Abstract
Mass spectrometry has rapidly evolved to become the platform of choice for proteomic analysis. While CID remains the major fragmentation method for peptide sequencing, electron transfer dissociation (ETD) is emerging as a complementary method for characterization of peptides and post-translational modifications (PTMs). Here, we review the evolution of ETD and some of its newer applications including characterization of PTMs, non-tryptic peptides and intact proteins. We will also discuss some of the unique features of ETD such as its complementarity with CID and the use of alternating CID/ETD along with issues pertaining to analysis of ETD data. The potential of ETD for applications such as multiple reaction monitoring and proteogenomics in the future will also be discussed.
Keywords: ETD, proteogenomics, non-tryptic peptide, O-GlcNAc, MRM
1 Introduction
The extensive use of CID in proteomics experiments over the last two decades has established it as a robust method for MS/MS fragmentation whose characteristics are relatively well understood from both experimental as well as computational perspectives [1]. In particular, CID is now well optimized for sequencing of small peptides obtained by protease digestion. However, in the case of modified peptides with labile PTMs such as O-GlcNAc, CID often fails to localize the site of modification. In 1998, Zubarev et al. reported electron capture dissociation (ECD) as a novel dissociation method to analyze peptide/protein structures [2]. Six years later, Syka et al. reported ETD as a new electron-based dissociation technique for fragmentation of gaseous ions [3]. In both ETD and ECD, an electron reacts with positively charged peptides followed by rapid neutralization of the charged site by an electron leading to generation of a radical, which in turn induces dissociation at the N-Cα bond [3].
In this review, we focus on the evolution of ETD in the context of instrumentation, analysis of various PTMs using ETD, top-down analysis using ETD and analysis of ETD data. Fundamentals of ETD and instrument-related aspects of ETD have been recently covered well elsewhere and will not be discussed here [4, 5].
2 Evolution of ETD
Chemistry of gaseous ions has long been investigated in the field of physical chemistry and chemical physics. ETD reactions resulting from electron transfer, a type of ion chemistry, include several steps, i.e. 1) generation of analyte cation and reagent anion, 2) appropriate delivery of the cation and the anion into the same space in time, 3) mass analysis of resultant ions. Each of these steps is discussed below.
2.1 Preparation of radical anions
In ion/ion reactions, electron transfer from an anion to a cation always competes with proton transfer from a cation to an anion [6, 7]. Thus, radical anions that prefer electron transfer are the key chemical moieties in ETD and their generation is, therefore, a key step. In the earliest ETD experiments, anthracene (C14H10) [3] molecules were introduced into the chemical ionization (CI) source to generate reactive anthracene radical anions (m/z 177 and m/z 179). Coon et al. also tested several other potential reagents for carrying out ETD including several polycyclic aromatic hydrocharbon molecules (e.g. fluoranthene) [8–10]. As a result, when reacted with triply charged peptides, some prefer proton transfer, whereas others are useful for ETD (e.g. fluoranthene radical anion (m/z 202)). Currently, fluoranthene is preferred because, of the molecules tested thus far, it works best [10], although the efficiency of electron transfer with fluoranthene is only ~40%. Thus, it is still desirable for ETD to find additional molecules that have low electron affinity and are structurally less hindered (e.g., a planar-like shape) to facilitate transfer of an electron to an analyte cation.
As an alternative, ESI modalities other than the CI, have been employed to generate precursor reagent molecules with CID for converting them into reagent anions [11]. From such experiments, among the tested compounds, the most attractive molecule for ETD was 2-(fluoranthene-8-carbonyl) benzoic acid (the efficiency of electron transfer, ~26%). The advantages of the ESI-based methods to generate reagent anions are that 1) it is relatively simple, 2) no additional ion source necessary, and 3) efficient and robust compared to CI-based methods. However, it is a slower method because additional time is required for polarity switching to and from negative mode, as discussed in the next section.
2.2 Injection of radical anions
The initial setup developed by Don Hunt’s group for ETD employed a linear ion-trap in which a negative CI source produced reagent anions from the back [3] (refer to as ‘back-end’). In this back-end method, only a relatively short time (~300 milliseconds (msec)) was spent to obtain a single ETD spectrum. This time scale fits well with that of liquid chromatography (LC), implying the feasibility of a high throughput analysis. However, this earlier back-end configuration was not readily applicable to hybrid instruments (e.g. LTQ-Orbitrap and quadrupole time-of-flight (QTOF)) owing to the location of high resolution analyzer at the rear. To circumvent this, a front-end method was introduced, for example, by utilizing a pulsed nanoESI/APCI, or ESI of precursor reagents and subsequent CID to generate reactive reagent ions on ion-trap and QTOF mass spectrometers [11–13]. The dual and pulsed method that used two ESI emitters to generate both analyte cation and reagent anion was also introduced on an LTQ-Orbitrap. A relative longer time of ~800 msec (~200 msec for anion injection and ~300 msec for each polarity switching) was taken to generate reagent ions for ETD [14]. Nonetheless, the important achievement of this configuration is that high resolution (60,000) and high accuracy (~2 ppm) ETD data were generated to better understand the performance characteristics of ETD MS. This front-end production of reagent ions was further improved by developing a dual electrospray ion source on LTQ-Orbitrap with reduction of the switching time upto ~30 ms [15]. The back-end ion source for ETD on the hybrid instrument (e.g. LTQ-Orbitrap ETD) has now been introduced with a systematic optimization for the efficient anion transfer from back to front [16]. The significant advance of this configuration was that it took only about 4–8 msec to inject a sufficient amount of uoranthene radical anion to the LTQ in the front from the rear in a stable and robust manner. As an alternative, reagent anions can be injected in between ion optics (i.e. rods) prior to ion-trap. This ‘in-between’ method has been implemented into a 3-D ion-trap (e.g. amaZon).
2.3 Optimization of ETD
Because the inefficiency of ETD is associated, at least in part, with non-specific interactions between ions produced by ETD, the use of additional energy required to dissociate non-dissociative charge reduced precursor ions has been evaluated. To increase the dissociation of non-specifically bound fragment ions, subsequent activation with vibrational collision energy has been shown to produce an additional set of odd-electron c•- and even-electron z- type ions (instead of c- and z•-type ions from ETD) from the non-specifically bound ion complex [17]. However, a 1-Da shift that results from a hydrogen rearrangement from c- to z-type ions makes the isotopic envelope of a fragment ion more complex [17, 18]. As an alternative approach, activated ion (AI) ETD methodology, which increases the internal energy of peptide ions by irradiating them with infrared photons during ion/ion reactions has been tried [19, 20]. This configuration resembles AI ECD MS [21] that was introduced earlier for increasing ECD efficiency by loosening secondary structure of a peptide/protein with increased internal energy by irradiation. The AI ETD method greatly improves the efficiency of peptide fragmentation, increasing the number of peptide identifications up to that obtained by combining both CID and ETD [20]. Another advantage of the AI ETD method is the lack of a requirement for additional time because of simultaneous in-space and in-time ion/ion reactions.
The instrument that is the most actively developed thus far for ETD is an ion-trap mass spectrometer while more technical developments with respect to ion/ion chemistry have been introduced on a QTOF mass spectrometer. It is possible that introduction of commercially available high accuracy and high resolution hybrid instruments such as QTOF (e.g. Synapt G2 HDMS) and quadrupole Fourier transformation ion cyclotron resonance (e.g. solariX) equipped with ETD could open new opportunities in ETD-based researches including top-down proteomics. The reaction time (~100 msec for small peptides and ~10 msec for larger peptides) for ETD has to be further optimized to minimize unwanted ion/ion reactions (e.g. the production of internal fragments and/or the neutralization of singly charged fragments), especially in top-down ETD experiments.
3 Analysis of PTMs by ETD
One of major advantages of ETD in MS-based proteomics is the ability to localize the exact site of PTMs that can be missed by CID. ETD MS-based PTM analysis has been reviewed [4, 5, 22]. In this section, we review the application of ETD for analysis of certain PTMs including phosphorylation, O-GlcNAc, N-linked glycosylation, O-linked glycosylation, glycation, isoaspartic acid formation, arginine methylation, lysine ubiquitination and ADP-ribosylation and its application to large-scale analysis. As listed in table 1, there are several studies that have employed ETD for large-scale PTM analyses such as phosphorylation, acetylation, methylation and glycosylation.
Table 1.
List of global PTM analyses employing ETD
| PTM | Sample | Method | Number | Mass spectrometer | Search algorithm | Reference |
|---|---|---|---|---|---|---|
| Phosphorylation | Human 293T cells | CID alone1; ETD alone2; alternating CID/ETD5 | 1435 sites | Agilent 6340 ion trap | Spectrum Mill | [91] |
| Phosphorylation | Drosophila melanogaster Kc167 cells | ETD alone; ET-CID8; alternating CID/ETD | 297 sites | LTQ XL | SEQUEST | [92] |
| Phosphorylation | Yeast | ETD alone | 1252 sites | Modified LTQ | OMSSA | [93] |
| Phosphorylation | Human embryonic stem cells (H1) | CID alone; ETD alone | 10844 sites | Modified LTQ Orbitrap | OMSSA | [94] |
| Phosphorylation | Medicago truncatula Jemalong A17 roots | ETD alone; conditional CID/ETD4 | 3404 sites | Modified LTQ-Orbitrap | OMSSA | [95] |
| Phosphorylation | Human pancreatic cancer P198 cells | CID alone; ETD alone; conditional CID/ETD; alternating CID/ETD | 2221 unique peptides | LTQ-Orbitrap XL | SEQUEST; Mascot; X!Tandem | [27] |
| N-linked glycosylation | Campylobacter jejuni | Alternating CID/HCD6; alternating CID/ETD | 81 sites with glycan structures | LTQ-Orbitrap XL and HCT Ultra ion trap | Manual interpretation | [31] |
| GlcNAc | Human 293T cells | HCD alone3; alternating HCD/ETD7 | 83 sites | LTQ-Orbitrap Velos | SEQUEST, followed by manual validation | [32] |
| GlcNAc | Mouse brain; HeLa cells | HCD alone; CID alone; ETD alone | 14 peptides from mouse brain; 30 from HeLa nuclei | LTQ-Orbitrap XL; LTQ- Orbitrap Velos | Mascot and SEQUEST, followed by manual validation | [96] |
| GlcNAc | HeLa cells | CID alone; ETD alone | 180 sites | LTQ-Orbitrap XL; LTQ-FT | OMSSA | [97] |
| Glycation | Human plasma | ETD alone | 726 unique peptides | LTQ | SEQUEST | [41] |
| Phosphorylation; Acetylation; Methylation | Suz12-deficient mouse embryonic stem cells | CID alone; ETD alone | 46 sites from H3.2 and H3.3 | LTQ-Orbitrap XL | Mascot | [98] |
| Acetylation; Methylation | HeLa S3 cells | ETD alone | 200 sites from H3.2 and 70 sites from H4 | LTQ and LTQ-Orbitrap XL | Mixed integer linear optimization computational framework | [99] |
CID alone: experiments where CID was only employed as a fragmentation method.
ETD alone: experiments where ETD was only employed as a fragmentation method.
HCD alone: experiments where HCD was only employed as a fragmentation method.
conditional CID/ETD: experiments where either CID or ETD was chosen based on the m/z value of a precursor ion.
alternating CID/ETD: experiments where a precursor ion was fragmented alternatively and successively by CID and ETD.
alternating CID/HCD: experiments where a precursor ion was fragmented alternatively and successively by CID and HCD.
alternating HCD/ETD: experiments where a precursor ion was fragmented alternatively and successively by HCD and ETD.
ET-CID: experiments where a charge reduced precursor ion during electron transfer (ET) was further fragmented by CID.
3.1 Serine/threonine phosphorylation
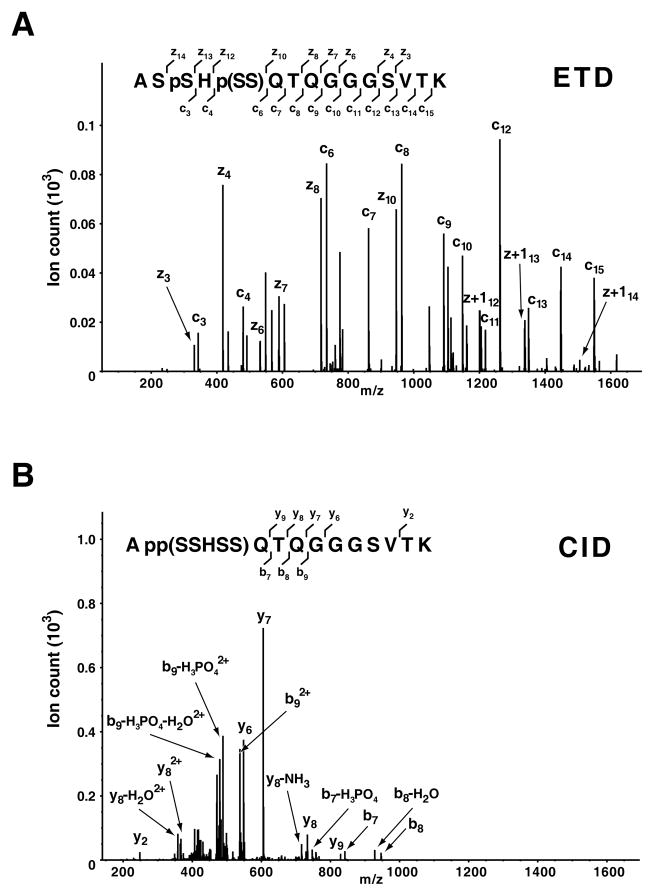
Phosphorylation is perhaps the most widely studied PTM in eukaryotes. The low stoichiometry of phosphopeptides coupled to their being negatively charged makes it difficult to identify them from very crude mixtures in the positive ion mode. Further, phosphoserine and phosphothreonine residues are labile under CID fragmentation conditions while phosphotyrosine is quite stable [23–25]. Fragmentation by ETD allows peptides to retain the labile phospho moiety on serine and threonine residues, which helps to accurately localize the phosphorylation sites [24]. Figure 1A shows an example of a phosphoserine containing peptide where the peptide was identified and the one of the phosphorylation sites localized by ETD. The MS/MS spectrum of the same peptide (Figure 1B) showed poor fragmentation using CID and did not lead to identification of the peptide by the search algorithm. In this instance, manual interpretation of the CID MS/MS spectrum could provide data on some of the peptide sequence but could still not help in the localization of either of the two phosphorylation sites. Note that the ETD spectrum (Figure 1A) was processed as described previously [26, 27].
Figure 1. ETD and CID spectra of a phosphorylated peptide.
A doubly phosphorylated peptide was fragmented by ETD (A) or CID (B) on an LTQ-Orbitrap ETD mass spectrometer. ETD generates a more informative MS/MS spectrum through a more uniform generation of c- and z-type ions while CID leads to a predominant neutral loss of phosphate moiety (H3PO4) with few other backbone fragments. Phosphorylation of serine-3 and either serine-5 or serine-6 was confirmed by ETD, while the exact phosphorylation sites could not be discerned from the CID spectrum. Note that ‘p(SS)’ indicates phosphorylation of one of the two serine residues within parentheses.
3.2 Glycosylation
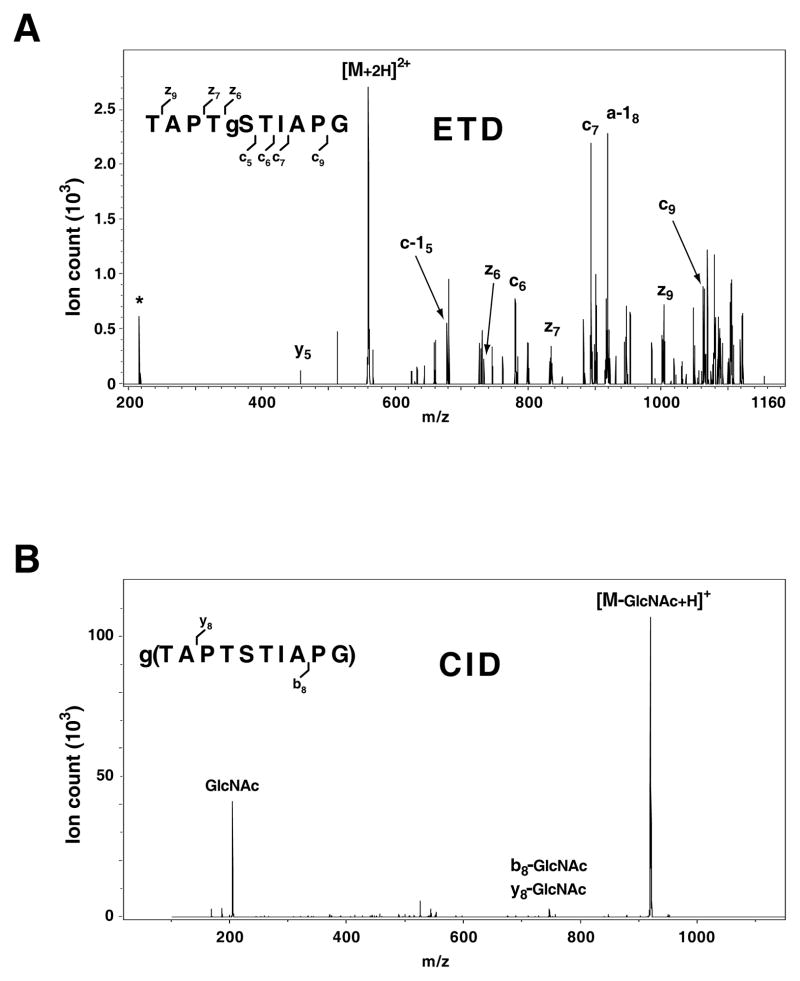
There are three major types of glycosylation: N-linked glycosylation at asparagine, O-linked glycosylation at serine and threonine and O-GlcNAc at serine and threonine. MS/MS analysis of N- and O-linked glycopeptides by CID often leads to a loss of the glycan (and some of its fragments) from the peptide with poor fragmentation of the peptide backbone [28]. ETD results in a more complete fragmentation of the peptide backbone which helps localize the site of the glycan attachment [28, 29]. In fact, it has been demonstrated that using both ETD and CID, it is possible to simultaneously elucidate both the glycan structure and peptide sequence of N- and O-linked glycopeptides [28–31]. On the other hand, peptides modified by GlcNAc on serine/threonine/asparagine residues undergo backbone fragmentation by CID but undergo a loss of GlcNAc as well [32]. In this situation, although one can discern the peptide sequence, it is still difficult to localize the site of modification as the loss of GlcNAc moiety does not alter mass of the amino acid residue where GlcNAc is attached. In many reports [32–35], ETD has been used as the primary fragmentation method both to identify the peptide sequence and to localize GlcNAc. Figure 2 shows an example of CID and ETD of a synthetic peptide (TAPTSTIAPG) that was modified by O-GlcNAc on the serine. ETD of this peptide yields an informative series that lead to correct identification of O-GlcNAc modification site at the serine residue (Figure 2A). In contrast, loss of GlcNAc from the peptide is a major fragmentation pathway under CID (Figure 2B), resulting in the most intense peaks arising from the doubly charged modified peptide, i.e. the singly charged GlcNAc moiety (m/z 204.1) and the singly charged unmodified peptide TAPTSTIAPG.
Figure 2. ETD and CID spectra of an O-GlcNAcylated peptide.
A synthetic O-GlcNAcylated peptide, TAPTgSTIAPG, was fragmented by ETD (A) or CID (B) on a 3D ion-trap mass spectrometer. A series of informative fragments was generated by ETD enabling localization of the site of modification while loss of the GlcNAc moiety (m/z 204.1) was predominant under CID. ‘gS’ refers to O-GlcNAc modified serine ‘*’ indicates N2 adduct-containing fluoranthene radical anion.
3.4 ADP-ribosylation
Mono- or poly-adenosine diphosphate (ADP)-ribosylation is enzyme mediated and can occur on arginine and lysine. Fragmentation of ADP-ribosylated arginine containing peptides by CID produces a few backbone fragments with predominant dissociation of the pyrophosphate bond, resulting in adenosine monophosphate (AMP, m/z 348) and the peptide ion that lost AMP [36]. ETD of ADP-ribosylated peptides shows a more informative backbone fragmentation with retention of the intact ADP-ribose moiety [36, 37]. Non-enzymatic modification of lysine residues (glycation) can also occur by ADP-ribose and is discussed below.
3.3 Glycation
Glycation is the non-enzymatic glycosylation of free amine groups on proteins and lipids by simple sugars such as fructose, glucose, ribose etc. Using arrays containing immobilized dipeptides, it has been shown that both glucose and fructose are highly reactive to the primary amino groups (lysine), thiol groups (cysteine), and the imidazolyl group (histidine) [38]. A complex series of reactions such as Amadori reaction, Schiff base reaction and Maillard reaction occur following glycation, leading to advanced glycation end products. For example, the level of glycated hemoglobin (also known as HbA1c) is measured to monitor how blood sugar levels are maintained in diabetics. ETD of glycated lysine containing peptides by D-glucose, for example, results in almost complete sequence coverage, while CID predominantly generates various neutral losses (2H2O, 3H2O, 4H2O, 3H2O+HCHO and C6H10O5) from glycated peptides [39–42]. In addition, fragmentation patterns of the glycated peptide with ADP-ribose at lysine residue by both CID and ETD have been observed similarly to that of ADP-ribosylated peptides by CID and ETD [43].
3.5 Isoaspartic acid
Deamidation of asparagine and isomerization of aspartic acid in proteins occurs through a succinimide intermediate, which in turn, is converted into a mixture of aspartic and isoaspartic acids. Thus, isoaspartic acid is an isomer of aspartic acid and leads to lengthening of the protein backbone by one methylene unit. ETD allows differentiation of isoaspartic acid from aspartic acid using diagnostic ions such as c + 57 and z − 57 peaks [44, 45] that are also observed under ECD (i.e. c + 58 and z − 57) [46, 47]. These diagnostic ions can define the presence and the location of isoaspartic acid. ETD analysis shows that for doubly charged peptide ions, supplemental activation is essential to obtain the diagnostic c + 57 or z − 57 ions needed to differentiate aspartic and isoaspartic acids from deamidated, digested peptide samples [44]. This method has been applied to a large scale analysis of blood samples from Alzheimer disease patients [48].
3.6 Arginine methylation
Methylation of arginine residues on proteins requires the methyl donor S-adenosyl methionine that is converted into S-adenosyl homocysteine. Arginine methylation can be mono-methylation or di-methylation (asymmetric or symmetric) and frequently occurs in an arginine-rich context such as RGG, GRG, RXR or RG motifs [49]. It has been demonstrated that arginine methylation containing peptides undergo peptide backbone dissociation by ETD providing localization information, while characteristic neutral losses (73.064 Da from monomethylated arginine and 87.087 Da from dimethylated arginines) are predominant when CID is used [50]. Thus, CID of the methylated arginine containing peptides could be a useful screen only to identify peptides that are modified, while ETD of the methylated peptide could provide information regarding the peptide sequence and the type of modification and its location within the peptide.
3.7 Ubiquitination
Ubiquitination is the attachment of ubiquitin, a 76 amino acid long protein, on proteins. The linkage occurs between the carboxylic acid of the C-terminal glycine in ubiquitin and the epsilon amine of lysine in the substrate protein. A characteristic di-glycine moiety on lysine residue is generated upon digestion of the ubiquitinated protein by trypsin, the standard protease used in MS–based proteomics. Even though CID is a promising fragmentation method to identify the di-glycine modified peptides [51–53], it is still desirable to use ETD as a complementary fragmentation method for these di-glycine modified peptides (which are higher charged than unmodified tryptic peptides because the modified lysine is not cleaved by trypsin) [51]. ETD has been used for the analysis of di-glycine containing peptides, while similarly informative series of fragment ions were produced by CID and ETD [54]. Thus, it might be a good example for utilizing both complementary fragmentation methods (CID and ETD) in an alternative acquisition mode. It should be noted that two alkylation moieties (C4H6N2O2, 114.0429 Da) by iodoacetamide on lysine can give rise to an identical mass tag as a di-glycine residue [55]. Thus, researchers might have to pay more attention about this potential artifact.
3.8 Endogenous peptides
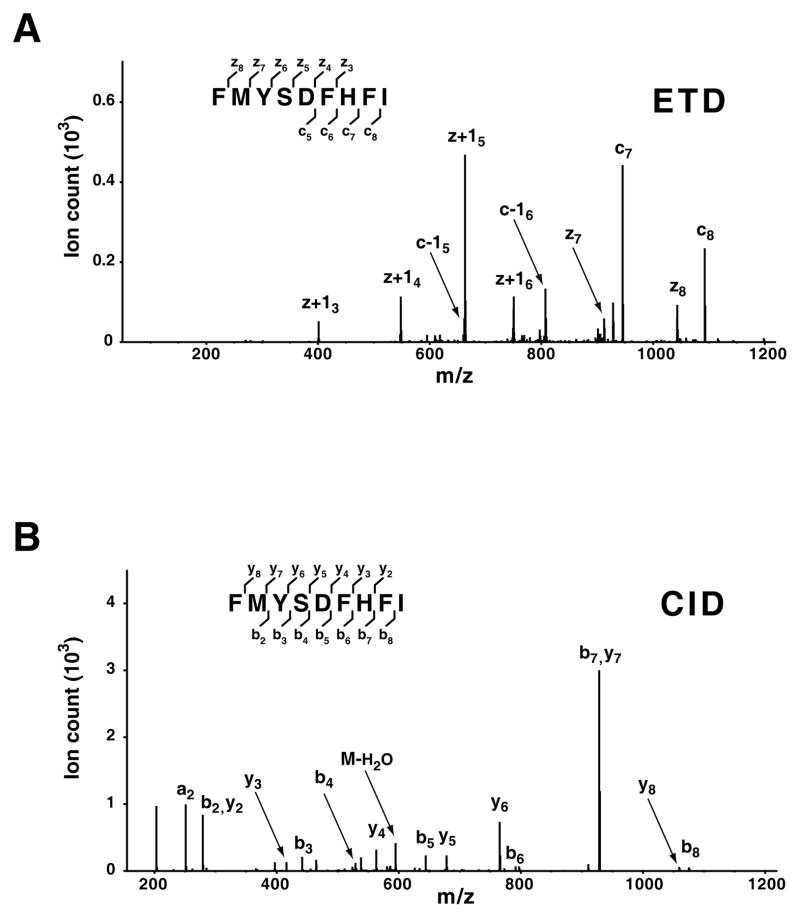
Although trypsin has been widely used as the standard protease in MS-based proteomics, other enzymes such as Lys-C and Lys-N are also being tried for proteomic applications [56]. However, there are biological situations where endogenous peptides need to be analyzed without any further protease digestion. For example, substance P (RPKPQQFFGLM-NH2), one of neuropeptides is an 11 amino acid long peptide containing a C-terminal amide modification which is structurally unusual. It is very important to elucidate both peptide sequences and structures of neuropeptides in the field of neuroscience. In addition, there are peptides (9~12 amino acids) presented by MHC class I/II molecules, to T-lymphocytes. Discovery of these endogenous immunogenic peptides is an important step in the field of immunoproteomics. Here, we discuss the use of multiple fragmentation methods for analysis of endogenous peptides. In this regard, Smith and co-workers have recently described an alternating fragmentation regimen to identify non-tryptic peptides [57]. As shown in Figure 3, a presenting peptide, FMYSDFHFI, derived from RNA polymerase from the influenza A virus, was fragmented by CID or ETD. Both patterns of fragments are comparably informative for identification of the peptide sequence. This example shows cross-validation of a non-tryptic peptide that was dissociated by two complementary fragmentation methods, i.e. CID and ETD. Two peaks i.e. m/z 279.1 and m/z 928.4 in Figure 3A are assigned to b2/y2 and b7/y7, respectively, because the fragment ions were detected on an ion-trap. However, all assigned fragment ions (c and z-ions) observed under ETD were distinguishable (Figure 3B), thereby increasing the confidence of identification. Thus, ETD as an additional fragmentation method or alternative acquisition of CID and ETD spectra would be of great interest for analysis of endogenous peptides.
Figure 3. ETD and CID spectra of a non-tryptic peptide.
A non-tryptic peptide, FMYSDFHFI, was fragmented by ETD (A) or CID (B) on an LTQ-Orbitrap ETD mass spectrometer. In this example, both fragmentation methods provide informative patterns of product ions, illustrating cross-validation of a non-tryptic peptide in an alternating CID/ETD approach.
4 Top-down proteomics
Currently, the most common pipeline in proteomics is the bottom-up approach in which protease-digested peptides that are ~7–25 amino acids in length are analyzed by LC-MS/MS. The top-down approach is slowly gaining popularity because intact proteins as well as protein complexes can be analyzed, which can lead to better elucidation of protein isoforms and PTMs. However, the top-down approach ideally requires mass spectrometers with very high resolution and mass accuracy (e.g. FT-ICR). Most studies to date where top down analysis has been carried out have employed CID/ECD not ETD [58–60]. In contrast, although ETD is superior to CID because of the more complete fragmentation and to ECD because of its compatibility with the LC-time scale, it has not yet been explored for top-down analysis. In the limited number of studies where ETD was used for a top-down analysis, only small number of proteins have been identified on a chromatographic time scale from E. coli samples [61, 62].
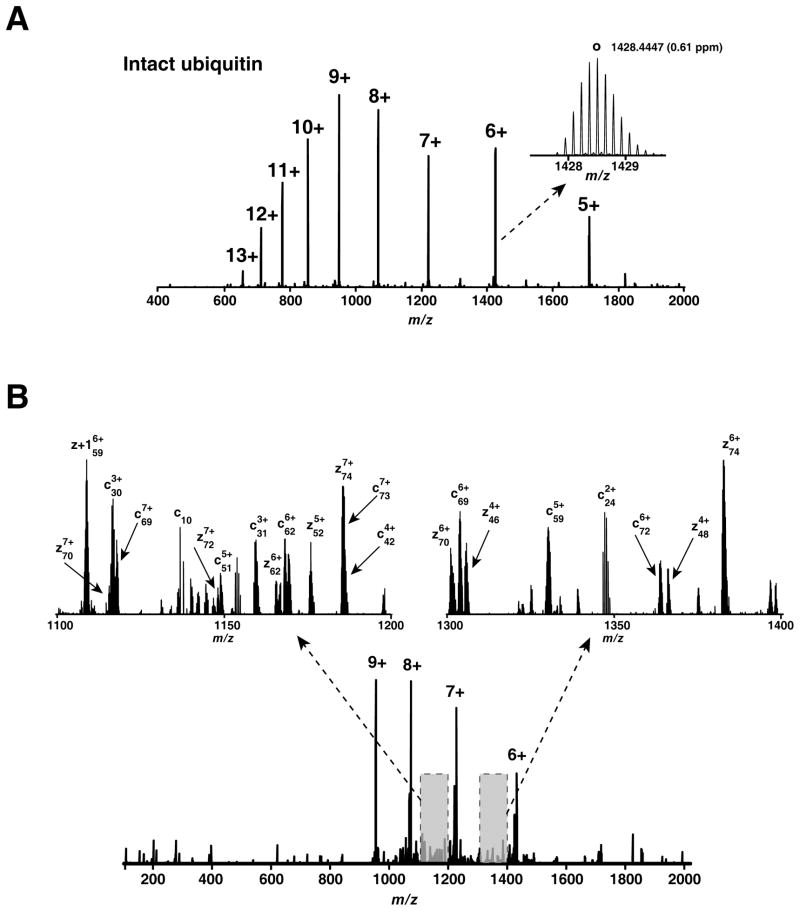
Regardless of the method of fragmentation employed, there are still a number of outstanding issues that limit the widespread applicability of top-down proteomics in a traditional LC-MS/MS setting. The low signal-to-noise ratio of high mass proteins is one of the main challenges in top-down proteomics [63]. What is not generally appreciated is that when intact proteins are analyzed in top-down experiments, a single intact protein, could be detected as various ion species because it exists as differently modified forms (e.g. oxidation), various charge species, and multiple isotopes leading to “splitting of the signal” and thereby lowering the signal-to-noise ratio. Other problems include the complexity of proteome both in terms of the number of proteins/peptides as well as the enormous dynamic range (e.g. 10–12 orders of magnitude for serum proteins). Finally, the relative lack of automated data analysis tools makes top-down analyses even more challenging although some dedicated tools have been developed for top-down analysis (for ECD/ETD and PTM types of analysis) [64]. Figure 4 shows an example of intact protein analysis by ETD. Figure 4A shows how the total signal of intact ubiquitin is distributed among several ion species (about 10 different charged ions, each of which is composed of ~10 isotopic peaks). Upon fragmentation of ubiquitin (z = 9) by ETD, a series of c- and z-type ions with non-dissociated ubiquitin ion species (indicated as +9, +8, +7, and +6) are observed (Figure 4B). In this case, manual assignment could be used to enrich the sequence information (see magnified regions of the spectrum) although many of the fragment ions could not be assigned using commercial software. This example illustrates how ETD-based top-down analysis is feasible but not yet optimal for automated data interpretation.
Figure 4. Intact ubiquitin analysis.
Intact ubiquitin was detected on an Orbitrap mass analyzer at a resolution of 100,000 (A). The inset in Figure 4A shows the isotopic distribution of a ubiquitin precursor (m/z = 1048; z = 6) at a mass accuracy of 0.61 ppm. Ubiquitin (m/z 953) was fragmented by ETD (B) and fragments were detected in the Orbitrap mass analyzer at a resolution of 60,000. A magnified view of two regions in the spectrum is shown along with manually assigned fragment ions.
5 ETD and CID as complementary technologies
ETD predominantly generates c- and z-type ions, while CID produces b- and y-type ions. Because of the nature of these fragment ions, the fragmentation data from ETD is complementary to that derived by CID. However, the two fragmentation methods are biased with regard to m/z values and amino acids. For example, CID works best for doubly charged smaller peptides, while ETD does best for higher charged peptides for sequencing analysis. The peptide bond at the amino-terminal end of proline (i.e. X-P, where X is an any amino acid) is the preferred dissociation site under CID, whereas the fact that two atomic bonds have to be broken to generate fragment ions on either side makes the dissociation in X-P almost impossible under ETD. Although neutral losses such as loss of H2O, NH3 and phosphate have been described in CID, an unusually large variety of neutral losses have been observed in ETD [65]. Overall, our predictive ability for fragmentation events, regardless of the dissociation method used remains poor. This unpredictability, in turn, makes de novo interpretation quite challenging. Although a statistical correlation of ETD behavior on peptide bond dissociation has been described [66], a comprehensive analysis of fragmentation events during ETD/CID by statistical or machine learning methods has not yet been carried out. Such an analysis has the potential to not only improve our understanding of the fragmentation process but also our ability to carry out de novo sequencing.
The orthogonal nature of ETD can be exploited by coupling it to CID in a number of ways. One can carry out separate CID and ETD experiments or experiments where CID of a given peptide alternates with ETD of the same peptide. Yet another strategy involves conditional selection of either ETD or CID during the run based on certain preset parameters related to physicochemical properties of peptide ions (e.g. choosing ETD for a peptide ion with a high charge state). In separate CID and ETD or conditional CID/ETD experiments, mass spectral data is separately processed and then the results are compared [66–68]. However, in alternating experiments, the same peptide ion is fragmented by CID and ETD in succession to generate a pair of CID and ETD spectra within the same LC-MS/MS run [27, 57]. Here, it is possible to process the CID and ETD mass spectral data separately or after merging [27, 69]. Both approaches (conditional CID/ETD and alternating CID/ETD) have been shown to provide a greater identification than a single fragmentation method (i.e. CID alone or ETD alone). In the case of alternating CID/ETD method, one can merge both CID and ETD spectra prior to database searching to take an advantage of complementarity of CID and ETD [27, 69]. Note that high energy collision dissociation (HCD) [70] that was recently introduced is also complementary to ETD [68].
As discussed in the examples above, CID and ETD are indeed complementary for deducing amino acid sequences or PTMs of peptides. Thus, these two are orthogonal datasets of the same peptide, which can be used to cross-validate the results of a database search making the identification even more confident. Thus, it is preferable to utilize multiple fragmentation methods for a single peptide to produce the maximum information, whenever analysis time and sample amount are not limiting.
6 Analysis of ETD data
Early studies employing ETD as a fragmentation method mainly relied on manual interpretation of MS/MS spectra to annotate c- and z-type ions. As discussed below, even now, the existing search algorithms for analysis of ETD data are not as advanced as they are for CID. To fully realize the potentials of ETD, it is still desirable to investigate the behavior of ETD and further, to understand ETD data.
Initial efforts for analysis of ETD data has focused on finding of characteristics of ETD spectra, resulting in the observation of highly intense charge-reduced precursor peptide ions and fragment ions associated with either various neutral losses or hydrogen rearrangements. This complexity of ETD products often diminishes the performance of searches, resulting in errors and a lower number of identifications [71]. It has been shown that search algorithms can improve peptide identification by deleting these interfering product ions as well as fragments associated with neutral losses such as H2O and NH3 affects [26]. Even though only marginal increase in peptide identification has been achieved by this post-acquisition data processing, there has been significant improvement in our understanding of the ETD behavior. It has reported that optimization of database search algorithms for ETD dataset can improve peptide identification [72]. Overall, it is clear that owing to the peculiarities of fragmentation induced by ETD, the search algorithms are far from being optimized for analysis of ETD data. In this regard, it should be pointed out that not all search algorithms are even capable of handling ETD data. Of the programs that can use ETD data, the performance is somewhat variable as demonstrated by a recent study [73] comparing multiple search algorithms such as OMSSA [74], Mascot [75], Spectrum Mill and X!Tandem [76]. Although Spectrum Mill and Mascot performed best for ETD data of phosphopeptides, non-phosphorylated peptides and known standard peptides [73], Protein Prospector, which was not evaluated by the above mentioned study, reported an improved peptide identification when its scoring system was modified for ETD data by weighting the precursor ion charge state and peptide sequence [77]. As discussed in the previous section, hydrogen rearranged fragment ions by supplemental activation after ETD makes spectra complex. Findings from statistical analyses of the behavior of ETD with supplemental activation [18, 65] have been implemented into pFind 2.1, which has been shown to perform slightly better than Mascot for ETD data [18]. Kim et al. have developed MS-GFDB [69], which employs the generating function approach using MS-GF [78]. A significant increase in identification of tryptic as well as Lys-N peptides from ETD data was observed in MS-GFDB over Mascot.
The Lys-N peptides, when fragmented by ETD, led to generation of predominantly c-type ions [79]. This characteristic tendency to display more c-type ions in ETD spectra of Lys-N peptides has been implemented into LysNDeNovo [80], an algorithm enabling de novo sequencing of Lys-N generated peptides fragmented by ETD. One database search algorithm that was specifically developed for analysis of ETD spectra is Z-core [81]. This algorithm is based on preprocessing of the data in addition to a probabilistic modeling approach and reported a minor increase in peptide identification as compared to OMSSA [81].
Although database search (e.g. Z-core, MS-GFDB, pFind 2.1) and de novo algorithms (e.g. CompNovo, Lys-NDeNovo) have been developed for analysis of ETD data, we feel that there is still adequate scope for optimizing their performance. One situation that has not been dealt with adequately pertains to scenarios when consecutive CID and ETD fragmentation is employed in an alternating mode for bottom up or top-down analyses [66, 82].
7 Future prospects
As described in the sections above, ETD has matured quite rapidly for MS-based proteomics analyses. Here, we will discuss the use of ETD in some areas where its potential has not yet been fully explored.
CID-based MS/MS is now a routine method for targeted analysis by multiple reaction monitoring (MRM). In these experiments, a certain number of transitions (many investigators use 3 or more transitions) from more than one unique peptide are considered as ‘requirements’ to characterize and quantify the corresponding proteins of interest. This poses problems when using the standard CID-based MRM pipeline for identifying and quantitating peptides/PTMs from complex protein mixtures. For example, it is not feasible to have multiple proteotypic peptides from every protein. It may also be difficult to generate three or more useful transitions from every proteotypic peptide. Also, when peptides containing labile PTMs (e.g. O-GlcNAc) are analyzed by CID, the site of modification is readily lost. Finally, not all peptides generated by proteases other than trypsin are optimal candidates for CID. In the above-mentioned scenarios, utilization of ETD should be helpful for MRM experiments because generation of complementary c- and z-type ions by ETD will extend the total number of useful transitions and also preserve labile PTMs. In the case of longer peptides with a high number of charges, ETD is better suited for MRM experiments. Even though ETD has been restricted to profiling type of experiments in proteomics thus far, we feel that application of ETD for MRM analysis has great potential and needs to be explored further.
In proteogenomics, MS-based proteomics data is correlated with genomic and transcriptomic data to better characterize genomes (for reviews, see [83–85]). For instance, the results from a proteogenomic analysis can include identification of novel genes that were missed by gene prediction algorithms. Using data from high resolution and high accuracy mass spectrometers [86] for searching against a six-frame-translation database [87], proteogenomics can be used for annotation of genomes. These types of studies rely greatly on the extent of coverage of the proteins and analyzing samples with both CID and ETD as orthogonal techniques can be carried out to maximize the sequence coverage obtained. In addition, the use of the alternating CID and ETD fragmentation in the same LC-MS run can also provide cross-validation of identified peptides by the two fragmentation methods. Thus, although CID is the routinely used fragmentation method, we anticipate that ETD will be employed more routinely for proteogenomic and similar applications in the future.
In typical LC-MS/MS experiments, the emphasis is on protein identification and not necessarily on the depth or comprehensiveness of the analysis. In a recent study, it was reported that only ~16% of all ions detected in a mass spectrometer were sampled for MS/MS (of which only ~50–60% were eventually identified) [88]. Although an increased depth can be achieved through the use of multiple proteases and fractionation methods, advances in instrumentation can achieve the same goal without any additional sample manipulation. This can happen in two different ways: introduction of newer fragmentation methods and development of newer mass spectrometers capable of multiple traps operating simultaneously. For example, negative ETD has been demonstrated as a new method in which a- and x-type ions are produced though free-radical chemistry [89] which is similar to electron detachment dissociation [90]. Therefore, it is conceivable that a complete set of complementary fragments (i.e. a, b, c, x, y, and z-type ions) from peptide ions can be acquired by employing negative ETD along with CID and ETD. Availability of newer types of mass spectrometers with multiple traps/analyzers that potentially permit multiple fragmentation methods in parallel would not only be attractive for increasing the depth of coverage but also help create new and imaginative solutions for protein/peptide analysis.
With continued advances in instrumentation and software for ETD analysis, we are positive that ETD will become an essential component of any proteomics platform in the future.
Acknowledgments
We would like to thank Raghothama Chaerkady, Derese Getnet and Robert O’Meally for helpful suggestions. A. P. is supported in part by a grant S10RR023025 from the High End Instrumentation Program of the National Institutes of Health, an NIH roadmap grant for Technology Centers of Networks and Pathways (U54RR020839) and a contract (HHSN268201000032C) from the National Heart, Lung, and Blood Institute.
Abbreviations
- CID
collision induced dissociation
- ETD
electron transfer dissociation
- ECD
electron capture dissociation
- CI
chemical ionization
- MS
mass spectrometry
- MS/MS
tandem mass spectrometer
- PTM
post-translational modification
- O-GlcNAc
O-linked beta-N-acetylglucosamine
- QTOF
quadrupole time-of-flight
- ESI
electrospray ionization
- msec
millisecond
- AI
activated ion
- HCD
high energy collision dissociation
- MRM
multiple reaction monitoring
- LC
liquid chromatography
- ADP
adenosine diphosphate
- AMP
adenosine monophosphate
Footnotes
Authors have declared no conflict of interest.
References
- 1.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 2.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 3.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, et al. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8:4466–4483. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 5.Mikesh LM, Ueberheide B, Chi A, Coon JJ, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang TY, McLuckey SA. Gas-phase chemistry of multiply charged bioions in analytical mass spectrometry. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:365–385. doi: 10.1146/annurev.anchem.111808.073725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia Y, McLuckey SA. Evolution of instrumentation for the study of gas-phase ion/ion chemistry via mass spectrometry. J Am Soc Mass Spectrom. 2008;19:173–189. doi: 10.1016/j.jasms.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, et al. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. International Journal of Mass Spectrometry. 2004;236:33–42. [Google Scholar]
- 9.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt DF, Coon JJ, Syka JEP, Marto JA. Electron transfer dissociation for biopolymer sequence analysis. 2005/0199804 A1. US patent. 2005 Sep 15;
- 11.Huang TY, Emory JF, O’Hair RA, McLuckey SA. Electron-transfer reagent anion formation via electrospray ionization and collision-induced dissociation. Anal Chem. 2006;78:7387–7391. doi: 10.1021/ac061409v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang X, Xia Y, McLuckey SA. Alternately pulsed nanoelectrospray ionization/atmospheric pressure chemical ionization for ion/ion reactions in an electrodynamic ion trap. Anal Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Y, Chrisman PA, Erickson DE, Liu J, et al. Implementation of ion/ion reactions in a quadrupole/time-of-flight tandem mass spectrometer. Anal Chem. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAlister GC, Phanstiel D, Good DM, Berggren WT, et al. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams DK, Jr, McAlister GC, Good DM, Coon JJ, et al. Dual electrospray ion source for electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:7916–7919. doi: 10.1021/ac071444h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAlister GC, Berggren WT, Griep-Raming J, Horning S, et al. A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, et al. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun RX, Dong MQ, Song CQ, Chi H, et al. Improved peptide identification for proteomic analysis based on comprehensive characterization of electron transfer dissociation spectra. J Proteome Res. 2010;9:6354–6367. doi: 10.1021/pr100648r. [DOI] [PubMed] [Google Scholar]
- 19.Ledvina AR, Beauchene NA, McAlister GC, Syka JE, et al. Activated-ion electron transfer dissociation improves the ability of electron transfer dissociation to identify peptides in a complex mixture. Anal Chem. 82:10068–10074. doi: 10.1021/ac1020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledvina AR, McAlister GC, Gardner MW, Smith SI, et al. Infrared photoactivation reduces peptide folding and hydrogen-atom migration following ETD tandem mass spectrometry. Angew Chem Int Ed Engl. 2009;48:8526–8528. doi: 10.1002/anie.200903557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn DM, Ge Y, McLafferty FW. Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 22.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Pandey A, Andersen JS, Mann M. Use of mass spectrometry to study signaling pathways. Sci STKE. 2000;2000:pl1. doi: 10.1126/stke.2000.37.pl1. [DOI] [PubMed] [Google Scholar]
- 24.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44:861–878. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 25.Harsha HC, Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Good DM, Wenger CD, McAlister GC, Bai DL, et al. Post-acquisition ETD spectral processing for increased peptide identifications. J Am Soc Mass Spectrom. 2009;20:1435–1440. doi: 10.1016/j.jasms.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MS, Zhong J, Kandasamy K, Delanghe B, et al. Systematic evaluation of alternating CID and ETD fragmentation for phosphorylated peptides. Proteomics. 2011;11:2568–2572. doi: 10.1002/pmic.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4:628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catalina MI, Koeleman CA, Deelder AM, Wuhrer M. Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun Mass Spectrom. 2007;21:1053–1061. doi: 10.1002/rcm.2929. [DOI] [PubMed] [Google Scholar]
- 30.Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, et al. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics. 2007;7:4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- 31.Scott NE, Parker BL, Connolly AM, Paulech J, et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics. 10:M000031–MCP000201. doi: 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao P, Viner R, Teo CF, Boons GJ, et al. Combining High-Energy C-Trap Dissociation and Electron Transfer Dissociation for Protein O-GlcNAc Modification Site Assignment. J Proteome Res. 2011;10:4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khidekel N, Ficarro SB, Clark PM, Bryan MC, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, et al. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zee BM, Garcia BA. Electron transfer dissociation facilitates sequencing of adenosine diphosphate-ribosylated peptides. Anal Chem. 2010;82:28–31. doi: 10.1021/ac902134y. [DOI] [PubMed] [Google Scholar]
- 37.Messner S, Altmeyer M, Zhao H, Pozivil A, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munch G, Schicktanz D, Behme A, Gerlach M, et al. Amino acid specificity of glycation and protein-AGE crosslinking reactivities determined with a dipeptide SPOT library. Nat Biotechnol. 1999;17:1006–1010. doi: 10.1038/13704. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Frolov A, Tang N, Hoffmann R, et al. Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides. Rapid Commun Mass Spectrom. 2007;21:661–666. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Tang N, Brock JW, Mottaz HM, et al. Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J Proteome Res. 2007;6:2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Schepmoes AA, Brock JW, Wu S, et al. Improved methods for the enrichment and analysis of glycated peptides. Anal Chem. 2008;80:9822–9829. doi: 10.1021/ac801704j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Tang N, Schepmoes AA, Phillips LS, et al. Proteomic profiling of nonenzymatically glycated proteins in human plasma and erythrocyte membranes. J Proteome Res. 2008;7:2025–2032. doi: 10.1021/pr700763r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedorova M, Frolov A, Hoffmann R. Fragmentation behavior of Amadori-peptides obtained by non-enzymatic glycosylation of lysine residues with ADP-ribose in tandem mass spectrometry. J Mass Spectrom. 2010;45:664–669. doi: 10.1002/jms.1758. [DOI] [PubMed] [Google Scholar]
- 44.Chan WY, Chan TW, O’Connor PB. Electron transfer dissociation with supplemental activation to differentiate aspartic and isoaspartic residues in doubly charged peptide cations. J Am Soc Mass Spectrom. 21:1012–1015. doi: 10.1016/j.jasms.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor PB, Cournoyer JJ, Pitteri SJ, Chrisman PA, et al. Differentiation of aspartic and isoaspartic acids using electron transfer dissociation. J Am Soc Mass Spectrom. 2006;17:15–19. doi: 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 46.Yang H, Fung EY, Zubarev AR, Zubarev RA. Toward proteome-scale identification and quantification of isoaspartyl residues in biological samples. J Proteome Res. 2009;8:4615–4621. doi: 10.1021/pr900428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cournoyer JJ, Pittman JL, Ivleva VB, Fallows E, et al. Deamidation: Differentiation of aspartyl from isoaspartyl products in peptides by electron capture dissociation. Protein Sci. 2005;14:452–463. doi: 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H, Lyutvinskiy Y, Soininen H, Zubarev RA. Alzheimer’s Disease and Mild Cognitive Impairment are Associated with Elevated Levels of Isoaspartyl Residues in Blood Plasma Proteins. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2011-110626. [DOI] [PubMed] [Google Scholar]
- 49.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Straubinger RM, Aletta JM, Cao J, et al. Accurate localization and relative quantification of arginine methylation using nanoflow liquid chromatography coupled to electron transfer dissociation and orbitrap mass spectrometry. J Am Soc Mass Spectrom. 2009;20:507–519. doi: 10.1016/j.jasms.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner SA, Beli P, Weinert BT, Nielsen ML, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim W, Bennett EJ, Huttlin EL, Guo A, et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobott F, Watt SJ, Smith J, Edelmann MJ, et al. Comparison of CID versus ETD based MS/MS fragmentation for the analysis of protein ubiquitination. J Am Soc Mass Spectrom. 2009;20:1652–1659. doi: 10.1016/j.jasms.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 56.Raijmakers R, Neerincx P, Mohammed S, Heck AJ. Cleavage specificities of the brother and sister proteases Lys-C and Lys-N. Chem Commun (Camb) 2010;46:8827–8829. doi: 10.1039/c0cc02523b. [DOI] [PubMed] [Google Scholar]
- 57.Shen Y, Tolic N, Xie F, Zhao R, et al. Effectiveness of CID, HCD, and ETD with FT MS/MS for Degradomic-Peptidomic Analysis: Comparison of Peptide Identification Methods. J Proteome Res. 2011;10:3929–3943. doi: 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth MJ, Forbes AJ, Boyne MT, 2nd, Kim YB, et al. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Cui W, Wen J, Blankenship RE, et al. Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Anal Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin S, Loo JA. Top-Down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int J Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi A, Bai DL, Geer LY, Shabanowitz J, et al. Analysis of intact proteins on a chromatographic time scale by electron transfer dissociation tandem mass spectrometry. Int J Mass Spectrom. 2007;259:197–203. doi: 10.1016/j.ijms.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, et al. Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal Chem. 2008;80:1459–1467. doi: 10.1021/ac7018409. [DOI] [PubMed] [Google Scholar]
- 63.Compton PD, Zamdborg L, Thomas PM, Kelleher NL. On the scalability and requirements of whole protein mass spectrometry. Anal Chem. 2011;83:6868–6874. doi: 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, et al. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 2007;35:W701–706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia Q, Lee MV, Rose CM, Marsh AJ, et al. Characterization and diagnostic value of amino acid side chain neutral losses following electron-transfer dissociation. J Am Soc Mass Spectrom. 2011;22:255–264. doi: 10.1007/s13361-010-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal Chem. 2008;80:4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frese CK, Altelaar AF, Hennrich ML, Nolting D, et al. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 10:2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 69.Kim S, Mischerikow N, Bandeira N, Navarro JD, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2011;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olsen JV, Macek B, Lange O, Makarov A, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 71.Good DM, Wenger CD, Coon JJ. The effect of interfering ions on search algorithm performance for electron-transfer dissociation data. Proteomics. 10:164–167. doi: 10.1002/pmic.200900570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leinenbach A, Hartmer R, Lubeck M, Kneissl B, et al. Proteome analysis of Sorangium cellulosum employing 2D-HPLC-MS/MS and improved database searching strategies for CID and ETD fragment spectra. J Proteome Res. 2009;8:4350–4361. doi: 10.1021/pr9004647. [DOI] [PubMed] [Google Scholar]
- 73.Kandasamy K, Pandey A, Molina H. Evaluation of several MS/MS search algorithms for analysis of spectra derived from electron transfer dissociation experiments. Anal Chem. 2009;81:7170–7180. doi: 10.1021/ac9006107. [DOI] [PubMed] [Google Scholar]
- 74.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 75.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 76.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 77.Baker PR, Medzihradszky KF, Chalkley RJ. Improving software performance for peptide electron transfer dissociation data analysis by implementation of charge state- and sequence-dependent scoring. Mol Cell Proteomics. 9:1795–1803. doi: 10.1074/mcp.M110.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taouatas N, Drugan MM, Heck AJ, Mohammed S. Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat Methods. 2008;5:405–407. doi: 10.1038/nmeth.1204. [DOI] [PubMed] [Google Scholar]
- 80.van Breukelen B, Georgiou A, Drugan MM, Taouatas N, et al. LysNDeNovo: an algorithm enabling de novo sequencing of Lys-N generated peptides fragmented by electron transfer dissociation. Proteomics. 10:1196–1201. doi: 10.1002/pmic.200900405. [DOI] [PubMed] [Google Scholar]
- 81.Sadygov RG, Good DM, Swaney DL, Coon JJ. A new probabilistic database search algorithm for ETD spectra. J Proteome Res. 2009;8:3198–3205. doi: 10.1021/pr900153b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horn DM, Zubarev RA, McLafferty FW. Automated de novo sequencing of proteins by tandem high-resolution mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2010;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 84.Krug K, Nahnsen S, Macek B. Mass spectrometry at the interface of proteomics and genomics. Mol Biosyst. 7:284–291. doi: 10.1039/c0mb00168f. [DOI] [PubMed] [Google Scholar]
- 85.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 73:2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, et al. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res. 2011 doi: 10.1101/gr.127951.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yates JR, 3rd, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 88.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 89.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. Electron transfer dissociation of peptide anions. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 90.Zubarev RA, Budnik BA, Haselmann KF. Electron detachment dissociation of peptide di-anions: an electron-hole recombination phenomenon. Chem Phys Lett. 2001;342:299–302. [Google Scholar]
- 91.Molina H, Horn DM, Tang N, Mathivanan S, et al. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Domon B, Bodenmiller B, Carapito C, Hao Z, et al. Electron transfer dissociation in conjunction with collision activation to investigate the Drosophila melanogaster phosphoproteome. J Proteome Res. 2009;8:2633–2639. doi: 10.1021/pr800834e. [DOI] [PubMed] [Google Scholar]
- 93.Chi A, Huttenhower C, Geer LY, Coon JJ, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grimsrud PA, den Os D, Wenger CD, Swaney DL, et al. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parker BL, Gupta P, Cordwell SJ, Larsen MR, et al. Purification and identification of O-GlcNAc-modified peptides using phosphate-based alkyne CLICK chemistry in combination with titanium dioxide chromatography and mass spectrometry. J Proteome Res. 2011;10:1449–1458. doi: 10.1021/pr100565j. [DOI] [PubMed] [Google Scholar]
- 97.Wang Z, Udeshi ND, Slawson C, Compton PD, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung HR, Pasini D, Helin K, Jensen ON. Quantitative mass spectrometry of histones H3.2 and H3. 3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics. 2010;9:838–850. doi: 10.1074/mcp.M900489-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, et al. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]