Abstract
Human Proteinpedia (http://www.humanproteinpedia.org) is a publicly available proteome repository for sharing human protein data derived from multiple experimental platforms. It incorporates diverse features of human proteome including protein-protein interactions, enzyme-substrate relationships, PTMs, subcellular localization, expression of proteins in various human tissues and cell lines in diverse biological conditions including diseases. Through a public distributed annotation system developed especially for proteomic data, investigators across the globe can upload, view and edit proteomic data even before they are published. Inclusion of information on investigators and laboratories that generated the data, visualization of tandem mass spectra, stained tissue sections, protein/peptide microarrays, fluorescent micrographs and Western blots ensure quality of proteomic data assimilated in Human Proteinpedia. Many of the protein annotations submitted to Human Proteinpedia have also been made available to the scientific community through Human Protein Reference Database (http://www.hprd.org), another resource developed by our group. In this protocol, we describe how to submit, edit and retrieve proteomic data in Human Proteinpedia.
Key terms: mass spectrometry, tissue microarrays, biomarkers, disease proteomics, HPRD, proteotypic peptides, multiple reaction monitoring
Introduction
Human Proteinpedia (Kandasamy et al., 2009; Mathivanan et al., 2008) is a unified protein resource with a large collection of experimentally-derived protein data obtained from multiple experiments, which include co-immunoprecipitation, mass spectrometry, fluorescence-based experiments, protein and peptide microarrays, Western blotting and yeast two-hybrid assays. Currently, the resource contains data derived from over 2,710 experiments gathered from 249 laboratories worldwide. Human Proteinpedia uses controlled vocabularies from the Gene Ontology (UNIT 7.2; Ashburner et al., 2000) for subcellular localization, eVOC (Kelso et al., 2003) for the human tissues, RESID (Garavelli, 2004) for post-translational modifications (PTMs), PSI-MS (Martens et al., 2011) for mass spectrometry data and interpretation of MS/MS spectra and PSI-MI (Kerrien et al., 2007) for protein-protein interactions (PPIs). Human Proteinpedia incorporates a Public Distributed Annotation System (PDAS), developed by our group for handling protein data, which enables scientific community to submit and edit their protein data.
Human Proteinpedia contains data derived from several experiments that include, 354 experiments for tissue expression (332 of them are mass spectrometry-derived expression studies), 192 cell line-based expression studies, 19 disease specific expression studies, 22 experiments for the detection of PPIs and 5 studies on subcellular localization. We have also included mass spectrometry derived data from HUPO initiatives such as human plasma proteome project (Omenn et al., 2011), human brain proteome project (Hamacher et al., 2004) and human liver proteome project (He, 2005). In order to ensure easy access for proteomic investigators, we describe the protocol of accessing and handling protein data submission in this resource, which will facilitate data sharing in the biomedical community.
We describe five basic protocols for utilizing Human Proteinpedia. Basic Protocol 1 describes the query system in Human Proteinpedia. Basic Protocol 2 describes PDAS for annotating data from both high-throughput and targeted investigations. Basic Protocol 3 explains integration of Human Proteinpedia data with HPRD (Keshava Prasad et al., 2009). Basic Protocol 4 explains the various ways to access and download the entire data available in the Human Proteinpedia. Finally, basic Protocol 5 describes protein data for diverse protein features in this resource.
Basic Protocol 1
QUERYING HUMAN PROTEINPEDIA
The query page in Human Proteinpedia (Fig. 1) can be accessed through the URL: http://www.humanproteinpedia/query. The query system includes search by gene symbol or protein name, protein database accession numbers, type of protein feature or the type of experimental platform annotated in Human Proteinpedia. When a query is made through a free text search in a field of interest, the other text boxes are automatically deactivated.
Figure 1.
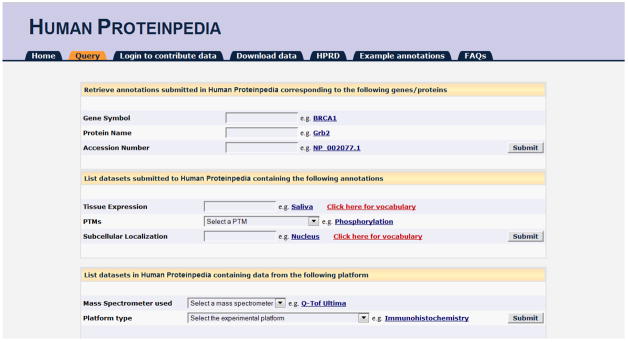
The Human Proteinpedia query page. It provides options to search by gene symbol, protein name, external database accession number, tissue types, types of PTM and subcellular localization. It can also be queried by types of mass spectrometer or various experimental platforms.
Necessary Resources
Hardware
A computer with internet connection.
Software
Javascript compatible web browsers, such as Mozilla Firefox, Internet Explorer or Google Chrome and an e-mail account.
Files
None
Querying Human Proteinpedia using gene symbol, protein name or protein accession numbers
-
1
Open the Human Proteinpedia query page located at http://www.humanproteinpedia.org/query
The query system contains three independent levels of query to fetch data from Human Proteinpedia (Fig. 1). In the first level, one can query Human Proteinpedia using gene symbol/protein name (e.g., BRCA1) or external database identifiers such as RefSeq (e.g., NP_002077.1) (Pruitt et al., 2009) or UniProt (e.g., P01860) (UniProt, 2012). -
2
Type gene symbol of a protein in the text box provided for gene symbol (e.g., BRCA1).
-
3
Click the ‘Submit’ button to reach query results page.
The query fetched data from 19 different mass spectrometry-based experiments, which included 16 studies aimed at PTMs and three experiments of tissue expression studies. The results fetched and displayed include, experimental platform, experiment type, experiment description, name and laboratory details of Principal Investigators (PIs), and data submitters, publication status of the data, sample details such as tissue or cell line and source organisms. The mass spectrometry-based experiments provide ionization method, fractionation technique, and name of the MS/MS search engine used along with the peptide and PTM data. These include the peptide sequence, modifications, peptide score/probability, precursor mass, charge state, sequence identifier and MS/MS spectrum. MS/MS spectrum is created for each peptide by PRIDE viewer (UNIT 13.8; Medina-Aunon et al., 2011) from the MS/MS peak lists. The entire dataset can also be downloaded. The multiple results are displayed one below the other and a unique Human Proteinpedia identifier is provided on the top of each result. -
4
Perform a new search by typing a protein name (e.g., ‘Grb2’) in the text box provided for protein name search.
-
5
Click the ‘Submit’ button to reach the query results page.
The search for the protein name ‘Grb2’ fetched 21 immunohistochemistry-based experiments carried out on ‘Grb2.’ The results include the details such as Human Proteinpedia identifier, experiment type, description of the experiment, names and laboratory details of PIs and data providers, status of publication, sample sources and source organisms. It also provides additional protein details such as protein name, tissue name, cell type, antibody reliability score fetched from the Human Protein Atlas (HPA) (Uhlen et al., 2010) and a link to HPA and the images showing protein’s staining pattern.
Query by datasets
Query by datasets allows the users to query using protein features such as tissue expression (e.g., saliva), PTMs (e.g., phosphorylation) or subcellular localization (e.g., nucleus). The controlled vocabularies obtained from the standard vocabulary consortia for each category are provided as a popup page near the text box provided for each search.
-
6
Choose tissue of interest from the list of controlled vocabularies for expression by clicking the ‘click here for vocabulary’ link. (e.g., brain).
-
7
Click the ‘Submit’ button to fetch the results.
Fig. 2 shows multiple experiments carried out for tissue expression studies on ‘Brain.’ The query fetched five mass spectrometry-based tissue expression experiments of which four of them are from the recently submitted ‘unpublished’ studies. The results page provides Human Proteinpedia accession number hyperlinked to its corresponding experimental details, brief description of the experiment, status of publication, annotation category (e.g., tissue expression), experimental platform and names and laboratory details of contributing group. Similar results are fetched for the query by PTMs and subcellular localization.
Figure 2.
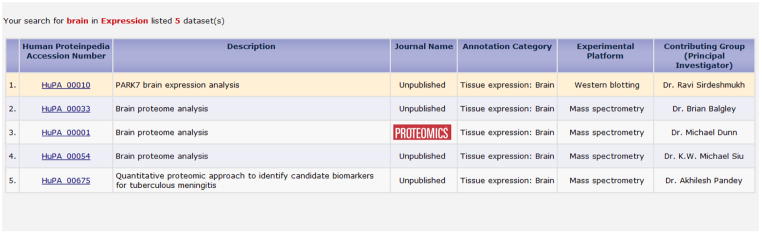
Query results of tissue expression (e.g., brain). The table displays the types of experiments carried out to check expression of proteins in brain. It also provides the number of datasets fetched.
Query by experimental platform
-
8
The query by experimental platform can be carried out either by the name of the mass spectrometer (e.g., LTQ-Orbitrap) or by the type of experimental platform (e.g., immunohistochemistry). The standard terms are given in the dropdown menus for both the categories. The search by either of these two categories fetches the results in the format as given in Fig. 2.
Basic Protocol 2
ANNOTATION OF PROTEIN FEATURES THROUGH COMMUNITY PARTICIPATION
A large fraction of the available data in Human Proteinpedia is driven by the contribution by the scientific community. Users can submit protein information obtained from their investigations through the Public Distributed Annotation System (PDAS). PDAS is a set of computational protocols made available for the scientific community to share their protein-related data. PDAS for protein data is a novel feature developed for Human Proteinpedia. Researchers could contribute their data in four different ways: 1) enter protein data using the web interface, 2) upload high-throughput data in batch mode using the web interface, 3) send data through FTP or e-mail; or, 4) set up PDAS servers at their own location. The users need to register and login to annotate, upload or manage their data in PDAS through web interface.
Necessary Resources
Hardware
A computer with internet connection.
Software
Javascript compatible web browsers, such as Mozilla Firefox, Internet Explorer or Google Chrome and an e-mail account.
Files
None
Login and registration
-
1
Click on the ‘Login to contribute data’ tab located at http://pdas.hprd.org/
-
2
If you are a new user, click on the link provided below the ‘Login’ button ‘New user click here to register’.
The registration page requires users to enter their contact and laboratory details. The required information includes full name of the contributor, email address, title, name of the organization, address, country, lab URL, whether PI or not. If the data provider is not a PI, the PI details need to be submitted. Also, the user needs to contain username and password in the lowest panel. -
3
Click the ‘Submit’ button to register.
-
4
Once registration is completed, the user is taken back to the login page.
-
5
Type the user name and password and click the ‘Submit’ button.
-
6
Now you are in the PDAS annotation system.
The web-based annotation of protein features obtained from experiments can be carried out through PDAS, which is located at http://pdas.hprd.org/loggedin. This page can be accessed only by registered users. The protein features include PTMs, PPIs, tissue expression, substrates and subcellular localization. The data submitted through this portal are subsequently manually verified, formatted and uploaded into Human Proteinpedia.
The PDAS annotation page provides access to the users who would like to contribute their experimental data into Human Proteinpedia. Users can enter their high-throughput or low-throughput data using this annotation system. The panel on the left side provides the user details along with an option to edit their registration details. The targeted protein data could be annotated by selecting the database name and typing the protein identifier from that database. High-throughput data can be uploaded by clicking the ‘Batch upload’ button. This page serves as a portal where the user can view/edit their own submitted data.
Annotation of high-throughput data through ‘Batch upload’
The data obtained from high-throughput experiments can also be shared through the PDAS annotation system using the portal for ‘Batch upload’ located at http://pdas.hprd.org/Batch_Upload
High-throughput data of PTMs, PPIs, tissue expression and substrates can be annotated through this annotation system.The data can also be sent through e-mail or through FTP in the prescribed format as indicated at the following URLs:
Tissue expression: http://pdas.hprd.org/expression_file_format
Substrates: http://pdas.hprd.org/substrates_file_format
-
7
Click the ‘Batch upload’ button provided in the PDAS annotation page.
The tab delimited data file in the prescribed format can be uploaded using this portal. If the experiment carried out is to detect a PTM using mass spectrometry, the data providers need to provide the peak lists files obtained from mass spectrometer to link the peptide data with its MS/MS spectrum and also to provide them for data download. -
8
Click on the Submit button.
The uploaded data files will be sent to the Human Proteinpedia annotation team to manually verify and upload the data into Human Proteinpedia. The user is permitted to opt for keeping the data private until their manuscript on the submitted dataset(s) is published. -
9
Standard annotation file formats were created for each of the protein features. The data need to be uploaded in those standard formats where the mandatory annotation fields are indicated by an asterisk. Annotation files must be provided in tab delimited text format. The sample annotation files can be accessed by clicking on the hyperlinks provided at the bottom of the table on the PDAS annotation page. The data provider must mention the external database used for the protein identification.
The required fields for the annotation of PTMs are provided at http://pdas.hprd.org/ptm_file_format provide the protein details such as protein identifiers, name of protein database used for searches, type of PTM, site (position at which the amino acid gets modified in the protein sequence), modified amino acid, experiment type, upstream enzyme(s), score given by the search algorithm(s), database search engine(s) used (e.g., Mascot), the mass spectrum file name and the status of publication. Peptide score, algorithm used and mass spectrum name are the fields to be annotated only for the mass spectrometry-based experiments.The required fields for the annotation of PPIs are provided at the URL: http://pdas.hprd.org/interaction_file_formatDirect PPIs which involve two molecules require the protein database identifiers for protein A and protein B, experiment name and the status of publication. Complex interactions, which involve multiple proteins, require the protein database identifier for each protein in the complex and the experimental method used to isolate the complex. When multiple complexes are annotated at a time, a common identifier for all the proteins in each complex needs to be provided to distinguish them. Multiple complexes can be sent in a single file by providing the different identifiers for each complex.The required fields for the annotation of tissue expression are provided at http://pdas.hprd.org/expression_file_format. It requires protein database identifier with the mention of database name, site of expression (normal tissue/cancer tissue/cell line), type of the experiment used to detect the expression and a PubMed identifier.The required fields for the annotation of substrates are provided at http://pdas.hprd.org/substrates_file_format. The annotation of substrates requires the protein database identifier with the mention of database name, modification type, protein sequence position at which the protein is modified, modified amino acid, name of the experiment carried out to identify the substrate, sequence identifier for the substrate, database search algorithm used, peptide score obtained from database search algorithm, name of the mass spectrum file from which the peptide is identified and PubMed identifier.
Annotation of low-throughput data
Annotation of a targeted protein can be carried out by the community through a web-based interface located on the PDAS annotation page. The protein identifier provided by the user is used to match the HPRD accession numbers for fetching the protein name and gene symbol from HPRD and they are displayed on the top of the low-throughput data annotation page. The low-throughput protein features include PTMs, PPIs, tissue expression, substrates and subcellular localization. The earlier annotation contributed by the logged in users can be viewed in the lower bottom table with the heading ‘Existing annotation using Human Proteinpedia by the <username>’.
-
10
Choose HGNC gene symbol from the dropdown menu and type any gene symbol approved by the gene nomenclature committee (e.g., ‘VIM’) in the text box in the PDAS annotation page.
-
11
Click the ‘Enter’ button.
You are directed to the URL (http://pdas.hprd.org/annotation) where different types of protein feature can be entered for using PDAS. It also provides the protein name and gene symbol of the queried protein hyperlinked to HPRD. If the query did not fetch the protein which user is looking for, the query could be modified by clicking on the link, ‘annotate a different protein’.
Annotation of PTMs
-
12
Click the ‘PTM’ button.
Annotation of site directed mutagenesis or mass spectrometry-based detections could be carried out using this link. -
13
Select site directed mutagenesis.
Fig. 3 shows the annotation page for PTM detected using the site directed mutagenesis. The data fields include, the detailed description of the experiment, sample source (normal tissue, cancer tissue or cell lines), status of publication, availability of data, a text box to enter the name of the tissue/cell lines, type of PTM from the standard menu provided as a dropdown, protein accession number, position of the amino acid, amino acid and type of experiment. The public data in Human Proteinpedia is accessible to anonymous users whereas the private data can be viewed only by the data contributor and their collaborators. Controlled vocabularies were given for the normal human tissues and can be added in the textbox by clicking on ‘Click here to add the normal tissue’. -
14
To annotate PTMs identified by mass spectrometry-based experiments, choose the type of experiment as ‘mass spectrometry detection’.
Along with the list of annotation fields for site-directed mutagenesis-based PTM annotation, the mass spectrometry detection methods have additional data fields which include labeling technique, protease, whether the sample is from SDS-PAGE, name of the mass spectrometer used, instrument vendor, ionization method, activation method, mass tolerance used for database searching (MS and MS/MS), database used for searching, database search algorithm used, type of experiment, peptide sequence, peptide score/probability, precursor mass, charge state and provision to upload the mass spectrum.
Figure 3.
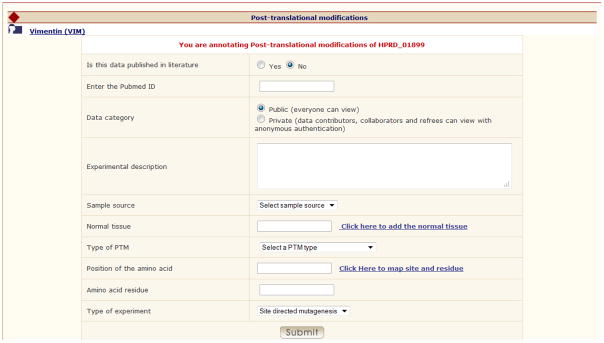
Targeted protein annotation page for PTMs.
Annotation of PPIs
Direct as well as the complex PPIs can be submitted using PDAS. Direct interaction means the interaction between two interactors, whereas a complex interaction means multiple proteins have been detected as the part of a complex and the exact topology of interaction between the individual interactors is not known.
-
15
Select protein-protein interactions from PDAS annotation page.
-
16
Select ‘Direct’ from the drop down menu for choosing the type of interaction. This page displays the fields to be annotated for direct PPIs.
The annotation fields include PubMed identifier for the article in which the interaction was described, data category (public or private), brief description about the experiment, sample source (normal tissue, cancer tissue or cell line), interacting molecule and experiment type. To make the process simple, the users can provide any external database protein identifiers for the interactors. The entered PPI dataset would be sent to Human Proteinpedia core annotation team to verify and upload the data into Human Proteinpedia. -
17
Select ‘Complex’ as interaction type.
The complex interaction annotation fields are the same as that of direct PPI with the exception that multiple text fields were provided to enter multiple interactors. Complex interactions could also be sent to Human Proteinpedia team by email, FTP or through batch upload.
Annotation of tissue expression
Data derived from experiments such as antibody array detection, Western blot detection, immunohistochemistry, ELISA and mass spectrometry-based detection can be annotated for tissue expression.
-
18
Click on the Tissue expression button from the PDAS annotation page.
-
19
Select ‘Western blot detection’ from the dropdown menu.
The required annotation fields include status of publication, description of experiment, sample type (normal tissue/disease tissue/cell line), site of expression as given in the pull down menu, type of experiment, species of primary antibody, source of antibody (whether commercial), name of the vendor if commercial, primary antibody dilution and image obtained from the experiment. All the other types of experiments for the annotation of tissue expression other than mass spectrometry-based detection are the same as that of detection by Western blot. In case of mass spectrometry-based detection, the mass spectrometry specific parameters such as labeling technique, protease used, name of the mass spectrometer used (as given in the pull down menu), instrument vendor (as given in the pull down menu), mass tolerance used for database searching, database used for searching (e.g., RefSeq), search algorithm used (e.g., Sequest), peptide sequence, peptide score or probability, precursor mass, charge state and image of the mass spectrum needs to be provided. -
20
Click on the ‘Submit’ button.
The annotated information would reach the Human Proteinpedia core annotation team to validate and upload the data into Human Proteinpedia.
Annotation of Substrates
Substrates are proteins which are modified by enzymes. The annotation fields are same as that of the PTMs. Enzyme and substrate names need to be added as additional fields.
Annotation of ‘Subcellular localization’
Subcellular localization can be annotated from different types of experiments that include fluorescence-based experiments, immunohistochemistry and mass spectrometry-based detection.
-
21
Click ‘Subcellular localization’ from the PDAS annotation page.
-
22
Choose the type of experiment from the drop down menu (e.g., fluorescence-based experiments).
The required fields for subcellular localization annotation include the PubMed ID if published, description of the experiment, experimental sample (normal tissue, disease tissue or cell line), name of the sample source, name of subcellular localization (as selected from the pull down menu), experiment type, species of the primary antibody (selected from a pull down menu), source of antibody whether commercial or not, name of the vendor if commercial, primary antibody dilution and the image obtained from the experiment. For the immunohistochemistry-based expression studies, the same annotation fields need to be annotated as that of fluorescence-based experiments. For the mass spectrometry-based detection, the additional mass spectrometry specific fields must be provided as described in the previous section. -
23
Click the ‘Submit’ button.
The annotated information would reach the Human Proteinpedia core annotation team to validate and upload into the database.
Basic Protocol 3
INTEGRATION OF HUMAN PROTEINPEDIA INTO HPRD
Human protein reference database (HPRD) is a repository hosting over 30,000 human proteins with the manually curated protein features such as tissue expression, PPIs, PTMs, subcellular localization and substrates. Protein features submitted to Human Proteinpedia are also made available to the scientific community through HPRD by integrating the data in Human Proteinpedia into HPRD (Kandasamy et al., 2009; Mathivanan et al., 2008). The integrated protein features are PTMs, subcellular localization and tissue expression. The volume of data in HPRD increases by incorporating additional data from Human Proteinpedia as Human Proteinpedia considers all the conditions of human samples.
Necessary Resources
Hardware
A computer with internet connection.
Software
Javascript compatible web browsers, such as Mozilla Firefox, Internet Explorer or Google Chrome and an e-mail account.
Files
None
Open HPRD query page located at http://www.hprd.org/query
-
Type any gene symbol (e.g., ‘VIM’) in the text box provided for the gene symbol and click ‘submit’ button.
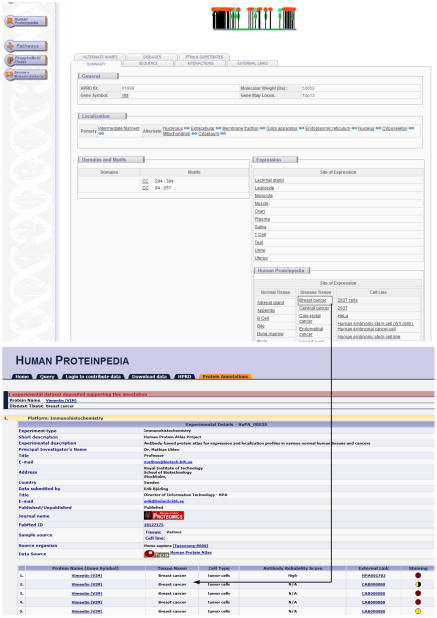
The query fetches the vimentin molecule page in HPRD. In the summary tab, under the expression panel, the manually literature curated normal tissue expression were provided from HPRD annotation. The Human Proteinpedia tissue expression data for vimentin is provided below the HPRD expression panel with the side heading ‘Human Proteinpedia’. Human Proteinpedia provides expression details of disease as well as cell line expression data which is not available in HPRD. The names of the tissues or cell lines from Human Proteinpedia are hyperlinked to the annotation pages of vimentin in Human Proteinpedia (Fig. 4). HPRD provides only normal tissue expression whereas incorporation of Human Proteinpedia into HPRD gains cancer and cell line expression as additional details for HPRD. -
Click on the ‘PTMS & SUBSTRATES’ from the HPRD molecule page to view PTMs in HPRD, which are submitted through Human Proteinpedia.
The PTM annotations in Human Proteinpedia are made available in HPRD under the tab PTMs with the heading of ‘Human Proteinpedia’. It provides, site (sequence position at which amino acid is modified), amino acid residue, type of PTM, upstream enzymes details and the sequence of the identified peptide. The position of amino acid residue, which is post-translationally modified, is hyperlinked to its corresponding annotation in Human Proteinpedia. -
To view an example of subcellular localization submitted through Human Proteinpedia into HPRD, open HPRD query page and type ‘ATP13A1’ in the text box provided for gene symbol.
This query fetches the molecule page of the gene ‘ATP13A1’ in HPRD. The summary tab in the molecule page provides the subcellular localization from HPRD as well as from Human Proteinpedia. Subcellular localization of ‘endoplasmic reticulum’ was fetched for ‘ATP13A1’ from Human Proteinpedia, which was not curated in HPRD clarifying the potential use of integrating Human Proteinpedia into HPRD.
Figure 4.
Integration of Human Proteinpedia data into HPRD. It describes the incorporation Human Proteinpedia data for the molecule ‘Vimentin’. Along with the normal tissue expression, Human Proteinpedia data provides cancer and cell line expression also. All the expression data displayed in HPRD are hyperlinked to the dataset description page of Human Proteinpedia.
Basic Protocol 4
DATA DOWNLOAD INTO HUMAN PROTEINPEDIA
Large amount of experimental data are being deposited into Human Proteinpedia, which can be used by the community for many meta-analysis studies. Human Proteinpedia data are made freely available for the community to download for any further analysis. The contributor uploads the mass spectrometry-derived raw files into Tranche server (Smith et al., 2011). Tranche is a distributed data repository to host proteomic data. The link to download the datasets from Tranche is provided in the experiment description page for the mass spectrometry-derived datasets.
Necessary Resources
Hardware
A computer with internet connection.
Software
Javascript compatible web browsers, such as Mozilla Firefox, Internet Explorer or Google Chrome and an e-mail account.
Files
None
-
Click on the ‘Download data’ tab located at the top panel of Human Proteinpedia portal: http://www.humanproteinpedia.org/download
The download tab provides list of publicly available datasets for download. For each of the datasets, this page provides the hyperlinked Human Proteinpedia accession number, which is linked to the detailed description of the dataset. It also provides brief description of the dataset, publication status, annotation category, experimental platform, download link and name of PI and name of the contributing laboratory. The entire dataset available in Human Proteinpedia has been made available for download on the top of this page. -
Click on Human Proteinpedia accession number for more details about the dataset.
This page provides detailed information about the selected dataset and the fields varies slightly based on the type of experiment and the type of platform are being used as explained in the example annotation files in Basic Protocol 5. -
Click on download data link provided for each of the dataset.
This link takes us to the page where the raw data files (link to Tranche server), processed protein identification files and meta-annotation data which provide brief description on the experiment could be downloaded for the dataset of interest.
Basic Protocol 5
EXAMPLE ANNOTATION FILES
Example annotation files are provided to make the users aware of different kinds of experimental data available in Human Proteinpedia. The display formats for Tissue expression and PTMs are discussed in Basic Protocol 1.
Necessary Resources
Hardware
A computer with internet connection
Software
Javascript compatible web browsers, such as Mozilla Firefox, Internet Explorer or Google Chrome and an e-mail account.
Files
None
-
Click on ‘Example annotation’ link provided on the top of the Human Proteinpedia portal located at http://www.humanproteinpedia.org/example
This page provides the example annotation for various annotation features as well as for the types of experimental platforms. The hyperlinked ‘click here’ buttons would take us to the example annotation pages. -
Click on the ‘click here’ link provided for the subcellular localization.
Protein name and its subcellular localization ‘Golgi apparatus’ are given in the top table followed by the experimental description. The experimental details include, experiment type, short and detailed descriptions of the experiment, PI and laboratory details, PubMed ID, sample source and source organism. The source organism is provided with the NCBI taxonomy ID. The lower bottom table describes the name of the protein, subcellular localization and external links to HPA. It also provides the immunofluorescence images describing the subcellular localization of KIAA2013 in Golgi apparatus. -
Click on the ‘click here’ link provided for the PPIs located at http://www.humanproteinpedia.org/Experimental_details?comp_id=3
This page shows a co-immunoprecipitation experiment carried out to study PPI. This page shows a complex interaction involving 11 molecules fetched from this experiment. Along with the experiment details as provided in the subcellular localization experiments, the interacting proteins and the name of the experiment is provided in the lower bottom table. -
Click on the ‘click here’ link for the Enzyme substrates located at http://www.humanproteinpedia.org/Experimental_details?ptm_id=13284
This page shows the casein kinase II substrates analysis using peptide microarray. RBM10 is the protein identified as substrate for the enzyme casein kinase II using peptide microarray. The peptide microarray image for the identification of substrates is provided in the lower bottom table along with the contributor’s details and experimental details.
GUIDELINES FOR UNDERSTANDING RESULTS
Human Proteinpedia is a portal which integrates and disseminates published and unpublished protein data pertaining to PTMs, PPIs, subcellular localization and tissue expression. The data available in Human Proteinpedia can be accessed either by query system or by downloading files. The query for a protein fetches all the experimental data for the protein by one or more laboratories. The data derived from mass spectrometry-based experiments provides most of the details obtained from the experiment that include one or more peptides identified from that protein, the database search engine score for the confidence of identification and it also provides the mass spectrum for each of the peptide identification allowing the users to check the assigned peaks for each of the identified amino acid. The mass spectra could be viewed by PRIDE viewer using the peak list data of each identified peptide. It is available for those peptides where peak list files were provided by the contributors. Multiple experiments on a single protein by different laboratories provide more evidence for a given reaction. Identification of a PTM in the same site by multiple groups provides the confidence on the site of modification. Also, the annotated mass spectrum and the peptide score derived from database search engines for the mass spectrometry-based experiments could be used as the confidence measure for the identification of protein.
COMMENTARY
Background information
The Human Proteinpedia was developed in 2009 as a unified resource to store and share the protein data derived from various experiments pertaining to human proteins. With the advent of recent proteomics technologies such as mass spectrometry, protein/peptide microarrays, yeast two hybrid systems, a large amount of data are being published every year. The data derived from these experiments take a while before they are disseminated to the biomedical community through publications in the form of either PDF documents or supplementary files. Human Proteinpedia accelerates the sharing of experimental data in different user-friendly formats and thus removes the inconvenience and delay caused by the time window required for publication. We have developed a separate PDAS dedicated to handle proteomic data to facilitate submission of proteomic data to Human Proteinpedia. The data can also be sent through FTP or e-mail. We also curate data from published literature and contact those authors to provide us the data with high resolution images and data from Human Proteinpedia is also made available into HPRD to reach out to a larger user community.
Troubleshooting
The data derived from multiple experiments are fetched and stored in Human Proteinpedia to make it easy for the scientific community to access the data by simple queries. The data displayed in the Human Proteinpedia website are obtained from the experimentalists and formatted according to the Human Proteinpedia database architecture. Human Proteinpedia provides the data associated with the experiment including images and contact details of the data provider. The users can contact the data provider for further information.
The protein names for each protein in Human Proteinpedia are fetched from HPRD. Although HPRD provides comprehensive information on the names and alternative names for each protein, in order to avoid multiple and non-specific hits, it is advisable to use the gene symbols from the approved gene nomenclature committee.
The images derived from the experiments are adjusted to accommodate in the Human Proteinpedia display. The original images as sent by the contributors can be obtained from the Human Proteinpedia core team upon request at http://pdas.hprd.org/help. Private data available in Human Proteinpedia is accessible only to the contributor and their collaborators, As soon as the data contributor releases it to the public, these data are made available to all users.
The protein assignments to the experimental data are mapped to HPRD proteins to have a unified identifier for each protein. HPRD annotation is based on the RefSeq protein database. The proteins which do not match the HPRD proteins are stored with the accession numbers given by the contributors. Those proteins which have accessions that do not match with HPRD proteins can be accessed only through download files.
Acknowledgments
A.P was partially funded for this project by a grant from National Institutes of Health Roadmap initiative U54 RR020839 and a contract N01-HV-28180 from the National Heart Lung and Blood Institute. We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics, Bangalore. B.M. is a recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Government of India. T.S.K.P. is supported by a research grant on “Development of Infrastructure and a Computational Framework for Analysis of Proteomic Data” from DBT.
Footnotes
Internet Resources
The Human Proteinpedia home page.
http://www.humanproteinpedia.org/download
The Human Proteinpedia download server.
The Human Proteinpedia public distributed annotation server.
The user’s queries and data request are answered through this link.
Literature Cited
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- Hamacher M, Klose J, Rossier J, Marcus K, Meyer HE. “Does understanding the brain need proteomics and does understanding proteomics need brains?”--Second HUPO HBPP Workshop hosted in Paris. Proteomics. 2004;4:1932–1934. doi: 10.1002/pmic.200400859. [DOI] [PubMed] [Google Scholar]
- He F. Human liver proteome project: plan, progress, and perspectives. Molecular & cellular proteomics: MCP. 2005;4:1841–1848. doi: 10.1074/mcp.R500013-MCP200. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, Patankar N, Shafreen B, Renuse S, Pawar H, Ramachandra YL, Acharya PK, Ranganathan P, Chaerkady R, Keshava Prasad TS, Pandey A. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic acids research. 2009;37:D773–781. doi: 10.1093/nar/gkn701. Update of Mathivanan et al., 2008 for the detailed explanation on various feature annotated in Human Proteinpedia and the potential uses of Human Proteinpedia data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, McCarthy MI, Hide T, Hide W. eVOC: a controlled vocabulary for unifying gene expression data. Genome research. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, Tyers M, Salama JJ, Moore S, Ceol A, Chatr-Aryamontri A, Oesterheld M, Stumpflen V, Salwinski L, Nerothin J, Cerami E, Cusick ME, Vidal M, Gilson M, Armstrong J, Woollard P, Hogue C, Eisenberg D, Cesareni G, Apweiler R, Hermjakob H. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC biology. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic acids research. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Rompp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, Tasman N, Coleman M, Reisinger F, Souda P, Hermjakob H, Binz PA, Deutsch EW. mzML--a community standard for mass spectrometry data. Molecular & cellular proteomics: MCP. 2011;10:R110 000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, Bjorling E, Blagoev B, Bose R, Brahmachari SK, Burlingame AS, Bustelo XR, Cagney G, Cantin GT, Cardasis HL, Celis JE, Chaerkady R, Chu F, Cole PA, Costello CE, Cotter RJ, Crockett D, DeLany JP, De Marzo AM, DeSouza LV, Deutsch EW, Dransfield E, Drewes G, Droit A, Dunn MJ, Elenitoba-Johnson K, Ewing RM, Van Eyk J, Faca V, Falkner J, Fang X, Fenselau C, Figeys D, Gagne P, Gelfi C, Gevaert K, Gimble JM, Gnad F, Goel R, Gromov P, Hanash SM, Hancock WS, Harsha HC, Hart G, Hays F, He F, Hebbar P, Helsens K, Hermeking H, Hide W, Hjerno K, Hochstrasser DF, Hofmann O, Horn DM, Hruban RH, Ibarrola N, James P, Jensen ON, Jensen PH, Jung P, Kandasamy K, Kheterpal I, Kikuno RF, Korf U, Korner R, Kuster B, Kwon MS, Lee HJ, Lee YJ, Lefevre M, Lehvaslaiho M, Lescuyer P, Levander F, Lim MS, Lobke C, Loo JA, Mann M, Martens L, Martinez-Heredia J, McComb M, McRedmond J, Mehrle A, Menon R, Miller CA, Mischak H, Mohan SS, Mohmood R, Molina H, Moran MF, Morgan JD, Moritz R, Morzel M, Muddiman DC, Nalli A, Navarro JD, Neubert TA, Ohara O, Oliva R, Omenn GS, Oyama M, Paik YK, Pennington K, Pepperkok R, Periaswamy B, Petricoin EF, Poirier GG, Prasad TS, Purvine SO, Rahiman BA, Ramachandran P, Ramachandra YL, Rice RH, Rick J, Ronnholm RH, Salonen J, Sanchez JC, Sayd T, Seshi B, Shankari K, Sheng SJ, Shetty V, Shivakumar K, Simpson RJ, Sirdeshmukh R, Siu KW, Smith JC, Smith RD, States DJ, Sugano S, Sullivan M, Superti-Furga G, Takatalo M, Thongboonkerd V, Trinidad JC, Uhlen M, Vandekerckhove J, Vasilescu J, Veenstra TD, Vidal-Taboada JM, Vihinen M, Wait R, Wang X, Wiemann S, Wu B, Xu T, Yates JR, Zhong J, Zhou M, Zhu Y, Zurbig P, Pandey A. Human Proteinpedia enables sharing of human protein data. Nature biotechnology. 2008;26:164–167. doi: 10.1038/nbt0208-164. This paper describes the multiple layer of contribution from the external laboratories and potential use of integrating Human Proteinpedia into HPRD. [DOI] [PubMed] [Google Scholar]
- Medina-Aunon JA, Carazo JM, Albar JP. PRIDEViewer: a novel user-friendly interface to visualize PRIDE XML files. Proteomics. 2011;11:334–337. doi: 10.1002/pmic.201000448. [DOI] [PubMed] [Google Scholar]
- Omenn GS, Baker MS, Aebersold R. Recent Workshops of the HUPO Human Plasma Proteome Project (HPPP): a bridge with the HUPO CardioVascular Initiative and the emergence of SRM targeted proteomics. Proteomics. 2011;11:3439–3443. doi: 10.1002/pmic.201100382. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic acids research. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BE, Hill JA, Gjukich MA, Andrews PC. Tranche distributed repository and ProteomeCommons.org. Methods in molecular biology. 2011;696:123–145. doi: 10.1007/978-1-60761-987-1_8. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- UniProt C. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic acids research. 2012;40:D71–75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]