (residues 496–583), RIP3 (residues 388–518), and the RIP1/RIP3 complex. Right, SDS-PAGE of gel filtration fractions of RIP1 and RIP3.
(B) EM images of RIP1 and RIP3.
(C and D) Cross-seeding in the polymerization of denatured RIP1 and the denatured RIP1/RIP3 complex by using ThT binding assays, respectively.
See also Figure S4.
Because RIP1 and RIP3 can each form fibrils on their own, we wondered whether the observed fibrils are similar to that of the RIP1/RIP3 complex. To address this question, we performed cross-fibril polymerization experiments to see whether the amyloids of RIP1, RIP3, or the RIP1/RIP3 complex can enhance the polymerization of each other. Because the purified recombinant proteins are already fibrillar, we needed to first denature the proteins. Consistent with the recognized high stability of amyloid structures (Balbirnie et al., 2001), we had to use harsh conditions: 8 M urea for RIP1 and 150 mM NaOH for the RIP1/RIP3 complex. When RIP1 was first denatured and then diluted 100-fold into the native ThT binding buffer to allow refolding, it showed minimal enhancement of ThT fluorescence as a function of time. When the same denatured RIP1 was subsequently diluted to allow renaturation in the presence of 10% native RIP1, RIP3, or the RIP1/RIP3 complex as polymerization seeds, ThT fluorescence increased much more quickly (Figure 4C). Similarly, denatured RIP1/RIP3 complex can be induced to polymerize much more efficiently in the presence of 10% native seeds of RIP1, RIP3, or the RIP1/RIP3 complex (Figure 4D). These experiments suggest that the fibrils of RIP1 and RIP3, as well as the RIP1/RIP3 complex, share a similar structural architecture.
Mutations of RIP1 and RIP3 Weaken Filament Formation
To elucidate the key determinants in RIP1/RIP3 interaction, we generated point mutations on residues spanning the core RHIMs of RIP1 and RIP3. Residues 533–548 of RIP1 and residues 451–466 of RIP3 were mutated to Asp for possible maximal disruptive effects, except that D461 of RIP3 was mutated to Lys (Figure 1B). The RHIM consensus sequences were also mutated to Pro (Figure S5). For RIP1 mutants, we used a coexpression construct of His-RIP1 (residues 496–583) and RIP3 (residues 388–518). For RIP3 mutants, a coexpression construct containing an untagged wild-type (WT) RIP1 and a His-tagged RIP3 was used.
All mutant His-RIP1 proteins could pull down coexpressed WT RIP3 with Ni-affinity beads. However, when the copurified samples were subjected to gel filtration chromatography, clear differences appeared (Figures 1B and 5A). Whereas RIP1 mutants flanking the core RHIM did not show any defects in RIP3 interaction or fibril formation, RIP1 mutants near the center of the RHIM (I539D, I539P, Q540D, Q540P, I541D, I541P, G542D, G542P, A543D, Y544D, and N545D) were the most defective (Figures 5A, S5A, and S5B). These RIP1 mutants dissociated from RIP3, as observed by the appearance of an additional RIP1 mutant peak at ~17 ml.
Figure 5. Mutagenesis of RIP1 and RIP3.
(A) Superimposed gel filtration profiles of complexes of mutant RIP1 and WT RIP3. Left, RIP1 mutants that dissociated from RIP3; right, RIP1 mutants that did not dissociate from RIP3 and migrated near the void position.
(B) Superimposed gel filtration profiles of complexes of mutant RIP3 and WT RIP1. Left, complexes of mutant RIP3 and WT RIP1 that migrated later than the void position; right, complexes of mutant RIP3 and WT RIP1 that migrated near the void position.
(C) ThT fluorescence of WT and mutant full-length RIP1 and RIP3. (D) EM images of negatively stained WT and mutant full-length RIP1 and RIP3.
See also Figure S5.
Similarly, all His-RIP3 point mutants could pull down coexpressed WT RIP1 with Ni-affinity beads. When these complexes were subjected to gel filtration chromatography, a range of effects occurred, including dissociation on gel filtration chromatography and a change in elution positions (Figures 1B, 5B, S5C, and S5D). Asp mutants on central RHIM residues of RIP3 “457-GVQVGD-462” eluted at around 12–14 ml in comparison with the void position of around 8 ml for the WT RIP1/RIP3 complex. S456D was also partially defective, as it eluted around 11 ml. The changes in the elution positions suggest that the mutant complexes have lower apparent molecular masses than the WT complex and exhibit weakened interaction between RIP1 and RIP3. A double mutant of the core RHIM of RIP3 (V458E and V460E) induced dissociation from WT RIP1 on gel filtration chromatography (Figure S5C). Pro mutants on central RHIM residues of RIP3 “458-VQVG-461” resulted in partial dissociation with coexpressed WT RIP1, and V460P and G461P were the most defective. In contrast, mutations on more peripheral residues “451-NIYNC-455” and “463-NNYL-466” migrated in a manner similar to the WT complex.
Similar to the results for the RHIM constructs of RIP1 and RIP3, mutations on full-length RIP1 and RIP3 also weakened or disrupted filament formation. ThT staining of insect cell-expressed recombinant RIP1-FL mutant I539D and RIP3-FL mutant V460P showed that both proteins exhibited close to background levels of ThT fluorescence in comparison to the WT proteins (Figure 5C). Upon subtilisin digestion, EM of WT RIP1-FL and RIP3-FL showed filamentous structures, though they were apparently less regular than the RIP1/RIP3 complex (Figures 5D and 1D). In contrast, the RIP1-FL mutant I539D and the RIP3-FL mutant V460P were mostly degraded by subtilisin and showed only residual aggregates (Figure 5D).
Amyloidal Nature and Ultrastability of the Endogenous RIP1/RIP3 Complex
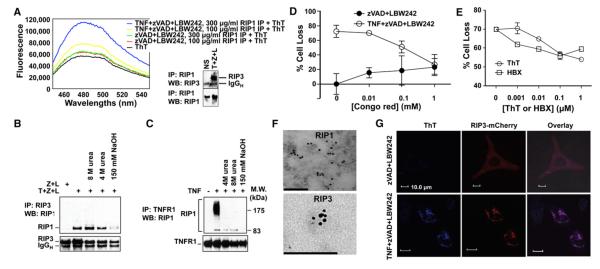
To determine whether the fibrils of endogenous RIP1/RIP3 complex isolated from necrotic cells that we observed under EM (Figure 1D) are amyloidal in nature, we first used ThT binding. Because the amount of endogenous complexes isolated from cells was much lower than that obtained from recombinant proteins, we measured ThT fluorescence by using a more sensitive instrument. HT-29 cells stimulated with TNF, zVAD-fmk, and LBW242 underwent RIP1/RIP3-dependent programmed necrosis (He et al., 2009). The RIP1-containing complex isolated from necrotic cells, but not from control cells, exhibited increased ThT fluorescence in a concentration-dependent manner (Figure 6A).
Figure 6. RIP1 and RIP3 Form Amyloidal Clusters In Vivo during Programmed Necrosis.
(A) Endogenous RIP1-containing complexes from necrotic HT-29 cells bind ThT. The right panel shows the specific pull-down of RIP3 by RIP1 upon TNF, zVADfmk, and LBW242 stimulation (T+Z+L).
(B) RIP3 complexes isolated by immunoprecipitation from HT-29 cells treated with T+Z+L after lysis in regular lysis buffer or buffer containing the indicated amount of urea or NaOH.
(C) TNFR1 complexes isolated by immunoprecipitation from MEFs treated with TNF after lysis in regular lysis buffer or buffer containing the indicated amount of urea or NaOH.
(D and E) Amyloid-binding compounds inhibit TNF-induced necrosis in HT-29 cells. Results shown are averages of triplicates ±SEM.
(F) Clustering of RIP1 and RIP3 in necrotic MEFs shown by immunogold EM. Scale bars, 200 nm (RIP1) and 100 nm (RIP3).
(G) Costaining of ThT with RIP3 puncta in necrotic HeLa cells as visualized by confocal microscopy.
See also Figure S6.
Given that the recombinant RIP1/RIP3 complex is ultrastable and requires 150 mM NaOH to be denatured, we tested the stability of the endogenous complex isolated from necrotic cells. We lysed cells with buffers containing 4 M urea, 8 M urea, or 150 mM NaOH. Thirty minutes after lysis, we diluted the lysates 10-fold with regular nondenaturing lysis buffer. Immunoprecipitation with anti-RIP3 antibody showed that 4 M urea or 8 M urea did not disrupt the interaction between RIP3 and RIP1, whereas 150 mM NaOH did (Figure 6B). This result is consistent with the stability of the recombinant complex and with the recognized stability of amyloidal structures in general (Balbirnie et al., 2001). In contrast, the interaction between polyubiquitinated RIP1 and TNFR1, which is crucial for TNF-induced NF-κB activation, was completely abolished in all conditions, including 4 M urea (Figure 6C).
Classical β-amyloid-binding dyes often inhibit amyloid oligomerization (Sánchez et al., 2003). Pretreatment of HT-29 cells with CR (Figure 6D), ThT, or another β-amyloid-binding compound, 2-(2-hydroxyphenyl)-benzoxazole (HBX) (Alavez et al., 2011), inhibited necrosis in a dose-dependent manner (Figure 6E). The inhibition by ThT and HBX is specific for necrosis because they had no effect in apoptotic HT-29 cells (Figure S6A). Clustering of RIP1 and RIP3 into punctate-like structures is a distinct feature of necrosis (Figures S6B–S6D). This was confirmed by immunogold EM (Figure 6F). When HeLa cells, which do not express endogenous RIP3, were transfected with RIP3-mCherry and stimulated with TNF, zVAD-fmk, and LBW242 to induce necrosis, they showed complete overlap of ThT staining with RIP3-mCherry puncta (Figure 6G). By contrast, ThT signal was undetectable in untreated cells (Figure 6G). Collectively, these results definitively show that RIP1 and RIP3 form amyloidal complexes during induction of programmed necrosis.
RHIM Residues of RIP1 and RIP3 Are Crucial for Cluster Formation, Kinase Activation, and Programmed Necrosis
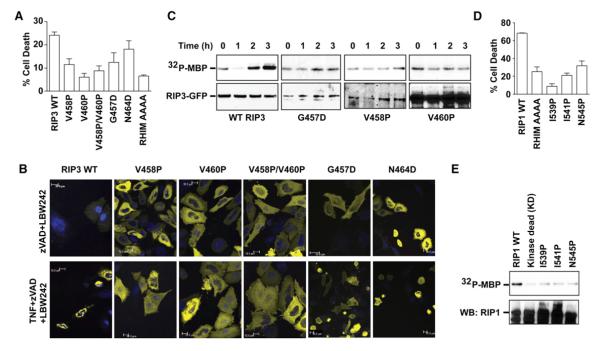
To determine the functional role of RHIMs in the assembly of the amyloidal RIP1/RIP3 complex in vivo, we introduced selected mutations within and flanking the RHIMs that were shown to cause disruption of filament formation and mutual interaction in vitro (Figure 1B). For RIP3, we stimulated HeLa cells transfected with WT and mutant RIP3-yellow fluorescent protein (YFP) with TNF, zVAD-fmk, and LBW242. As expected, WT RIP3-transfected cells underwent cell death, whereas the RHIM AAAA mutant was highly protected. Importantly, RIP3 mutants V458P, V460P, and G457D protected cells from TNF-induced necrosis, whereas the mutant N464D that did not show significant defects in vitro behaved most similar to the WT (Figure 7A). In addition, mutants V458P and V460P completely abolished puncta formation in response to necrosis stimulation, whereas G457D partially disrupted RIP3 clustering (Figure 7B). On the other hand, N464D still formed puncta (Figure 7B). Expression of RIP3-YFP in 293T cells showed that the mutations V458P and V460P severely compromised RIP3 kinase activation by using myelin basic protein (MBP) as the substrate (Figure S7A). To further validate the results from transient expression, we stably reconstituted RIP3−/− fibroblasts with WT and mutant RIP3. In agreement with results in 293T cells, WT RIP3 showed robust kinase activity upon induction of necrosis, whereas the V458P, V460P, and G457D mutants were dramatically impaired in kinase activation (Figure 7C). Notably, there is a close correlation between in vitro and in vivo experiments. For instance, V460P is the most defective in fibril formation and RIP1 interaction in vitro (Figure S5D; note the weakest RIP3 band in the fibril fraction) and is the most defective in cell death induction and kinase activation. Similarly, G457D is a less defective mutant in cell death induction and is also less impaired in kinase activation and cluster formation (Figures 7A–7C).
Figure 7. The RIP1/RIP3 Interaction Is Crucial for Kinase Activation, Clustering, and Cell Death.
(A) Effects of RIP3 mutations on TNF-induced necrosis in HeLa cells transfected with WTor the indicated RIP3 mutants fused to YFP. AAAA, quadruple Ala mutant of RIP3 at the VQVG RHIM sequence. Results shown are averages of triplicates ±SEM.
(B) Effects of RIP3 mutations on puncta formation in HeLa cells transfected with the indicated RIP3-YFP plasmids as examined by confocal microscopy.
(C) Effects of RIP3 mutations on kinase activity of the anti-RIP3 immunoprecipitates in RIP3−/− fibroblasts stably expressing the indicated RIP3-GFP mutants using MBP as the substrate. Bottom panel shows the RIP3 western blot of the same membrane.
(D) Effects of RIP1 mutations on TNF-induced necrosis in RIP1−/− fibroblasts transfected with the indicated RIP1 fused to GFP. Results shown are averages of triplicates ±SEM.
(E) Effects of RIP1 mutations on kinase activity of the anti-GFP immunoprecipitates in 293T cells transfected with the indicated RIP1-GFP constructs using MBP as the substrate. The same membrane was probed for RIP1 on western blot in the lower panel.
See also Figure S7.
To assess RIP1 mutational phenotypes in vivo, we transfected RIP1−/− fibroblasts with WT and mutant RIP1-green fluorescent protein (GFP). Cell death upon necrosis induction was enumerated in the GFP+ population by using flow cytometry. Like the RHIM AAAA mutant, the RIP1 mutants I539P, I541P, and N545P were much weaker in mediating necrosis than WT RIP1 (Figure 7D), recapitulating the in vitro interaction data. Similar to evaluation of the kinase activity of RIP3 mutants, we expressed RIP1-GFP constructs in 293T cells. After immunoprecipitation with anti-GFP antibody, the immune complexes were subjected to in vitro kinase assay using MBP as the substrate. All three RIP1 mutants, I539P, I541P, and N545P, showed defective kinase activation (Figure 7E). We attempted to stably reconstitute RIP1−/− fibroblasts with WT and mutant RIP1; however, because RIP1 is generally expressed at much lower levels than RIP3, likely due to toxicity, we could not evaluate the kinase activities in these cells. Similar defects in clustering in mutations within the core RHIM residues were observed with truncated RIP1 (residues 496–583) and RIP3 (residues 388–518) lacking the KDs and DD (Figure S7B). Thus, these results define a critical role for RHIM-mediated amyloidal RIP1/RIP3 fibrils in the activation of RIP1/RIP3 kinase activity and the induction of programmed necrosis.
DISCUSSION
Amyloids are fibrous protein aggregates composed of cross-β structures and associated with many neurodegenerative (Chiti and Dobson, 2006) and infective prion diseases (Uptain and Lindquist, 2002). Amyloids can also perform normal cellular functions, such as host interaction, hazard protection, and memory storage (Chiti and Dobson, 2006). However, this aspect of the function of amyloids is less defined, especially in mammals. Here, we show that RIP1 and RIP3 form a functional, hetero-oligomeric amyloidal signaling complex that mediates programmed necrosis. The discovery of cross-β amyloid structures in protein complexation and signal transduction provides new insights into both the amyloid field and the signaling field.
How is the assembly of the RIP1/RIP3 necrosome regulated? It has been shown that both RIP1 and RIP3 kinase activities are required for complex formation and cell death (Cho et al., 2009). Consistent with a role for amyloid assembly in RIP1 and RIP3 kinase activation, hyperphosphorylated RIP1 and RIP3 are found predominantly in NP-40 insoluble fractions (Figures S6C and S6D). Interestingly, abnormal phosphorylation of tau and α-synuclein by multiple kinases is known to be involved in the formation of neurofibrillary tangles in Alzheimer’s and Parkinson’s diseases, respectively, as well as in other neurodegenerative tauopathies and synucleinopathies (Avila et al., 2010; Oueslati et al., 2010). Our data are consistent with a feed-forward, gain-of-function mechanism in which kinase activation and RIP1/RIP3 necrosome formation are mutually reinforcing. In their inactive states, the core RHIM sequences may be hidden by long-range interactions in the unstructured flanking sequences of RIP1 and RIP3 and possibly by RIP1 ubiquitination. Indeed, expression of RIP1 and RIP3 RHIM fragments, but not full-length RIP1 and RIP3, induced spontaneous clustering in cells without stimulation (Figure S7B). In line with the observation of autoinhibition, it was reported that large proteins have a low propensity to form amyloid fibrils, although they possess a great tendency to form β-sheet-rich aggregates that are weak binders of ThT and CR (Ramshini et al., 2011). In contrast, the RIP1/RIP3 complex binds ThT and CR robustly, even though the folded KDs and the DD in these proteins are primarily α-helical. Kinase activation and the resultant hyperphosphorylation may be the key events that reduce this autoinhibition, perhaps as a result of charge repulsion to expose the RHIM core, leading to enhanced complex formation. In turn, complex formation further potentiates kinase activation through autophosphorylation and cross-phosphorylation, propagating the pronecrotic signal.
Is it possible that the amyloid structures per se also have toxicity to cells and contribute to cell death? It is well known that amyloids are toxic, likely as a consequence of their mass and induction of inflammation or disruption of membrane integrity (Eisenberg and Jucker, 2012). However, given that specific RIP3 kinase substrates have been identified and are crucial for induction of programmed necrosis (Sun et al., 2012; Zhao et al., 2012), kinase activation seems to be the key consequence of the formation of the RIP1/RIP3 amyloid scaffold. Drawing a parallel with the structural model of the designed amyloid of ribonuclease A (Sambashivan et al., 2005), the KDs of RIP1 and RIP3 would extend from the central amyloid spine to the periphery and find space to retain their globular structures and functions.
Previous structural studies have revealed several arrangements of cross-β amyloid spines in the formation of dry steric zippers, including those from both parallel and antiparallel β sheets (Eisenberg and Jucker, 2012; Sawaya et al., 2007). The fiber diffraction pattern of RIP1/3-RHIM (Figure 2F) showed that each β sheet in the fibrils consists of parallel β strands rather than antiparallel β strands, as evidenced by the strong 4.7 Å spacing along the fibril axis (meridional direction). Structures of related self-complementing steric zippers have also led to insights regarding complementary molecular surfaces in RIP1 and RIP3 (Sawaya et al., 2007). The core RHIM sequences of RIP1 and RIP3 and IQIG and VQVG, respectively, suggest that the hydrophobic Ile and Val residues pack to form the double β sheet in the steric zipper. This type of packing is remarkably similar to the structure of the VQIVYK sequence from tau (PDB 2ON9) (Sawaya et al., 2007), a microtubule-associated protein that plays an important role in stabilizing axonal microtubules. Particularly, the first 3 residues of the RHIM core sequences and the tau sequence are basically identical. We built two alternative models of the RHIM steric zipper based on the tau structure (Figure S7C). In both models, heterosteric zippers (Eisenberg and Jucker, 2012) for the β-amyloid spine are proposed. Notably, the hydrophobic packing between Ile and Val residues is reminiscent of tau. It is likely that packing in RIP1 or RIP3 homo-oligomeric fibrils (with Ile-Ile or Val-Val contacts) is less optimal than the Ile-Val contacts in the hetero-oligomeric complex. This preference may potentiate the unusual assembly of the RIP1/RIP3 heterodimeric complex and explain the apparent 1:1 stoichiometry between RIP1 and RIP3.
A number of additional proteins, including the cytoplasmic DNA sensor DAI, the Toll-like receptor signaling adaptor TRIF, and the murine cytomegalovirus protein M45, have been shown to contain RHIMs (Figure S7D). Both DAI and TRIF recruit RIP1 and/or RIP3 through their RHIMs to activate NF-κB or to induce cell death (Kaiser and Offermann, 2005; Meylan et al., 2004; Takaoka et al., 2007). Indeed, we show that DAI and RIP1 also form filamentous structures (Figure S7E). M45 binds to RIP1 and functions as a viral inhibitor in DNA sensing, Toll-like receptor signaling, and the TNF receptor pathway (Mack et al., 2008; Rebsamen et al., 2009; Upton et al., 2010). In all of these signaling processes, the high-order oligomeric scaffold of amyloids may be the key feature that brings signaling proteins into proximity to allow their activation. Similarly, yet distinctly, the oligomeric scaffold of the DD superfamily mediates caspase activation and apoptosis (Wang et al., 2010), as well as MyD88-dependent Toll-like receptor signaling (Lin et al., 2010). In addition, this DD superfamily scaffold exhibits functional characteristics akin to amyloids and prions in their abilities to seed and to propagate (Hou et al., 2011). For both the amyloid scaffold and the DD superfamily scaffold, the slow seeding phase and the fast growing phase in the assembly of these complexes may dictate a highly cooperative process and a digital threshold response mechanism. Assembly of highly oligomeric signalosomes may be an emerging principle in signal transduction.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, Purification, and Mutagenesis
The RIP1 (residues 496–583)/RIP3 (residues 388–518) complex and its truncation complexes were polycistronically subcloned into the pET28a (Novagen) or pDB-His-MBP vector (Berkeley Structural Genomics Center). The RIP1 (residues 496–583) and RIP3 (residues 388–518) constructs were subcloned into the pET28a or pSMT3 vector with a His-Sumo tag. All proteins were expressed in E. coli BL21 (DE3) RIPL cells (Novagen) and were purified by Ni-affinity and gel filtration chromatography.
The full-length RIP1(1–671)/RIP3(1–518) complex was subcloned into pFastBacDual vector (Invitrogen) with an N-terminal His-tag on RIP1 and an N-terminal MBP-tag on RIP3. The WT and mutant RIP1 and RIP3 (RIP1-FL, RIP3-FL, RIP1-FL-I539D, and RIP3-FL-V460P) were subcloned into pFast-BacHTA vector (Invitrogen) with an N-terminal His-tag. The proteins were expressed in Hi5 cells and were purified with either Ni-NTA or amylose resin followed by gel filtration chromatography.
Electron Microscopy and Image Processing
RIP1/RIP3-RHIM, RIP1-RHIM, and RIP3-RHIM were negatively stained with uranyl formate and imaged with a 1K × 1K charge-coupled device (CCD) camera (Gatan) on a CM10 electron microscope (FEI). For subsequent image processing, images were collected on imaging plates with a Tecnai T12 electron microscope. The particles were subjected to ten cycles of multireference alignment, each followed by K-means classification into 100 classes.
For full-length RIP1, RIP3, RIP1/RIP3 complex, and endogenous RIP1/3 complexes, the samples were stained using uranyl acetate and imaged with a Veleeta 2K × 2K cooled CCD camera (Olympus-Soft Imaging Solutions, Munster, Germany) on a JEM-1400 (JEOL, Ltd., Tokyo, Japan) electron microscope.
X-Ray Diffraction from Fibrils
The RIP1/RIP3-RHIM complex was dried and partially aligned by using a SpeedVac System (Savant) and mounted in a cryoloop. The diffraction image was collected by using a Rigaku RU-H3R X-ray generator on a Rigaku R-Axis IV imaging plate detector.
Congo Red Binding
CR absorption spectra were recorded from 430–600 nm in 96-well plates by using a SpectraMax M2 plate reader (Molecular Devices).
Thioflavin T Fluorescence
Fluorescence measurements were performed in 96-well plates on a Spectra-Max M2 plate reader (Molecular Devices) with an excitation wavelength of 430 nm.
Circular Dichroism Spectroscopy
CD spectra of RIP1, RIP3, and the RIP1/RIP3 complex were measured on an Aviv Model 410 spectropolarimeter at a cell length of 0.1 cm at 20°C with five scans per measurement. Buffer control was subtracted from each sample spectrum.
Fourier Transform Infrared Spectroscopy
Infrared spectra were recorded by using a Bruker Tensor spectrometer (Bruker Optik, Germany) at room temperature and with a resolution of 4 cm−1. The spectra represent the sum of 40 scans after reference subtraction.
Renaturation and Seeding Experiments Using ThT Fluorescence
RIP1 was denatured in 8 M urea with heating at 95°C for 10 min. The RIP1/RIP3 complex was denatured in 150 mM NaOH. The denatured proteins were diluted 100-fold into a native buffer and 50 μM ThT. The final concentration for RIP1 or the RIP1/RIP3 complex was 5μM. Seeding experiments were performed as described above with the exception that RIP1, RIP3, or RIP1/RIP3 complex seeds were added to the reaction to a final concentration of 0.5 μM. All reactions were measured kinetically at 25°C in 96-well plates with excitation and emission wavelengths of 430 nm and 485 nm, respectively.
Solid-State NMR Experiments
2D DARR spectra were recorded on a Bruker Advance III 400 spectrometer. The 2D 13C-13C DARR and DREAM spectra used for the resonance assignment, as well as the 1D 13C spectra, were recorded on an Advance II 900 spectrometer. Data were collected at ~10°C if not mentioned otherwise. All triple resonance spectra used for the assignment were recorded on an Advance I 750 spectrometer. All spectra were processed by using Topspin 3.1 and analyzed using the program CARA.
Confocal Microscopy
For ThT staining, HeLa cells were plated on coverslips and transfected with RIP3-mCherry. After 15–18 hr, cells were treated with zVAD-fmk, LBW242, and TNF for 3 hr. 20 μM ThT was added to cells 1 hr prior to fixation and imaging with a Leica SP5 confocal microscope. For microscopy of WT and mutant RIP1-CFP, RIP1-GFP, or RIP3-YFP, transfected NIH 3T3 fibroblasts or HeLa cells were induced to undergo necrosis and visualized by confocal microscopy.
Cell Death Assays
For inhibition of programmed necrosis, CR, ThT, or HBX were used at the indicated concentrations on HT-29 cells treated with TNF, LBW242, and zVAD-fmk. Cell death was determined by lactate dehydrogenase (LDH) release or propidium iodide (PI) exclusion using flow cytometry. For induction of apoptosis, HT-29 cells were pretreated with the indicated concentrations of inhibitors prior to stimulation with IFN-γ and anti-Fas antibody.
For effects of RIP3 mutations, HeLa cells were transfected with WT or mutant RIP3 fused to YFP. Cells were treated with zVAD-fmk, LBW242, and TNF, and cell death was determined in the YFP-positive population with PI by flow cytometry. For effects of RIP1 mutations, RIP1−/− fibroblasts were transfected with WT or mutant RIP1 fused to GFP. Cells were similarly treated, and cell death was determined in the GFP-positive population by PI flow cytometry.
ThT Binding and Immunogold EM of the Endogenous RIP1/RIP3 Complex
RIP1 immunoprecipitates were purified from necrotic HT-29 cells, and ThT fluorescence was measured from 450–550 nm by using a Spex Fluorolog-3 spectrofluorometer with excitation at 430 nm. For immunogold EM analyses, necrotic MEFs were fixed in 4% paraformaldehyde. RIP1 (BD PharMingen) and RIP3 (ProSci) antibodies were applied at 1:50 dilutions. Secondary immunogold conjugated antibodies (15 nm for RIP1 and 6 nm for RIP3) were used at 1:50 dilution.
Stability Assessment of the Endogenous RIP1/RIP3 Complex
HT-29 cells were lysed in lysis buffer supplemented with 4 M urea, 8 M urea, or 150 mM NaOH. Thirty minutes after lysis, cell lysates were diluted 10-fold with regular lysis buffer and immunoprecipitated with anti-RIP3 antibody. For TNFR1 complexes, WT MEFs were treated with TNF for 2 min. Cell lysis was performed as for the RIP1/RIP3 complexes.
In Vitro Kinase Assays
293T cells were transfected with indicated RIP1 or RIP3 mutants fused to GFP or YFP, respectively. The fusion proteins were immunoprecipitated 24 hr later with anti-GFP antibody (Roche). For RIP3 stable fibroblasts, RIP3 complexes were immunoprecipitated after necrosis induction. The resulting immune complexes were used for in vitro kinase assay using myelin basic protein (MBP, Stressgen) as substrate.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Cohen-Gould for help with EM imaging at the Weill Cornell Microscopy Facility, S.M. Damo for initial work on this project, Q. Li for insect cell expression, V. Kumar for ThT measurement of endogenous complexes, and D. Porter (Novartis) for the generous gift of LBW242. This work was supported by grants to F.K.C. (AI083497 and AI088502) and H.W. (AI045937). F.K.C. is a member of the UMass DERC (DK32520). J.N. is an Irvington Institute postdoctoral fellow of the Cancer Research Institute, and D.M. is supported by NIH training grant T32 AI07349. T.W. is an investigator in the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.06.019.
REFERENCES
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila J, Gómez de Barreda E, Engel T, Lucas JJ, Hernández F. Tau phosphorylation in hippocampus results in toxic gain-of-function. Biochem. Soc. Trans. 2010;38:977–980. doi: 10.1042/BST0380977. [DOI] [PubMed] [Google Scholar]
- Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa S, Woelfel M, Guildford M, Moquin D, Chan FK. Viral cell death inhibitor MC159 enhances innate immunity against vaccinia virus infection. J. Virol. 2010;84:10467–10476. doi: 10.1128/JVI.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J. Biol. Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, McQuade T, Zhang H, Zhang J, Chan FK. RIP1-dependent and independent effects of necrostatin-1 in necrosis and T cell activation. PLoS ONE. 2011;6:e23209. doi: 10.1371/journal.pone.0023209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detken A, Hardy EH, Ernst M, Meier BH. Simple and efficient decoupling in magic-angle spinning solid-state NMR: the XiX scheme. Chem. Phys. Lett. 2002;356:298–304. [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell. Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Häcker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gaither A, Porter D, Yao Y, Borawski J, Yang G, Donovan J, Sage D, Slisz J, Tran M, Straub C, et al. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-alpha signaling. Cancer Res. 2007;67:11493–11498. doi: 10.1158/0008-5472.CAN-07-5173. [DOI] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- He S, Liang Y, Shao F, Wang X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl. Acad. Sci. USA. 2011;108:20054–20059. doi: 10.1073/pnas.1116302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Pettegrew JW, Abraham DJ. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- LeVine H., III Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- Li Z, Hite RK, Cheng Y, Walz T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J. Electron Microsc. (Tokyo) 2010;59:53–63. doi: 10.1093/jmicro/dfp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mack C, Sickmann A, Lembo D, Brune W. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. USA. 2008;105:3094–3099. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Abedini A, Plesner A, Verchere CB, Raleigh DP. The flavanol (−)-epigallocatechin 3-gallate inhibits amyloid formation by islet amyloid polypeptide, disaggregates amyloid fibrils, and protects cultured cells against IAPP-induced toxicity. Biochemistry. 2010;49:8127–8133. doi: 10.1021/bi100939a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Moquin D, Chan FK. The molecular regulation of programmed necrotic cell injury. Trends Biochem. Sci. 2010;35:434–441. doi: 10.1016/j.tibs.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati A, Fournier M, Lashuel HA. Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein: implications for Parkinson’s disease pathogenesis and therapies. Prog. Brain Res. 2010;183:115–145. doi: 10.1016/S0079-6123(10)83007-9. [DOI] [PubMed] [Google Scholar]
- Ramshini H, Parrini C, Relini A, Zampagni M, Mannini B, Pesce A, Saboury AA, Nemat-Gorgani M, Chiti F. Large proteins have a great tendency to aggregate but a low propensity to form amyloid fibrils. PLoS ONE. 2011;6:e16075. doi: 10.1371/journal.pone.0016075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K, Vazquez J, Benedict CA, Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- Sánchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- Sun X, Lee J, Navas T, Baldwin DT, Stewart TA, Dixit VM. RIP3, a novel apoptosis-inducing kinase. J. Biol. Chem. 1999;274:16871–16875. doi: 10.1074/jbc.274.24.16871. [DOI] [PubMed] [Google Scholar]
- Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 2012;11:290–297. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verel R, Ernst M, Meier BH. Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. J. Magn. Reson. 2001;150:81–99. doi: 10.1006/jmre.2001.2310. [DOI] [PubMed] [Google Scholar]
- Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol. Rev. 2011;244:9–28. doi: 10.1111/j.1600-065X.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernández-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van Loo G, Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32(Web Server issue):W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu PW, Huang BC, Shen M, Quast J, Chan E, Xu X, Nolan GP, Payan DG, Luo Y. Identification of RIP3, a RIP-like kinase that activates apoptosis and NFkappaB. Curr. Biol. 1999;9:539–542. doi: 10.1016/s0960-9822(99)80239-5. [DOI] [PubMed] [Google Scholar]
- Zandomeneghi G, Krebs MR, McCammon MG, Fändrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl. Acad. Sci. U S A. 2012 doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.